Abstract
The clinical efficiency of everolimus, an mammalian target of rapamycin (mTOR) inhibitor, is palliative as sequential or second-line therapy for renal cell carcinoma (RCC). However, the limited response of everolimus in RCC remains uncertain. In the present study, everolimus-resistant RCC models were established to understand the mechanisms and to seek combination approaches. Consequently, the activation of ERK was found to contribute toward everolimus-acquired resistance and poor prognosis in patients with RCC. In addition, the efficacy and mechanism of combination treatment underlying RCC using everolimus and ERK inhibitors was investigated. The ERK inhibitor in combination with everolimus synergistically inhibited the proliferation of RCC cells by arresting the cell cycle in the G1 phase. The combination treatment markedly attenuated the deoxyribonucleoside triphosphate (dNTP) pools by downregulating the mRNA expression of RRM1 and RRM2 through E2F1. The overexpression of E2F1 or supplementation of dNTP rescued the anti-proliferation activity of the everolimus-SCH772984 combination. The antitumor efficacy of combination therapy was reiterated in RCC xenograft models. Thus, the current findings provided evidence that the everolimus-ERK inhibitor combination is a preclinical therapeutic strategy for RCC.
Keywords: renal cell carcinoma, ERK inhibitor, everolimus, dNTP, ribonucleotide reductase
Introduction
Renal cell carcinoma (RCC) is the second leading urogenital malignancy in China, accounting for 2%–3% of all adult cancers. Although an increasing number of patients are detected early with the development of advanced diagnostic technology, approximately 30% of the patients are diagnosed with metastatic RCC (mRCC) or subsequently developed tumor recurrence after surgery.1 Since mRCC is typically unresponsive to chemotherapy and radiotherapy, immunotherapy consisting of interleukin-2 (IL-2) and interferon-alpha (IFN-α) served as an adjuvant treatment in the 1990s and 2000s, despite low response rates (5%–20%).2 In recent years, the advent of targeted therapies that inhibit the vascular endothelial growth factor (VEGF) and mammalian target of rapamycin (mTOR) pathways led to a considerable improvement in progression-free survival (PFS) and overall survival (OS) of mRCC patients.
However, monotherapy rarely brings in durable disease control, as the benefits of these agents may be undermined by their toxicity and suboptimal tolerability as well as resistance to these agents. Furthermore, because carcinogenesis and the progression of cancer are a biologically complex system, the inhibition of a single target might not lead to thorough disease control. Based on the results from a phase 3 clinical trial, everolimus was only marginally effective in improving the PFS as compared to the placebo (median, 4.9 versus 1.9 months).3 Multiple clinical trials investigating the efficacy and safety of everolimus and the currently registered drugs (i.e., imatinib, sorafenib, sunitinib, dovitinib, and bevacizumab) for renal cancer combination treatment have shown several adverse effects and limited efficacy.4 Until 2016, the first successful combination therapy of everolimus and lenvatinib in patients with advanced or metastatic RCC was approved by the FDA,5, 6 thereby rendering it as the potential treatment for mRCC.
In mammals, ribonucleotide reductase (RNR) is a unique enzyme that catalyzes the rate-limiting step of de novo synthesis of deoxyribonucleoside triphosphates (dNTPs).7, 8 Mammalian RNR consists of two homodimer subunits: the large catalytic dimer RRM1 and the small regulatory dimer RRM2 or RRM2B.9 Another study demonstrated that MEK1/2 inhibitor pimasertib enhanced the efficacy of gemcitabine in pancreatic cancer by MDM2-mediated polyubiquitination of RRM1 via proteasomal degradation.10 Nonetheless, tumors could exert resistance to BRAF-MEK inhibitors through a strong reactivation of ERK protein caused by several different mechanisms.11, 12, 13 As a result, a novel, selective, and ATP-competitive ERK1/2 inhibitor, SCH772984, has been developed. Despite negative feedback activation up to and including phospho-ERK, SCH772984 has a long-lasting ability to inhibit the catalytic activity of ERK and block the signal transduction between ERK and RSK.14 Targeting ERK is more effective than targeting MEK,15 because ERK inhibitors can effectively overcome the resistance of tumor cells to MEK inhibitors.16 A previous study found that biopsy-accessible breast tumor samples from patients treated with everolimus displayed an increased activation of the mitogen-activated protein kinase (MAPK) pathway in an S6K-PI3K-Ras feedback loop-dependent manner.17 We aimed to investigate whether ERK activation contributes to the everolimus resistance in RCC and whether the inhibition of ERK signal extends the efficacy of everolimus, and we aimed to highlight the molecular mechanisms underlying the synergistic efficacy of everolimus-ERK inhibitor combination for the treatment of RCC.
Results
Activation of ERK Signal Contributes to Everolimus Resistance and Poor Prognosis for RCC
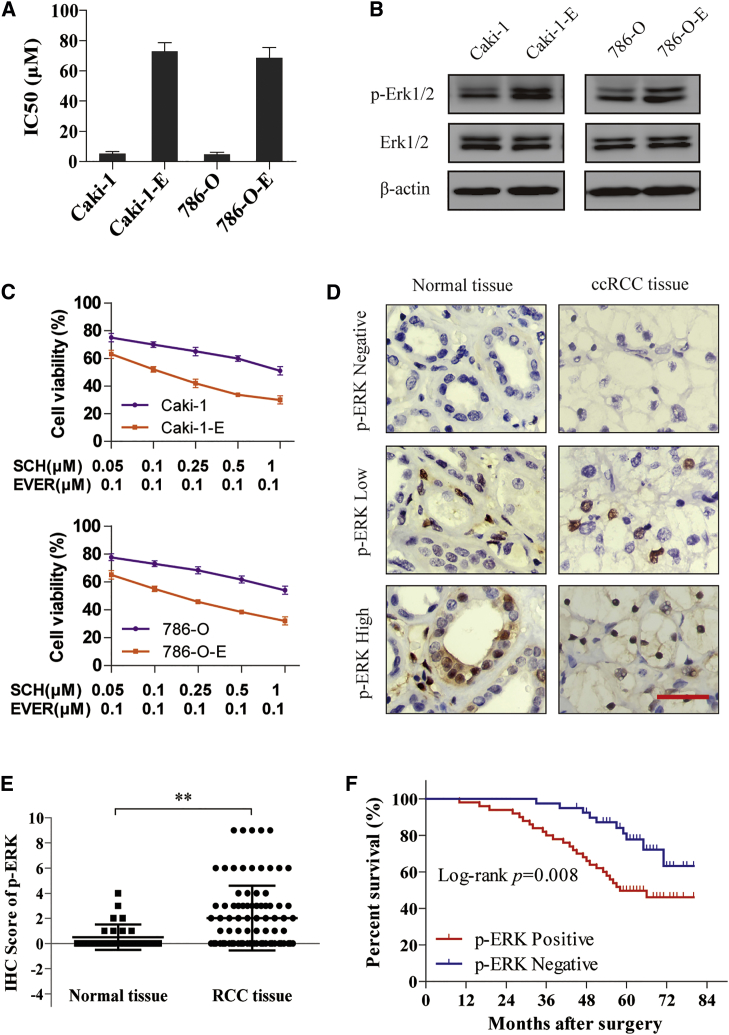
To understand whether ERK activation contributes to everolimus resistance in RCC, we treated the RCC cell lines with chronic (>3 months) everolimus therapy to establish the everolimus-resistant cell lines (Caki-1-E and 786-O-E). Compared to parental cells, Caki-1-E and 786-O-E cells exhibited poor response to everolimus, as demonstrated by an increased 50% inhibitory concentration (IC50) (Figure 1A) and constitutively activated ERK signal (Figure 1B). Moreover, the everolimus-resistant cells were more sensitive to the combination of everolimus and SCH772984 compared with parental cells (Figure 1C), which substantiated the hypothesis that ERK activation is the underlying mechanism.
