In the absence of an effective vaccine to prevent HCMV infections, alternative interventions must be developed. Prevention of viral entry into susceptible cells is an attractive alternative strategy. Here we report that heparan sulfate-binding peptides effectively inhibit entry into fibroblasts of in vitro-derived CMVs and partially inhibit in vivo-derived CMVs. This includes the inhibition of urine-derived HCMV (uCMV), which is highly resistant to antibody neutralization. While these antiviral peptides are highly effective at inhibiting cell-free virus, they do not inhibit MCMV cell-to-cell spread. This underscores the need to understand the mechanism of cell-to-cell spread and differences between in vivo-derived versus in vitro-derived CMV entry to effectively prevent CMV’s spread.
KEYWORDS: HCMV, MCMV, antiviral peptides, cytomegalovirus, entry, heparan sulfate
ABSTRACT
Human cytomegalovirus (HCMV) is a ubiquitous betaherpesvirus that can cause severe disease following in utero exposure, during primary infection, or latent virus reactivation in immunocompromised populations. These complications lead to a 1- to 2-billion-dollar economic burden, making vaccine development and/or alternative treatments a high priority. Current treatments for HCMV include nucleoside analogues such as ganciclovir (GCV), foscarnet, and cidofovir. Recently, letermovir, a terminase complex inhibitor, was approved for prophylaxis after stem cell transplantation. These treatments have unwanted side effects, and HCMV is becoming resistant to them. Therefore, we sought to develop an alternative treatment that targets a different stage in viral infection. Currently, small antiviral peptides are being investigated as anti-influenza and anti-HIV treatments. We have developed heparan sulfate-binding peptides as tools for preventing CMV infections. These peptides are highly effective at stopping infection of fibroblasts with in vitro-derived HCMV and murine cytomegalovirus (MCMV). However, they do not prevent MCMV infection in vivo. Interestingly, these peptides inhibit infectivity of in vivo-derived CMVs, albeit not as well as tissue culture-grown CMVs. We further demonstrate that this class of heparan sulfate-binding peptides is incapable of inhibiting MCMV cell-to-cell spread, which is independent of heparan sulfate usage. These data indicate that inhibition of CMV infection can be achieved using synthetic polybasic peptides, but cell-to-cell spread and in vivo-grown CMVs require further investigation to design appropriate anti-CMV peptides.
IMPORTANCE In the absence of an effective vaccine to prevent HCMV infections, alternative interventions must be developed. Prevention of viral entry into susceptible cells is an attractive alternative strategy. Here we report that heparan sulfate-binding peptides effectively inhibit entry into fibroblasts of in vitro-derived CMVs and partially inhibit in vivo-derived CMVs. This includes the inhibition of urine-derived HCMV (uCMV), which is highly resistant to antibody neutralization. While these antiviral peptides are highly effective at inhibiting cell-free virus, they do not inhibit MCMV cell-to-cell spread. This underscores the need to understand the mechanism of cell-to-cell spread and differences between in vivo-derived versus in vitro-derived CMV entry to effectively prevent CMV’s spread.
INTRODUCTION
Human cytomegalovirus (HCMV) is a significant pathogen within immunocompromised groups. Disease in these populations can result from primary infection or spontaneous latent virus reactivation (1, 2). As 60 to 90% of adults are latently infected with HCMV, there is a substantial population at risk for complications if their immune system becomes compromised (3, 4). HCMV infection/reactivation in immunocompromised persons can result in mononucleosis-like symptoms, interstitial pneumonia, gastroenteritis, retinitis, or organ transplant rejection in transplant patients (1, 3). HCMV is also the leading cause of congenital disease (5, 6). In utero infection may result in fetal abnormalities such as microcephaly or severe sequelae that can evolve over time in the form of progressive deafness, mental retardation, or learning disabilities (7, 8). HCMV infections impose a yearly 1- to 2-billion-dollar economic burden; therefore, development of effective treatment and preventive strategies is a high priority (5, 9). Because there is no effective vaccine, treatment of infected immunocompromised patients primarily consists of nucleoside analogs such as ganciclovir (GCV), foscarnet, or cidofovir which inhibit DNA replication (10 – 12). Unfortunately, GCV treatment can be myelosuppressive, while foscarnet and cidofovir are nephrotoxic (13). All DNA polymerase inhibitors select for resistant HCMV mutants, and cases of GCV-resistant HCMV infections are on the rise (1, 14, 15). This has led to the development of novel treatments such as the recently FDA-approved terminase inhibitor, letermovir (16).
Antiviral peptides (APs) are an attractive alternative treatment for inhibiting viral infections. Indeed, peptide therapeutics are being investigated for respiratory viruses and HIV (17 – 19). APs have different mechanisms for virus inhibition from inhibiting viral attachment, entry, replication, or egress (20). HCMV attaches to a host cell via heparan sulfate proteoglycans (HSPGs) (21). Viral glycoproteins gB and gM/gN initially interact with negatively charged sulfate moieties, which serve to “dock” the HCMV virion to the host cell (21). Docking triggers a signal cascade within the cell allowing for subsequent viral entry. HSPGs are ubiquitously expressed on most host cells, supporting the idea that HCMV can infect almost any human cell type (22).
HSPGs have a myriad of functions, including binding chemokines and cytokines and serving as scaffolds for ligand receptors, growth factors, and other cell adhesion molecules (23). Cell surface HSPGs are also major components of host-mediated endocytosis and cell membrane fusion processes. HSPG functions have been exploited for malarial and viral infections, including HCMV and herpes simplex virus 1 (24 – 26). Because of their major role in the early stages of HCMV replication, heparan sulfates (HSs) are an attractive target for intervention. HS-binding peptides effectively inhibit HCMV infection (27). However, these peptides were not tested against the more virulent in vivo-derived virus or in an in vivo setting (28).
We have previously reported that synthetic heparin-binding peptides bind pathological amyloid deposits in vitro and in vivo (29, 30). As HCMV attaches to cells via HS, we investigated whether these peptides could inhibit virus attachment. In this study, we demonstrate that these synthetic polybasic peptides are efficient at inhibiting viral entry of tissue culture-derived HCMV and murine cytomegalovirus (MCMV). We also provide evidence of effectively inhibiting an HCMV clinical isolate obtained from infected bodily secretions. However, these peptides could not prevent cell-to-cell spread of MCMV, potentially explaining the need to further investigate additional antiviral peptides for efficiency in vivo.
RESULTS
Peptide characteristics.
Three polybasic peptides, designated p5(coil), p5(coil)D, and p5 + 14(coil), were synthesized using a glycine-rich backbone to enhance flexibility of the peptide chain (Table 1). The p5(coil) peptide is the parental peptide (31, 32) from which the derivative p5(coil)D and p5 + 14(coil) peptides were designed. p5(coil)D is the D form of p5(coil). Because D-form peptides are more proteolytically stable and are equally effective as L-form peptides at inhibiting HCMV entry, we focused on p5(coil)D for the majority of this study (33). We also utilized the peptide p5 + 14(coil), which is p5(coil) with an additional repeat of the last 14 amino acids. The addition of 14 amino acids has been shown to increase the efficacy of peptide-induced HCMV inhibition (27). At a peptide concentration of 50 µM, both p5(coil)D and p5 + 14(coil) inhibited HCMV and MCMV infection of fibroblasts (Table 1). We chose 50 µM as the initial concentration for the screening the peptides because another polybasic D-form peptide could inhibit MCMV in vivo at this dose (33). All three peptides were predicted to adopt a flexible coil secondary structure, which is different from previously published peptides and may increase their efficacy (34, 35).
