Abstract
Background
Cancer is a complex disease with profound genomic alterations and extensive heterogeneity. Recent studies on large-scale genomics have shed light on the impact of core oncogenic pathways, which are frequently dysregulated in a wide spectrum of cancer types. Aberrant activation of the hepatocyte growth factor (HGF) signaling axis has been associated with promoting various oncogenic programs during tumor initiation, progression, and treatment resistance. As a result, HGF-targeted therapy has emerged as an attractive therapeutic approach. However, recent clinical trials involving HGF-targeted therapies have demonstrated rather disappointing results. Thus, an alternative, in-depth assessment of new patient stratification is necessary to shift the current clinical course.
Methods
To address such challenges, we have evaluated the therapeutic efficacy of YYB-101, an HGF-neutralizing antibody, in a series of primary glioblastoma stem cells (GSCs) both in vitro and in vivo. Furthermore, we performed genome and transcriptome analysis to determine genetic and molecular traits that exhibit therapeutic susceptibility to HGF-mediated therapy.
Results
We have identified several differentially expressed genes, including MET, KDR, and SOX3, which are associated with tumor invasiveness, malignancy, and unfavorable prognosis in glioblastoma patients. We also demonstrated the HGF-MET signaling axis as a key molecular determinant in GSC invasion, and we discovered that a significant association in HGF expression existed between mesenchymal phenotype and immune cell recruitment.
Conclusions
Upregulation of MET and mesenchymal cellular state are essential in generating HGF-mediated therapeutic responses. Our results provide an important framework for evaluating HGF-targeted therapy in future clinical settings.
Keywords: glioblastoma, HGF, MET, mesenchymal subtype, tumor-associated macrophages
Importance of the study
The HGF-MET signaling axis regulates multiple signaling networks that govern essential cellular functions. Aberrant activation of the HGF-MET signaling axis has also been implicated across various cancer types. However, recent clinical trials involving HGF-targeted therapy have demonstrated disappointing results, thus prompting exploration of alternative biomarkers that could shift the current clinical course. Toward this goal, we have evaluated the therapeutic efficacy of YYB-101, an HGF-neutralizing antibody that is currently enrolled in a phase I clinical trial. In the present study, we have identified differentially expressed genes that show sensitivity toward HGF-mediated therapy and are associated with worse prognosis in glioblastoma patients. We have identified the HGF-MET signaling axis as a key molecular determinant in GSC invasion. Furthermore, we discovered that a significant association exists between HGF transcriptome in relation to mesenchymal transcriptional subtype and tumor-associated immune cell enrichment. Collectively, our work provides an important conceptual groundwork for future HGF-targeted clinical therapies.
Glioblastoma (GBM) is the most common primary, malignant brain tumor.1,2 Despite aggressive therapeutic intervention, the current standard regimen provides only palliation with a median survival of less than 15 months.2,3 Prominent histopathological features of GBM consist of rapid vascularization, infiltrative growth, and pleomorphic vessels.4–6 Due to its highly infiltrative nature, complete surgical resection remains challenging. Recent advancement in the field of molecular genetics has revealed prevalent abnormality in the structure and orientation of proto-oncogenes. Proto-oncogenes are considered ideal therapeutic targets due to their innate capabilities in promoting tumor initiation and progression.7–9
Hepatocyte growth factor (HGF) and its binding receptor, c-MET, are frequently dysregulated in multiple neoplasms. This then leads to malignant phenotypic states and modulates essential intracellular functions, including cellular proliferation, stemness, angiogenesis, and therapeutic resistance.10–12 Upon HGF autocrine activation, MET facilitates cellular invasion via activation of matrix metalloproteinase-2 (MMP2) and growth factor receptor bound protein 2–associated-binding protein 1 (GAB1). Aberrant activation of the HGF-MET signaling axis has been associated with dismal prognosis across multiple tumor types, including ovarian, breast, gastric, and lung carcinomas.13–15 Given its profound effects on tumor malignancy, disruption of the HGF-MET signaling axis would be a prime therapeutic target in GBM treatment. Additionally, recent studies have highlighted the essential role of HGF in tumor cell and tumor microenvironment interactions, including recruitment of tumor-associated macrophages and neutrophils and poor responses to checkpoint blockade therapies.16–19 However, most HGF targeting clinical trials have shown disappointing results. Anti-HGF antibodies, including rilotumumab and ficlatuzumab, have been terminated in phase III and II clinical trials, respectively, because the overall or progression-free survival of patients with either gastroesophageal or lung adenocarcinomas was considered insignificant. The current limitation on HGF-targeted therapy necessitates assessment of additional biomarkers to achieve successful clinical outcomes.20 Toward this goal, we have employed YYB-101, which is currently being tested in solid tumors as a potential therapeutic agent in a phase I clinical trial,21,22 in a series of primary glioma stem cells (GSCs) to identify genomic and transcriptomic traits that exhibit targeted vulnerabilities to HGF-mediated therapy.
Materials and Methods
Patient-Derived GBM Specimens and Primary Cell Culture
With appropriate approval from the institutional review board, all GBM specimens were obtained from patients undergoing surgery at the Samsung Medical Center. The study protocol was approved by our institution’s ethical committees, and written informed consents were received from all patients. Surgical samples were enzymatically dissociated into single cells and cultured in Neurobasal medium. DNA and RNA were extracted from a portion of single cells and were then subjected to DNA and RNA sequencing.
Targeted-Panel, GliomaSCAN
GliomaSCAN is a targeted panel that covers a range of exonic regions of specific genes that are associated with glioma progression. An Agilent SureSelect kit was used to capture the exonic DNA fragments. The Illumina HiSeq 2000 instrument was used for sequencing and generated 2 × 101-bp paired-end reads.
Somatic Mutation
The sequenced reads in the FASTQ files were aligned to the human genome assembly (hg19) using the Burrows-Wheeler aligner version 0.6.2. The initial alignment BAM files were subjected to conventional preprocessing before mutation calling: sorting, removing duplicated reads, locally realigning reads around potential small indels, and recalibrating base quality score using SAMtools, Picard version 1.73, and Genome Analysis Toolkit (GATK) v2.5.2. We used MuTect (v1.1.4) and Somatic IndelDetector (GATK v2.2) to make high-confidence predictions on somatic mutations from neoplastic and nonneoplastic cell/tissue pairs. Variant Effect Predictor v73 was used to annotate the called somatic mutations.
