Abstract
In this survey we investigated a population of small mammals in Eastern Croatia in order to determine Leptospira carriage rates and identify circulating serovars. Out of 67 trapped animals, 20 (29.9%) isolates were obtained. Identification of isolates using microscopic agglutination test, pulsed field gel electrophoresis and multi locus sequence typing revealed that 10 (50.0%) isolates belong to serogroup Pomona, serovar Mozdok, 6 (30.0%) isolates to serogroup Australis, serovar Jalna, 2 (10.0%) isolates to serogroup Sejroe, serovar Saxkoebing, and 1 (5.0%) isolate to serogroup Grippotyphosa, serovar Grippotyphosa. One isolate from serogroup Bataviae was unable to be identified to the serovar level. Amplification of a 331-bp region of the locus LA0322 using real-time polymerase chain reaction determined that 12 (60.0%) isolates belong to L. kirschneri, 6 (30.0%) isolates to L. interrogans, and 2 (10.0%) isolates to L. borgpetersenii. Leptospira carriage rate was high (29.9%), which corresponds to a high incidence of human and domestic animal leptospirosis in Eastern Croatia. Furthermore, 90.0% of the isolates belong to serogroups Pomona, Australis and Sejroe which are also the most prevalent serogroups in humans in this area. These findings suggest that small mammals might be an important source of Leptospira spp. infection in Eastern Croatia.
Keywords: Leptospira, Leptospirosis, Reservoirs, Small mammals, Molecular, Zoonosis
1. Introduction
Leptospirosis is a zoonosis of worldwide distribution, caused by pathogenic spirochetes of the genus Leptospira (Levett, 2001). It affects all mammals, including humans, livestock and wildlife. Leptospires are very heterogenous and are classified phenotypically and genetically. Phenotypic classification with the serovar as the basic taxon is based on antigenic differences determined by the cross agglutination-absorption test (CAAT). So far, more than 300 serovars have been identified and grouped into 29 serogroups. Genetic classification is based on DNA homology and divides the genus Leptospira into 20 species (Smythe et al., 2013). Due to very demanding requirements and difficult implementation of CAAT, in the last few decades various molecular methods for identification of isolates to the serovar level have been developed. For that purpose pulsed field gel electrophoresis (Galloway and Levett, 2008, 2010), restriction fragment length polymorphism-based methods (Perolat et al., 1994), arbitrarily primed polymerase chain reaction (Ralph et al., 1993), variable number of tandem repeat analysis (Majed et al., 2005; Slack et al., 2005) and multi locus sequence typing (Thaipadungpanit et al., 2007) have been used. Genomic species of Leptospira spp. can be determined with various molecular methods, mostly based on 16S rDNA analysis. In the last decade, efforts have been made to develop real-time PCR for detection of pathogenic Leptospira from cultures and clinical materials (Levett et al., 2005; Merien et al., 2005; Fearnley et al., 2007; Ahmed et al., 2009). However, the combination of both molecular and serological identification methods still provides the best results in most cases.
Identification of Leptospira spp. isolates from small mammals is required to make a connection between human and animal disease and small mammal reservoirs because most infections are acquired from an environment contaminated with infected urine. Since clinical manifestations and disease outcome, among other things, might depend on the infecting Leptospira serovar, it is also of interest for clinicians to know what the circulating serovars in particular areas are.
Surveys conducted in the past decades revealed that Croatia is an endemic area of leptospirosis, especially in valleys of big rivers that flow from west to east through most of the lowland (Borcic et al., 1982; Milas et al., 2002; Turk et al., 2003). The importance of leptospirosis in Croatia is also highlighted by the official data from the Croatian National Institute of Health, according to which the mean yearly incidence of human leptospirosis from 1990 to 2007 was 1.83/100,000 inhabitants, with an incidence >2.5/100,000 inhabitants recorded approximately every 3–4 years (Balen Topic et al., 2010). These data make Croatia one of the countries with the highest incidence of human leptospirosis in Europe and in the world as well (Pappas et al., 2008). In the majority of previous reservoir surveys in Croatia, Leptospira carriage rates were investigated by serology and renal culture. Unfortunately, more thorough serological or molecular identification methods were unavailable so obtained isolates were not identified to the serovar level. Only in the latest study (Turk et al., 2003) were isolates identified to the serovar level, which set the ground for further epizootiological surveys.
The aim of this study was to determine Leptospira spp. carriage rates in a small mammal population and identify circulating serovars in order to correlate these data with incidence of human leptospirosis in Eastern Croatia.