TABLE 1.
Polybasic peptide descriptions and characteristicsa
| Peptide | No. of aa | Primary structure | Property | Net charge (positive) |
% of infection inhibition |
|
|---|---|---|---|---|---|---|
| MCMV | HCMV | |||||
| p5(coil) | 31 | GGGYS KGGKG GGKGG KGGGK GGKGG GKGGK G | Flexible coil | 8 | ||
| p5(coil)D | 31 | [GGGYS KGGKG GGKGG KGGGK GGKGG GKGGK G]D | D form of p5(coil) | 8 | 68 | 89 |
| p5 + 14(coil) | 45 | GGGYS KGGKG GGKGG KGGGK GGKGG GKGGK GGGKG GKGGG KGGKG | Flexible coil | 12 | 72 | 96 |
Peptide secondary structures were predicted via ITASSER software. Peptide inhibition of infection was determined at 50 μM. Positively charged residues are underlined.
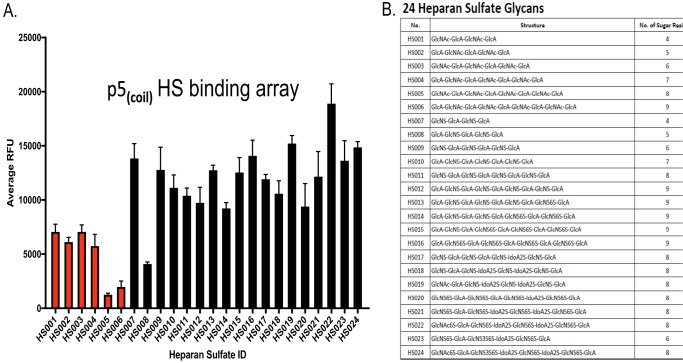
As p5(coil) was the peptide from which the others were generated, we tested the binding of a biotinylated variant to a panel of HS moieties using a synthetic glycoarray (Fig. 1A). p5(coil) bound significantly more effectively to sulfated glycans (black bars) than to unsulfated glycans (red bars) (Fig. 1A), with the exception of the 5-sugar HS008 glycan. Statistical analysis showed significantly enhanced binding of p5(coil) to almost all sulfated glycans relative to nonsulfated species (see Table S1 in the supplemental material). In general, no significant difference was observed between levels of peptide binding to the structurally different sulfated HSs. Figure 1B lists the structure of the glycans used in the glycan array. These results highlight that p5(coil) preferentially binds sulfated glycans.
FIG 1.
Binding of peptide p5(coil) to an array of synthetic HS glycans. (A) A 0.5-mg/ml aliquot of biotinylated p5(coil) peptide was incubated with a heparan sulfate glycan array, and the binding was visualized using a streptavidin-conjugated fluorophore. Nonsulfated glycans are shown in red. Each bar represents the mean and SD from 5 replicates. Statistical analysis data are presented in Table S1. (B) Composition of the HS glycans used in panel A.
Statistical analysis of the glycan array. Statistical significance was determined by an ordinary one-way ANOVA with Tukey’s multiple comparison of means. ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. Analysis was carried out using GraphPad Prism. Download Table S1, DOCX file, 0.2 MB (214.3KB, docx) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Efficacy of inhibition of CMV infection.
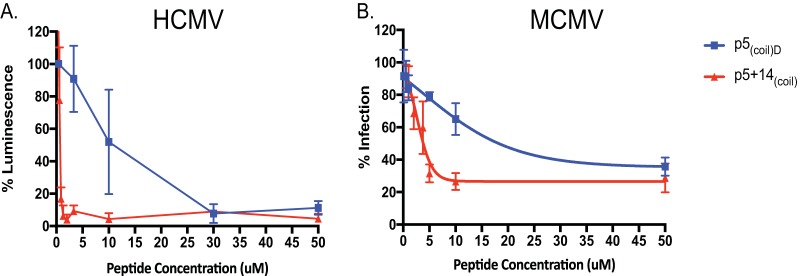
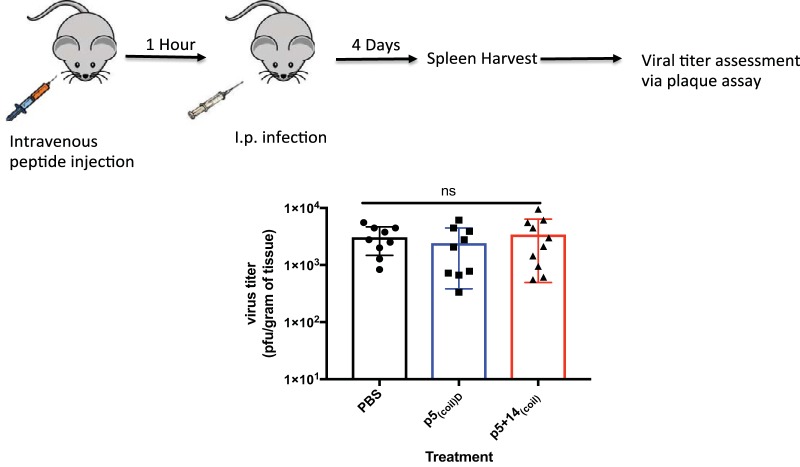
The blockade of HCMV and MCMV attachment to cells was studied in the presence of increasing concentrations of peptide p5(coil)D or p5 + 14(coil) (Fig. 2). The estimated 50% inhibitory concentrations (IC50s) of p5(coil)D and p5 + 14(coil) for blocking HCMV (TB40/E) were 9.98 and 0.6 μM, respectively (Fig. 2A), and those for MCMV were 22.6 and 2.97 μM, respectively (Fig. 2B). These data indicate that our peptides are capable of inhibiting both HCMV and MCMV; however, MCMV is inhibited to a lesser extent. Peptide inhibition of infection of mice was evaluated using the MCMV mouse model (Fig. 3). BALB/c mice were pretreated with p5(coil)D or p5 + 14(coil) at 250 μg per mouse 1 h prior to infection. Evaluation of the viral titer in spleens harvested 4 days postinfection (dpi) indicated no significant difference in viral burden (Fig. 3). In order to confirm peptide was present at the time of infection and following infection, we evaluated peptide biodistribution postadministration (see Table S2 in the supplemental material). Our biodistribution assay confirms that p5(coil)D or p5 + 14(coil) is present within the host at the time of infection and following infection. While there is a difference in biodistributions between the two peptides, there is no difference in levels of viral dissemination to the spleen. We have previously reported the inability of p5RD, another antiviral polybasic peptide, which is significantly more stable in vivo, to substantially inhibit infection of any primary dissemination organs (e.g., spleen, liver, and lung) (33). The inability of these peptides to reduce viral load in vivo could be due in vivo dosage/timing effect, but an alternative explanation is that the peptides differ in their ability to block in vivo-derived virus versus in vitro-derived viruses. Previous studies have reported differences between MCMVs grown in culture compared with those harvested in vivo, which was related to their HSPG usage for entry (36 – 40).