Figure 1.
Activation of ERK Signal Contributes to Everolimus Resistance and Poor Prognosis of RCC
(A) Caki-1 and 786-O cells were treated with chronic (>3 months) everolimus therapy to establish everolimus-resistant cell line models (Caki-1-E and 786-O-E). IC50 of everolimus in parental and resistant cells was detected. (B) Immunoblotting was performed with the parental and resistant cell lysates for phospho-ERK expression. (C) Cells were treated with ERK inhibitor SCH772984 and everolimus for 72 h. Cell viability was assessed by CCK-8 assay. (D) IHC staining was performed in 90 cases of ccRCC tissues and 30 cases of normal tissues. The representative images of phospho-ERK expression localized in the nucleus are shown (×400). Scale bar, 50 μm. (E) IHC score of phospho-ERK in the above samples. **p < 0.01. (F) Survival analysis of RCC patients related to phospho-ERK expression was analyzed by Kaplan-Meier survival curves. Error bars represent mean ± SD of three independent experiments.
Next, we explored the clinical role of ERK signal in patients by immunohistochemistry (IHC) staining in 90 cases of clear-cell RCC (ccRCC) tissues and 30 cases of normal tissues from patients who underwent surgery. As shown in Figure 1D, the relative expression of phospho-ERK was scored by the product of the intensity and percentage of staining. In comparison with the normal tissues, the expression of phospho-ERK in ccRCC tissues was significantly higher (p < 0.01; Figure 1E). Then the tumor tissues were divided into two groups based on the expression of phospho-ERK: positive expression (n = 50) and negative expression (n = 40). The prognostic implication of ERK signal was subsequently analyzed by Kaplan-Meier survival curves, indicating that patients with activated ERK signal showed a significantly poor OS (p < 0.01; Figure 1F).
Combination of ERK Inhibitor and Everolimus Synergistically Reduces the Viability of RCC Cells
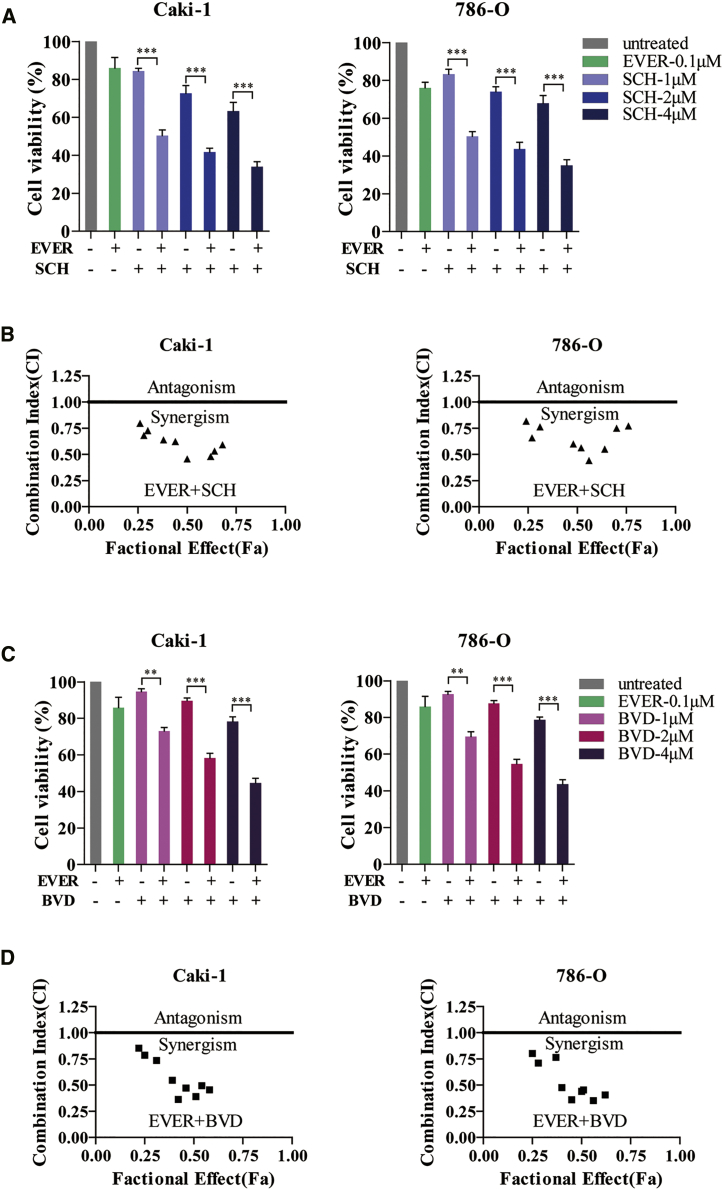
In a previous study, we demonstrated that 0.1 μM everolimus blocks the signal transduction from mTOR to P70S6K in a dose-independent manner.18 Subsequently, we used an everolimus dose at 0.1 μM, and in combination with 1, 2, and 4 μM SCH772984 or BVD-523, respectively, it was used to evaluate the synergistic effect on human RCC cell lines Caki-1 and 786-O. Overall, the proliferation of Caki-1 or 786-O cells was significantly suppressed when everolimus was combined with SCH772984 as compared to the monotherapy (Figure 2A); a similar effect was observed using another ERK inhibitor, BVD-523 (Figure 2C). Furthermore, the isobologram analysis demonstrated that the combination of everolimus with ERK inhibitor (whether SCH772984 or BVD-523) exhibited a synergistic antitumor effect on both Caki-1 and 786-O cells (Figures 2B and 2D).
Figure 2.
Combination of ERK Inhibitor and Everolimus Synergistically Reduces the Viability of RCC Cells
(A) Caki-1 and 786-O cells were treated with everolimus alone or in combination with ERK inhibitor SCH772984 for 72 h. The cell viability was assessed by CCK-8 assay. ***p < 0.001. (B) Synergistic effect of everolimus and SCH772984 in Caki-1 and 786-O cells was analyzed using isobologram. The horizontal line indicates the fractional effect of the combination. CI values <1 indicate synergism whereas >1 indicate antagonism. (C) Caki-1 and 786-O cells were treated with everolimus alone or in combination with ERK inhibitor BVD-523 for 72 h. The cell viability was assessed by CCK-8 assay. **p < 0.01; ***p < 0.001. (D) Synergistic effect of everolimus and BVD-523 in Caki-1 and 786-O cells was analyzed using isobologram. The horizontal line indicates the fractional effect of the combination. CI values <1 indicate synergism whereas >1 indicate antagonism. Error bars represent mean ± SD of three independent experiments.
