Abstract
Hydrogen sulfide (H2S), a known inhibitor of cytochrome c oxidase (CcOX), plays a key signaling role in human (patho)physiology. H2S is synthesized endogenously and mainly metabolized by a mitochondrial sulfide-oxidizing pathway including sulfide:quinone oxidoreductase (SQR), whereby H2S-derived electrons are injected into the respiratory chain stimulating O2 consumption and ATP synthesis. Under hypoxic conditions, H2S has higher stability and is synthesized at higher levels with protective effects for the cell. Herein, working on SW480 colon cancer cells, we evaluated the effect of hypoxia on the ability of cells to metabolize H2S. The sulfide-oxidizing activity was assessed by high-resolution respirometry, measuring the stimulatory effect of sulfide on rotenone-inhibited cell respiration in the absence or presence of antimycin A. Compared to cells grown under normoxic conditions (air O2), cells exposed for 24 h to hypoxia (1% O2) displayed a 1.3-fold reduction in maximal sulfide-oxidizing activity and 2.7-fold lower basal O2 respiration. Based on citrate synthase activity assays, mitochondria of hypoxia-treated cells were 1.8-fold less abundant and displayed 1.4-fold higher maximal sulfide-oxidizing activity and 2.6-fold enrichment in SQR as evaluated by immunoblotting. We speculate that under hypoxic conditions mitochondria undergo these adaptive changes to protect cell respiration from H2S poisoning.
1. Introduction
Hydrogen sulfide (H2S) has been increasingly recognized as a key signaling molecule in human (patho)physiology. While being able to regulate cell redox homeostasis and other crucial physiological functions at low (nM) concentrations [1–4], at higher (μM) levels, H2S exerts toxicity both inhibiting O2 consumption by cytochrome c oxidase (CcOX) in the mitochondrial electron transport chain [5] and impairing O2 transport/storage through covalent modification of the heme porphyrin ring in globins (reviewed in [6]). It is therefore crucial that cells tightly control H2S bioavailability to prevent toxicity.
In humans, at least three enzymes are directly involved in H2S synthesis (reviewed in [1, 7, 8]): cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE), belonging to the transulfuration pathway, and 3-mercaptopyruvate sulfurtransferase (MST). Of these, CBS is inhibited with relatively high affinity by nitric oxide (NO) and carbon monoxide (CO), particularly in the presence of the allosteric stimulator S-adenosyl-L-methionine [9–13]. H2S breakdown is instead mostly accomplished by a mitochondrial enzymatic pathway that couples the oxidation of H2S into thiosulfate (S2O32-) and sulfate (SO42-) to ATP synthesis [14]. The first step of sulfide breakdown is catalyzed by the membrane-associated sulfide:quinone oxidoreductase (SQR). This flavoprotein transfers electrons from H2S to coenzyme Q in the mitochondrial electron transfer chain, thus making H2S the first inorganic substrate that is able to sustain mitochondrial respiration [15]. Concomitantly, SQR transfers the H2S sulfur atom to an acceptor, leading to the formation of glutathione persulfide (GSSH) [16, 17] or, less likely, S2O32- [18, 19]. Differences in the SQR substrate specificity were recently reported comparing the soluble with the nanodisc-incorporated enzyme [20]. Three additional enzymes, persulfide dioxygenase (ETHE1), thiosulfate sulfurtransferase, and sulfite oxidase, cooperate with SQR in the mitochondrial sulfide oxidation pathway, to oxidize H2S into SO42- and S2O32-. To process 1 H2S molecule, mitochondria overall consume ~0.75 O2 molecules (0.25 by CcOX plus 0.5 by ETHE1, [21]). Besides being metabolized through the mitochondrial sulfide-oxidizing pathway, H2S can be oxidized by several metalloproteins such as globins, heme-based sensors of diatomic gaseous molecules, catalase, and peroxidases (see [8] and references therein) or be catabolized by the cytosolic thiol methyltransferase [22].
In vivo, H2S can therefore exert a dual effect on cell bioenergetics, at lower concentrations stimulating via SQR mitochondrial respiration and thus ATP synthesis or causing a reversible inhibition of CcOX at higher concentrations (reviewed in [23–26]). Notably, the sulfide-oxidizing activity varies considerably between different cell types and tissues, spanning from undetectable, as e.g., in neuroblastoma cells, to high, as observed in colonocytes [15, 21, 27]. The high H2S-detoxifying ability of colonocytes is perhaps not surprising as these cells are physiologically exposed to the fairly high H2S levels produced by the gut microbiota (reviewed in [28]).
Among other diseases, cancer has been increasingly associated with alterations of H2S metabolism [29–31]. In particular, CBS has been shown to be overexpressed in cell lines and samples of colorectal cancer [32] and other cancer types [33–36]. In colorectal cancer cell lines, CBS-derived H2S was proposed to promote cell proliferation and angiogenesis and to sustain cellular bioenergetics by stimulating both oxidative phosphorylation and glycolytic ATP synthesis. The enzyme is therefore currently recognized as a drug target [29, 31, 37]. CSE and CSE-derived H2S have been recognized as key elements in melanoma progression [38]. All three H2S-synthesizing enzymes have been posited to contribute to the correlation between increased H2S production and tumor stage and grade in bladder urothelial cell carcinoma [39]. Moreover, Szczesny et al. [36] observed higher expression levels of all three H2S-generating enzymes and increased H2S-producing activity in lung adenocarcinoma samples as compared to the adjacent normal lung tissue. A link between H2S production and mitochondrial DNA repair was proposed, and the inhibition of CBS and CSE by aminooxyacetic acid or siRNA-mediated depletion of CBS, CSE, or MST in the lung adenocarcinoma A549 cell line resulted in compromised integrity of mitochondrial DNA. Irrespectively of the downstream mechanisms linking increased H2S levels and cell proliferation and/or tumor progression, it remains to be established how cancer cells circumvent the potentially toxic effects of increased H2S.
Hypoxia is a common factor in the microenvironment of solid tumors that has been recognized to be associated to drug resistance and promotion of cancer progression, metastasization, and angiogenesis (see [40] for a review). The effect of hypoxia on cancer metabolism has been extensively investigated (reviewed in [41–43]). Among other changes, hypoxic cells undergo a reduction in mitochondrial mass, resulting from reduced biogenesis of this organelle and enhanced mitophagy [44–46]. Because mitochondria are the main site of sulfide oxidation, in the absence of compensatory mechanisms, hypoxic cells are expected to display a reduced ability to detoxify sulfide. The intricate interplay between H2S and O2 has been extensively investigated (reviewed in [47, 48]). As O2 facilitates both the chemical and enzymatic oxidative decomposition of H2S into persulfides and polysulfides, at low O2 tension a higher stability of H2S is expected. Furthermore, hypoxic/ischemic conditions have been reported to enhance H2S synthesis, through upregulation or stimulation of the sulfide-synthesizing enzymes [49, 50], accumulation of CBS in mitochondria, likely augmenting the H2S mitochondrial levels [51], and release of CO-mediated inhibition of CBS and CSE [52, 53]. Hypoxia is thus expected to increase H2S bioavailability, a condition that can have opposite physiological consequences. Indeed, while H2S has been shown to be protective against ischemic injuries [54, 55], the enhanced biosynthesis and chemical stability of H2S, combined with the reduced content in mitochondria (the main sites of sulfide disposal), may increase the risk of H2S toxicity in hypoxic cells.