2. Materials and methods
2.1. Animal trapping, sampling of kidneys and kidney culturing
During the relative abundance estimation of rodents in October 2005, at three sites in Eastern Croatia (Mikanovci, Cerna and Ilok), animals were trapped using 238 common snap traps positioned along transect lines (Fig. 1).
Fig. 1.
Localities in Eastern Croatia where animals were trapped.
Species of animals was determined based on morphological characteristics. Animals were aseptically dissected. Kidney tissue was immediately inoculated into the homemade Korthof’s medium and sampled for DNA extraction. Positive cultures were subcultured in Korthof’s medium until they reached stable growth, then subcultured to Ellinghausen-McCullough-Johnson-Harris (EMJH) liquid medium and cultivated up to a density of 2–4 × 108 leptospires per millilitre, suitable for serological and molecular identification procedures.
2.2. Microscopic agglutination test (MAT) with rabbit antisera
To identify the obtained isolates to the serogroup level, the microscopic agglutination test (MAT) was performed following the standard procedure using a panel of 18 rabbit anti-Leptospira reference antisera. Reference rabbit antisera used in this study were from the Koninklijk Instituut voor de Tropen (KIT), Amsterdam, The Netherlands (Table 1).
Table 1.
Reference strains used in production of rabbit antisera.
| Serogroup | Serovar | Strain |
|---|---|---|
| Australis | Australis | Ballico |
| Autumnalis | Autumnalis | Akiyami A |
| Bataviae | Bataviae | Van Tienen |
| Ballum | Castellonis | Castellon 3 |
| Canicola | Canicola | Hond Utrecht IV |
| Cynopteri | Cynopteri | 3522C |
| Grippotyphosa | Grippotyphosa | Moskva V |
| Hebdomadis | Hebdomadis | Hebdomadis |
| Icterohaemorrhagiae | Icterohaemorrhagiae | RGA |
| Icterohaemorrhagiae | Copenhageni | M 20 |
| Panama | Panama | CZ 214K |
| Pomona | Pomona | Pomona |
| Pyrogenes | Pyrogenes | Salinem |
| Semaranga | Patoc | Patoc I |
| Sejroe | Hardjo | Hardjoprajitno |
| Sejroe | Saxkoebing | Mus 24 |
| Sejroe | Sejroe | M 84 |
| Tarassovi | Tarassovi | Mitis Johnson |
2.3. DNA extraction from Leptospira cultures and animal kidneys
Reference strains and isolates were grown at 30°C in EMJH medium and harvested by centrifugation during the late logarithmic phase. Genomic DNA from animal kidneys and Leptospira isolates was extracted using QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions and stored at −20 °C.
2.4. Polymerase chain reaction (PCR)
Polymerase chain reaction was performed on all obtained isolates and kidneys of related animals using three primer pair sets. PCR with primers LeptoA (5′-GGCGGCGCGTCTTAAACATG-3′) and LeptoB (5′-TTCCCCCCATTGAGCAAGATT-3′) as described by Merien et al. (1992) was used to confirm the presence of Leptospira DNA in cultures and kidney tissue. Semi-nested PCR with two primer sets, L3 (5′-TGAGGGTTAAAACCCCCAAC-3′) and L4 (5′-GATTTTTCGGGTAAAGATT-3′) followed by L4 and Lepat2 (5′-TCACAT(CT)GCTGCTTATTTT-3′) as described by Gravekamp et al. (1993) was performed to confirm pathogenicity of Leptospira DNA present in cultures and kidney tissue. All products were electrophoresed in 1% agarose gel and compared to a molecular size marker.
2.5. Pulsed-field gel electrophoresis (PFGE)
Subsequent typing of the isolates to the serovar level was performed by pulsed-field gel electrophoresis (PFGE). Preparation of agarose plugs was performed as described by Galloway and Levett (2008). Genomic DNA of isolates from serogroups Pomona, Saxkoebing, Bataviae and Grippotyphosa was restricted with the endonuclease NotI and subjected to PFGE for 18 h with circulating 0.5 × TBE buffer. Electrophoresis conditions were as follows: switch times of 2.16 and 35.07 s, angle of 120 °, gradient of 6 V/cm, temperature of 14 °C, and linear ramping factor. Genomic DNA of isolates from serogroup Australis was restricted with endonuclease SgrAI and subjected to PFGE for 22 h with circulating 0.5 × TBE buffer. Electrophoresis conditions were as follows: switch times of 5 and 30 s, angle of 120°C, gradient of 6 V/cm, temperature of 14°C, and linear ramping factor. Gels were stained with ethidium bromide and analysed with Gel Doc 2000 System (Bio-Rad Laboratories, Richmond, CA, USA).