Concurrent Inhibition of mTOR and ERK Exhibited a Synergistic Effect in the Treatment of RCC by Enhancing Everolimus-Induced G1 Arrest
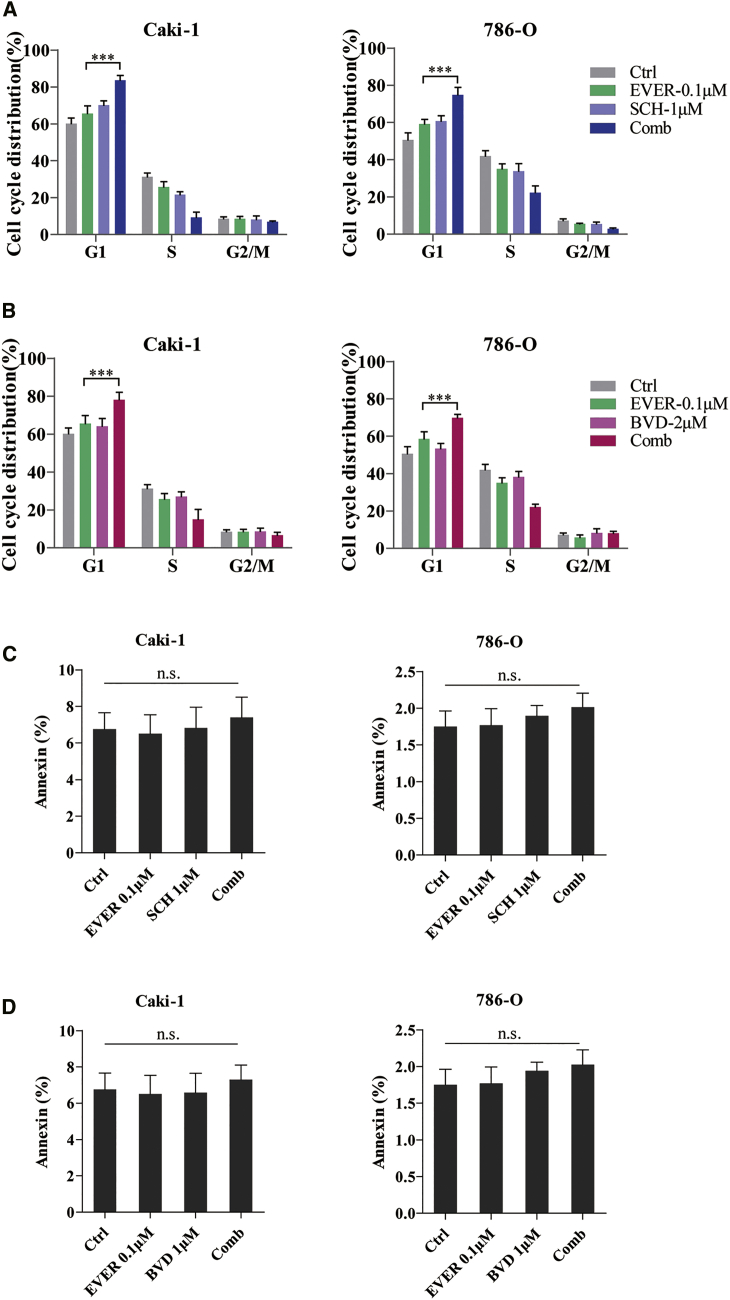
To further explore the mechanism underlying the significantly decreased cell proliferation as a result of the combination of everolimus and ERK inhibitor, we performed the cell cycle distribution and apoptosis assay on Caki-1 and 786-O cells. As expected, the increased sensitivity of Caki-1 and 786-O cells to everolimus when treated with SCH772984 was attributed to the enhanced G1 cell-cycle arrest, as evaluated by propidium iodide (PI) staining (Figure 3A); a similar effect was observed using another ERK inhibitor, BVD-523 (Figure 3B), while the combination of everolimus and ERK inhibitor did not show a significant efficacy on cell apoptosis (Figures 3C and 3D). Together, these findings suggested that everolimus and ERK inhibitor acted synergistically in inhibiting the growth of RCC cells by enhancing the everolimus-induced G1 cell-cycle arrest.
Figure 3.
ERK Inhibitor and Everolimus Synergistically Reduce the Viability of RCC Cells by Inducing G1 Cell-Cycle Arrest
(A and B) Caki-1 and 786-O cells were treated with everolimus alone or in combination with ERK inhibitor SCH772984 (A) or BVD-523 (B) for 24 h. The cell cycle distribution was detected by PI staining and is shown in the bar graph as the percentage of the cells. ***p < 0.001. (C and D) Caki-1 and 786-O cells were treated with everolimus alone or in combination with ERK inhibitor SCH772984 (C) or BVD-523 (D) for 48 h. The apoptotic cells were detected by PI and Annexin V dual staining and are shown in the bar graph as the percentage of cells. n.s. indicates not significant. Error bars represent mean ± SD of three independent experiments.
Cell-Cycle Arrest Induced by the Combination Therapy of Everolimus and ERK Inhibitor Results from Impaired dNTP Synthesis
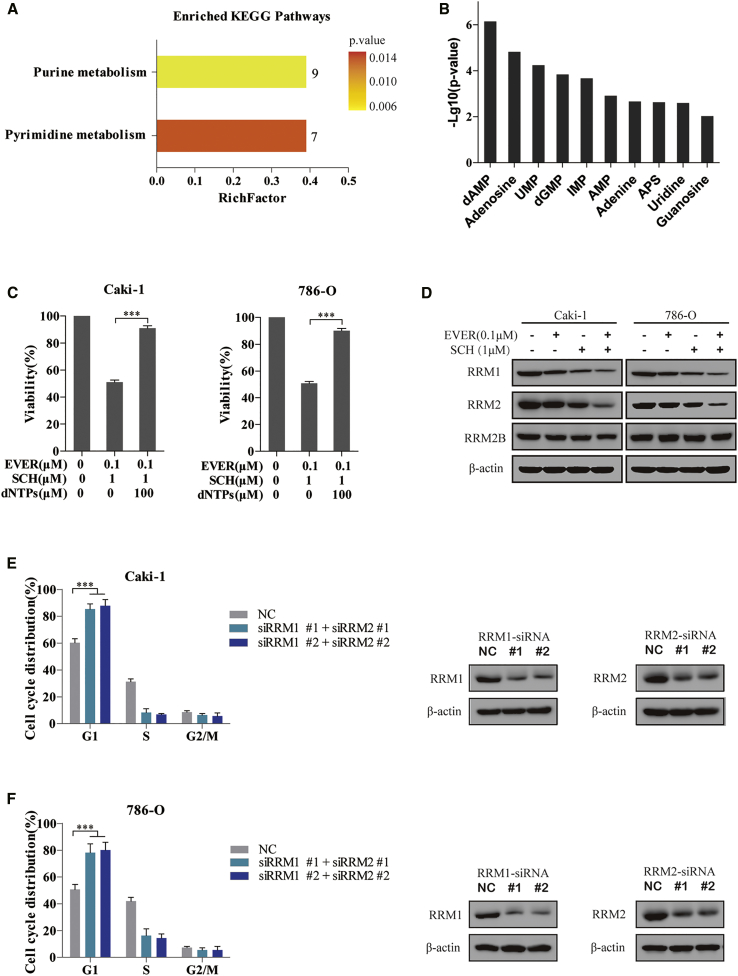
RCC is essentially a metabolic disease.19 mTORC1, the target of everolimus, participates in lipid, nucleotide, and glucose metabolism, which are required for cell proliferation and growth.20 MEK inhibitor regulates the RRM1 degradation in pancreatic cancer.10 Thus, to explore whether ERK inhibitor could enhance the effect of everolimus on the metabolism of RCC, we performed a targeted metabolomic assay in Caki-1 cells exposed to single or combination treatment, and we measured the amount of 200 metabolites among different groups. As expected, the combination of everolimus and SCH772984 led to the alteration of a total of 35 metabolites as compared to monotherapy (Figure S1). Furthermore, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of these changed metabolites revealed that purine and pyrimidine metabolisms were highly affected by combination therapy (Figure 4A). After one-way ANOVA analysis, the top 10 decreased metabolites involved in nucleotide synthesis were ranked by −Log10(p value) (Figure 4B).
Figure 4.