This information prompted us to investigate in the present study the effect of hypoxia on the mitochondrial sulfide-oxidizing activity and SQR expression in colorectal cancer cells.
2. Materials and Methods
2.1. Materials
The human colon cancer cell line SW480 was purchased from the American Type Culture Collection (ATCC no. CCL228™). Sodium sulfide nonahydrate (Na2S·9H2O, 431648), 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), acetyl coenzyme A, oxaloacetate, CelLytic™ MT cell lysis reagent, protease inhibitor cocktail (P8340), and rabbit polyclonal antibody against human SQR (HPA017079) were purchased from Sigma. The bicinchoninic acid assay (BCA) kit was from Thermo Fisher Scientific. Cell culture media and antibiotics were from Sigma, EuroClone, or Gibco. Mini-PROTEAN TGX Stain-Free Precast Gels, the Clarity Western ECL Substrate, and the Laemmli protein sample buffer were purchased from Bio-Rad. Bovine serum albumin was from AppliChem.
2.2. Preparation of Sulfide Stock Solutions
Stock solutions of Na2S were prepared by quickly washing the surface of a crystal of sodium sulfide nonahydrate with degassed ultrapure (Milli-Q®) water and then dissolving it in degassed Milli-Q water under N2 atmosphere, as reported in [56]. The concentration of Na2S in solution was measured spectrophotometrically using 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) according to Nashef et al. [57] in a Cary 60 UV-VIS spectrophotometer. The concentration of Na2S was then adjusted to 3-5 mM by dilution with degassed ultrapure (Milli-Q®) water in a gas-tight glass syringe.
2.3. Cell Culture
The human colon cancer cell line SW480 was maintained in Dulbecco's Modified Eagle Medium (DMEM) containing 4.5 g·L−1 glucose, supplemented with 2 mM l-glutamine, 10% (v/v) heat-inactivated fetal bovine serum (FBS), 100 U·mL−1 penicillin, and 100 μg·mL−1 streptomycin. Cells at 37°C and 5% CO2 in 25 cm2 or 75 cm2 flasks were grown under normoxic conditions (air O2) or incubated for 24 h under hypoxic conditions (1% O2) in a Galaxy 14 S incubator (Eppendorf) designed to maintain cell cultures at controlled O2 tension. After trypsinization, the cells were washed in the culture medium, counted using the trypan blue dye exclusion test, centrifuged at 1000 ×g for 5 min, and resuspended in fresh medium at a final density of 8 × 106 cells·mL−1. Trypan blue-positive cells were always less than 5%. Cells grown under air conditions or exposed to hypoxia are, respectively, referred to as “normoxic” and “hypoxia-treated” cells.
2.4. Measurements of the Mitochondrial Sulfide-Oxidizing Activity
The mitochondrial sulfide-oxidizing activity of tested cells was evaluated as described in [25], by measuring the stimulatory effect of sulfide on cellular O2 consumption. Measurements were carried out at 37°C, using a high-resolution respirometer (Oxygraph-2k, Oroboros Instruments, Innsbruck, Austria), equipped with two 1.5 mL chambers and a micropump (TIP-2k) allowing for steady injections of relatively small amounts of sulfide into the chambers. According to Abou-Hamdan et al. [25], in these assays, sulfide is injected into a cell suspension at increasing flux (determined by the pump rate) and the mitochondrial sulfide-detoxifying activity is evaluated from the observed stimulation of cellular O2 consumption. Indeed, upon increasing the rate of sulfide injection, the concentration of sulfide in solution and, in turn, the sulfide-sustained cellular O2 consumption increase until the concentration of injected sulfide becomes inhibitory for CcOX. In colorectal cancer cells, SQR-mediated sulfide detoxification was shown to promote both forward electron transfer to O2 via quinol:cytochrome c reductase (complex III)/cytochrome c/CcOX and reverse electron transfer through complex I [21]. Therefore, measurements were herein carried out in the presence of rotenone, a known inhibitor of complex I, to prevent electrons derived from SQR-mediated sulfide oxidation to be partially diverted from O2 reduction with consequent underestimation of the mitochondrial sulfide-oxidizing activity. Herein, the assays were typically conducted in FBS-supplemented cell medium under stirring as follows. A suspension of four million cells was added into the respirometer chamber, and the basal respiration was measured for ~10 min. Afterwards, following the addition of 5 μM rotenone resulting in O2 consumption inhibition, a solution of 3-5 mM sulfide was injected for time intervals of 180 s at increasing rates (10 nL·s−1, 20 nL·s−1, 40 nL·s−1, 80 nL·s−1, and 160 nL·s−1) and the effect on O2 consumption was measured. Control experiments were carried out in the presence of both rotenone (5 μM) and antimycin A (5 μM), an inhibitor of complex III. The latter assays allowed us to evaluate the effect of sulfide on extramitochondrial and nonenzymatic O2 consumption and thus obtain by subtraction (from the experiments performed in the absence of antimycin A) the genuine mitochondrial O2 consumption activity due to sulfide oxidation and from it an estimate of the H2S-oxidizing activity, considering that ~1.33 molecules of H2S per O2 molecule are reportedly consumed by the mitochondrial sulfide-oxidizing pathway [21].
2.5. Evaluation of Mitochondrial Content by the Citrate Synthase Assay
Cells were harvested and lysed using the CelLytic™ MT cell lysis reagent and protease inhibitor cocktail from Sigma according to the manufacturer's instructions. Cell extracts were assayed spectrophotometrically for citrate synthase in 100 mM Tris-HCl, 0.3 mM acetyl-CoA, 0.1 mM DTNB and 0.1 mM oxaloacetate, as described in [58].
2.6. Immunoblotting Assays
Cells were harvested and lysed as described in the previous section, and after total protein content determination by the bicinchoninic acid method, proteins (20 μg per lane) were separated by SDS-PAGE using Mini-PROTEAN TGX Stain-Free Precast Gels (Bio-Rad). The formulation of these gels includes trihalo compounds which lead to UV fluorescence emission upon reaction with proteins [59], allowing estimation of the total protein load in a gel lane, using a ChemiDoc MP imaging system (Bio-Rad) without resorting to staining procedures or housekeeping proteins for normalization purposes. Proteins commonly used as housekeepers, such as glyceraldehyde 3-phosphate dehydrogenase and β-actin, indeed are known to change their expression levels under hypoxia [60, 61]. Afterwards, the proteins separated by SDS-PAGE were transferred onto a polyvinylidene difluoride membrane using a Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (from Bio-Rad) at 180 mA for 30 min. The membrane was blocked with PBS-T (phosphate-buffered saline with 0.1% Tween 20 (v/v)) containing 3% bovine serum albumin (BSA, w/v) and then incubated overnight at 4°C with the antibody against human SQR (1 : 150, in PBS-T with 3% BSA (w/v)). After three washing steps with PBS-T (15 min), the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (1 : 5000, in PBS-T with 3% BSA (w/v)), followed by three washing steps with PBS-T (15 min) and detection by enhanced chemiluminescence (Clarity Western ECL Substrate, Bio-Rad). Finally, the blotted membrane was subjected to densitometric analysis using the Image Lab software (Bio-Rad), followed by the normalization of the target protein band intensity to the total protein load determined as described above.