2.6. Multi locus sequence typing (MLST)
Subsequent typing of the isolates from serogroup Australis to the serovar level was performed by multi locus sequence typing (MLST). MLST was performed by amplifying and sequencing seven housekeeping genes as previously described (Thaipadungpanit et al., 2007). Sequence types (STs) were determined from the resulting allelic profiles of the seven sequenced genes and compared to an established Internet database to obtain serovar identification (http://www.mlst.net).
2.7. Real-time PCR with Tm determination
Real-time PCR was applied as described by Merien et al. (2005) with one primer set, LEB1-F (5′-CATTCATGTTTCGAATCATTTCAAA-3′) and LEB1-R (5′-GGCCCAAGTTCCTTCTAAAAG-3′), that amplifies a 331-bp of the locus LA0322 obtained from the complete genome sequence of L. interrogans serovar Lai. Melting temperature (Tm) of PCR product was used to distinguish particular Leptospira genomic species. Real-time PCR was performed on all Leptospira isolates and kidneys of Leptospira spp. positive animals to determine genomic species of Leptospira. All specimens were tested ten times and mean melting temperatures were calculated.
3. Results
3.1. Identification of trapped animals and isolation of Leptospira spp. from kidneys
The study included 67 animals, 30 (44.8%) from Ilok, 28 (41.8%) from Mikanovci, and 9 (13.4%) from Cerna. Out of 67 trapped animals, 29 (43.3%) were identified as Apodemus agrarius, 20 (29.8%) as A.flavicollis, 9 (13.4%) as A. sylvaticus, 6 (8.9%) as S. araneus, 1 (1.5%) as Muscardinus avellanarius, 1 (1.5%) as Microtus arvalis and 1 (1.5%) as Myodes glareolus. Kidney cultures from 20/67 (29.9%) animals were positive for Leptospira spp.: 10/20 from A. agrarius (M613, M615, M616, M619, M621, M634, M635, M639, M640, M656), 5/20 from A. flavicollis (M664, M666, M675, M676, M678), 3/20 from A. sylvaticus (M641, M644, M649), 1/20 from M. avellanarius (M628) and 1/20 from M. arvalis. Results are shown in Table 2.
Table 2.
Localities where animals were trapped, identified species of animals and numbers of animals with positive renal cultures.
| Locality/species | A. agrarius | A. flavicollis | A. sylvaticus | M. avellanarius | M. arvalis | S. araneus | M. glareolus | Total |
|---|---|---|---|---|---|---|---|---|
| Mikanovci | 9/18 | 0/2 | 0/0 | 1/1 | 1/1 | 0/6 | 0/0 | 11/28 |
| Cerna | 1/9 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 1/9 |
| Ilok | 0/2 | 5/18 | 3/9 | 0/0 | 0/0 | 0/0 | 0/1 | 8/30 |
| Total | 10/29 | 5/20 | 3/9 | 1/1 | 1/1 | 0/6 | 0/1 | 20/67 |
3.2. Microscopic agglutination test (MAT) with rabbit antisera
Serogroup identification of 20 isolates with 18 rabbit antisera revealed that 10/20 isolates belong to serogroup Pomona (M613, M615, M616, M619, M621, M628, M634, M635, M639, M656), 6/20 to serogroup Australis (M641, M644, M649, M666, M675, M678), 2/20 isolates to serogroup Sejroe (M664, M676), 1/20 isolate to serogroup Bataviae (M640) and 1/20 to serogroup Grippotyphosa (M631).