Attenuation of dNTP Pools Caused by the Downregulation of RRM1 and RRM2 Is Required for G1 Arrest Induced by the Combination of Everolimus and ERK Inhibitor
(A and B) A targeted metabolomic assay was performed in Caki-1 cells exposed to 0.1 μM everolimus, 1 μM SCH772984, the combination of both, or an equivalent volume of DMSO for 72 h. After one-way ANOVA analysis, KEGG pathway enrichment analysis of the altered 35 metabolites revealed that the purine and pyrimidine metabolisms were highly affected by the combination therapy. The number at the top of the bar indicates the number of altered metabolites; the RichFactor indicates the percentage ratio of the altered metabolites to all metabolites in this pathway (A). The top 10 altered metabolites involved in purine and pyrimidine metabolism pathways were ranked by −Log10(p value) (B). (C) 100 μM dNTP was added to the everolimus-SCH772984 combination system for 72 h in Caki-1 and 786-O cells. The cell viability was measured using CCK-8 assay. ***p < 0.001. (D) Caki-1 and 786-O cells were treated with everolimus and SCH772984 for 24 h. Then, immunoblotting analysis was performed on the cell lysates to detect the essential enzymes involved in purine metabolism. (E and F) Caki-1 (E) and 786-O (F) cells were transfected with the indicated siRNAs to concurrently knock down RRM1 and RRM2 for 48 h. The knockdown efficiency was evaluated by immunoblotting analysis. The cell cycle distribution was detected in the cells by PI staining. ***p < 0.001. Error bars represent mean ± SD of three independent experiments.
To demonstrate whether deficient nucleotide synthesis was responsible for the synergistic effect of the combination therapy, cells were treated with exogenous dNTPs, which rescued the proliferation inhibition induced by everolimus combined with SCH772984 (Figure 4C). In agreement with this result, the synthesis of nucleotides is required for DNA replication and ribosome biogenesis in G1-S transition cells. A remaining quandary was how the dNTP pools were suppressed by the everolimus-SCH772984 combination. As deoxyadenosine monophosphate (dAMP), adenosine, uridine monophosphate (UMP), and deoxyguanosine monophosphate (dGMP) are the significantly decreased metabolites induced by combination therapy, we focused on these critical enzymes of the de novo purine and pyrimidine synthesis pathway. Immunoblotting analysis revealed that the levels of RRM1 and RRM2 proteins were remarkably decreased in everolimus-SCH772984-treated cells as compared to monotherapy-treated cells (Figure 4D). Furthermore, the knockdown of RRM1 and RRM2 by small interfering RNA (siRNA) in Caki-1 cells indeed induced G1 cell-cycle arrest (Figure 4E), which was reiterated in 786-O cells (Figure 4F). These results suggested that the combination of everolimus and ERK inhibitor perturbed the dNTP pools by reducing the expression of RRM1 and RRM2, which, in turn, induced G1 cell-cycle arrest via impaired DNA replication.
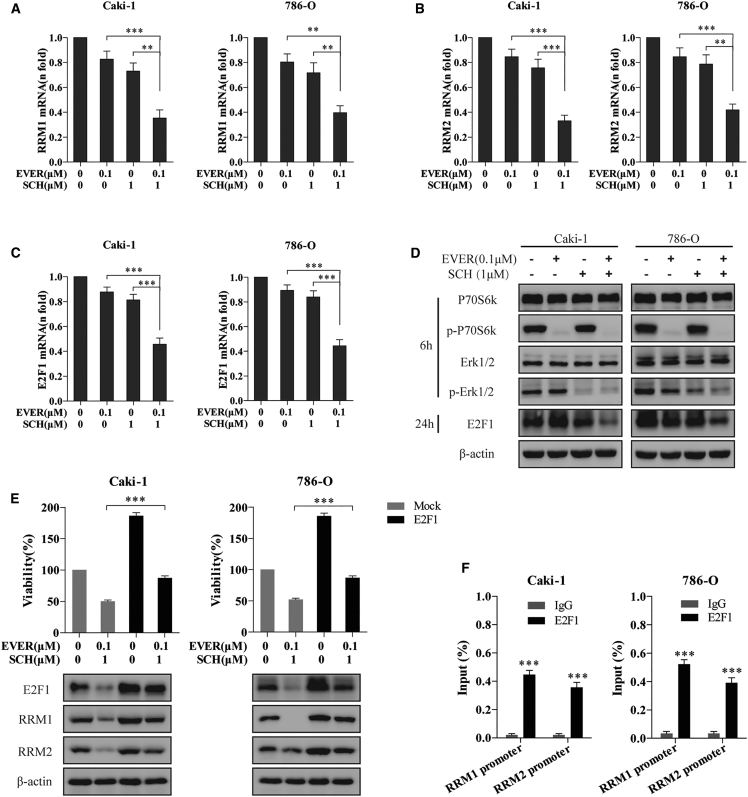
Everolimus and ERK Inhibitor Synergistically Suppressed the RRM1 and RRM2 Transcription via E2F1 Inhibition
Next, we explored the mechanism underlying the everolimus- and ERK inhibitor-mediated downregulation of RRM1 and RRM2 in RCC cells. A majority of the substrates of mTOR and ERK transfers into the nucleus to play a key role in cell proliferation and nutrient synthesis. We confirmed the transcriptional downregulation of RRM1 (Figure 5A) and RRM2 (Figure 5B) as a result of the everolimus-SCH772984 combination using qRT-PCR assay in Caki-1 and 786-O cells. Interestingly, a previous study suggested that hepatitis B virus (HBV) induced the RRM2 expression via the Chk1-E2F1 axis as a DNA damage response.21 Another study showed that gambogic acid sensitizes the pancreatic carcinoma to gemcitabine by inhibiting the ERK-E2F1-RRM2-signaling pathway.22 Hence, we speculated that the combination of everolimus and ERK inhibitor might impair the RRM1 and RRM2 transcription by decreasing the E2F1 expression levels in RCC cells. To test this hypothesis, we examined the expression of E2F1 post-treatment with everolimus and SCH772984 using both real-time qPCR and an immunoblotting assay. Strikingly, the combination therapy overtly reduced the mRNA and protein levels of E2F1 when the signaling pathways (p-P70S6K or p-ERK1/2) were blocked by everolimus or SCH772984, respectively, in Caki-1 and 786-O cells (Figures 5C and 5D).
Figure 5.
E2F1 Downregulation Is Required for the Transcriptional Inhibition of RRM1 and RRM2 Induced by the Combination of Everolimus and ERK Inhibitor
(A–C) Caki-1 and 786-O cells treated with everolimus alone or in combination with SCH772984 for 72 h were subjected to qRT-PCR analysis to elucidate the mRNA levels of RRM1 (A), RRM2 (B), and E2F1 (C). (D) Phospho-p70S6k, phospho-Erk1/2, and E2F1 protein levels were detected by immunoblotting analysis when Caki-1 and 786-O cells were treated with everolimus and SCH772984 for the indicated time points. (E) Caki-1 and 786-O cells were transfected with E2F1 overexpression plasmid or empty plasmid (mock) for 48 h, followed by everolimus-SCH772984 combination treatment for another 24 h. The overexpression efficiency and the alterations of RRM1 and RRM2 were detected by immunoblotting analysis. Also, the proliferation of cells was measured by CCK-8 assay. ***p < 0.001. (F) Caki-1 and 786-O cells were subjected to ChIP assay using E2F1 antibody, followed by qRT-PCR analysis using primers targeting the promoter regions of RRM1 and RRM2. ***p < 0.001. Error bars represent mean ± SD of three independent experiments.
The proliferation inhibition induced by RRM1 and RRM2 depletion appeared to be dependent on the decline in the level of E2F1, while the overexpression of E2F1 partially restored the RRM1 and RRM2 protein levels accompanied by the viability in the everolimus-SCH772984 combination-treated cells (Figure 5E). To confirm this finding, chromatin immunoprecipitation (ChIP) assays using E2F1 antibody followed by qPCR analysis were performed in Caki-1 and 786-O cells to examine the recruitment of E2F1 to the promoter of RRM1 and RRM2 genes. As shown in Figure 5F, E2F1 binding was enhanced in the promoter regions of RRM1 and RRM2 in Caki-1 and 786-O cells. In addition, the higher levels of RRM1, RRM2, and E2F1 in everolimus-resistant cells as compared to parental cells (Figure S2) reconfirmed our hypothesis. Together, these results suggested that the combination of everolimus and ERK inhibitor reduced the recruitment of E2F1 to the promoter of RRM1 and RRM2, which led to trans-inhibition of RRM1 and RRM2, followed by dNTP deprivation in cancer cells.