Copy Number Alterations
ONCOCNV was used to generate estimated copy number alterations in tumor cells.
RNA Sequencing
RNA-seq libraries were prepared using the Illumina TruSeq RNA Sample Prep kit. Sequenced reads were trimmed to include 30 nucleotides from the 5ʹ end of each read for mRNA analysis. Afterward, the trimmed reads were mapped onto hg19 using GSNAP, not allowing mismatch, indels, or splicing. The resulting alignments were sorted and summarized into BED files using SAMtools and bedTools. The BED files were used to calculate values of RPKM (reads per kilobase of transcript per million reads) for each gene, using the DEGseq package.
Invasion Assay
Twenty-eight patient-derived GBM cells (3 × 105 cells/mL) were plated onto the upper wells of a Matrigel-coated 24-well Transwell chamber (353383, BD Bioscience) containing Neurobasal-A medium. Lower wells of the Transwell chamber contained exogenous HGF (80 ng/mL, R&D) and/or YYB-101 (10 μg/mL). Invading cells were fixed with methanol and stained with hematoxylin and eosin, and cell numbers were recorded with the Aperio analysis program (Algorithm; Nuclear 9).
Proliferation Assay
Cell proliferation was measured using the ATPlite one-step Luminescence ATP Detection Assay System kit (PerkinElmer) according to the manufacturer’s instructions. Briefly, the cells were seeded in 384-well plates at a density of 5 × 102 cells/well, and incubated for 6 days. The viable cells were measured using the EnVision Multilabel Reader (PerkinElmer).
Immunoblot Assay
Tumor cells were lysed in radioimmunoprecipitation assay buffer supplemented with proteinase and phosphatase inhibitors. Total proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. The blots were blocked for 1 hour in 5% bovine serum albumin and incubated overnight with the following primary antibodies: phosphorylated (p-)MET (Y1234/1235) (3077s, Cell Signaling), MET (370100, Life Technologies), p-GAB1 (Y307) (3234p, Cell Signaling), GAB1 (3232p, Cell Signaling), p-Akt (S473, D9E, 4060s, Cell Signaling), Akt (9272, Cell Signaling), p-Erk (T202/Y204) (9102s, Cell Signaling), or Erk (9101, Cell Signaling). After washing with Tris-buffered saline and 0.05% Tween 20, the blots were incubated with horseradish peroxidase‒conjugated secondary antibody for 1 hour at room temperature. Detection was performed using the chemiluminescence method.
Flow Cytometry
GBM patient-derived cells (GBM102) were dissociated into single cells and labeled with the following anti-human MET allophycocyanin (APC)-conjugated antibody (FAB3582A, R&D Systems) at a concentration of 5 μL per 1 × 106 cells for 20 min. Antibodies against mouse immunoglobulin conjugated to APC were used as the isotype control (17-4714-42, eBioscience). According to MET expression, GBM102 isolates were sorted into 2 subpopulations. Stained cells were analyzed using the FACS Aria III instrument (BD Biosciences).
Lentivirus Production and Transduction
293FT cells were transfected using Lipofectamine 2000 (Life Technologies) with lentivirus packaging plasmid and MET expression plasmid (37560, Addgene) or pLenti cytomegalovirus green fluorescent protein Puro (17448, Addgene). Forty-eight hours after the transfection, viral supernatants were collected using Lenti-X concentrator (631232, Clontech). Resulting products were added into the culture medium for 2 days, and puromycin selection was performed to eliminate non-infected cells.
Orthotopic GBM Xenograft Models
Animal experiments were conducted in accordance with the current regulations and standards of the Laboratory Animal Research Center at the Samsung Medical Center. To establish human GBM orthotopic xenografts, 6-week-old female BALB/c nude mice (Orient Bio) were used. Patient-derived GBM cells (2 × 105 cells/5 μL in Hanks Balanced Salt Solution; Gibco) were directly injected into the brains of anesthetized mice by using a rodent stereotactic frame. Treatment with the anti-HGF antibody, YYB-101 (5 mg/kg or 20 mg/kg twice a week, intraperitoneal injection), began a week after the tumor cell injection. Mice were sacrificed when either 20% of total body weight loss or neurological symptoms such as lethargy, ataxia, and seizures were observed.
Immunohistochemistry
Brain tissue specimens were fixed with formalin and embedded into paraffin. Tissue sections of paraffin-embedded specimens were stained with the following primary antibodies: p-MET (Y1234/1235) (E9P0077, Enogene), p-GAB1 (Y307) (3234p, Cell Signaling), p‒focal adhesion kinase (FAK) (Y397) (ab4803, Abcam), MMP2 (6E3F8) (ab86607, Abcam), and urokinase-type plasminogen activator (uPA)/urokinase (AF1310, R&D system).
CIBERSORT
We used normalized bulk tumor gene expression data to infer relative proportion of 22 types of infiltrating immune cell using the CIBERSORT algorithm.23 The 22 immune cell types evaluate immune cell populations of T cells, B cells, macrophages, natural killer cells, eosinophils, dendritic cells, neutrophils, etc. CIBERSORT is a deconvolution algorithm that utilizes set-of-reference gene expression data from each representative cell type and evaluates cell type proportions from bulk tumor samples. Gene expression data were uploaded onto the CIBERSORT web portal (http://cibersort.stanford.edu/; Accessed July 6, 2018) with the parameter set at a default of 1000 permutations. The output results were normalized based on tumor purity levels that were derived from the ESTIMATE results.
ESTIMATE
To evaluate tumor purity level from bulk tumor samples, we employed ESTIMATE (Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data).24 ESTIMATE is an algorithm tool for predicting tumor purity and the presence of infiltrating immune/stromal cells from bulk tumor samples using gene expression data. The ESTIMATE algorithm is based on a single-sample Gene Set Enrichment Analysis and generates corresponding scores for stromal, immune, and tumor cells.