3.3. Pulsed-field gel electrophoresis and multi locus sequence typing
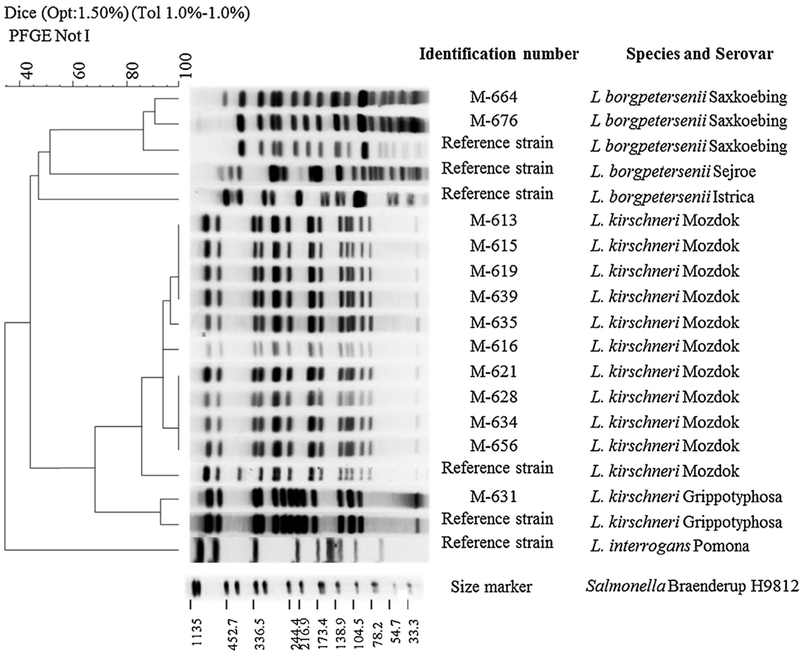
Patterns of ten isolates from serogroup Pomona (M613, M615, M616, M619, M621, M628, M634, M635, M639, M656) coincided with electrophoretic profile of reference strain L. kirschneri serovar Mozdok.
Patterns of two isolates from serogroup Saxkoebing (M664, M676) coincided with electrophoretic profile of reference strain L. borgpetersenii serovar Saxkoebing.
The pattern of the isolate from serogroup Grippotyphosa (M631) coincided with electrophoretic profile of reference strain L. kirschneri serovar Grippotyphosa.
The pattern of the isolate from serogroup Bataviae (M640) did not coincide with electrophoretic profile of any of the reference strains from L. kirschneri genomic species from serogroup Bataviae and remained undetermined. Selected PFGE patterns are shown in Fig. 2.
Fig. 2.
Dendrogram and electrophoretic profiles of NotI-restriction of whole genome of selected Leptospira isolates and reference strains.
Patterns of six isolates from serogroup Australis (M641, M644, M649, M666, M675, M678) coincided with electrophoretic profiles of reference strains L. interrogans serovar Jalna and L. interrogans serovar Lora. Therefore, isolates from serogroup Australis were further analysed with MLST and identified as sequence type (ST) 24, which is consistent with L. interrogans serovar Jalna but different from L. interrogans serovar Lora (ST 25).
3.4. Polymerase chain reaction (PCR)
PCRs with three primer pairs (LeptoA–LeptoB, L3–L4 and L4–Lepat2), performed on DNAs extracted from all obtained Leptospira isolates and kidneys of animals from which Leptospira spp. were isolated, resulted in products with the expected size of 331 bp, 643 bp and 270 bp, respectively.
3.5. Real-time PCR with Tm determination
Out of 20 isolates, 12 had mean melting temperatures characteristic for L. kirschneri (M613, M615, M616, M619, M621, M628, M631, M634, M635, M639, M640, M656), 6 forL. interrogans (M641, M644, M649, M666, M675, M678) and two for L. borgpetersenii (M664 and M676). Results of real-time PCR performed on 20 kidneys of animals from which Leptospira spp. were isolated, coincided with results of real-time PCR performed on isolated Leptospira spp. cultures. Results of genomic species determination are shown in Table 3.
Table 3.
PCR based determination of melting temperatures (Tm) fo.
| Isolate | Tm range/mean | Determined genomic species |
|---|---|---|
| M 613 | (84.31–84.76)/84.59 | L. kirschneri |
| M 615 | (84.32–84.74)/84.49 | L. kirschneri |
| M 616 | (84.23–84.84)/84.55 | L. kirschneri |
| M 619 | (84.21–84.73)/84.47 | L. kirschneri |
| M 621 | (84.12–84.70)/84.48 | L. kirschneri |
| M 628 | (84.12–84.69)/84.48 | L. kirschneri |
| M631 | (84.05–84.79)/84.53 | L. kirschneri |
| M 634 | (84.08–84.79)/84.22 | L. kirschneri |
| M 635 | (84.10–84.80)/84.55 | L. kirschneri |
| M 639 | (84.02–84.74)/84.41 | L. kirschneri |
| M 640 | (84.44–84.84)/84.62 | L. kirschneri |
| M 641 | (83.39–83.71)/83.60 | L. interrogans |
| M 644 | (83.39–83.70)/83.56 | L. interrogans |
| M 649 | (83.29–83.77)/83.60 | L. interrogans |
| M 656 | (84.22–84.72)/84.54 | L. kirschneri |
| M 664 | (86.00–86.51)/86.35 | L. borgpeterseni |
| M 666 | (83.25–83.69)/83.61 | L. interrogans |
| M 675 | (83.19–83.72)/83.61 | L. interrogans |
| M 676 | (85.94–86.30)/86.24 | L. borgpeterseni |
| M 678 | (83.18–83.63)/83.58 | L. interrogans |
Results of serogroup, serovar and genomic species determination as well as methods used are summarised in Table 4.