2.7. Data Analysis
Oxygen consumption rates (OCR) were calculated using the software DatLab4 (Oroboros Instruments, Austria). Data are reported as mean ± standard error of the mean (SEM). Statistical significance (P) was estimated using Student's t-test in Microsoft Excel. ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001 were considered significant.
3. Results
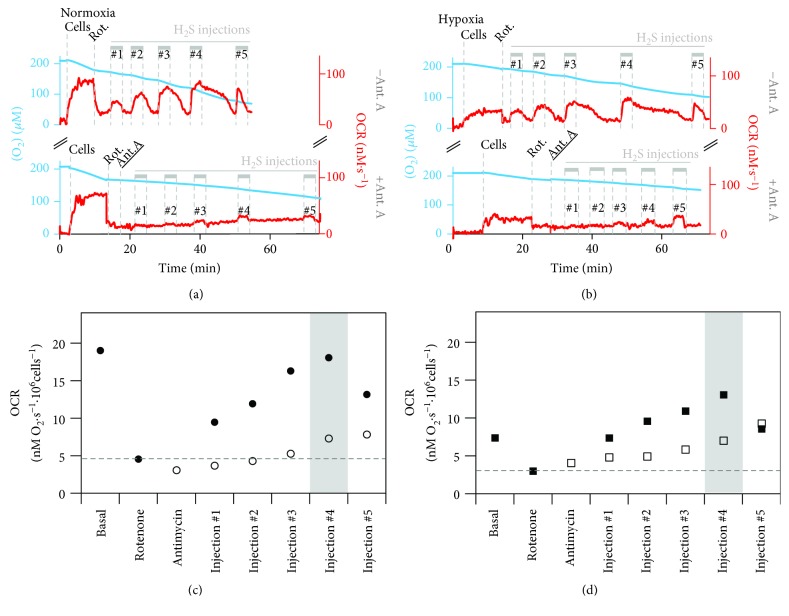
Colorectal cancer SW480 cells were either grown under normoxic (air O2) conditions or exposed for 24 h to hypoxia (1% O2), and their sulfide-oxidizing activity was assayed by high-resolution respirometry, according to Abou-Hamdan et al. [25], as described in Materials and Methods. A representative oxygraphic trace acquired with untreated (“normoxic”) cells is shown in Figure 1(a). The trace shows that ~80% of oxygen consumption was blocked by the addition of the complex I inhibitor rotenone, added to prevent sulfide oxidation through reversal of complex I activity, as described in [21, 62]. Sulfide was then injected five times at increasing rates into the oxygraphic chamber via a micropump. The first four injections led to the stimulation of O2 consumption, pointing to a fully operative mitochondrial sulfide-oxidizing pathway in the tested cells (Figures 1(a) and 1(b)). The stimulation persisted for the entire duration (3 minutes) of sulfide injection, after which the O2 consumption rate (OCR) declined back to the value measured in the absence of sulfide. The decline took a few minutes, as if some sulfide persisted in solution, sustaining cell respiration even after the injection was stopped. The extent of O2 consumption stimulation by sulfide increased with the rate of sulfide injection (up to 80 nL·s−1, Figures 1(a) and 1(c)). However, upon further increasing the injection rate (to 160 nL·s−1), a decline in OCR was observed already before sulfide injection was stopped, likely due to CcOX inhibition by sulfide, as suggested previously [25].
Figure 1.
Stimulation of O2 consumption by sulfide. Representative oxygen consumption traces (blue) and corresponding O2 consumption rate (OCR, red traces) acquired with normoxic (a) or hypoxia-treated SW480 cells (b), following the addition of cells (4 × 106), rotenone (Rot., 5 μM) either alone (top traces) or plus antimycin A (Ant. A, 5 μM, bottom traces), and subsequent injection of a sulfide solution (3-5 mM) at increasing rates (10 nL·s−1, 20 nL·s−1, 40 nL·s−1, 80 nL·s−1, and 160 nL·s−1, corresponding, respectively, to injections #1 to #5). (c, d) OCR values obtained from the oxygraphic traces, respectively shown in (a) and (b), measured at basal condition and upon sulfide injection at increasing rates after addition of rotenone alone (full symbols) or rotenone plus antimycin A (hollow symbols). Mitochondrial H2S consumption in normoxic cells was calculated by determining the OCR measured at the highest non-inhibitory H2S injection rate (highlighted with grey bar in (c)) and subtracting the OCR measured after the addition of rotenone (horizontal dashed line in (c)), yielding ΔOCR(-Ant). Then, the ΔOCR at the corresponding sulfide injection in the antimycin A-containing measurement was calculated in the same manner, yielding ΔOCR(+Ant). By calculating ΔOCR(‐Ant) − ΔOCR(+Ant), the genuine mitochondrial H2S-dependent OCR (OCRmitH2S) was determined. Finally, OCRmitH2S was multiplied by 1.33 to account for the number of H2S molecules consumed per O2 molecule, yielding an estimated sulfide oxidizing activity of 12.7 nM H2S·s−1·106 cells−1. Employing the same procedure for cells exposed to hypoxia (b, d), an activity of 9.5 nM H2S·s−1·106 cells−1 was estimated.
For comparison, the measurements described above were carried out on the same cells after 24 h exposure to hypoxic conditions. A representative oxygraphic trace is shown in Figure 1(b). Hypoxia-treated cells displayed a lower basal respiratory activity compared to untreated cells (6.3 ± 0.5 nM O2·s−1 vs. 17.1 ± 1.1 nM O2·s−1 per million cells). Yet, as observed for normoxic cells, after rotenone addition a progressive stimulation of cell respiration was observed upon injecting sulfide at an increasing rate (Figures 1(b) and 1(d)), until the amount of injected sulfide exceeded the detoxifying activity of the cells, and CcOX inhibition occurred, leading to impairment of cell respiration (see last sulfide injection in Figure 1(b), top).