FIG 2.
The p5(coil) family of peptides prevent HCMV and MCMV infection. Peptides were serially diluted and assayed in an HCMV (TB40/E UL18Luc) luciferase assay (A) or an MCMV plaque reduction assay (B). IC50s for HCMV of 9.98 μM for p5(coil)D and 0.6 μM for p5 + 14(coil) and IC50s for MCMV of 22.6 μM for p5(coil)D and 2.97 μM for p5 + 14(coil) were calculated with GraphPad Prism. Each point represents an average of 3 or 4 replicates ± SD from 3 experiments.
FIG 3.
Peptide efficacy in vivo. BALB/c mice were treated with peptide (250 μg/mouse) i.v. and infected with 1 × 106 PFU of MCMV i.p. 1 h later. Bars represent the average of 4 or 5 mice per group from 2 experiments. One-way ANOVA with Tukey’s multiple comparison of means was used to determine statistical significance. ns, not significant.
p5(coil)D or p5 + 14(coil) biodistribution in BALB/c mice. Mice were injected with 250 μg/mouse of 125I-labeled peptide. Mice were euthanized at 1 and 4 h postinjection (hpi), and the indicated organs were harvested. The amount of 125I present in each organ was measured. Data represent the percentage of injected dose (ID)/g of tissue. Download Table S2, DOCX file, 0.3 MB (263.3KB, docx) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Peptide inhibition of in vivo- and in vitro-derived MCMV.
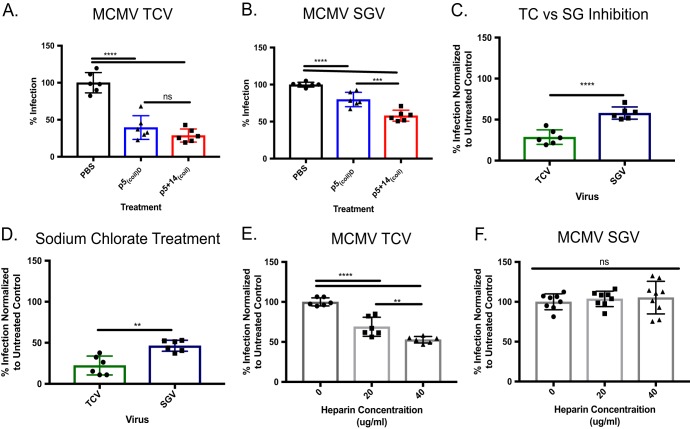
To evaluate the possibility of differential inhibition between in vitro-derived virus and in vivo-derived virus, we performed plaque reduction assays using MCMV salivary gland-isolated virus (SGV) and MCMV passaged on cultured cells (tissue culture-derived virus [TCV]) (Fig. 4). Both p5(coil)D and p5 + 14(coil) significantly inhibited infection of both murine TCV (Fig. 4A) and SGV (Fig. 4B) relative to untreated cells (phosphate-buffered saline [PBS]). Additionally, in both cases p5 + 14(coil) was significantly more efficacious than p5(coil)D. When used at 50 μM, p5 + 14(coil) inhibited murine TCV (75%) more effectively than SGV (40%) (Fig. 4C). Our data support observations made by Ravindranath and Graves (36) and indicate SGV and TCV use different entry strategies. One entry mechanism is inhibited by peptides (i.e., TCV), and the other is only partially blocked (i.e., SGV).
FIG 4.
Differential effects of peptide treatment on in vivo- and in vitro-derived MCMV. Peptide at a 50 µM concentration was used to inhibit (A) tissue culture-derived (TCV) or (B) salivary gland-derived (SGV) MCMV. (C) Data from panels A and B to compare the efficacy of p5 + 14(coil) inhibition of TCV and SGV. (D) MEF 10.1 cells were treated with 50 mM sodium chlorate to remove 2-O- and 6-O-linked sulfations. Treated cells were infected with ∼100 PFU of TCV or SGV. Either TCV (E) or SGV (F) was incubated with various concentrations of heparin and used to infect MEF 10.1 cells (∼100 PFU/well). Data were normalized to untreated (PBS) controls. Bars represent the average ± SD from 2 experiments with 3 replicates per experiment. Statistical significance was determined by one-way ANOVA with a Tukey’s multiple comparison of means. ns, not significant; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
To further investigate the differences in SGV and TCV entry identified by the peptide inhibition studies, mouse embryonic fibroblasts (MEFs) were treated with 50 mM sodium chlorate prior to infection to remove 2-O- and 6-O-linked HS sulfations (41). We focused on these sulfation patterns based on observations from HCMV, which indicated that these O-linked sulfations were important for viral attachment (28). This treatment resulted in inhibition of infection of both SGV and TCV, with the latter being significantly more impacted (Fig. 4D). It is known that incubation of MCMV with heparin blocks cellular entry; therefore, we studied the effect of increasing heparin concentration on infection efficiency of TCV (Fig. 4E) and SGV (Fig. 4F). Pretreatment of TCV with heparin resulted in a dose-dependent decrease in infection, with 50% loss of efficiency in the presence of 40 µg/ml heparin (Fig. 4E). In contrast, there was no significant decrease in the infectivity of murine SGV following pretreatment with 40 µg/ml heparin (Fig. 4F).
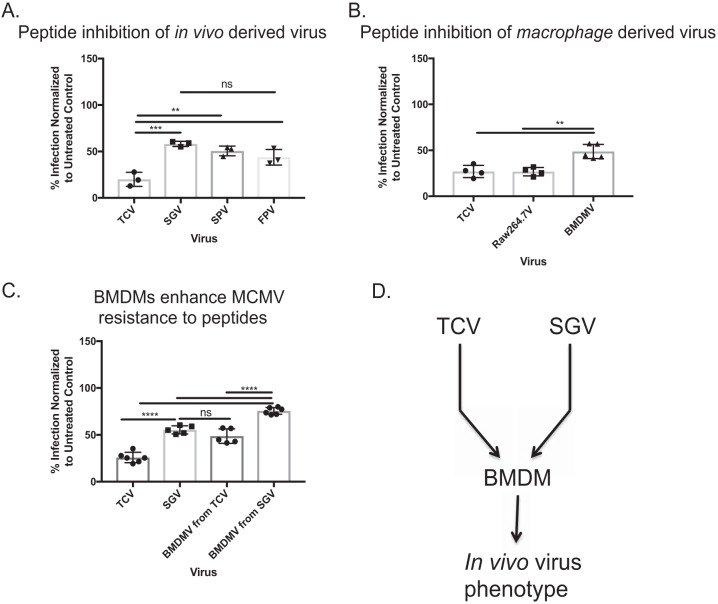
Because viruses derived from different tissues vary in their susceptibility to antibody neutralization (37), we speculated that perhaps not all in vivo-derived MCMVs would be resistant to peptide inhibition. Therefore, we infected mice subcutaneously in the footpad, and SGV was isolated on day 14 postinfection, splenic virus (SPV) at day 5, and the footpad virus (FPV) at day 3. The different time points were necessary to maximize viral load in the given organ. Once MCMV organ titers were determined, whole-organ homogenates were plated at ∼100 PFU on peptide-treated MEF 10.1 cells (Fig. 5A). As expected there was some inhibition of in vivo-grown virus, but not to the same levels as TCV. Interestingly, there was no difference between the different organs in regard to peptide inhibition susceptibility. Because there is no difference between viruses isolated from different organs, the SGV phenotype is not the result of SGV sample preparation.