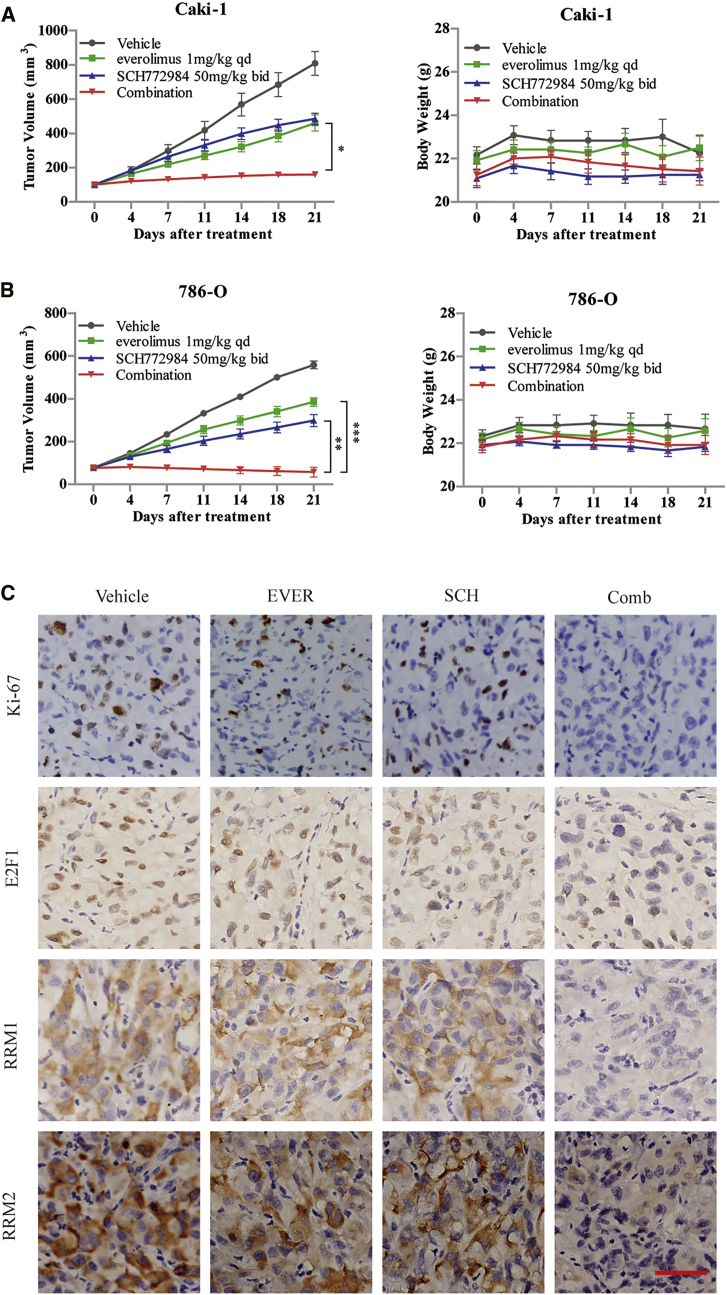
SCH772984 Significantly Enhances the Antitumor Efficacy of Everolimus in RCC Xenograft Tumors
To assess the therapeutic efficacy of combined everolimus and ERK inhibitor in vivo, two human RCC xenograft Caki-1 and 786-O models were established. In both models, single treatment with either everolimus at 1 mg/kg/day or SCH772984 50 mg/kg twice daily led to a slight reduction in tumor size after 21 days of treatment. Conversely, the combination therapy provided a synergistic antitumor efficacy, as tumor growth was almost completely blunted at the endpoint of the treatment. Moreover, no significant loss in body weight or death was observed in either group of mice during the whole study, which indicated controlled toxicity of the combination therapy (Figures 6A and 6B). Consistent with the mechanisms discovered in vitro, IHC staining on Caki-1 xenograft tumor tissues showed that the combination of everolimus and SCH772984 remarkably inhibited the expressions of E2F1, RRM1, and RRM2 as compared to monotherapy (Figure 6C). These observations suggested that the combination therapy of everolimus and ERK inhibitor is promising for RCC.
Figure 6.
ERK Inhibitor Sensitized the RCC Tumors to Everolimus in Xenograft Models
(A and B) Caki-1 (A) and 786-O (B) xenograft models were treated with everolimus (1 mg/kg/day) and SCH772984 (50 mg/kg, twice daily) alone or in combination daily for 21 days. The inhibition rate of the tumor growth (left panel) and body weights of the mice (right panel) were assessed by two-way ANOVA analysis. The error bars represent means ± SEM (n = 6 mice/group). *p < 0.05; **p < 0.01; ***p < 0.001. (C) Tumor tissues from Caki-1 xenografts were resected after the last dose and immunostained with Ki-67, E2F1, RRM1, and RRM2 antibodies to detect the intratumoral molecular alteration. Magnification, ×200; scale bar, 100 μm.
Discussion
Everolimus, similar to other PI3K or mTOR inhibitors, has been approved by the FDA for clinical therapy, as the PI3K-AKT-mTOR pathway is frequently dysregulated in a broad spectrum of cancers. However, the antitumor activities of these inhibitors are limited due to the pro-tumorigenic aberrations in other signaling networks.23 Hence, a rational combination therapy is required to optimize the efficacy of everolimus. Thus, some clinical trials tend to concurrently block the mTOR and MAPK pathways in order to combat the adaptive and innate resistance.24, 25, 26 Regardless of different efficiencies of combination strategies, convincing mechanism-based evidence is lacking to conclude the concurrent blockade of mTOR and MAPK signaling. In this study, we identified a novel combination strategy for RCC, in which everolimus and an ERK inhibitor synergistically inhibit the growth of cancer through the attenuation of dNTP pools by downregulating the levels of RRM1 and RRM2 transcripts.
Furthermore, mTORC1, the target of everolimus, promotes glucose, lipid, and nucleotide metabolism in mammals. A recent study established that mTORC1 increases the transcriptional expression of MTHFD2 in an ATF4-dependent manner, thereby inducing the mitochondrial tetrahydrofolate cycle that provides one-carbon units for de novo purine synthesis.27 Nevertheless, only a few studies have addressed the impact on the metabolism of the tumor cells. This study explored the mechanism underlying the synergistic effect of everolimus-ERK inhibitor combination with respect to tumor metabolism. As expected, the combination of everolimus and ERK inhibitor synergistically downregulates some of the dNTPs, such as dAMP, adenosine, UMP, and dGMP. Subsequently, the combination of everolimus and ERK inhibitor decreases the recruitment of E2F1 to the promoter of RRM1 and RRM2, which, in turn, results in E2F1-mediated transcriptional inhibition of RRM1 and RRM2. Consequently, the downregulated RRM1 and RRM2 attenuate the dNTP pools and inhibit the proliferation of RCC cells by suppressing the G1-S transition.