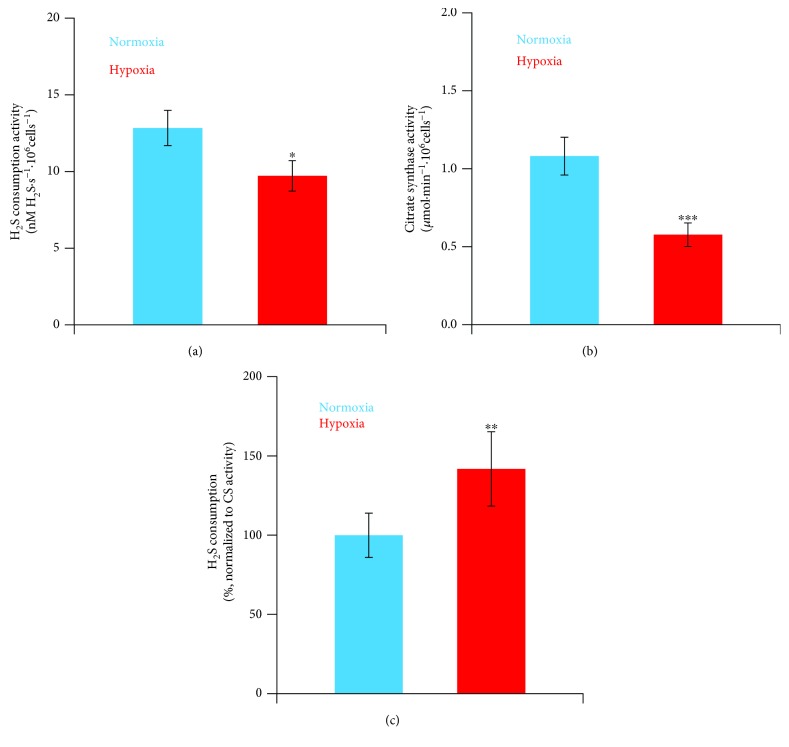
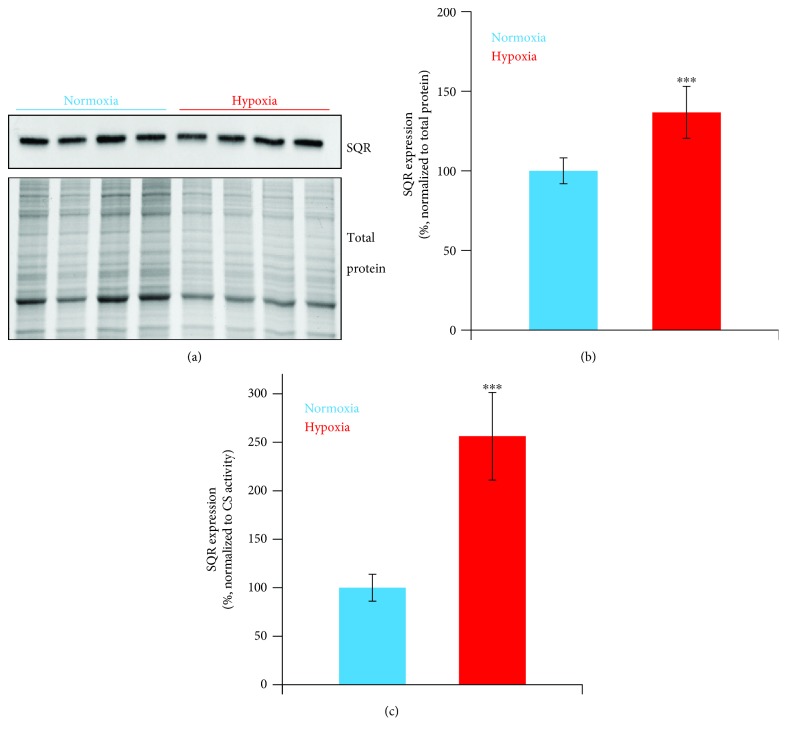
To evaluate the contribution of mitochondria to the observed sulfide-oxidizing activity, we used antimycin A, a known inhibitor of complex III that blocks quinol oxidation in the respiratory chain and thus prevents sulfide oxidation by mitochondria [25]. As shown in Figures 1(a) and 1(b) (bottom traces), in the presence of rotenone, antimycin A considerably prevented O2 consumption stimulation by sulfide in both normoxic and hypoxia-treated cells, proving that under the tested conditions sulfide oxidation occurs mostly at the mitochondrial level. The effect of sulfide on mitochondrial O2 consumption was quantitatively evaluated by subtracting the OCR values measured during sulfide injection in the presence of both rotenone and antimycin A from those measured at identical sulfide injection rates in the presence of rotenone only (see legend of Figure 1 for more details). According to this analysis, at the highest non-inhibitory (for CcOX) injection rate sulfide sustained a mitochondrial O2 consumption of 9.7 ± 1.2 nM O2·s−1 and 7.3 ± 0.8 nM O2·s−1 per million cells, in normoxic and hypoxia-treated cells, respectively. Considering that the mitochondrial sulfide-oxidizing pathway overall was reported to consume ~1.33 molecules of H2S per O2 molecule [21], a mitochondrial sulfide-oxidizing activity of 12.8 ± 1.5 and 9.7 ± 1.1 nM H2S·s−1 per million cells was estimated for normoxic and hypoxia-treated cells, respectively (Figure 2(a)). To evaluate the mitochondrial content in the tested cells, we carried out citrate synthase activity assays, a validated surrogate biomarker of mitochondrial content ([63] and references therein). Normoxic and hypoxia-treated cells displayed, respectively, a citrate synthase activity of 1.1 ± 0.1 μmol·min−1·106 cells−1 and 0.6 ± 0.1 μmol·min−1·106 cells−1 (Figure 2(b)), consistent with a reduction in the mitochondrial content upon exposure to hypoxia [44–46]. The measured citrate synthase activity was used to normalize the calculated mitochondrial sulfide-oxidizing activity, which proved to be in hypoxia-treated cells ~1.4-fold higher than in normoxic cells (Figure 2(c)). Finally, we have assayed by immunoblotting combined with “stain-free” imaging technology the SQR expression level in the tested cells (Figure 3(a)) and found that hypoxia-treated cells display 1.4-fold higher SQR protein levels than normoxic cells (Figure 3(b)). Considering that hypoxia-treated cells have a lower mitochondrial content (based on citrate synthase activity assays, Figure 2(b)), we estimate that the mitochondria of hypoxia-treated cells contain 2.6-fold more SQR than those of normoxic cells (Figure 3(c)).
Figure 2.
Effect of hypoxia on mitochondrial sulfide consumption. (a) Mean values of maximal estimated sulfide consumption activity (calculated as described in the legend of Figure 1(c)), measured in normoxic (n = 9, blue bar) and hypoxia-treated (n = 8, red bar) cells. (b) Citrate synthase activity in normoxic (n = 13, blue bar) and hypoxia-treated (n = 10, red bar) cell lysates. (c) Maximal sulfide consumption activity normalized to the citrate synthase activity, as measured in normoxic (blue bar) and hypoxia-treated (red bar) cells. ∗P ≤ 0.05; ∗∗P ≤ 0.01; ∗∗∗P ≤ 0.001.
Figure 3.