FIG 5.
Generation of peptide-resistant MCMV in vitro. (A) Virus was harvested from salivary gland (SGV), spleen (SPV), and footpads (FPV) of mice infected with MCMV. MEF 10.1 cells were treated with 50 µM p5 + 14(coil) and then infected with ∼100 PFU of each virus. The percentage of infection inhibition was determined by comparison to untreated controls. (B) TCV was grown on RAW 264.7 macrophages and BMDMs. Progeny virus was then subjected to a plaque reduction assay. The percentage of virus inhibition was determined by comparison to untreated controls. (C) Comparison of peptide inhibition of MEF 10.1 cells infected with TCV, SGV, or BMDM-derived virus (BMDMV) originally from TCV or SGV. Bars represent the average ± SD from 2 experiments with 2 or 3 replicates per experiment. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison of means with a two-tailed t test. ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. (D) Schematic of method used to generate in vivo-like virus in vitro.
We speculated that the differences in susceptibility to the peptides correlated with differences in entry complexes that determine cellular tropism (42). MCMVs grown on macrophages contain different entry complexes compared to MCMVs grown on fibroblasts (42). Therefore, we tested whether MCMVs derived from macrophages in vitro would mimic peptide inhibition of SGV or TCV. In vitro-derived MCMV was grown on the macrophage cell line RAW 264.7 or bone marrow-derived macrophages (BMDMs). The progeny viruses were subjected to a plaque reduction assay (Fig. 5B). Peptide treatment of cells blocked RAW 264.7-grown virus (∼75% inhibition) but only partially blocked BMDM virus (∼50% inhibition). Not only are BMDMs capable of generating “in vivo-like” virus, but when SGV virus was grown on BMDMs, the peptide was even less efficient in blocking MCMV infection (Fig. 5C). Regardless of the initial MCMV input into BMDMs, they produced “in vivo-like” virus (Fig. 5B to D). Additionally, these results indicate that the phenotype observed with SGV and virus isolated from other tissues is not an artifact of sample preparation.
Peptide inhibition of in vivo-derived HCMV.
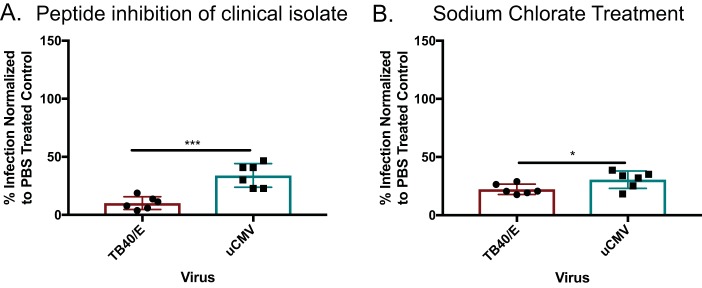
We sought to determine whether p5 + 14(coil) could inhibit infection of an HCMV clinical isolate. This virus was used directly from a patient’s urine (11). This is important because the tropism/entry complex can mutate after a single passage in vitro (43, 44). Peptide p5 + 14(coil) significantly inhibited the entry of a clinical isolates of HCMV into human fibroblasts (∼70% inhibition [Fig. 6A]), but was significantly more effective when tissue culture-grownTB40/E virus was used (∼90% inhibition). Because these results mimicked MCMV’s peptide inhibition, we tested which HS moieties were important for viral entry. MRC-5 fibroblasts were treated with 50 mM sodium chlorate to remove 2-O and 6-O sulfations (Fig. 6B). Despite the small but significant difference between HCMVs from in vitro- or in vivo-grown viruses, removing the 2-O and 6-O sulfations prevented infection of both HCMVs, albeit more inhibition of tissue culture-grown HCMVs than the clinical isolate was observed. These results corroborate the MCMV data. Both viruses highly rely on sulfated surface glycans for entry.
FIG 6.
Peptide and sodium chlorate treatment prevents urine-derived CMV (uCMV) infection. (A) MCR-5 human fibroblasts were treated with p5 + 14(coil) at 50 µM and infected with the TB40/E strain or uCMV. (B) MRC-5 cells were treated with 50 mM sodium chlorate to remove 2-O- and 6-O-linked sulfations. All cells were infected with ∼100 PFU of TB40/E or clinically derived HCMV (uCMV). The percentage of infection was normalized to PBS-treated controls. Bars are the averages of replicates from two experiments ± SD. Statistical significance was determined by unpaired t test. *, P ≤ 0.05; ***, P ≤ 0.001.
Do polybasic peptides prevent MCMV cell-to-cell spread?
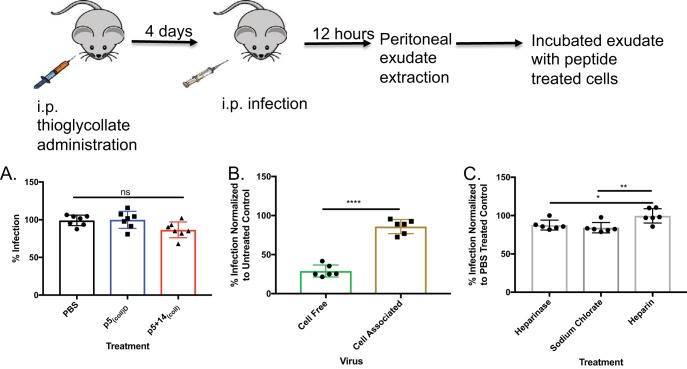
Cell-to-cell spread in vivo is important for viral dissemination within the host (11, 45, 46). We evaluated the effectiveness of p5(coil)D and p5 + 14(coil) on cell-to-cell spread. Infected peritoneal exudate cells (PECs) were harvested from MCMV-infected mice and coincubated with MEF 10.1 cells, treated with 100 μM p5(coil)D, p5 + 14(coil), or PBS as a control. Inhibition of MCMV infection was measured via plaque reduction assay. Peptide treatment has no effect on cell-to-cell spread (Fig. 7A). Because most of the PEC population consists of immune cells (data not shown), we tested our peptide’s ability to inhibit cell-to-cell spread from infected to uninfected MEF 10.1 cells in a plaque reduction assay. As seen with the PECs, peptide treatment did not inhibit cell-to-cell spread regardless of cell type (Fig. 7B). To evaluate whether or not HS mediates cell-to-cell spread, we treated murine fibroblasts with heparinase, sodium chlorate, and heparin. Cell-associated virus spread was marginally affected in the absence of HS, pointing to a different entry mechanism from cell-free MCMV, as heparin treatment was ineffective at inhibiting infection (Fig. 7C). These data highlight the complexity of CMV entry whether the virus enters from cell-to-cell spread or cell-free virus.
FIG 7.