Table 4.
Results of identification and methods used.
| Isolate | Serogroup determined by MAT | Serovar determined by PFGE | Serovar determined by MLST | Genomic species determined by real time PCR(Tm) |
|---|---|---|---|---|
| M 613 | Pomona | Mozdok | L. kirschneri | |
| M 615 | Pomona | Mozdok | L. kirschneri | |
| M 616 | Pomona | Mozdok | L. kirschneri | |
| M 619 | Pomona | Mozdok | L. kirschneri | |
| M 621 | Pomona | Mozdok | L. kirschneri | |
| M 628 | Pomona | Mozdok | L. kirschneri | |
| M631 | Grippotyphosa | Grippotyphosa | L. kirschneri | |
| M 634 | Pomona | Mozdok | L. kirschneri | |
| M 635 | Pomona | Mozdok | L. kirschneri | |
| M 639 | Pomona | Mozdok | L. kirschneri | |
| M 640 | Bataviae | Undetermined | Undetermined | L. kirschneri |
| M 641 | Australis | Undetermined | Jalna | L. interrogans |
| M 644 | Australis | Undetermined | Jalna | L. interrogans |
| M 649 | Australis | Undetermined | Jalna | L. interrogans |
| M 656 | Pomona | Mozdok | L. kirschneri | |
| M 664 | Sejroe | Saxkoebing | L. borgpetersenii | |
| M 666 | Australis | Undetermined | Jalna | L. interrogans |
| M 675 | Australis | Undetermined | Jalna | L. interrogans |
| M 676 | Sejroe | Saxkoebing | L. borgpetersenii | |
| M 678 | Australis | Undetermined | Jalna | L. interrogans |
4. Discussion
In this survey we investigated distribution and abundance of a mouse-like rodent population and their Leptospira carriage rates in Eastern Croatia. Moreover, we determined genetic and antigenic variability of circulating Leptospira spp. serovars.
Species identification of trapped animals and numbers of animals with positive renal cultures is presented in Table 2. Prevalence of A. agrarius can probably be explained by the fact that two of three investigated sites were small wooded areas surrounded by fields, a common habitat of this species. Increased abundance of A. agrarius in woods is also attributable to reduction of food sources in fields during autumn, when trapping was conducted. This is, to our knowledge, the first report of Leptospira spp. isolation from M. avellanarius. The carriage rate detected in this investigation (29.9%) and the carriage rate of 29.5% detected in another investigation conducted in Croatia by Tadin et al. (2012) are the highest reported carriage rates in small mammals. In Europe, carriage rate of 12.1% in Switzerland (Adler et al., 2002) was reported. From other parts of the world, carriage rates of 0% in Madagaskar (Ralaiarijaona et al., 2001), 10.5% in Japan (Koizumi et al., 2008), 11% in Tanzania (Mgode et al., 2005), 12.6% in Korea (Cho et al., 1998) and 15% in Thailand (Doungchawee et al., 2005) were reported. A higher carriage rate than in our study (82.9%) was reported in domestic mice in Terceira island (Collares-Pereira et al., 2000). The highest Leptospira carriage rates among small mammals are reported in rats worldwide (90.5% in USA by Vinetz et al., 1996, 27.1% in Turkey by Sunbul et al., 2001, 63% in Thailand by Niwetpathomwat and Doungchawee, 2005, 50% in India by Priya et al., 2007, 80% in Brazil by Faria et al., 2008). The high carriage rate detected in this investigation further shows that Croatia is an endemic area of leptospirosis, especially in valleys of big rivers Sava and Drava (Borcic et al., 1982; Milas et al., 2002).
Serogrouping of isolates revealed that 10/20 belong to serogroup Pomona, 6/20 to serogroup Australis, 2/20 to serogroup Sejroe, 1/20 to serogroup Bataviae and 1/20 to serogroup Grippotyphosa. These results correlate to the latest serological surveys on human leptospirosis in Eastern Croatia which reports Pomona, Australis and Sejroe as the most prevalent serogroups (Peric et al., 2005; Cvitkovic, 2007). In this study 9/10 (90.0%) of L. kirschnerii serovar Mozdok serogroup Pomona were isolated from A. agrarius, which confirms this species as natural host of L. interrogans serogroup Pomona (Borcic et al., 1986).