Interestingly, our study identified RRM1 and RRM2 as the funnel molecules that overlap in the mTOR- and ERK-signaling pathways. The RRM1 and RRM2 enzyme are crucial for RNR in normoxia to maintain DNA replication due to their critical role in the homeostasis of dNTPs.28 The dNTPs are required for DNA synthesis in the G1 to S phase. Similarly, E2F1 plays a pivotal role in the transition of G1 to S phase, and it regulates the expression of several genes required for passage into the S phase. Consistent with the current insights on RCC cells, CG-5, a pan-Glut inhibitor, abrogated the resistance to gemcitabine by decreasing the expression of RRM2 through E2F1-mediated transcriptional inhibition in pancreatic cancer cells.29 In addition to these mechanistic insights, high RRM1 and RRM2 expressions were correlated with poor prognosis in patients with pancreatic cancer30 and lung adenocarcinoma31 treated with gemcitabine, and the downregulation of these molecules increased the chemosensitivity of pancreatic cancer cells to gemcitabine.32, 33 Also, RRM1 and RRM2 predict the prognosis and act as potential therapeutic targets in patients with multiple myeloma34 and glioblastoma.35
Therefore, this study complemented the current understanding of RRM1 and RRM2 in RCC by proving their role in the everolimus-ERK inhibitor combination-targeted therapies. Since the ERK inhibitors, SCH772984 and BVD-523, are in the phase of clinical trials, the everolimus-ERK inhibitor combination strategy in RCC might seem promising.
Materials and Methods
Clinical Samples
Clinical samples and data were collected after obtaining informed consent according to an established protocol approved by the Ethics Committee of Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine. Human ccRCC and normal tissues were obtained from patients who underwent nephrectomy in the Urology Department of Shanghai Ninth People’s Hospital between 2005 and 2010. The patients were followed up from the date of surgery to the day of death or the day of study completion. The median OS was 60 months (range: 10–80 months).
Cell Lines and Inhibitors
The human RCC cell lines Caki-1 and 786-O were purchased from the American Type Culture Collection (ATCC, USA). Caki-1 cells were cultured in McCoy’s 5A medium, while 786-O cells were maintained in RPMI 1640 medium. Both media were supplemented with 10% fetal bovine serum (FBS), at 37°C in a humidified environment with 5% CO2. Everolimus (10 mM, Selleck Chemicals) was diluted in the cell culture medium to a final concentration of 100 nM. SCH772984 and BVD-523 (5 mg, Selleck Chemicals) was solubilized in DMSO as a 10-mM stock solution, stored at −20°C, and used at final concentrations of 1–4 μM.
Cell Viability Assay and Criteria for Synergism
Cells were seeded overnight in 96-well plates at a density of 2,000 cells/well. After 72-h treatment with the indicated inhibitors, 10 μL CCK-8 reagent (Life Technologies) was added to each well and incubation continued for 1–2 h at 37°C. Then, the absorbance (optical density [OD]) was measured at 450 nm on a SoftMax pro plate reader. Cell viability (%) = [OD (dosing) − OD (blank)]/[OD (control) − OD (blank)] × 100%. Isobologram analysis was performed to assess the synergistic effect. Combination index (CI) was calculated by the CompuSyn software (Combo Syn, Paramus, NJ, USA) according to the Chou-Talalay method36 (CI < 1, synergism; CI = 1, additive effect; CI > 1, antagonism).
Cell Cycle Assay
After treatment with the indicated inhibitors for 24 h, approximately 5 × 105 tumor cells were harvested, washed with cold PBS, and fixed with 70% ethanol for 24 h at 4°C. Then, the cells were stained with PI (Sigma-Aldrich, USA) and incubated in the dark for 30 min at room temperature. The cell cycle distribution was analyzed by a FACScan flow cytometer (FACS Canto II, Becton Dickinson).
Apoptosis Assay
After treatment with the indicated inhibitors for 48 h, at least 5 × 105 tumor cells were collected, washed with cold PBS, and resuspended in 100 μL binding buffer. Then, the cells were stained with PI and Annexin V- fluorescein isothiocyanate (FITC) using the Apoptosis Detection Kit (BD Pharmingen, Germany), according to the manufacturer’s instructions. Finally, at least 1 × 104 cells were analyzed by flow cytometry.
Targeted Metabolomic Assay
After treatment with the indicated inhibitors for 72 h, 1 × 107 Caki-1 cells (6 replicates) were extracted using 1 mL cold MeOH:ACN:H2O (2:2:1, v/v) solvent mixture in liquid nitrogen for 1 min. After three freeze-thaw cycles, the samples were incubated for 1 h at −20°C, followed by centrifugation at 14,000 × g for 15 min to precipitate the proteins. The supernatants were reconstituted in 100 μL ACN:H2O (1:1, v/v) solvent after drying in a vacuum concentrator. The chromatographic separation of samples was performed on an Agilent 1260 HPLC system (Agilent Technologies). Then, a triple quadrupole mass spectrometry (QqQ-MS)-based multiple reaction monitoring (MRM) technique was used for liquid chromatography-tandem MS (LC-MS/MS) analysis on an Agilent 6460 QqQ mass spectrometer (Agilent Technologies). For targeted metabolomic analysis, 200 metabolites standard (100 μg/mL) were first analyzed in both ESI-positive and -negative modes in a single 15-min LC run using the MassHunter Optimizer software (Agilent Technologies) to obtain the optimal MRM transitions. For each metabolite, the retention time (marked as rtsingle) was measured by the corresponding MRM transition. The acquired MRM raw data were converted and analyzed by MRMAnalyzer (R) program, as described previously.37
Immunoblotting Analysis
Total protein extracted by cell lysis using RIPA buffer was quantified by a bicinchoninic acid (BCA) assay (Thermo Scientific). An equivalent of 20 μg total protein/sample was subjected to SDS-PAGE and subsequently transferred to nitrocellulose membranes (Millipore). After blocking with 3% BSA for 1 h at room temperature, the membranes were probed overnight with specific primary antibodies at 4°C. Then, the membranes were incubated with secondary antibodies for 1 h after washing with tris buffered saline + Tween (TBST) three times. Then, immunoreactive bands were detected using enhanced chemiluminescence assay (Thermo Scientific). β-actin was used as a loading control. In this study, p70S6K, p-p70S6K(Thr389), Erk1/2, p-Erk1/2(Thr202/Tyr204), and RRM1 antibodies were obtained from Cell Signaling Technology and E2F1, RRM2, RRM2B, and β-actin antibodies were from Abcam.
qRT-PCR Analysis
Total RNA isolated using RNeasy Mini Kit (QIAGEN) from cells was reverse transcribed to cDNA using the QuantiTect Reverse Transcription Kit (QIAGEN). Then, real-time qPCR was performed using SYBR Green PCR Master Mix (Applied Biosystems), according to the manufacturer’s instructions on a ViiA 7 Real-Time PCR System (Applied Biosystems). Ct values were obtained for β-actin and the indicated genes during the log phase of the cycle. The expression of the indicated genes was normalized to that of β-actin [ΔCt = Ct (indicated genes) − Ct (β-actin)] and compared to the values obtained for a control using the following formula: 2−ΔΔCt [ΔΔCt = ΔCt (test) − ΔCt (control)]. The sequences of primers used for RT-qPCR analysis were as follows:
RRM1: 5′-CAGTGATGTGATGGAAGA-3′ and 5′-CTCGGTCATAGATAATAGCA-3′,
RRM2: 5′-AGACTTATGCTGGAACTG-3′ and 5′-TCTGATACTCGCCTACTC-3′,
E2F1: 5′-CAGAGCAGATGGTTATGG-3′ and 5′-CTGAAAGTTCTCCGAAGA-3′, and
β-actin: 5′-CATGTACGTTGCTATCCAGGC-3′ and 5′-CTCCTTAATGTCACGCACGAT-3′.
siRNA and Plasmid Transfection
Cells were seeded in a 6-well plate overnight to achieve 50%–60% confluency, and then they were transfected with a specific siRNA duplex or plasmid for 48 h using Lipofectamine RNAiMAX or 2000 Transfection Reagent (Invitrogen), according to the manufacturer’s protocol. A nonspecific oligonucleotide was used as a negative control. siRNAs and plasmid were purchased from GenePharma (Shanghai, China). The sequences were as follows:
siRRM1 1: 5′-GCUUUGUUAUGGACUCAAUTT-3′,
siRRM1 2: 5′-GACCACAACAUAUGUUGAUTT-3′,
siRRM2 1: 5′-CCCAUCGAGUACCAUGAUATT-3′, and
siRRM2 2: 5′-CGUCGAUAUUCUGGCUCAATT-3′.