Results
Neutralization of HGF via YYB-101 Treatment Inhibits Tumor Cell Invasion and Growth in Glioblastoma
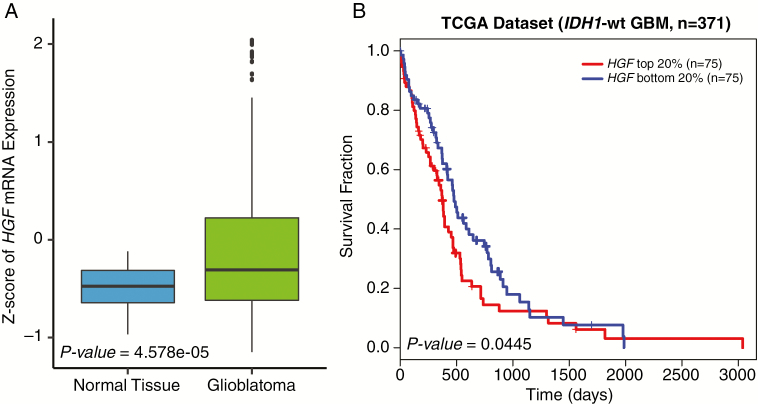
To assess clinical relevance of HGF, we first surveyed transcriptome expression levels of HGF between normal tissue and glioblastoma specimens. Interestingly, HGF mRNA level was significantly upregulated in GBM compared with nonneoplastic brain tissues (Fig. 1A). Additionally, when we stratified isocitrate dehydrogenase 1 (IDH1) wild-type GBM patients according to HGF mRNA expression levels, patients with high HGF portrayed unfavorable survival outcomes compared with those with low HGF expression levels, thus demonstrating the clinical importance of HGF in GBM prognosis (Fig. 1B).
Fig. 1.
Clinical relevance of HGF in GBM. (A) TCGA microarray data analysis for HGF expression in GBM vs normal brain tissues. P-value was calculated using the Wilcoxon rank sum test. (B) TCGA IDH1 wild-type GBM patient survival analysis based on HGF expression levels. P-value was calculated using the log-rank test.
Cancer stem cells are functionally defined by their enhanced abilities for perpetual self-renewal and govern essential cellular functions that modulate tumor survivability. Previous studies have demonstrated that GBMs harbor a subpopulation of highly tumorigenic GSCs.25–28 GSCs are responsible for conventional treatment resistance and tumor relapse, emphasizing the need for an alternative therapeutic avenue.
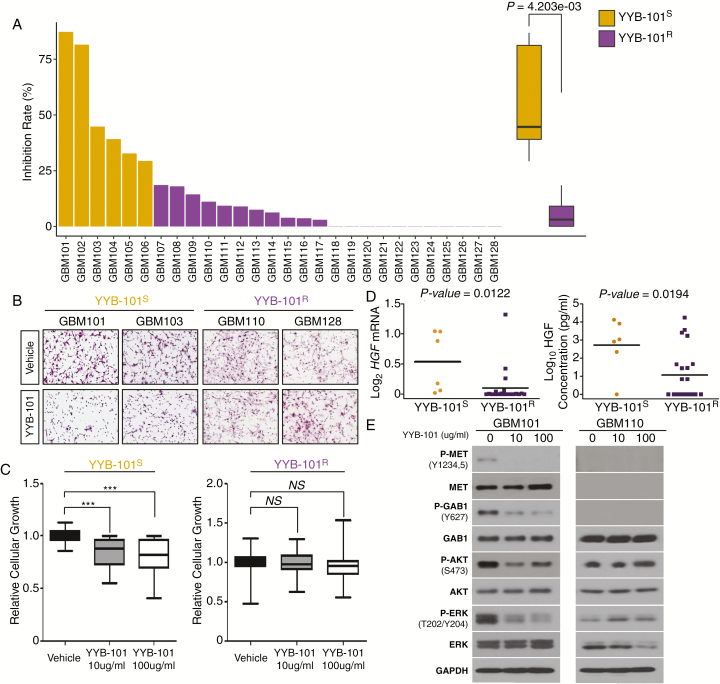
In order to evaluate the significance of HGF neutralization in GSC invasion and growth, we have employed YYB-101, a humanized version of the anti-HGF antibody, in a series of GSCs.29–35 Twenty-eight GSCs were treated with either 10 μg/mL of YYB-101 or vehicle and assessed for their invasive inhibitory effects. Six GBM tumor isolates exhibited significant reduction in tumor invasive capability in response to YYB-101, termed “YYB-101 sensitive,” or YYB-101S, while the rest of the tumors showed no notable inhibitory effects, termed “YYB-101 resistant,” or YYB-101R (Fig. 2A, B). We also investigated YYB-101’s ability to regulate cellular proliferation. Short-term proliferation kinetics of YYB-101S isolates showed a significant reduction in cellular growth compared with those of the YYB-101R group (Fig. 2C). When we compared mRNA and protein expression levels of HGF between the 2 groups, YYB-101S tumors showed significantly higher levels (Fig. 2D). To investigate the downstream molecular events of HGF signaling in GBM, we surveyed essential molecular effectors that are frequently activated during tumor progression. Immunoblot analysis revealed significant attenuation of phosphorylation levels in MET, GAB1, Akt, and extracellular signal-regulated kinase (ERK) of YYB-101S cells in response to YYB-101 treatment, whereas YYB-101R cells exhibited no significant changes (Fig. 2E). Furthermore, significant differences in the basal expression levels of these molecular effectors did not exist (Supplementary Figure S1). Collectively, these results support the notion that the neutralization of HGF inhibits the invasive and proliferative properties of GSCs in vitro.
Fig. 2 .
Invasive inhibitory effect of YYB-101 on GSC invasion and growth. (A) Invasion assays to determine the effect of YYB-101 treatment on GSCs. P-value was calculated using the Wilcoxon rank sum test. (B) Representative immunohistochemical images of cellular invasion assays using patient-derived GBM cells treated with or without YYB-101. (C) Relative cellular growth on GBM cells under the effect of YYB-101 treatment. P-values were calculated using paired Student’s t-tests. *P < 0.05; **P < 0.01; ***P < 0.001. (D) HGF mRNA (left panel) and protein (right panel) expression comparison between YYB-101S and YYB-101R tumors. P-values were calculated using the Wilcoxon rank sum test. (E) Immunoblots of important cellular modulators: p-MET, MET, p-GAB1, GAB1, p-Akt, Akt, p-ERK, and ERK. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control.