Discrepancies between humoral response and actual carriership confirmed by renal culture or PCR were reported by several authors (Sunbul et al., 2001; Mgode et al., 2005; Priya et al., 2007). Due to evolutionary adaptation of rodent reservoirs to Leptospira spp. infection, serology is very limited in determination of carrier status. Low or absent antibody production in response to infection results in poor correlation between humoral response and actual carriership in reservoirs. That means that serological testing of reservoir animals certainly cannot be used for investigation of carriage rates and especially not for predicting the leptospiral serovars responsible for the infection. Knowing that, it is obvious that data on Leptospira carriage rates in most parts of the world are very insufficient. It is also unreliable to determine the infecting serovar or even serogroup based on serological testing.
Even in humans, where a stronger correlation of humoral response and infecting serovar is expected, it is often not possible to infer the infecting Leptospira serovar from the results of serological testing, especially in the early stage of infection (Levett, 2003; Murray et al., 2011). Identification of isolates to the serovar level is very important for complete understanding of epizootiology and epidemiology of the disease. We used pulsed-field gel electrophoresis (PFGE) and multi locus sequence typing (MLST) to identify 20 Leptospira isolates to the serovar level. Real time polymerase chain reaction (real-time PCR) with determination of melting temperature (Tm) was used to determine genomic species of Leptospira from cultures and kidney tissues.
PCR based Tm revealed that 12/20 (60.0%) isolates belong to L. kirschneri (10 to serovar Mozdok, one to serovar Grippotyphosa and one to undetermined serovar), 6/20 (30.0%) isolates to L. interrogans (serovar Jalna) and 2/20 (10.0%) isolates to L. borgpetersenii (serovar Saxkoebing).
Those results differ from a previously conducted study of small mammals in different parts of Croatia which reports 62.5% of isolates as L. borgpetersenii serovar Istrica, 31.3% as L. kirschneri serovar Tsaratsovo and 6.3% as L. interrogans serovar Lora (Turk et al., 2003). The variety of serovars observed in these two studies illustrates the overall diversity of Leptospira serovars present in Croatia.
Real-time PCR detected Leptospira spp. DNA in the kidneys of all animals from which Leptospira spp. isolates were obtained. At the same time, genomic species of Leptospira in tissue was determined. Results of genomic species determination from kidney tissue correlated with results obtained for Leptospira isolates cultures. The method we used is standardized for determination of Leptospira genomic species from cultures but is not widely used for determination and identification of Leptospira from tissues. Our investigation suggests that there is no interference of rodent DNA or any other limitations for using this method in reservoir surveys when culturing of Leptospira is not feasible due to either field or laboratory conditions as well as personnel skills. Due to the high carriage rate found, we can conclude that Croatia remains an endemic area of leptospirosis. Accordance of serogroups previously detected in humans and herein in animals indicates that small mammals might be a main source of infection for humans and animals. Analysis of isolated Leptospira to the serovar level revealed great genetic diversity of Leptospira circulating in Croatia.
Acknowledgement
This work was supported by Ministry of science, education and sports of the Republic of Croatia, scientific project number 053-1430115-211.
References
- Adler H, Vonstein S, Deplazes P, Steiger C, Frei R, 2002. Prevalence of Leptospira spp. in various species of small mammals caught in an inner-city area in Switzerland. Epidemiol. Infect 128, 107–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A, Engelberts MF, Boer KR, Ahmed N, Hartskeerl RA, 2009. Development and validation of a real-time PCR for detection of pathogenic Leptospira species in clinical materials. PLoS ONE 4, e7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BalenTopic M, Habus J, Milas Z, Tosev EC, Stritof Z, Turk N, 2010. Human leptospirosis in Croatia: current status of epidemiology and clinical characteristics. Trans. R. Soc. Trop. Med. Hyg 104, 202–206. [DOI] [PubMed] [Google Scholar]