ChIP Assay
ChIP assay was performed using Magna ChIP HiSens Chromatin Immunoprecipitation Kit (17-10460, Merck Millipore), according to the manufacturer’s instructions. The DNA fragments isolated in the complex with the target protein were identified by qPCR using primers for RRM1 and RRM2 promoters. The antibody used for ChIP was anti-E2F1 (ab4070, Abcam). The sequences of primers used for qPCR were as follows:
RRM1: 5′-GTTCCCGCCGGTTAGGTTTT-3′ and 5′-CTGCTCTCCTGCTACCATGT-3′, and
RRM2: 5′-TTTGGAAGTCGCGCTAACCT-3′ and 5′-GCGCGTTTTGACATCTGCAT-3′.
Xenograft Model and Treatments
Animal experiments were conducted in compliance with the institutional ethical guidelines on animal care and approved by the Animal Care and Welfare Committee of Shanghai Jiao Tong University. 4- to 6-week-old male nude mice (BALB/c nu/nu), purchased from Charles River (Beijing, China), were housed under pathogen-free conditions. Approximately 5 × 106 Caki-1 or 786-O cells were injected subcutaneously into the flank region of nude mice to establish the xenograft models. The mice were randomized into four groups (n = 6) and administered the drug when the tumor volume reached approximately 100 mm3. Everolimus (1 mg/kg) was administered orally daily, and SCH772984 (50 mg/kg) was administered by intraperitoneal injection two times per day. For the combination treatment, both drugs were administered concurrently. The tumor volume and body weight were measured two times per week using calipers. The tumor volume was calculated using the following formula: V = π (length × width2)/6. After 3 weeks of treatment, the mice were euthanized, and the tumor tissues were harvested, weighed, and fixed in formalin for IHC staining.
IHC Analysis
Paraffin-embedded tumor tissue specimens were sliced into 5-μm-thick sections and mounted onto slides. Then, the slices were deparaffinized, rehydrated, subjected to antigen retrieval, and incubated with specific primary antibodies overnight at 4°C. Subsequently, the sections were incubated with secondary antibodies for 1 h after washing three times with PBS. After staining with diaminobenzidine (DAB), the sections were visualized under a microscope. In this study, the patient tumor sections were stained with phospho-p44/42 Erk1/2 (Thr202/Tyr204) (4370, Cell Signaling Technology) to assess the expression of the protein in RCC. The quantification of phospho-Erk1/2 was scored by the product of intensity and percentage of staining. The xenograft tumor sections were stained with anti-Ki67 (ab15580, Abcam), anti-E2F1 (ab4070, Abcam), anti-RRM1 (8637, Cell Signaling Technology), and RRM2 (ab57653, Abcam) to verify the molecular mechanisms detected in vitro.
Statistical Analysis
Data were presented as mean ± SD of three independent experiments. Two-tailed Student’s t test or one-way ANOVA, followed by Dunnett’s post hoc test, was performed to assess the statistical difference between the indicated groups with respect to phospho-Erk1/2 expression, cell viability, cycle distribution, and mRNA expression. Two-way ANOVA, followed by Bonferroni post hoc test was used to analyze the synergistic effect on the size of animal tumors and therapy response over a period. Log-rank test (Kaplan-Meier method) was used to analyze the correlation between phospho-Erk1/2 expression and OS. p < 0.05 was considered as a statistically significant difference. Statistical analyses were performed using GraphPad Prism version 7 (GraphPad, CA, USA).
Author Contributions
Y.Z. and W.L. conducted the experiments, analyzed the data, and wrote the manuscript. J. Zhou and J. Zhang collected clinical samples and data and analyzed the data. Y.H. and Z.W. conceived the project, supervised the research, and revised the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This work was sponsored by the National Natural Science Foundation of China (81802517), the Integrated Traditional Chinese and Western Medicine of Shanghai (ZHYY-ZXYJHZX-1-03), the Clinical Research Program of 9th People’s Hospital, Shanghai Jiao Tong University School of Medicine (JYLJ005), and the Program for Outstanding Medical Academic Leader.
Footnotes
Supplemental Information includes two figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2019.01.001.
Contributor Information
Yiran Huang, Email: hyrrenji2@aliyun.com.
Zhong Wang, Email: zhongwang2000@sina.com.
Supplemental Information
References
- 1.Fisher R., Gore M., Larkin J. Current and future systemic treatments for renal cell carcinoma. Semin. Cancer Biol. 2013;23:38–45. doi: 10.1016/j.semcancer.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Li G., Ci W., Karmakar S., Chen K., Dhar R., Fan Z., Guo Z., Zhang J., Ke Y., Wang L. SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer. Cancer Cell. 2014;25:455–468. doi: 10.1016/j.ccr.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motzer R.J., Escudier B., Oudard S., Hutson T.E., Porta C., Bracarda S., Grünwald V., Thompson J.A., Figlin R.A., Hollaender N., RECORD-1 Study Group Phase 3 trial of everolimus for metastatic renal cell carcinoma : final results and analysis of prognostic factors. Cancer. 2010;116:4256–4265. doi: 10.1002/cncr.25219. [DOI] [PubMed] [Google Scholar]
- 4.Czarnecka A.M., Kornakiewicz A., Lian F., Szczylik C. Future perspectives for mTOR inhibitors in renal cell cancer treatment. Future Oncol. 2015;11:801–817. doi: 10.2217/fon.14.303. [DOI] [PubMed] [Google Scholar]
- 5.Motzer R.J., Hutson T.E., Glen H., Michaelson M.D., Molina A., Eisen T., Jassem J., Zolnierek J., Maroto J.P., Mellado B. Lenvatinib, everolimus, and the combination in patients with metastatic renal cell carcinoma: a randomised, phase 2, open-label, multicentre trial. Lancet Oncol. 2015;16:1473–1482. doi: 10.1016/S1470-2045(15)00290-9. [DOI] [PubMed] [Google Scholar]
- 6.Colwell J. FDA Approves Drug Combo for Kidney Cancer. Cancer Discov. 2016;6:687–688. doi: 10.1158/2159-8290.CD-NB2016-067. [DOI] [PubMed] [Google Scholar]
- 7.Kolberg M., Strand K.R., Graff P., Andersson K.K. Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta. 2004;1699:1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Stubbe J. Ribonucleotide reductases in the twenty-first century. Proc. Natl. Acad. Sci. USA. 1998;95:2723–2724. doi: 10.1073/pnas.95.6.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aye Y., Li M., Long M.J., Weiss R.S. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene. 2015;34:2011–2021. doi: 10.1038/onc.2014.155. [DOI] [PubMed] [Google Scholar]
- 10.Vena F., Li Causi E., Rodriguez-Justo M., Goodstal S., Hagemann T., Hartley J.A., Hochhauser D. The MEK1/2 Inhibitor Pimasertib Enhances Gemcitabine Efficacy in Pancreatic Cancer Models by Altering Ribonucleotide Reductase Subunit-1 (RRM1) Clin. Cancer Res. 2015;21:5563–5577. doi: 10.1158/1078-0432.CCR-15-0485. [DOI] [PubMed] [Google Scholar]