Genomic and Transcriptomic Characterization of YYB-101S and YYB-101R Tumors
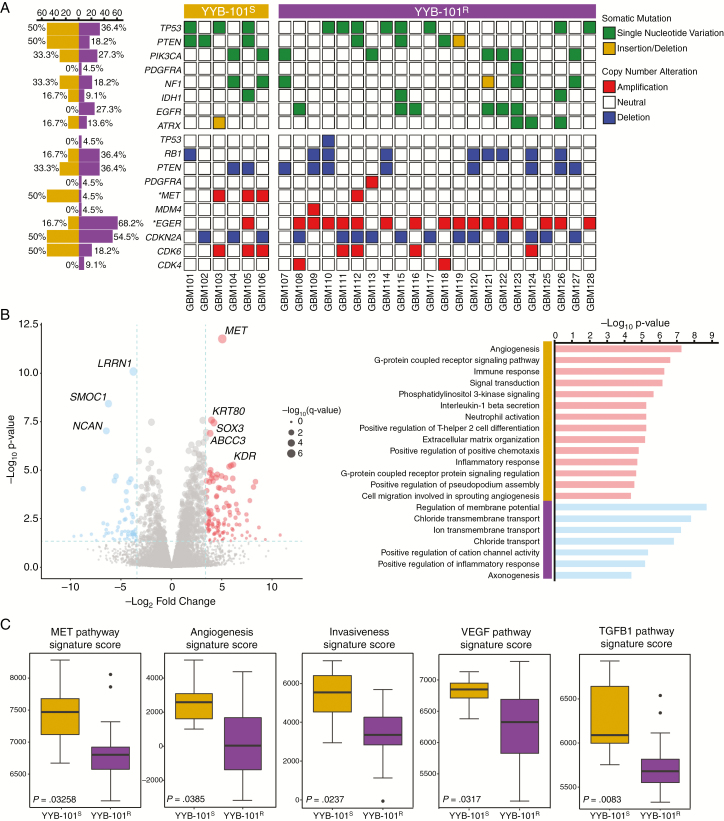
To assess differential genomic and transcriptomic traits that drive YYB-101S and YYB-101R tumors, 28 GSCs were subjected to whole-transcriptome sequencing and targeted panel sequencing called “GliomaSCAN.” Both groups harbored multiple somatic alterations in the core oncogenic pathways that are frequently dysregulated in GBM, including the p53, receptor tyrosine kinase, and phosphoinositide 3-kinase signaling pathways. Interestingly, YYB-101R isolates comprised mainly epidermal growth factor receptor (EGFR) amplified tumors, while YYB-101S tumors harbored higher frequency of MET amplification (Fig. 3A). Consistently, YYB-101R tumors were enriched with EGFR-associated pathways (Supplementary Figure S2). These results suggest that YYB-101R tumors did not exhibit any phenotypic response to YYB-101 because they are mediated by EGFR-dependent cellular growth and invasion.
Fig. 3.
Genomic and transcriptomic analysis of YYB-101S and YYB-101R tumors. (A) Mutational landscape of YYB-101S and YYB-101R tumors. Somatic mutations, including single-nucleotide variations (SNVs) and small insertions/deletions and copy number alterations, are shown. All somatic mutations with allele frequency of >5% are shown. An asterisk indicates statistical significance (P < 0.05) using Fisher’s exact test. (B) Left panel: A volcano plot representation of DEGseq analysis between YYB-101S and YYB-101R isolates. Genes with >2.5 log2 fold change and <0.05 P-value are colored in red, and those with <−2.5 log2 fold change and <0.05 P-value are colored in blue. Right panel: Gene Ontology (GO) analysis of differentially expressed genes from DEGseq output results. (C) Single-sample Gene Set Enrichment Analysis of corresponding signatures between YYB-101S and YYB-101R tumors. P-values were calculated using the Wilcoxon rank sum test.
Transcriptomic analysis of YYB-101S and YYB-101R revealed a set of differentially expressed genes, including MET, KDR, and SOX3. Of the 3 genes, MET demonstrated the most robust difference (Fig. 3B). The YYB-101S enriched transcriptomes, the “YYB-101S signature,” were associated with several biological mechanisms, including angiogenesis and cell migration based on Gene Ontology analysis (Fig. 3B). Notably, the YYB-101S signature demonstrated a significant correlation with GBM prognosis, as patients with higher expressions of the YYB-101S signature displayed worse survival probability (Supplementary Figure S3). On the contrary, YYB-101 treatment did not downregulate expression levels of the signature genes except for MET in YYB-101S tumors (Supplementary Figure S4). YYB-101S tumors also showed enrichment of MET, angiogenesis, invasiveness, vascular endothelial growth factor, and transforming growth factor–β1 associated pathways. These results corroborate with our previous in vitro evaluation of HGF-neutralizing effects on tumor invasiveness. Collectively, our results highlight the inhibitory effects of YYB-101 via combined genome and in vitro phenotypic analyses.