- Borcic B, Kovacic H, Sebek Z, Aleraj B, Tvrtkovic N, 1982. Smallterrestrial mammals as reservoir of leptospires in the Sava Valley (Croatia). Folia Parasitol. 29, 177–182. [PubMed] [Google Scholar]
- Borcic B, Kovacic H, Sebek Z, Aleraj B, Tvrtkovic N, 1986. Field mouse (Apodemus agrarius Pall.) our natural reservoir of Leptospira serotype pomona. Vet. Arhiv 56, 169–178. [Google Scholar]
- Cho MK, Kee SH, Song HJ, Kim KH, Song KJ, Baek LJ, Kim HH, Oh HB, Kim YW, Chang WH, 1998. Infection rate of Leptospira interrogans in the field rodent Apodemus agrarius in Korea. Epidemiol. Infect 121, 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collares-Pereira M, Mathias ML, Santos-Reis M, Ramalhinho MG, Duarte-Rodrigues P, 2000. Small mammals and Leptospira transmission risk in Terceira island (Azores). Eur. J. Epidemiol 16, 1151–1157. [DOI] [PubMed] [Google Scholar]
- Cvitkovic A, 2007. Human leptospirosis in Slavonski Brod, 1995–2005. Acta Med. Croatica 61 (4), 349–353. [PubMed] [Google Scholar]
- Doungchawee G, Phulsuksombat D, Naigowit P, Khoaprasert Y, Sangjun N, Kongtim S, Smyth L, 2005. Survey of leptospirosis of small mammals in Thailand. Southeast Asian J. Trop. Med. Public Health 36 (6), 1516–1522. [PubMed] [Google Scholar]
- Faria MT, Calderwood MS, Athanazio DA, McBride AJA, Hartskeerl RA, Pereira MM, Ko AI, Reis MG, 2008. Carriage of Leptospira interrogans among domestic rats from an urban setting highly endemic for leptospirosis in Brazil. Acta Tropica 108, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnley C, Wakeley P, Gallego-Beltran J, Dalley C, Williamson S, Gaudie C, Woodward M, 2007. The development of a real-time PCRto detect pathogenic Leptospira species in kidney tissue. Res. Vet. Sci 85 (1), 8–16. [DOI] [PubMed] [Google Scholar]
- Galloway R, Levett PN, 2008. Evaluation of a modified pulsed-field gel electrophoresis approach for the identification of Leptospira serovars. Am. J. Trop. Med. Hyg 78, 628–632. [PubMed] [Google Scholar]
- Galloway R, Levett PN, 2010. Application and validation of PFGE for serovar identification of Leptospira clinical isolates. PLoS Negl. Trop. Dis 4 (9), 824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravekamp C, Van de Kemp H, Franzen M, Carrington D, Schoone GJ, Van Eys GJ, Everard CO, Hartskeerl RA, Terpstra WJ, 1993. Detection of seven species of pathogenic leptospires by PCRusing two sets of primers. J. Gen. Microbiol 139, 1691–1700. [DOI] [PubMed] [Google Scholar]
- Koizumi N, Muto M, Yamamoto S, Baba Y, Kudo M, Tamae Y, Shimomura K, Takatori I, Iwakiri A, Ishikawa K, Soma H, Watanabe H, 2008. Investigation of reservoir animals of Leptospira in the northern part of Miyazaki prefecture. Jpn. J. Infect Dis 61 (6), 465–468. [PubMed] [Google Scholar]
- Levett PN, 2001. Leptospirosis. Clin. Microbiol. Rev 14, 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levett PN, 2003. Usefulness of serologic analysis as a predictor of the infecting serovar in patients with severe leptospirosis. Clin. Infect. Dis 36, 447–452. [DOI] [PubMed] [Google Scholar]
- Levett PN, Morey RE, Galloway RL, Turner DE, Steigerwalt AG, Mayer LW, 2005. Detection of pathogenic leptospires by real-time quantitative PCR. J. Med. Microbiol 54, 45–49. [DOI] [PubMed] [Google Scholar]
- Majed Z, Bellenger E, Postic D, Pourcel C, Baranton G, Picardeau M, 2005. Identification of variable-number tandem-repeat loci in Leptospira interrogans sensu stricto. J. Clin. Microbiol 43 (2), 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merien F, Amouriaux P, Perolat P, Baranton G, Saint Girons I, 1992. Polymerase chain reaction for detection of Leptospira spp. in clinical samples. J. Clin. Microbiol 30, 2219–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merien F, Portnoi D, Bourhy P, Charavay F, Berlioz-Arthaud A, Baranton G, 2005. A rapid and quantitative method for the detection of Leptospira species in human leptospirosis. FEMS Microbiol. Lett 249 (1), 139–147. [DOI] [PubMed] [Google Scholar]