Peptide inhibition of MCMV cell-to-cell spread. (A) MEF 10.1 cells were treated with PBS, p5(coil), or p5 + 14(coil) at 100 μM 1 h prior to incubation with 1 × 105 MCMV-infected peritoneal exudate cells. (B) MEF10.1 cells were treated with p5 + 14(coil) at 50 μM 1 h prior to incubation with either cell-free MCMV or 300 MEF 10.1 cells infected with MCMV. (C) MEF 10.1 cells were treated with 12 U/μl of heparinase for 1 h, 50 mM sodium chlorate overnight, or 40 µg/ml of heparin and then infected with cell-associated MCMV. Bars represent the average from 2 experiments with 3 or 4 replicates per experiment. Statistical significance was determined by an ordinary one-way ANOVA with a Tukey’s multiple comparison of means or an unpaired t test. ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001.
DISCUSSION
Decades of investment and innovation have failed to generate a protective HCMV vaccine (10, 11). Without a vaccine, treatment of HCMV relies on drugs that target two different stages of HCMV replication. First, nucleoside analogues (e.g., GCV, foscarnet, and cidofovir) have been the standard treatment for HCMV disease, but these compounds are myelosuppressive and nephrotoxic. The recently approved letermovir (trade name Prevymis), which inhibits the terminase complex, is not myelosuppressive or nephrotoxic (16). All of these agents potentially select for resistant viruses (15, 47). Therefore, a multifaceted approach may be required to inhibit additional stages during HCMV replication, and this could include APs that block viral entry. Other anti-CMV APs inhibit viral entry of in vitro-derived CMV, but none have been shown to inhibit a clinical isolate of HCMV (27, 28, 33, 48). We demonstrated that our p5(coil) peptides are capable of inhibiting infection of both HCMV and MCMV and that these peptides can partially inhibit infection of in vivo-derived CMVs. This is the first report of an effective inhibition of HCMV isolated directly from urine. Clinical isolates have previously been shown to be resistant to anti-HCMV antibody neutralization (49). Although inhibition of in vivo-derived virus was not as efficient as the blockade of tissue culture-derived virus, greater than 50% reduction in infectivity was achieved, which is comparable to that observed following sodium chlorate treatment of cells. These results support previous observations and point to the important role of 2-O- or 6-O-sulfated cell surface glycans in viral entry via HS moieties (28).
Our data indicate that the p5(coil) peptides, which presumably inhibit the interactions of viral attachment proteins with HS, were unable to inhibit cell-to-cell spread. Also, heparin treatment of infected cells did not inhibit cell-to-cell spread. These data provide insights into why polyclonal antibodies generated in response to the gB vaccine are of limited efficacy (50) (i.e., the mechanism of HCMV cell-to-cell spread may be independent of HS interactions involving the viral gB protein). The studies of peptide-mediated inhibition of viral attachment and infection have elucidated several important aspects of CMV infection pathways. First, these peptides, which bind preferentially to sulfated HS, effectively inhibited TCV entry into cultured cells. There was one HS exception, HS008, to which the peptide did not bind. This glycan consisted of GlcA-GlcNS-ClcA-GlcNS-GlcA, while HS007 is one sugar moiety shorter but binds to the peptide very well. The full set of features that are important for viral entry, (i.e., length, number of repeating sugar units, etc.) and whether it depends on in vivo- or cell culture-grown virus remain to be determined. Interestingly, another AP, p5 + 14, also did not bind to HS008 (data not shown) (27). Could the lack of binding to HS008 be the reason that the peptides cannot block 100% of the infection? Without knowing the composition of the HS on the cell surface, we can only speculate the lack of complete inhibition of infectivity (i.e., 5% remaining infectivity for HCMV and 30% for MCMV). Entry differences could be due to differences in entry complexes, glycosylations of the virion proteins, or other factors during replication in vivo. Because we have demonstrated that virus grown on BMDMs recapitulates the “in vivo” virus phenotype, this will allow an in-depth investigation of these possibilities. Our data demonstrate the significant biochemical differences between in vivo-grown virus and virus cultured in vitro. Previously, differences between in vivo-derived and tissue culture-grown MCMVs were defined as “virulent” for in vivo-grown virus or “attenuated” when grown in vitro. These differences were not due to selection of mutated viruses, but rather differential usage of sugar moieties for entry (36, 39, 40), which could explain why one may be more virulent than the other. Interestingly, it appears that all in vivo-derived viruses, regardless of the tissue from which they were isolated, utilize discrete surface HS compared to tissue culture-derived virus. These differences in HS usage may explain the lack of peptide-mediated inhibition of in vivo-grown MCMV. Furthermore, it is noteworthy that cell attachment by in vivo-grown virus did not depend on 2-O or 6-O glycans, as indicated by the differential inhibition following sodium chlorate treatment of the cells (Fig. 6).
Perhaps most notably, we have shown that the HS-binding peptides do not inhibit cell-to-cell spread of CMV, which represents the main mechanism of cellular transmission in vivo (45, 51 – 53). Our data indicate that cell-to-cell spread does not require HS (i.e., heparinase and sodium chlorate treatment of uninfected cells prior to being cocultured with infected fibroblasts only achieved approximately 15% inhibition). These data once again indicate that our current understanding of CMV entry remains incomplete. Further understanding of the mechanism of cell-to-cell spread may provide a path to improve treatment strategies for HCMV.
MATERIALS AND METHODS
Cells and viruses.
All experiments were performed with low-passage cells (<20 passages). MRC-5 human lung fibroblasts were cultured in modified Eagle’s medium (MEM; Lonza, Rockland, ME) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA), 1% penicillin–streptomycin, and 1% l-glutamine. Cells of the mouse embryonic fibroblast (MEF) line 10.1 (54) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Lonza, Rockland, ME) supplemented with 10% Fetalclone III serum (HyClone, Logan, UT), 1% penicillin–streptomycin, and 1% l-glutamine. RAW 264.7 cells were cultured in DMEM (Lonza, Rockland, ME) with 10% Fetalclone III (HyClone, Logan, UT), 1% penicillin–streptomycin, and 1% l-glutamine. Primary bone marrow-derived macrophages (BMDMs) were cultured in RPMI 1640 (Lonza, Rockland, ME) supplemented with 10% Fetalclone III (HyClone, Logan, UT), 1% penicillin–streptomycin, and 1% l-glutamine.
The HCMV TB40/E and MCMV K181 (55) were used in this study. HCMV TB40/E expressing luciferase under the control of the UL18 promoter was a gift from Christine O’Connor and Eain Murphy (University of Buffalo and Forge Life Science, LLC). TB40/E viruses were propagated on HUVECs. MCMV was produced in vitro using MEF 10.1 cells. All viruses were stored at −80°C until use. Viral titer was assessed by plaque assay (described below) on MEF 10.1 cells (MCMV) or MRC-5 cells (HCMV). The HCMV clinical isolates were collected at Johns Hopkins University without any identifiers that can link them to a specific patient. To generate BMDM virus, differentiated BMDMs were infected with K181 and virus was harvested after a 100% cytopathic effect (CPE) was observed. Salivary gland-derived virus was harvested from mouse salivary glands at 14 dpi with K181 via the intraperitoneal (i.p.) route. Spleen-derived virus was obtained by harvesting the organ 5 dpi after i.p. infection, while footpad-derived virus was obtained by harvesting footpads 3 dpi following footpad inoculation with K181. All organs were homogenized, and titers were determined via plaque assay.