Invasive Inhibitory Effect of YYB-101 Is Mediated via the HGF-MET Signaling Axis
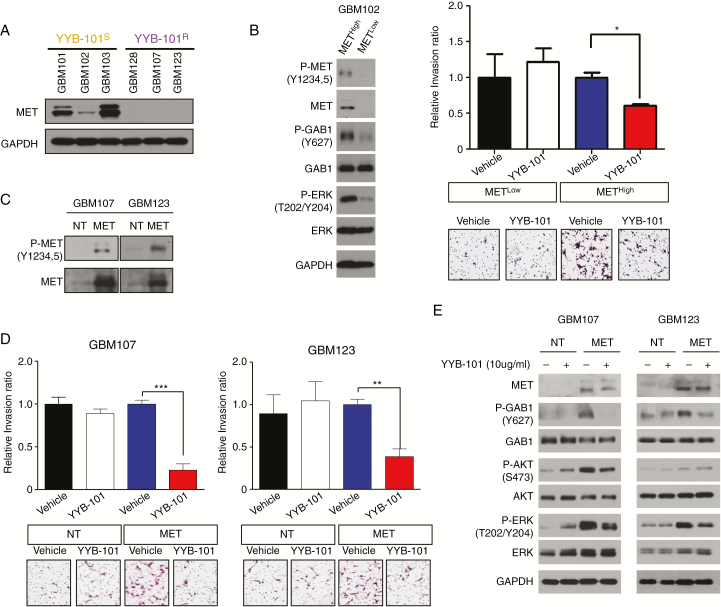
Previous results have demonstrated upregulation of MET in YYB-101S tumors. This suggests that aberrant activation of the HGF-MET signaling axis contributes to the invasive kinetics of GSCs. Consistently, immunoblot assay demonstrated enrichment of MET protein expressions in the YYB-101S isolates (Fig. 4A). To evaluate the HGF-MET signaling axis in GBM, we employed fluorescence-activated cell sorting technology to isolate METHigh and METLow subpopulations (Fig. 4B). Notably, METHigh tumor cells demonstrated higher phosphorylation levels of MET, GAB1, and ERK compared with METLow tumor cells. After confirming the purity and viability of the sorted populations, we determined the invasive kinetics of each subpopulation in the presence of YYB-101. As expected, METHigh cells exhibited significant invasive inhibitory response to YYB-101, in contrast to the static response of METLow cells (Fig. 4B). As we presumed MET to be the molecular determinant of YYB-101 response, we transduced YYB-101R GSCs with MET-expressing lentivirus and examined whether previous phenotypic response would fluctuate (Fig. 4C). As expected, MET transduction activated the HGF-MET signaling pathway, indicated by upregulation of p-GAB1, and showed substantially increased drug sensitivity to YYB-101 treatment (Fig. 4D, E). Consistently, YYB-101 treatment also attenuated phosphorylation levels of GAB1, Akt, and ERK in MET-overexpressed GSCs (Fig. 4E). Our results collectively demonstrate that the HGF-MET signaling axis is an essential component of the tumor invasive property of GSCs.
Fig. 4.
The HGF-MET signaling axis promotes cellular invasion in GBM. (A) immunoblots of MET and GAPDH in YYB-101S and YYB-101R isolates. (B) Left panel: Immunoblots of p-MET, MET, p-GAB1, GAB1, p-ERK, ERK, and GAPDH on METHigh and METLow subpopulations that were derived from fluorescence-activated cell sorting (FACS). Right panel: Relative cellular invasion ratio of METHigh and METLow tumor cells in presence of vehicle or YYB-101 treatments. P-values were calculated using Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001. (C) Immunoblots of p-MET and MET in nontarget (NT) or MET transduced cells. (D) Relative cellular invasion ratio of NT or MET transduced cells in presence of vehicle or YYB-101 treatments. P-values were calculated using Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001. (E) Immunoblots of MET, p-GAB1, GAB1, p-Akt, Akt, p-ERK, ERK, and GAPDH on NT or MET transduced cells in presence of vehicle or YYB-101 treatments.
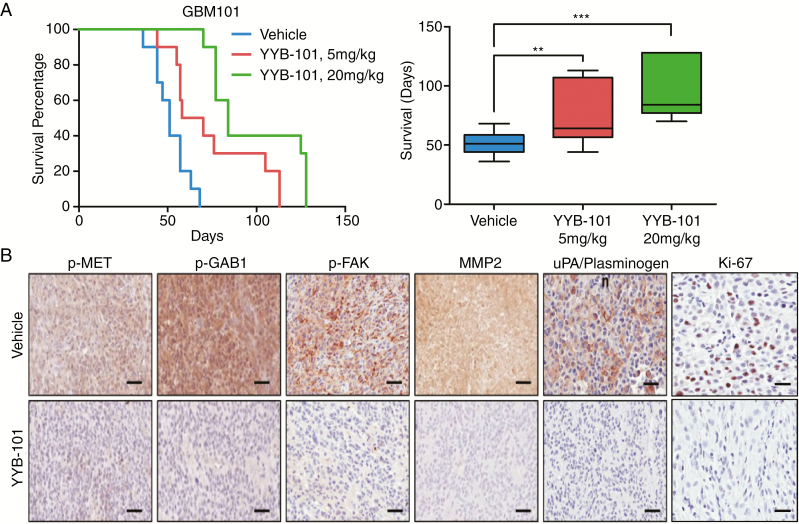
Targeting HGF Prolongs In Vivo Xenograft Survival in GBM
To investigate the functional role of HGF in tumor propagation in vivo, we generated patient-derived xenograft models using BALB/c nude mice. While a majority of the animals from the vehicle group died within 1–2 months (median survival: 51 days), animals that were treated with either YYB-101 (5 mg/kg body weight [mpk]) or YYB-101 (20 mpk) survived significantly longer (median survival: 58 or 84 days, respectively) (Fig. 5A). Based on immunohistochemical analysis where representative cellular proliferative and invasive markers such as Ki-67, MMP2, and uPA/plasminogen were used, significant reductions in cellular index were noted. Consistently, we also observed downregulated phosphorylation levels of MET, GAB1, and FAK in the YYB-101 treated group (Fig. 5B). Together, our results support HGF as a critical regulator of tumor growth in vivo.
Fig. 5 .
YYB-101 treatment impedes tumor growth in vivo and downregulates important cellular molecular effectors. (A) Kaplan–Meier survival curves and representative boxplot of vehicle vs YYB-101-treated (5 mpk, 20 mpk) groups in BALB/c nude mice. P-values were calculated using the log-rank test for survival curve and Student’s t-test for boxplot. (B) Representative immunohistochemical images of p-MET, p-GAB1, p-FAK, MMP2, uPA/plasminogen, and Ki-67 stainings. Scale bars: p-MET, p-GAB1, p-FAK, MMP2, and uPA/plasminogen, 200 μm; Ki-67, 100 μm.