- Mgode GF, Mhamphi G, Katakweba A, Paemelaere E, Willekens N, Leirs H, Machang’u RS, Hartskeerl RA, 2005. PCR detection of Leptospira DNA in small mammals and insectivores from Tanzania. Belg. J. Zool 135, 17–19. [Google Scholar]
- Milas Z, Turk N, Staresina V, Margaletic J, Slavica A, Zivkovic D, Modric Z, 2002. The role of myomorphous mammals as reservoirs of Leptospira in the pedunculate oak forests of Croatia. Vet. Arhiv 72 (3), 119–129. [Google Scholar]
- Murray CK, Gray MR, Mende K, Parker TM, Samir A, Rahman BA, Habashy EE, Hospenthal DR, Pimentel G, 2011. Use of patient-specific Leptospira isolates in the diagnosis of leptospirosis employing microscopic agglutination testing (MAT). Trans. R. Soc. Trop. Med. Hyg 105 (4), 209–213. [DOI] [PubMed] [Google Scholar]
- Niwetpathomwat A, Doungchawee G, 2005. An investigation of rodent leptospirosis in Bangkok, Thailand. J. Vet. Res 9, 95–100. [Google Scholar]
- Pappas G, Papadimitriou P, Siozopoulou V, Christoub L, Akritidisc N, 2008. The globalization of leptospirosis: worldwide incidence trends. Int. J. Infect. Dis 12 (4), 351–357. [DOI] [PubMed] [Google Scholar]
- Peric L, Simasek D, Barbic J, Peric N, Prus V, Sisljagic V, Zibar L, 2005. Human leptospirosis in eastern Croatia, 1969–2003: epidemiological, clinical, and serological features. Scand. J. Infect. Dis 37 (10), 738–741. [DOI] [PubMed] [Google Scholar]
- Perolat P, Merien F, Ellis WA, Baranton G, 1994. Characterization of Leptospira isolates from serovar hardjo by ribotyping, arbitrarily primed PCR and mapped restriction site polymorphism. J. Clin. Microbiol 32, 1949–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priya CG, Hoogendijk KT, Berg M, Rathinam SR, Ahmed A, Muthukkaruppan VR, Hartskeerl RA, 2007. Field rats form a major infection source of leptospirosis in and around Madurai, India. J. Postgrad. Med 53, 236–240. [DOI] [PubMed] [Google Scholar]
- Ralaiarijaona RL, Bellenger E, Chanteau S, Roger F, Pérolat P, Rasolofo Razanam-parany V, 2001. Detection of leptospirosis reservoirs in Madagascar using the polymerase chain reaction technique. Arch. Inst. Pasteur Madagascar 67, 34–36. [PubMed] [Google Scholar]
- Ralph D, McClelland M, Welsh J, Baranton G, Perolat P, 1993. Leptospira species categorized by arbitrarily primed polymerase chain reaction (PCR) and by mapped restriction polymorphisms in PCR-amplified rRNA genes. J. Bacteriol 175, 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack AT, Dohnt MF, Symonds ML, Smythe LD, 2005. Development of a multiple-locus variable number of tandem repeat analysis (MLVA) for Leptospira interrogans and its application to Leptospira interrogans serovar Australis isolates from Far North Queensland, Australia. Ann. Clin. Microbiol. Antimicrob 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe L, Adler B, Hartskeerl RA, Galloway RL, Turenne CY, Levett PN, 2013. Classification of Leptospira genomospecies 1, 3, 4 and 5 as Leptospira alstonii sp. nov. Leptospira vanthielii sp. nov., Leptospira terpstrae sp. nov. and Leptospira yanagawae sp. nov., respectively. Int. J. Syst. Evol. Microbiol 63, 1859–1862. [DOI] [PubMed] [Google Scholar]
- Sunbul M, Esen S, Leblebicioglu H, Hokelek M, Pekbay A, Eroglu C, 2001. Rattus norvegicus acting as reservoir of Leptospira interrogans in the Middle Black Sea region of Turkey as evidenced by PCR and presence of serum antibodies to Leptospira strain. Scand. J. Infect. Dis 33, 896–898. [DOI] [PubMed] [Google Scholar]
- Tadin A, Turk N, Korva M, Margaletic J, Beck R, Vucelja M, Habus J, Svoboda P, Zupanc TA, Henttonen H, Markotic A, 2012. Multiple co-infections of small mammals with hantaviruses, Leptospira and Babesia in Croatia. Vector-Borne Zoonot. Dis 12 (5), 388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, et al. , 2007. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl. Trop. Dis 1, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk N, Milas Z, Margaletic J, Staresina V, Slavica A, Riquelme-Sertour N, Bellenger E, Baranton G, Postic D, 2003. Molecular characterisation of Leptospira spp. strains isolated from small rodents in Croatia. Epidemiol. Infect 130, 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinetz JM, Glass GE, Flexner CE, Mueller P, Kaslow DC, 1996. Sporadic urban leptospirosis. Ann. Intern. Med 125, 794–798. [DOI] [PubMed] [Google Scholar]