BMDM differentiation.
Bone marrow from naive mice was harvested and incubated in RPMI 1640 for 4 h. The medium was then changed and supplemented with macrophage colony-stimulating factor (M-CSF; PeproTech, Rocky Hill, NJ) at 1 ng/μl for 7 days. The medium was changed every 3 days. Differentiated cells were infected with K181 at a multiplicity of infection (MOI) of ∼0.1. Virus was harvested 1 day after cells showed 100% CPE.
Treatment of cells and viruses.
Cells were washed with PBS prior to addition of treatment. Heparinase I (NEB, Ipswich, MA) was used at 12 U/μl in medium. Cells were pretreated for 1 h at 37°C before addition of virus or cells. MEF 10.1 or MRC-5 cells were treated for 1 h with 50 mM sodium chlorate to remove 2-O- and 6-O-linked HS as previously described (41). Heparin at various concentrations was preincubated with virus or infected fibroblasts for 30 min at 4°C. Following pretreatment, viral infectivity was assayed by plaque assay or cell-to-cell transfer assay as described below.
Peptides.
Peptides were purchased and purified as previously described (27). Briefly, peptides were purified by high-performance liquid chromatography (HPLC), and purity was confirmed by mass spectrometry (MS). Purified peptides were lyophilized and resuspended in PBS prior to use.
Plaque reduction assay.
Lyophilized peptides were resuspended in PBS and stored at 4°C until use. Fibroblasts were seeded into 24-well dishes. After cells reached ∼80% confluence, medium was removed and cells were washed with PBS. Peptide in PBS plus 10% FBS or PBS plus 10% FBS alone as the control was added, and the mixture was incubated for 30 min at 37°C. Virus was added to treated and untreated control wells (∼100 PFU/well) and incubated for an additional hour. Following virus incubation, medium was removed and the overlay was added. Overlays consisted of 0.75% carboxymethyl cellulose (CMC; Sigma-Aldrich, St. Louis, MO) for MCMV-infected wells and 0.5% SeaKem agarose (Lonza, Rockland, ME) in complete medium for HCMV-infected wells. For MCMV assays, plates were incubated for 5 days, at which point they were stained with Coomassie stain and plaques counted using a dissecting microscope. Reduction in viral infectivity was expressed as a percentage of infectivity of PBS-treated wells. Data were analyzed using Prism 7 (GraphPad Software, La Jolla, CA).
Cell-to-cell transfer of virus.
To examine fibroblast-mediated cell-to-cell spread, MEF 10.1 cells were infected with K181 at an MOI of 3. Following 16 h of incubation, cells were trypsinized, washed, and counted. Approximately 300 potentially infected cells were added to an uninfected 100% confluent monolayer of MEF 10.1 cells. The uninfected monolayer was pretreated with peptide or PBS. Infected cells were given an hour to transfer infection, medium was removed, and a CMC overlay was added. In order to evaluate immune cell-mediated cell-to-cell spread, mice were injected i.p. with 3% thioglycollate (56). After 4 days, animals were infected i.p. with MCMV K181 at 1 × 106 PFU, and peritoneal exudate cells (PECs) were collected 12 h postinfection. PECs were added to an uninfected monolayer either treated with p5 + 14(coil) or untreated. PECs were incubated for an hour, medium was removed, and a CMC overlay was added. Analysis was performed as described for the plaque reduction assay.
HCMV infectivity assay.
HCMV TB40/E infectivity was assessed using a luciferase reporter assay as previously described (33). Briefly, MRC-5 fibroblasts were seeded into a 24-well dish. After reaching ∼80% confluence, cells were washed once with PBS and peptide with PBS plus 10% FBS or the PBS-plus-10% FBS control was incubated at 37°C for 30 min. Virus was added at ∼1,000 relative light units (RLU) and incubated at 37°C for 1 h. Cells were washed, medium was replaced, and then the mixture was incubated at 37°C for 3 days. On day 3, cells were washed with PBS and lysed using passive lysis buffer (Promega, Madison, WI), and the cell lysates were pelleted. Luciferase reagent (Gaussian luciferase) was combined 1:1 with cell lysate in a clear-bottom 96-well plate. Luminescence was measured using a Synergy 2 plate reader (BioTek, Winooski, VT) and recorded as RLU. The RLU from untreated uninfected samples was subtracted as background, and results were normalized to untreated infected wells to 100% infection. Data were analyzed using Prism 7 (GraphPad Software, La Jolla, CA). The data were expressed as the percentage of luminescence of the untreated infected well.
Statistical analysis and IC50 determination.
Each experiment represents two or more independent experiments with at least three replicates per experimental group, unless otherwise indicated. Individual data points are shown for all graphs. Error bars represent the standard deviation (SD) from each data set. Statistical significance was determined by one-tailed Student's t test or one-way analysis of variance (ANOVA) with Tukey’s multivariance analysis when appropriate. IC50 values were calculated using a linear regression sigmoidal dose-dependent test. All statistical analysis was performed using Prism 7 (GraphPad Software, La Jolla, CA). Statistical significance was assigned to P values of <0.05. Significant values are labeled as follows: ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; and ****, P ≤ 0.0001.
Glycan array.
The glycan array was performed by Z-biotech, LLC. Briefly, biotinylated peptide was incubated with various heparan sulfate derivatives. Following incubation, an antibiotin, fluorescently labeled antibody was added, and a plate reader determined fluorescence. The data represent 6 technical replicates per HS residue.
Mice.
Animal use was approved under The University of Tennessee, Knoxville, IACUC protocol. The animals used were housed and bred at the Walters Life Science Laboratory Animal Facility at The University of Tennessee. BALB/c mice (6 to 12 weeks) were purchased from Jackson Laboratory (Bar Harbor, ME) and housed under specific-pathogen-free conditions.
In vivo analysis of peptide efficacy.
Mice were treated with intravenously via the retro-orbital route with the indicated peptide at 250 μg/mouse 1 h prior to infection. One hour posttreatment, animals were infected i.p. with 1 × 106 PFU of K181. Four days postinfection, animals were euthanized and spleens were harvested. Viral burden was determined by plaque assay as described above.
Peptide biodistribution.
To determine the distribution of our APs in vivo, mice were injected intravenously (i.v.) in the lateral tail vein with 125I-labeled peptides (<120 μCi, 20 μg of peptide). At 1 and 4 h postinjection, mice were euthanized with an isoflurane inhalation overdose, spleen, liver, lung, and nine other tissues were harvested, and the tissue radioactivity was measured as previously described (30, 33). The biodistribution of radiolabeled peptide was expressed as a percentage of the injected dose per gram of tissue.
ACKNOWLEDGMENTS
This work was supported by PHS grant R01DK079984 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) to J.S.W. The UT CMV Research Fund and the UTHSC Pot of Gold Fund provided additional funding for this work.