Transcriptomic Analysis Reveals Association Between HGF and Mesenchymal Cellular State
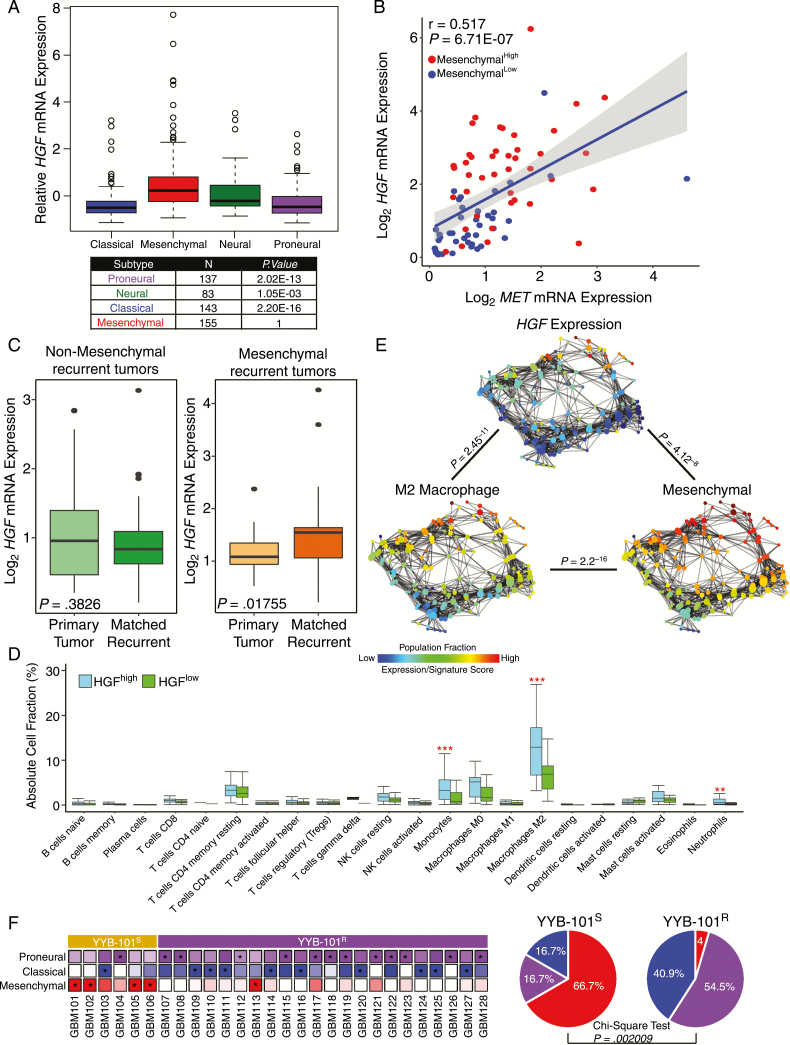
As previous genomic studies on large sets of GBM specimens portrayed distinct GBM subtypes based on transcriptome profiling,36,37 we determined potential correlations between expression levels of HGF and GBM subgroups by using the dataset of The Cancer Genome Atlas (TCGA). When we stratified GBM specimens into 4 distinct subtypes, HGF mRNA expression appeared to be the highest in the mesenchymal subgroup (Fig. 6A). Since the significance of the HGF-MET signaling pathway has been implicated in GBM progression, we further investigated the relationship between the HGF-MET signaling axis and the mesenchymal transcriptional subtype. As a surrogate indicator of mesenchymal activation, the metagene sets previously reported36 were used. Strikingly, mesenchymalHigh GBMs demonstrated significantly higher levels of both HGF and MET mRNA expressions in contrast to mesenchymalLow GBMs (Fig. 6B).
Fig. 6.
Association between HGF expression and mesenchymal transcriptional subtype in GBM. (A) TCGA GBM subtype analysis based on HGF mRNA expression. P-values were calculated using the Wilcoxon rank sum test. (B) Coexpression of HGF and MET mRNA in mesenchymalHigh and mesenchymalLow tumors. The P-value was calculated using Pearson’s correlation test. (C) HGF mRNA expression level between primary and matching recurrent tumors classified as nonmesenchymal (left panel) or mesenchymal (right panel). P-values were calculated using the Wilcoxon rank sum test. (D) Immune cell fraction analysis between HGFHigh and HGFLow tumors. Immune cell fractions were estimated using CIBERSORT and corrected using ESTIMATE purity scores for each sample. P-values were calculated using the Wilcoxon rank sum test. (E) Topological representation of transcriptome data for individual tumors. Each node represents a group of tumors with similar transcriptional profiles. A single tumor can appear in several nodes, and 2 nodes are connected by an edge if they share at least one tumor in common. P-values were calculated using Pearson’s correlation test between the fraction distribution of corresponding transcriptome expression, gene signature score, or population fraction over the nodes. (F) Left panel: Expression profiles of individual tumor isolates from YYB-101S and YYB-101R groups according to tumor-intrinsic subtypes. For each tumor, the subtype with the highest expression is marked with an asterisk. Right panel: Representative pie charts of mesenchymal, classical, and proneural glioma-intrinsic subtype frequencies in YYB-101S and YYB-101R isolates. P-value was calculated using the chi-square test.