Effect of hypoxia on SQR expression. Representative Western blot analyzing SQR expression in normoxic and hypoxia-exposed SW480 cells (a), with the corresponding total protein load quantitation by stain-free imaging technology (see Materials and Methods). SQR levels in normoxic (n = 4 in triplicate, blue bars) and hypoxia-treated cells (n = 4 in triplicate, red bars), as normalized to total protein (b) or citrate synthase activity (c). ∗∗∗P ≤ 0.001.
4. Discussion
O2 and H2S are key molecules in living systems, able to control each other's availability, and regulate numerous processes in human (patho)physiology. As reviewed in [47], the interplay between H2S and O2 is intricate and based on several mechanisms: (i) direct reaction between the two, (ii) O2-dependent H2S breakdown through the mitochondrial sulfide-oxidizing pathway, (iii) H2S-mediated stimulation or inhibition of mitochondrial O2 consumption, (iv) O2-dependent regulation of expression and cellular relocalization of the H2S-synthesizing enzymes, and (v) O2-dependent control of CO-mediated inhibition of H2S production by CBS. H2S has indeed been recognized as an O2 sensor [64]. Despite this, to our knowledge no studies have been conducted yet to explore the effect of prolonged exposure to hypoxia on the cell ability to dispose of H2S, which represented the main objective of the present study.
Under hypoxic conditions, H2S plays a key protective role against ischemia/reperfusion damages [54, 55] through only partly understood molecular mechanisms including induction of antioxidant and vasorelaxation effects on microcirculation. Moreover, H2S appears to mediate the repair of damaged mitochondrial DNA [36], occurring in ischemia/reperfusion, and to protect from hypoxia-induced proteostasis disruption, as demonstrated in Caenorhabditis elegans [65]. In knockdown experiments with Hepa1-6 cells, H2S-mediated protection during O2 deprivation was found to require SQR [66], pointing to a key role of H2S catabolism in the cellular protective responses to hypoxia. Consistently, under hypoxic conditions, thiosulfate, a major product of H2S oxidation, has been shown to exert protective effects against ischemia/reperfusion damage [66–68] and also to generate H2S [69]. In this context, it is noteworthy that H2S is able to mimic hypoxia-induced responses such as vasodilation [70], neoangiogenesis [71], and expression of the hypoxia-inducible factor (HIF-1α, [72]), a master gene regulator promoting cell survival under hypoxic conditions shown to stimulate CBS expression in hypoxia [49]. The occurrence of H2S under hypoxic conditions is therefore likely part of a more general adaptive response adopted by the cells to ensure survival and protection from damages resulting from O2 deprivation (and possible reoxygenation).
In hypoxic cells, H2S bioavailability therefore needs to be finely regulated for this gaseous molecule to occur at physiologically protective yet non-poisonous levels. In this regard, it seems relevant to gain insight into the regulation of H2S production and breakdown at low O2 tensions. Previous studies focused on the H2S-synthesizing enzymes have shown that, under hypoxic conditions, H2S synthesis is enhanced [47] through multiple mechanisms [49–53] (see Introduction). In addition, H2S breakdown via both chemical and enzymatic reaction pathways is negatively affected by low O2 tensions. Evidence for a lower mitochondrial sulfide-oxidizing activity at lower O2 concentrations was initially provided in [73] working on immortalized cells derived from alveolar macrophages and, then, corroborated by Abou-Hamdan et al. in a more recent investigation on CHO cells [74].
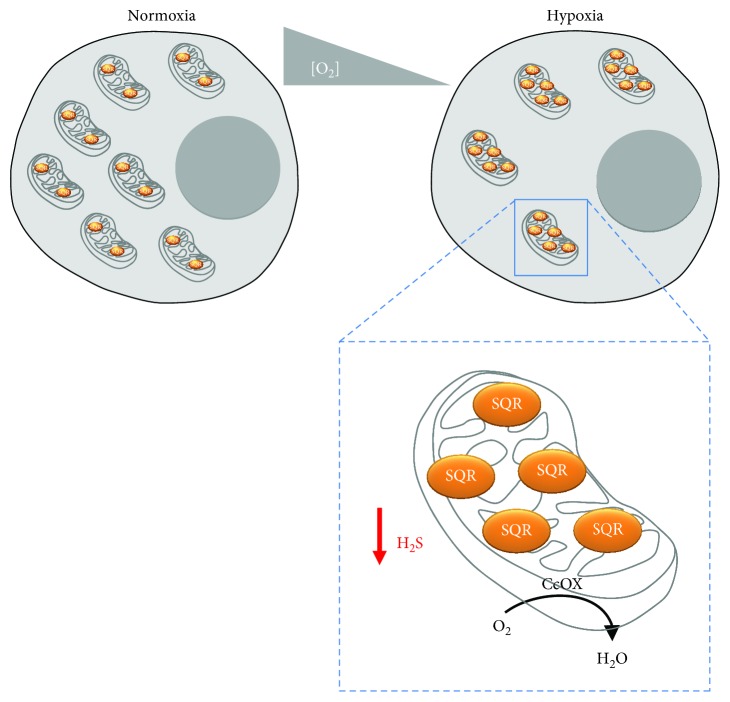
In the present study, using SW480 colorectal cancer cells as a model, we tested the effect of prolonged (24 h) exposure to 1% O2 on the cellular ability to dispose of sulfide at the mitochondrial level. Exposure to hypoxia leads to a notable (2.7-fold) reduction in basal respiration and to a marked (1.8-fold) decrease in the mitochondrial content (Figure 2(b)), as previously documented and suggested to result from enhanced mitophagic activity and reduced organelle biogenesis [44–46]. Hypoxia-treated cells also display a lower ability to dispose of H2S as compared to normoxic cells (Figure 2(a)). However, considering the above-mentioned decrease in mitochondrial content, the sulfide-detoxifying capacity of hypoxia-treated cells normalized to their minor mitochondrial content actually turned out to be 1.4-fold higher than that of untreated cells, pointing to an enhanced sulfide disposal capacity of mitochondria in hypoxia-treated cells. To gain further insight, we analyzed the SQR expression by immunoblotting, employing “stain-free” imaging technology for total protein quantitation and normalization purposes. Using this approach, we made the somewhat puzzling observation that hypoxia-treated cells, though displaying slightly reduced overall sulfide-oxidizing activity, have modestly (~1.4-fold) increased SQR levels. Interestingly, normalizing the SQR expression to the mitochondrial content revealed that, in line with their enhanced sulfide-oxidizing capacity, mitochondria of hypoxia-treated SW480 cells have ~2.6-fold higher levels of SQR than those of normoxic cells. Altogether, these results are intriguing in that they suggest that mitochondria in hypoxia-treated cells display lower mass but are enriched in SQR. The increased SQR levels could have a protective role in hypoxic cells preventing mitochondria to be poisoned by enhanced production of sulfide (Figure 4).
Figure 4.