- 11.Corcoran R.B., Ebi H., Turke A.B., Coffee E.M., Nishino M., Cogdill A.P., Brown R.D., Della Pelle P., Dias-Santagata D., Hung K.E. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brady D.C., Crowe M.S., Turski M.L., Hobbs G.A., Yao X., Chaikuad A., Knapp S., Xiao K., Campbell S.L., Thiele D.J., Counter C.M. Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature. 2014;509:492–496. doi: 10.1038/nature13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulikakos P.I., Persaud Y., Janakiraman M., Kong X., Ng C., Moriceau G., Shi H., Atefi M., Titz B., Gabay M.T. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris E.J., Jha S., Restaino C.R., Dayananth P., Zhu H., Cooper A., Carr D., Deng Y., Jin W., Black S. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 15.Carlino M.S., Todd J.R., Gowrishankar K., Mijatov B., Pupo G.M., Fung C., Snoyman S., Hersey P., Long G.V., Kefford R.F., Rizos H. Differential activity of MEK and ERK inhibitors in BRAF inhibitor resistant melanoma. Mol. Oncol. 2014;8:544–554. doi: 10.1016/j.molonc.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatzivassiliou G., Liu B., O’Brien C., Spoerke J.M., Hoeflich K.P., Haverty P.M., Soriano R., Forrest W.F., Heldens S., Chen H. ERK inhibition overcomes acquired resistance to MEK inhibitors. Mol. Cancer Ther. 2012;11:1143–1154. doi: 10.1158/1535-7163.MCT-11-1010. [DOI] [PubMed] [Google Scholar]
- 17.Carracedo A., Ma L., Teruya-Feldstein J., Rojo F., Salmena L., Alimonti A., Egia A., Sasaki A.T., Thomas G., Kozma S.C. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J. Clin. Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou Y., Wang J., Leng X., Huang J., Xue W., Zhang J., Huang Y. The selective MEK1 inhibitor Selumetinib enhances the antitumor activity of everolimus against renal cell carcinoma in vitro and in vivo. Oncotarget. 2017;8:20825–20833. doi: 10.18632/oncotarget.15346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linehan W.M., Ricketts C.J. The metabolic basis of kidney cancer. Semin. Cancer Biol. 2013;23:46–55. doi: 10.1016/j.semcancer.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saxton R.A., Sabatini D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ricardo-Lax I., Ramanan V., Michailidis E., Shamia T., Reuven N., Rice C.M., Shlomai A., Shaul Y. Hepatitis B virus induces RNR-R2 expression via DNA damage response activation. J. Hepatol. 2015;63:789–796. doi: 10.1016/j.jhep.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 22.Xia G., Wang H., Song Z., Meng Q., Huang X., Huang X. Gambogic acid sensitizes gemcitabine efficacy in pancreatic cancer by reducing the expression of ribonucleotide reductase subunit-M2 (RRM2) J. Exp. Clin. Cancer Res. 2017;36:107. doi: 10.1186/s13046-017-0579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janku F., Yap T.A., Meric-Bernstam F. Targeting the PI3K pathway in cancer: are we making headway? Nat. Rev. Clin. Oncol. 2018;15:273–291. doi: 10.1038/nrclinonc.2018.28. [DOI] [PubMed] [Google Scholar]
- 24.Farley J., Brady W.E., Vathipadiekal V., Lankes H.A., Coleman R., Morgan M.A., Mannel R., Yamada S.D., Mutch D., Rodgers W.H. Selumetinib in women with recurrent low-grade serous carcinoma of the ovary or peritoneum: an open-label, single-arm, phase 2 study. Lancet Oncol. 2013;14:134–140. doi: 10.1016/S1470-2045(12)70572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu T., Tolcher A.W., Papadopoulos K.P., Beeram M., Rasco D.W., Smith L.S., Gunn S., Smetzer L., Mays T.A., Kaiser B. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clin. Cancer Res. 2012;18:2316–2325. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- 26.Bedard P.L., Tabernero J., Janku F., Wainberg Z.A., Paz-Ares L., Vansteenkiste J., Van Cutsem E., Pérez-García J., Stathis A., Britten C.D. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin. Cancer Res. 2015;21:730–738. doi: 10.1158/1078-0432.CCR-14-1814. [DOI] [PubMed] [Google Scholar]
- 27.Ben-Sahra I., Hoxhaj G., Ricoult S.J.H., Asara J.M., Manning B.D. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foskolou I.P., Jorgensen C., Leszczynska K.B., Olcina M.M., Tarhonskaya H., Haisma B., D’Angiolella V., Myers W.K., Domene C., Flashman E., Hammond E.M. Ribonucleotide Reductase Requires Subunit Switching in Hypoxia to Maintain DNA Replication. Mol. Cell. 2017;66:206–220.e9. doi: 10.1016/j.molcel.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai I.L., Chou C.C., Lai P.T., Fang C.S., Shirley L.A., Yan R., Mo X., Bloomston M., Kulp S.K., Bekaii-Saab T., Chen C.S. Targeting the Warburg effect with a novel glucose transporter inhibitor to overcome gemcitabine resistance in pancreatic cancer cells. Carcinogenesis. 2014;35:2203–2213. doi: 10.1093/carcin/bgu124. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Fujita H., Ohuchida K., Mizumoto K., Itaba S., Ito T., Nakata K., Yu J., Kayashima T., Souzaki R., Tajiri T. Gene expression levels as predictive markers of outcome in pancreatic cancer after gemcitabine-based adjuvant chemotherapy. Neoplasia. 2010;12:807–817. doi: 10.1593/neo.10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Souglakos J., Boukovinas I., Taron M., Mendez P., Mavroudis D., Tripaki M., Hatzidaki D., Koutsopoulos A., Stathopoulos E., Georgoulias V., Rosell R. Ribonucleotide reductase subunits M1 and M2 mRNA expression levels and clinical outcome of lung adenocarcinoma patients treated with docetaxel/gemcitabine. Br. J. Cancer. 2008;98:1710–1715. doi: 10.1038/sj.bjc.6604344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhutia Y.D., Hung S.W., Krentz M., Patel D., Lovin D., Manoharan R., Thomson J.M., Govindarajan R. Differential processing of let-7a precursors influences RRM2 expression and chemosensitivity in pancreatic cancer: role of LIN-28 and SET oncoprotein. PLoS ONE. 2013;8:e53436. doi: 10.1371/journal.pone.0053436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan P., Liu L., Yin Y., Zhao Z., Zhang Y., Amponsah P.S., Xiao X., Bauer N., Abukiwan A., Nwaeburu C.C. MicroRNA-101-3p reverses gemcitabine resistance by inhibition of ribonucleotide reductase M1 in pancreatic cancer. Cancer Lett. 2016;373:130–137. doi: 10.1016/j.canlet.2016.01.038. [DOI] [PubMed] [Google Scholar]
- 34.Sagawa M., Ohguchi H., Harada T., Samur M.K., Tai Y.T., Munshi N.C., Kizaki M., Hideshima T., Anderson K.C. Ribonucleotide Reductase Catalytic Subunit M1 (RRM1) as a Novel Therapeutic Target in Multiple Myeloma. Clin. Cancer Res. 2017;23:5225–5237. doi: 10.1158/1078-0432.CCR-17-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasmussen R.D., Gajjar M.K., Tuckova L., Jensen K.E., Maya-Mendoza A., Holst C.B., Møllgaard K., Rasmussen J.S., Brennum J., Bartek J., Jr. BRCA1-regulated RRM2 expression protects glioblastoma cells from endogenous replication stress and promotes tumorigenicity. Nat. Commun. 2016;7:13398. doi: 10.1038/ncomms13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou T.C., Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 37.Cai Y., Weng K., Guo Y., Peng J., Zhu Z.-J. An integrated targeted metabolomic platform for high-throughput metabolite profiling and automated data processing. Metabolomics. 2015;11:1575–1586. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.