Recent large-scale longitudinal GBM studies have demonstrated potential association between mesenchymal subtype at recurrence and dismal prognosis. Therefore, we evaluated changes in the HGF mRNA expression levels of 65 paired gliomas between diagnosis and relapse. Interestingly, we discovered significantly elevated levels of HGF transcriptome expression in recurrent tumors that were classified as mesenchymal, while no notable differences were observed from non-mesenchymal recurrent tumors (Fig. 6C). Our results suggest that the functional role of HGF could be responsible for tumor malignancy in mesenchymal recurrent GBMs and that the employment of HGF-targeting agent could potentially prevent such transitions from occurring. As mesenchymal subtype has been associated with high infiltration of immune cell population, we further investigated the immune cell composition between the HGFHigh and HGFLow GBMs by using CIBERSORT. As anticipated, tumor-promoting M2 macrophages were highly populated in HGFHigh tumors compared with HGFLow tumors (Fig. 6D). In addition to M2 tumor-associated macrophage abundance, there was a significantly greater fraction of monocytes and neutrophils in the HGFHigh GBMs as well. To further explore the relationship between immune cell population, transcriptome expression, and GBM subtype enrichment, we employed topological data analysis (TDA). TDA utilizes gene expression profiles of corresponding tumor samples, and by using the Mapper algorithm, it generates a low-dimensional network where nodes represent sets of tumors with similar global transcriptional profiles and 2 nodes are connected by an edge if they have at least one tumor in common. TDA reduces high dimensionality of large datasets while retaining local high-dimensional structure. Notably, our results showed that HGF transcriptome, tumor-promoting M2 macrophage fraction, and mesenchymal transcriptional activity were all significantly associated with one another. Our results highlighted that infiltration of tumor-associated macrophages and mesenchymal cellular state have significant association with HGF expression in GBM malignancy (Fig. 6E). Since higher expression levels of HGF could be derived from immune cells rather than tumor cells, HGF expression levels with mesenchymal activities were evaluated in GSCs. Both conventional and glioma-intrinsic subtype analyses showed significant correlation (Supplementary Figure S4), indicating that tumor-derived HGF triggers the mesenchymal cellular state and therefore potentially recruits tumor-associated macrophages in the process. Interestingly, when we applied glioma-intrinsic transcriptional subtype analysis38 to YYB-101S and YYB-101R tumors, we discovered that the YYB-101S group mainly exhibited higher degrees of the mesenchymal cellular state, while a majority of the YYB-101R tumors were identified as either proneural or classical (Fig. 6F). Collectively, our findings suggest that HGF is an important contributing factor in the oncogenicity of the mesenchymal GBM subgroup.
Discussion
Recent advancement in the field of genetics has uncovered core oncogenic pathways that are frequently dysregulated. Aberrant activation of the HGF-MET signaling axis has been studied extensively for its prominent role in triggering a series of intracellular signaling that promotes cellular proliferation, invasion, angiogenesis, and survival.39–44 High levels of HGF expression have been associated with dismal prognosis in multiple cancer types, including GBM, which makes HGF a promising therapeutic target in the clinical perspective.
Toward this goal, we have evaluated the therapeutic efficacy of YYB-101, an HGF-targeting antibody, in hopes of facilitating a more progressive course within a clinical framework. Through this approach, we distinguished a subset of GSCs with specific targeted vulnerabilities to HGF treatment and identified a set of genes that were specifically upregulated, the YYB-101S signature. Notably, the YYB-101S signature was significantly associated with tumor cell angiogenesis and invasiveness and correlated with unfavorable survival outcomes. Interestingly, YYB-101 resistant tumors comprised mainly EGFR-amplified tumors, suggesting they are mediated by EGFR-dependent cellular growth and invasion. Therefore, YYB-101R tumors remained ineffective against HGF neutralization. On the contrary, the YYB-101S group harbored higher frequency of MET amplification and/or transcriptional upregulation. We also demonstrated disruption of the HGF-MET signaling axis as a key molecular determinant in inhibiting GSC invasion and downregulating important cellular molecular effectors, including p-GAB1, p-Akt, and p-ERK. HGF neutralization also impeded GBM tumor growth in a patient-derived xenograft model.
Finally, transcriptome analysis revealed the dominance of mesenchymal-intrinsic tumors in the YYB-101S cohort, while YYB-101R isolates comprised mainly proneural or classical tumors. Consistently, TDA revealed that a significant correlation exists between HGF expression level with mesenchymal cellular state and the infiltrative M2 macrophage population. Previous studies have postulated high degrees of GBM subtype plasticity, demonstrating dynamic subtype transitions from diagnosis to recurrence.9,38,45,46 In particular, mesenchymal transition has been generally associated with dismal prognosis and treatment resistance in GBM progression, largely due to infiltration of tumor-associated immune cells. Tumor cells release several chemoattractant factors, including CSF-1, MCP-3, and HGF, which mediate macrophage recruitment,16 and in such a context, HGF-mediated therapy will prove to be an appealing therapeutic strategy against tumor-associated macrophage recruitment. In addition, a recent study has highlighted the association between the HGF/c-MET pathway to increased neutrophil counts and poor response to checkpoint inhibitors.19 Consistently, we have found that patients with elevated HGF expressions also had greater fractions of neutrophils, suggesting co-treatment of HGF antibody may improve the response to immunotherapy. Mesenchymal transcriptional subtype has also been identified in various tumor classes, including colorectal cancer,47,48 which suggests the potential therapeutic benefits of HGF-targeted therapy that can be acquired from a wide spectrum of malignant tumors.
Conclusively, our results demonstrate the prevalent role of the HGF signaling axis in GBM molecular architecture. Based on genomic and molecular evaluation of the HGF-neutralizing antibody in GBM, we showed that MET and mesenchymal cellular state act as essential components of therapeutic response to HGF-targeted therapy. Furthermore, the results of this study also support a combinational therapeutic strategy of the HGF-targeting antibody with immunotherapy. These results will provide a constructive groundwork for the assessment of HGF-targeted therapies in future clinical trials.
Supplementary Material
Supplementary material is available online at Neuro-Oncology online (http://neuro-oncology.oxfordjournals.org/).
Acknowledgment
We would like to thank members of the Translational Research Organization (TRO) team for helping out with in vivo experiments. We appreciate the helpful discussions with Dr. Na Kyung Lee. The biospecimens for this study were provided by Samsung Medical Center BioBank.
Conflict of interest statement. The authors declare no competing financial interests.
Funding
This work was supported by the Korea Health Technology R&D project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C3418). Additional support was from the National OncoVenture/National Cancer Center (HI11C1191).