Adaptive changes occurring in mitochondria in response to hypoxia. Upon prolonged exposure to hypoxia, mitochondria become less abundant, but enriched in sulfide:quinone oxidoreductase (SQR). Consistently, their maximal sulfide-oxidizing activity increases, while overall decreasing in the cell. These changes are proposed to occur to prevent H2S inhibition of cytochrome c oxidase (CcOX) and thus protect cell respiration from H2S poisoning.
5. Conclusions
This is to our knowledge the first study in which the effect of prolonged cell exposure to hypoxia on the mitochondrial sulfide-oxidizing activity has been evaluated. The evidence collected here on SW480 colorectal cancer cells shows that hypoxia-treated cells metabolize sulfide with overall reduced maximal efficacy and have reduced mitochondrial content, but mitochondria are better equipped to dispose of H2S. Physiologically, this may represent a regulatory mechanism to ensure higher protective H2S levels, while protecting mitochondria from H2S toxicity.
Acknowledgments
This work was partially supported by the Ministero dell'Istruzione, dell'Universita e della Ricerca of Italy (PNR-CNR Aging Program 2012–2014 and PRIN 20158EB2CM_003 to AG). iNOVA4Health Research Unit (LISBOA-01-0145-FEDER-007344), which is cofunded by Fundação para a Ciência e Tecnologia/Ministério da Ciência e do Ensino Superior, through national funds, and by FEDER under the PT2020 Partnership Agreement, is acknowledged by JBV.
Abbreviations
- H2S:
Hydrogen sulfide
- SQR:
Sulfide:quinone oxidoreductase
- NO:
Nitric oxide
- CO:
Carbon monoxide
- CcOX:
Cytochrome c oxidase
- CBS:
Cystathionine β-synthase
- CSE:
Cystathionine γ-lyase
- MST:
3-Mercaptopyruvate sulfurtransferase
- SO42-:
Sulfate
- S2O32-:
Thiosulfate
- DTNB:
5,5′-Dithiobis-(2-nitrobenzoic acid)
- PBS-T:
Phosphate-buffered saline with 0.1% Tween 20 (v/v)
- OCR:
Oxygen consumption rate.
Contributor Information
João B. Vicente, Email: jvicente@itqb.unl.pt.
Alessandro Giuffrè, Email: alessandro.giuffre@uniroma1.it.
Data Availability
The data used to support the findings of this study are available from the corresponding authors upon request.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- 1.Kabil O., Banerjee R. Enzymology of H2S biogenesis, decay and signaling. Antioxidants & Redox Signaling. 2014;20(5):770–782. doi: 10.1089/ars.2013.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ono K., Akaike T., Sawa T., et al. Redox chemistry and chemical biology of H2S, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Radical Biology & Medicine. 2014;77:82–94. doi: 10.1016/j.freeradbiomed.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuevasanta E., Moller M. N., Alvarez B. Biological chemistry of hydrogen sulfide and persulfides. Archives of Biochemistry and Biophysics. 2017;617:9–25. doi: 10.1016/j.abb.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Filipovic M. R., Zivanovic J., Alvarez B., Banerjee R. Chemical biology of H2S signaling through persulfidation. Chemical Reviews. 2018;118(3):1253–1337. doi: 10.1021/acs.chemrev.7b00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper C. E., Brown G. C. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. Journal of Bioenergetics and Biomembranes. 2008;40(5):533–539. doi: 10.1007/s10863-008-9166-6. [DOI] [PubMed] [Google Scholar]
- 6.Pietri R., Roman-Morales E., Lopez-Garriga J. Hydrogen sulfide and hemeproteins: knowledge and mysteries. Antioxidants & Redox Signaling. 2011;15(2):393–404. doi: 10.1089/ars.2010.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olson K. R. H2S and polysulfide metabolism: conventional and unconventional pathways. Biochemical Pharmacology. 2018;149:77–90. doi: 10.1016/j.bcp.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Giuffrè A., Vicente J. B. Hydrogen sulfide biochemistry and interplay with other gaseous mediators in mammalian physiology. Oxidative Medicine and Cellular Longevity. 2018;2018:31. doi: 10.1155/2018/6290931.6290931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puranik M., Weeks C. L., Lahaye D., et al. Dynamics of carbon monoxide binding to cystathionine β-synthase. Journal of Biological Chemistry. 2006;281(19):13433–13438. doi: 10.1074/jbc.M600246200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gherasim C., Yadav P. K., Kabil O., Niu W. N., Banerjee R. Nitrite reductase activity and inhibition of H2S biogenesis by human cystathionine ß-synthase. PLoS One. 2014;9(1, article e85544) doi: 10.1371/journal.pone.0085544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vicente J. B., Colaço H. G., Mendes M. I. S., Sarti P., Leandro P., Giuffrè A. NO• binds human cystathionine β-synthase quickly and tightly. Journal of Biological Chemistry. 2014;289(12):8579–8587. doi: 10.1074/jbc.M113.507533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vicente J. B., Colaço H. G., Sarti P., Leandro P., Giuffrè A. S-Adenosyl-L-methionine modulates CO and NO• binding to the human H2S-generating enzyme cystathionine β-synthase. Journal of Biological Chemistry. 2016;291(2):572–581. doi: 10.1074/jbc.M115.681221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vicente J. B., Colaço H. G., Malagrinò F., et al. A clinically relevant variant of the human hydrogen sulfide-synthesizing enzyme cystathionine β-synthase: increased CO reactivity as a novel molecular mechanism of pathogenicity? Oxidative Medicine and Cellular Longevity. 2017;2017:13. doi: 10.1155/2017/8940321.8940321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hildebrandt T. M., Grieshaber M. K. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. The FEBS Journal. 2008;275(13):3352–3361. doi: 10.1111/j.1742-4658.2008.06482.x. [DOI] [PubMed] [Google Scholar]
- 15.Goubern M., Andriamihaja M., Nubel T., Blachier F., Bouillaud F. Sulfide, the first inorganic substrate for human cells. The FASEB Journal. 2007;21(8):1699–1706. doi: 10.1096/fj.06-7407com. [DOI] [PubMed] [Google Scholar]
- 16.Mishanina T. V., Yadav P. K., Ballou D. P., Banerjee R. Transient kinetic analysis of hydrogen sulfide oxidation catalyzed by human sulfide quinone oxidoreductase. Journal of Biological Chemistry. 2015;290(41):25072–25080. doi: 10.1074/jbc.M115.682369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landry A. P., Ballou D. P., Banerjee R. H2S oxidation by nanodisc-embedded human sulfide quinone oxidoreductase. Journal of Biological Chemistry. 2017;292(28):11641–11649. doi: 10.1074/jbc.M117.788547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson M. R., Melideo S. L., Jorns M. S. Human sulfide:quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry. 2012;51(34):6804–6815. doi: 10.1021/bi300778t. [DOI] [PubMed] [Google Scholar]