REFERENCES
- 1.Britt W. 2007. Virus entry into host, establishment of infection, spread in host, mechanisms of tissue damage In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 2.Liu L. 2014. Fields Virology, 6th ed. Clin Infect Dis 59:613. doi: 10.1093/cid/ciu346. [DOI] [Google Scholar]
- 3.Sinclair J, Sissons P. 2006. Latency and reactivation of human cytomegalovirus. J Gen Virol 87:1763–1779. doi: 10.1099/vir.0.81891-0. [DOI] [PubMed] [Google Scholar]
- 4.Staras SAS, Dollard SC, Radford KW, Flanders WD, Pass RF, Cannon MJ. 2006. Seroprevalence of cytomegalovirus infection in the United States, 1988–1994. Clin Infect Dis 43:1143–1151. doi: 10.1086/508173. [DOI] [PubMed] [Google Scholar]
- 5.Manicklal S, Emery VC, Lazzarotto T, Boppana SB, Gupta RK. 2013. The “silent” global burden of congenital cytomegalovirus. Clin Microbiol Rev 26:86–102. doi: 10.1128/CMR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demmler GJ. 1994. Congenital cytomegalovirus infection. Semin Pediatr Neurol 1:36–42. [PubMed] [Google Scholar]
- 7.Schleiss MR. 2013. Cytomegalovirus in the neonate: immune correlates of infection and protection. Clin Dev Immunol 2013:501801. doi: 10.1155/2013/501801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stagno S, Reynolds DW, Huang ES, Thames SD, Smith RJ, Alford CA. 1977. Congenital cytomegalovirus infection. Occurrence in an immune population. N Engl J Med 296:1254–1258. doi: 10.1056/NEJM197706022962203. [DOI] [PubMed] [Google Scholar]
- 9.Nassetta L, Kimberlin D, Whitley R. 2009. Treatment of congenital cytomegalovirus infection: implications for future therapeutic strategies. J Antimicrob Chemother 63:862–867. doi: 10.1093/jac/dkp083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths P, Plotkin S, Mocarski E, Pass R, Schleiss M, Krause P, Bialek S. 2013. Desirability and feasibility of a vaccine against cytomegalovirus. Vaccine 31(Suppl 2):B197–B203. doi: 10.1016/j.vaccine.2012.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui XH, Adler SP, Schleiss MR, Arav-Boger R, Harrison GJD, Mcvoy MA. 2017. Cytomegalovirus virions shed in urine have a reversible block to epithelial cell entry and are highly resistant to antibody neutralization. Clin Vaccine Immunol 24:e00024-17. doi: 10.1128/CVI.00024-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramanan P, Razonable RR. 2013. Cytomegalovirus infections in solid organ transplantation: a review. Infect Chemother 45:260–271. doi: 10.3947/ic.2013.45.3.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biron KK. 2006. Antiviral drugs for cytomegalovirus diseases. Antiviral Res 71:154–163. doi: 10.1016/j.antiviral.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Lurain NS, Chou S. 2010. Antiviral drug resistance of human cytomegalovirus. Clin Microbiol Rev 23:689–712. doi: 10.1128/CMR.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limaye AP, Corey L, Koelle DM, Davis CL, Boeckh M. 2000. Emergence of ganciclovir-resistant cytomegalovirus disease among recipients of solid-organ transplants. Lancet 356:645–649. doi: 10.1016/S0140-6736(00)02607-6. [DOI] [PubMed] [Google Scholar]
- 16.Marty FM, Ljungman P, Chemaly RF, Maertens J, Dadwal SS, Duarte RF, Haider S, Ullmann AJ, Katayama Y, Brown J, Mullane KM, Boeckh M, Blumberg EA, Einsele H, Snydman DR, Kanda Y, DiNubile MJ, Teal VL, Wan H, Murata Y, Kartsonis NA, Leavitt RY, Badshah C. 2017. Letermovir prophylaxis for cytomegalovirus in hematopoietic-cell transplantation. N Engl J Med 377:2433–2444. doi: 10.1056/NEJMoa1706640. [DOI] [PubMed] [Google Scholar]
- 17.Zhao H, Zhou J, Zhang K, Chu H, Liu D, Poon VK, Chan CC, Leung HC, Fai N, Lin YP, Zhang AJ, Jin DY, Yuen KY, Zheng BJ. 2016. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci Rep 6:22008. doi: 10.1038/srep22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scala MC, Sala M, Pietrantoni A, Spensiero A, Di Micco S, Agamennone M, Bertamino A, Novellino E, Bifulco G, Gomez-Monterrey IM, Superti F, Campiglia P. 2017. Lactoferrin-derived peptides active towards influenza: identification of three potent tetrapeptide inhibitors. Sci Rep 7:10593. doi: 10.1038/s41598-017-10492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chupradit K, Moonmuang S, Nangola S, Kitidee K, Yasamut U, Mougel M, Tayapiwatana C. 2017. Current peptide and protein candidates challenging HIV therapy beyond the vaccine era. Viruses 9:281. doi: 10.3390/v9100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulder KC, Lima LA, Miranda VJ, Dias SC, Franco OL. 2013. Current scenario of peptide-based drugs: the key roles of cationic antitumor and antiviral peptides. Front Microbiol 4:321. doi: 10.3389/fmicb.2013.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Compton T, Nowlin DM, Cooper NR. 1993. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology 193:834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- 22.Esko JD, Lindahl U. 2001. Molecular diversity of heparan sulfate. J Clin Invest 108:169–173. doi: 10.1172/JCI200113530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarrazin S, Lamanna WC, Esko JD. 2011. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol 3:a004952. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shieh MT, WuDunn D, Montgomery RI, Esko JD, Spear PG. 1992. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol 116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla D, Spear PG. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest 108:503–510. doi: 10.1172/JCI200113799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wadstrom T, Ljungh A. 1999. Glycosaminoglycan-binding microbial proteins in tissue adhesion and invasion: key events in microbial pathogenicity. J Med Microbiol 48:223–233. doi: 10.1099/00222615-48-3-223. [DOI] [PubMed] [Google Scholar]
- 27.Dogra P, Martin EB, Williams A, Richardson RL, Foster JS, Hackenback N, Kennel SJ, Sparer TE, Wall JS. 2015. Novel heparan sulfate-binding peptides for blocking herpesvirus entry. PLoS One 10:e0126239. doi: 10.1371/journal.pone.0126239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borst EM, Standker L, Wagner K, Schulz TF, Forssmann WG, Messerle M. 2013. A peptide inhibitor of cytomegalovirus infection from human hemofiltrate. Antimicrob Agents Chemother 57:4751–4760. doi: 10.1128/AAC.00854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin EB, Williams A, Heidel E, Macy S, Kennel SJ, Wall JS. 2013. Peptide p5 binds both heparinase-sensitive glycosaminoglycans and fibrils in patient-derived AL amyloid extracts. Biochem Biophys Res Commun 436:85–89. doi: 10.1016/j.bbrc.2013.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wall JS, Richey T, Stuckey A, Donnell R, Macy S, Martin EB, Williams A, Higuchi K, Kennel SJ. 2011. In vivo molecular imaging of peripheral amyloidosis using heparin-binding peptides. Proc Natl Acad Sci U S A 108:E586–E594. doi: 10.1073/pnas.1103247108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wall JS, Williams A, Wooliver C, Martin EB, Cheng X, Heidel RE, Kennel SJ. 2016. Secondary structure propensity and chirality of the amyloidophilic peptide p5 and its analogues impacts ligand binding—in vitro characterization. Biochem Biophys Rep 8:89–99. doi: 10.1016/j.bbrep.