References
- 1. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. [DOI] [PubMed] [Google Scholar]
- 3. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 4. Das S, Marsden PA. Angiogenesis in glioblastoma. N Engl J Med. 2013;369(16):1561–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tchaicha JH, Reyes SB, Shin J, Hossain MG, Lang FF, McCarty JH. Glioblastoma angiogenesis and tumor cell invasiveness are differentially regulated by β8 integrin. Cancer Res. 2011;71(20):6371–6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gilbertson RJ, Rich JN. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 2007;7(10):733–736. [DOI] [PubMed] [Google Scholar]
- 7. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee JK, Wang J, Sa JK, et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet. 2017;49(4):594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Y, Lal B, Kwon S, et al. The scatter factor/hepatocyte growth factor: c-met pathway in human embryonal central nervous system tumor malignancy. Cancer Res. 2005;65(20):9355–9362. [DOI] [PubMed] [Google Scholar]
- 11. Michieli P, Mazzone M, Basilico C, et al. Targeting the tumor and its microenvironment by a dual-function decoy Met receptor. Cancer Cell. 2004;6(1):61–73. [DOI] [PubMed] [Google Scholar]
- 12. Abounader R, Laterra J. Scatter factor/hepatocyte growth factor in brain tumor growth and angiogenesis. Neuro Oncol. 2005;7(4):436–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edakuni G, Sasatomi E, Satoh T, Tokunaga O, Miyazaki K. Expression of the hepatocyte growth factor/c-Met pathway is increased at the cancer front in breast carcinoma. Pathol Int. 2001;51(3):172–178. [DOI] [PubMed] [Google Scholar]
- 14. Aune G, Lian AM, Tingulstad S, et al. Increased circulating hepatocyte growth factor (HGF): a marker of epithelial ovarian cancer and an indicator of poor prognosis. Gynecol Oncol. 2011;121(2):402–406. [DOI] [PubMed] [Google Scholar]
- 15. Kasahara K, Arao T, Sakai K, et al. Impact of serum hepatocyte growth factor on treatment response to epidermal growth factor receptor tyrosine kinase inhibitors in patients with non-small cell lung adenocarcinoma. Clin Cancer Res. 2010;16(18):4616–4624. [DOI] [PubMed] [Google Scholar]
- 16. Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Riabov V, Gudima A, Wang N, Mickley A, Orekhov A, Kzhyshkowska J. Role of tumor associated macrophages in tumor angiogenesis and lymphangiogenesis. Front Physiol. 2014;5:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kennedy BC, Showers CR, Anderson DE, et al. Tumor-associated macrophages in glioma: friend or foe?J Oncol. 2013;2013:486912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glodde N, Bald T, van den Boorn-Konijnenberg D, et al. Reactive neutrophil responses dependent on the receptor tyrosine kinase c-MET limit cancer immunotherapy. Immunity. 2017;47(4):789–802 e789. [DOI] [PubMed] [Google Scholar]
- 20. Kim KH, Kim H. Progress of antibody-based inhibitors of the HGF-cMET axis in cancer therapy. Exp Mol Med. 2017;49(3):e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song SW, Lee SJ, Kim CY, et al. Inhibition of tumor growth in a mouse xenograft model by the humanized anti-HGF monoclonal antibody YYB-101 produced in a large-scale CHO cell culture. J Microbiol Biotechnol. 2013;23(9):1327–1338. [DOI] [PubMed] [Google Scholar]
- 22. Kim H, Hong SH, Kim JY, et al. Preclinical development of a humanized neutralizing antibody targeting HGF. Exp Mol Med. 2017;49(3):e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5): 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dirks PB. Brain tumor stem cells: the cancer stem cell hypothesis writ large. Mol Oncol. 2010;4(5):420–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. [DOI] [PubMed] [Google Scholar]
- 27. Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. [DOI] [PubMed] [Google Scholar]
- 28. Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100(25):15178–15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yin J, Oh YT, Kim JY, et al. Transglutaminase 2 inhibition reverses mesenchymal transdifferentiation of glioma stem cells by regulating C/EBPβ signaling. Cancer Res. 2017;77(18):4973–4984. [DOI] [PubMed] [Google Scholar]
- 30. Lee JK, Chang N, Yoon Y, et al. USP1 targeting impedes GBM growth by inhibiting stem cell maintenance and radioresistance. Neuro Oncol. 2016;18(1):37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sa JK, Yoon Y, Kim M, et al. In vivo RNAi screen identifies NLK as a negative regulator of mesenchymal activity in glioblastoma. Oncotarget. 2015;6(24):20145–20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee Y, Kim KH, Kim DG, et al. FoxM1 promotes stemness and radio-resistance of glioblastoma by regulating the master stem cell regulator Sox2. PLoS One. 2015;10(10):e0137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim KH, Seol HJ, Kim EH, et al. Wnt/β-catenin signaling is a key downstream mediator of MET signaling in glioblastoma stem cells. Neuro Oncol. 2013;15(2):161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim E, Kim M, Woo DH, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23(6):839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joo KM, Kim J, Jin J, et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 2013;3(1):260–273. [DOI] [PubMed] [Google Scholar]
- 36. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. [DOI] [PubMed] [Google Scholar]
- 38. Wang Q, Hu B, Hu X, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32(1):42–56 e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4(12):915–925. [DOI] [PubMed] [Google Scholar]
- 40. Jeffers M, Rong S, Vande Woude GF. Enhanced tumorigenicity and invasion-metastasis by hepatocyte growth factor/scatter factor-met signalling in human cells concomitant with induction of the urokinase proteolysis network. Mol Cell Biol. 1996;16(3):1115–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boccaccio C, Comoglio PM. Invasive growth: a MET-driven genetic programme for cancer and stem cells. Nat Rev Cancer. 2006;6(8):637–645. [DOI] [PubMed] [Google Scholar]
- 42. Boccaccio C, Andò M, Tamagnone L, et al. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature. 1998;391(6664):285–288. [DOI] [PubMed] [Google Scholar]
- 43. Holland EC. Gliomagenesis: genetic alterations and mouse models. Nat Rev Genet. 2001;2(2):120–129. [DOI] [PubMed] [Google Scholar]
- 44. Joo KM, Jin J, Kim E, et al. MET signaling regulates glioblastoma stem cells. Cancer Res. 2012;72(15):3828–3838. [DOI] [PubMed] [Google Scholar]
- 45. Bhat KPL, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang J, Cazzato E, Ladewig E, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. De Sousa E Melo F, Wang X, Jansen M, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–618. [DOI] [PubMed] [Google Scholar]
- 48. Sadanandam A, Lyssiotis CA, Homicsko K, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.