- 19.Augustyn K. D., Jackson M. R., Jorns M. S. Use of tissue metabolite analysis and enzyme kinetics to discriminate between alternate pathways for hydrogen sulfide metabolism. Biochemistry. 2017;56(7):986–996. doi: 10.1021/acs.biochem.6b01093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landry A. P., Ballou D. P., Banerjee R. Modulation of catalytic promiscuity during hydrogen sulfide oxidation. ACS Chemical Biology. 2018;13(6):1651–1658. doi: 10.1021/acschembio.8b00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagoutte E., Mimoun S., Andriamihaja M., Chaumontet C., Blachier F., Bouillaud F. Oxidation of hydrogen sulfide remains a priority in mammalian cells and causes reverse electron transfer in colonocytes. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2010;1797(8):1500–1511. doi: 10.1016/j.bbabio.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Weisiger R. A., Pinkus L. M., Jakoby W. B. Thiol S-methyltransferase: suggested role in detoxication of intestinal hydrogen sulfide. Biochemical Pharmacology. 1980;29(20):2885–2887. doi: 10.1016/0006-2952(80)90029-5. [DOI] [PubMed] [Google Scholar]
- 23.Bouillaud F., Blachier F. Mitochondria and sulfide: a very old story of poisoning, feeding, and signaling? Antioxidants & Redox Signaling. 2011;15(2):379–391. doi: 10.1089/ars.2010.3678. [DOI] [PubMed] [Google Scholar]
- 24.Szabo C., Ransy C., Modis K., et al. Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part I. Biochemical and physiological mechanisms. British Journal of Pharmacology. 2014;171(8):2099–2122. doi: 10.1111/bph.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abou-Hamdan A., Guedouari-Bounihi H., Lenoir V., Andriamihaja M., Blachier F., Bouillaud F. Oxidation of H2S in mammalian cells and mitochondria. Methods in Enzymology. 2015;554:201–228. doi: 10.1016/bs.mie.2014.11.042. [DOI] [PubMed] [Google Scholar]
- 26.Vicente J. B., Malagrinò F., Arese M., Forte E., Sarti P., Giuffrè A. Bioenergetic relevance of hydrogen sulfide and the interplay between gasotransmitters at human cystathionine β-synthase. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2016;1857(8):1127–1138. doi: 10.1016/j.bbabio.2016.03.030. [DOI] [PubMed] [Google Scholar]
- 27.Mimoun S., Andriamihaja M., Chaumontet C., et al. Detoxification of H2S by differentiated colonic epithelial cells: implication of the sulfide oxidizing unit and of the cell respiratory capacity. Antioxidants & Redox Signaling. 2012;17(1):1–10. doi: 10.1089/ars.2011.4186. [DOI] [PubMed] [Google Scholar]
- 28.Blachier F., Davila A. M., Mimoun S., et al. Luminal sulfide and large intestine mucosa: friend or foe? Amino Acids. 2010;39(2):335–347. doi: 10.1007/s00726-009-0445-2. [DOI] [PubMed] [Google Scholar]
- 29.Hellmich M. R., Coletta C., Chao C., Szabo C. The therapeutic potential of cystathionine β-synthetase/hydrogen sulfide inhibition in cancer. Antioxidants & Redox Signaling. 2015;22(5):424–448. doi: 10.1089/ars.2014.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hellmich M. R., Szabo C. Hydrogen sulfide and cancer. Handbook of Experimental Pharmacology. 2015;230:233–241. doi: 10.1007/978-3-319-18144-8_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao X., Ding L., Xie Z. Z., et al. A review of hydrogen sulfide synthesis, metabolism, and measurement: is modulation of hydrogen sulfide a novel therapeutic for cancer? Antioxidants & Redox Signaling. 2018 doi: 10.1089/ars.2017.7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szabo C., Coletta C., Chao C., et al. Tumor-derived hydrogen sulfide, produced by cystathionine-β-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(30):12474–12479. doi: 10.1073/pnas.1306241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharyya S., Saha S., Giri K., et al. Cystathionine beta-synthase (CBS) contributes to advanced ovarian cancer progression and drug resistance. PLoS One. 2013;8(11, article e79167) doi: 10.1371/journal.pone.0079167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakraborty P. K., Xiong X., Mustafi S. B., et al. Role of cystathionine beta synthase in lipid metabolism in ovarian cancer. Oncotarget. 2015;6(35):37367–37384. doi: 10.18632/oncotarget.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sen S., Kawahara B., Gupta D., et al. Role of cystathionine β-synthase in human breast cancer. Free Radical Biology & Medicine. 2015;86:228–238. doi: 10.1016/j.freeradbiomed.2015.05.024. [DOI] [PubMed] [Google Scholar]
- 36.Szczesny B., Marcatti M., Zatarain J. R., et al. Inhibition of hydrogen sulfide biosynthesis sensitizes lung adenocarcinoma to chemotherapeutic drugs by inhibiting mitochondrial DNA repair and suppressing cellular bioenergetics. Scientific Reports. 2016;6(1, article 36125) doi: 10.1038/srep36125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Druzhyna N., Szczesny B., Olah G., et al. Screening of a composite library of clinically used drugs and well-characterized pharmacological compounds for cystathionine β-synthase inhibition identifies benserazide as a drug potentially suitable for repurposing for the experimental therapy of colon cancer. Pharmacological Research. 2016;113(Part A):18–37. doi: 10.1016/j.phrs.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panza E., De Cicco P., Armogida C., et al. Role of the cystathionine γ lyase/hydrogen sulfide pathway in human melanoma progression. Pigment Cell & Melanoma Research. 2015;28(1):61–72. doi: 10.1111/pcmr.12312. [DOI] [PubMed] [Google Scholar]
- 39.Gai J. W., Qin W., Liu M., et al. Expression profile of hydrogen sulfide and its synthases correlates with tumor stage and grade in urothelial cell carcinoma of bladder. Urologic Oncology: Seminars and Original Investigations. 2016;34(4):166.e15–166.e20. doi: 10.1016/j.urolonc.2015.06.020. [DOI] [PubMed] [Google Scholar]
- 40.Muz B., de la Puente P., Azab F., Azab A. K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia. 2015;3:83–92. doi: 10.2147/HP.S93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masson N., Ratcliffe P. J. Hypoxia signaling pathways in cancer metabolism: the importance of co-selecting interconnected physiological pathways. Cancer & Metabolism. 2014;2(1):p. 3. doi: 10.1186/2049-3002-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie H., Simon M. C. Oxygen availability and metabolic reprogramming in cancer. Journal of Biological Chemistry. 2017;292(41):16825–16832. doi: 10.1074/jbc.R117.799973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samanta D., Semenza G. L. Metabolic adaptation of cancer and immune cells mediated by hypoxia-inducible factors. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2018;1870(1):15–22. doi: 10.1016/j.bbcan.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H., Bosch-Marce M., Shimoda L. A., et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. Journal of Biological Chemistry. 2008;283(16):10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Solaini G., Baracca A., Lenaz G., Sgarbi G. Hypoxia and mitochondrial oxidative metabolism. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2010;1797(6-7):1171–1177. doi: 10.1016/j.bbabio.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 46.Wu H., Chen Q. Hypoxia activation of mitophagy and its role in disease pathogenesis. Antioxidants & Redox Signaling. 2015;22(12):1032–1046. doi: 10.1089/ars.2014.6204. [DOI] [PubMed] [Google Scholar]