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wall JS, Kennel SJ, Martin EB. 2017. Dual-energy SPECT and the development of peptide p5 + 14 for imaging amyloidosis. Mol Imaging 16:1536012117708705. doi: 10.1177/1536012117708705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pitt EA, Dogra P, Patel RS, Williams A, Wall JS, Sparer TE. 2016. The D-form of a novel heparan binding peptide decreases cytomegalovirus infection in vivo and in vitro. Antiviral Res 135:15–23. doi: 10.1016/j.antiviral.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravindranath RMH, Graves MC. 1990. Attenuated murine cytomegalovirus binds to N-acetylglucosamine, and shift to virulence may involve recognition of sialic acids. J Virol 64:5430–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chong KT, Gould JJ, Mims CA. 1981. Neutralization of different strains of murine cytomegalo-virus (Mcmv)—effect of in vitro passage. Arch Virol 69:95–104. doi: 10.1007/BF01315153. [DOI] [PubMed] [Google Scholar]
- 38.Chong KT, Mims CA. 1981. Murine cytomegalovirus particle types in relation to sources of virus and pathogenicity. J Gen Virol 57:415–419. doi: 10.1099/0022-1317-57-2-415. [DOI] [PubMed] [Google Scholar]
- 39.Jordan MC, Takagi JL. 1983. Virulence characteristics of murine cytomegalovirus in cell and organ cultures. Infect Immun 41:841–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborn JE, Walker DL. 1971. Virulence and attenuation of murine cytomegalovirus. Infect Immun 3:228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Safaiyan F, Kolset SO, Prydz K, Gottfridsson E, Lindahl U, Salmivirta M. 1999. Selective effects of sodium chlorate treatment on the sulfation of heparan sulfate. J Biol Chem 274:36267–36273. doi: 10.1074/jbc.274.51.36267. [DOI] [PubMed] [Google Scholar]
- 42.Wagner FM, Brizic I, Prager A, Trsan T, Arapovic M, Lemmermann NAW, Podlech J, Reddehase MJ, Lemnitzer F, Bosse JB, Gimpfl M, Marcinowski L, MacDonald M, Adler H, Koszinowski UH, Adler B. 2013. The viral chemokine MCK-2 of murine cytomegalovirus promotes infection as part of a gH/gL/MCK-2 complex. PLoS Pathog 9:e1003493. doi: 10.1371/journal.ppat.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dargan DJ, Douglas E, Cunningham C, Jamieson F, Stanton RJ, Baluchova K, McSharry BP, Tomasec P, Emery VC, Percivalle E, Sarasini A, Gerna G, Wilkinson GW, Davison AJ. 2010. Sequential mutations associated with adaptation of human cytomegalovirus to growth in cell culture. J Gen Virol 91:1535–1546. doi: 10.1099/vir.0.018994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinzger C, Schmidt K, Knapp J, Kahl M, Beck R, Waldman J, Hebart H, Einsele H, Jahn G. 1999. Modification of human cytomegalovirus tropism through propagation in vitro is associated with changes in the viral genome. J Gen Virol 80:2867–2877. doi: 10.1099/0022-1317-80-11-2867. [DOI] [PubMed] [Google Scholar]
- 45.Silva MC, Schroer J, Shenk T. 2005. Human cytomegalovirus cell-to-cell spread in the absence of an essential assembly protein. Proc Natl Acad Sci U S A 102:2081–2086. doi: 10.1073/pnas.0409597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinzger C, Knapp J, Plachter B, Schmidt K, Jahn G. 1997. Quantification of replication of clinical cytomegalovirus isolates in cultured endothelial cells and fibroblasts by a focus expansion assay. J Virol Methods 63:103–112. doi: 10.1016/S0166-0934(97)02082-X. [DOI] [PubMed] [Google Scholar]
- 47.Li F, Kenyon KW, Kirby KA, Fishbein DP, Boeckh M, Limaye AP. 2007. Incidence and clinical features of ganciclovir-resistant cytomegalovirus disease in heart transplant recipients. Clin Infect Dis 45:439–447. doi: 10.1086/519941. [DOI] [PubMed] [Google Scholar]
- 48.Baldwin J, Maus E, Zanotti B, Volin MV, Tandon R, Shukla D, Tiwari V. 2015. A role for 3-O-sulfated heparan sulfate in promoting human cytomegalovirus infection in human iris cells. J Virol 89:5185–5192. doi: 10.1128/JVI.00109-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui XH, Freed DC, Wang D, Qiu P, Li FS, Fu TM, Kauvar LM, McVoy MA. 2017. Impact of Antibodies and strain polymorphisms on cytomegalovirus entry and spread in fibroblasts and epithelial cells. J Virol 91:e01650-16. doi: 10.1128/JVI.01650-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schleiss MR, Permar SR, Plotkin SA. 2017. Progress toward development of a vaccine against congenital cytomegalovirus infection. Clin Vaccine Immunol 24:e00268-17. doi: 10.1128/CVI.00268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eisenfeld L, Silver H, McLaughlin J, Klevjer-Anderson P, Mayo D, Anderson J, Herson V, Krause P, Savidakis J, Lazar A, Rosenkrantz T, Pisciotto P. 1992. Prevention of transfusion-associated cytomegalovirus-infection in neonatal patients by the removal of white cells from blood. Transfusion 32:205–209. doi: 10.1046/j.1537-2995.1992.32392213801.x. [DOI] [PubMed] [Google Scholar]
- 52.Gilbert GL, Hayes K, Hudson IL, James J. 1989. Prevention of transfusion-acquired cytomegalovirus infection in infants by blood filtration to remove leucocytes. Neonatal Cytomegalovirus Infection Study Group. Lancet i:1228–1231. doi: 10.1016/S0140-6736(89)92330-1. [DOI] [PubMed] [Google Scholar]
- 53.Jackson JW, Sparer T. 2018. There is always another way! Cytomegalovirus’ multifaceted dissemination schemes. Viruses 10:E383. doi: 10.3390/v10070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harvey DM, Levine AJ. 1991. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev 5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- 55.Hudson JB, Walker DG, Altamirano M. 1988. Analysis in vitro of two biologically distinct strains of murine cytomegalovirus. Arch Virol 102:289–295. doi: 10.1007/BF01310834. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, Goncalves R, Mosser DM. 2008. The isolation and characterization of murine macrophages. Curr Protoc Immunol Chapter 14:Unit 14.11. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical analysis of the glycan array. Statistical significance was determined by an ordinary one-way ANOVA with Tukey’s multiple comparison of means. ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. Analysis was carried out using GraphPad Prism. Download Table S1, DOCX file, 0.2 MB (214.3KB, docx) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
p5(coil)D or p5 + 14(coil) biodistribution in BALB/c mice. Mice were injected with 250 μg/mouse of 125I-labeled peptide. Mice were euthanized at 1 and 4 h postinjection (hpi), and the indicated organs were harvested. The amount of 125I present in each organ was measured. Data represent the percentage of injected dose (ID)/g of tissue. Download Table S2, DOCX file, 0.3 MB (263.3KB, docx) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.