- 47.Olson K. R. Hydrogen sulfide as an oxygen sensor. Antioxidants & Redox Signaling. 2015;22(5):377–397. doi: 10.1089/ars.2014.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu B., Teng H., Zhang L., et al. Interaction of hydrogen sulfide with oxygen sensing under hypoxia. Oxidative Medicine and Cellular Longevity. 2015;2015:9. doi: 10.1155/2015/758678.758678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takano N., Peng Y. J., Kumar G. K., et al. Hypoxia-inducible factors regulate human and rat cystathionine β-synthase gene expression. Biochemical Journal. 2014;458(2):203–211. doi: 10.1042/BJ20131350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang M., Guo Z., Wang S. Regulation of cystathionine γ-lyase in mammalian cells by hypoxia. Biochemical Genetics. 2014;52(1-2):29–37. doi: 10.1007/s10528-013-9624-7. [DOI] [PubMed] [Google Scholar]
- 51.Teng H., Wu B., Zhao K., Yang G., Wu L., Wang R. Oxygen-sensitive mitochondrial accumulation of cystathionine β-synthase mediated by Lon protease. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(31):12679–12684. doi: 10.1073/pnas.1308487110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morikawa T., Kajimura M., Nakamura T., et al. Hypoxic regulation of the cerebral microcirculation is mediated by a carbon monoxide-sensitive hydrogen sulfide pathway. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(4):1293–1298. doi: 10.1073/pnas.1119658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan G., Vasavda C., Peng Y. J., et al. Protein kinase G-regulated production of H2S governs oxygen sensing. Science Signaling. 2015;8(373, article ra37) doi: 10.1126/scisignal.2005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bos E. M., van Goor H., Joles J. A., Whiteman M., Leuvenink H. G. Hydrogen sulfide: physiological properties and therapeutic potential in ischaemia. British Journal of Pharmacology. 2015;172(6):1479–1493. doi: 10.1111/bph.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jensen A. R., Drucker N. A., Khaneki S., et al. Hydrogen sulfide: a potential novel therapy for the treatment of ischemia. Shock. 2017;48(5):511–524. doi: 10.1097/SHK.0000000000000894. [DOI] [PubMed] [Google Scholar]
- 56.Nagy P., Palinkas Z., Nagy A., Budai B., Toth I., Vasas A. Chemical aspects of hydrogen sulfide measurements in physiological samples. Biochimica et Biophysica Acta (BBA) - General Subjects. 2014;1840(2):876–891. doi: 10.1016/j.bbagen.2013.05.037. [DOI] [PubMed] [Google Scholar]
- 57.Nashef A. S., Osuga D. T., Feeney R. E. Determination of hydrogen sulfide with 5,5′-dithiobis-(2-nitrobenzoic acid), N-ethylmaleimide, and parachloromercuribenzoate. Analytical Biochemistry. 1977;79(1-2):394–405. doi: 10.1016/0003-2697(77)90413-4. [DOI] [PubMed] [Google Scholar]
- 58.Srere P. A. [1] Citrate synthase: [EC 4.1.3.7. Citrate oxaloacetate-lyase (CoA-acetylating)] Methods in Enzymology. 1969;13:3–11. doi: 10.1016/0076-6879(69)13005-0. [DOI] [Google Scholar]
- 59.Rivero-Gutierrez B., Anzola A., Martinez-Augustin O., de Medina F. S. Stain-free detection as loading control alternative to Ponceau and housekeeping protein immunodetection in Western blotting. Analytical Biochemistry. 2014;467:1–3. doi: 10.1016/j.ab.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 60.Zhong H., Simons J. W. Direct comparison of GAPDH, β-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochemical and Biophysical Research Communications. 1999;259(3):523–526. doi: 10.1006/bbrc.1999.0815. [DOI] [PubMed] [Google Scholar]
- 61.Heerlein K., Schulze A., Hotz L., Bartsch P., Mairbaurl H. Hypoxia decreases cellular ATP demand and inhibits mitochondrial respiration of A549 cells. American Journal of Respiratory Cell and Molecular Biology. 2005;32(1):44–51. doi: 10.1165/rcmb.2004-0202OC. [DOI] [PubMed] [Google Scholar]
- 62.Helmy N., Prip-Buus C., Vons C., et al. Oxidation of hydrogen sulfide by human liver mitochondria. Nitric Oxide. 2014;41:105–112. doi: 10.1016/j.niox.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 63.Larsen S., Nielsen J., Hansen C. N., et al. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. The Journal of Physiology. 2012;590(14):3349–3360. doi: 10.1113/jphysiol.2012.230185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olson K. R., Dombkowski R. A., Russell M. J., et al. Hydrogen sulfide as an oxygen sensor/transducer in vertebrate hypoxic vasoconstriction and hypoxic vasodilation. Journal of Experimental Biology. 2006;209(20):4011–4023. doi: 10.1242/jeb.02480. [DOI] [PubMed] [Google Scholar]
- 65.Fawcett E. M., Hoyt J. M., Johnson J. K., Miller D. L. Hypoxia disrupts proteostasis in Caenorhabditis elegans. Aging Cell. 2015;14(1):92–101. doi: 10.1111/acel.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hine C., Harputlugil E., Zhang Y., et al. Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell. 2015;160(1-2):132–144. doi: 10.1016/j.cell.2014.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marutani E., Yamada M., Ida T., et al. Thiosulfate mediates cytoprotective effects of hydrogen sulfide against neuronal ischemia. Journal of the American Heart Association. 2015;4(11) doi: 10.1161/JAHA.115.002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leskova A., Pardue S., Glawe J. D., Kevil C. G., Shen X. Role of thiosulfate in hydrogen sulfide-dependent redox signaling in endothelial cells. American Journal of Physiology-Heart and Circulatory Physiology. 2017;313(2):H256–H264. doi: 10.1152/ajpheart.00723.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Olson K. R., Deleon E. R., Gao Y., et al. Thiosulfate: a readily accessible source of hydrogen sulfide in oxygen sensing. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology. 2013;305(6):R592–R603. doi: 10.1152/ajpregu.00421.2012. [DOI] [PubMed] [Google Scholar]
- 70.Zhao W., Zhang J., Lu Y., Wang R. The vasorelaxant effect of H2S as a novel endogenous gaseous KATP channel opener. The EMBO Journal. 2001;20(21):6008–6016. doi: 10.1093/emboj/20.21.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Papapetropoulos A., Pyriochou A., Altaany Z., et al. Hydrogen sulfide is an endogenous stimulator of angiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(51):21972–21977. doi: 10.1073/pnas.0908047106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beaumont M., Andriamihaja M., Lan A., et al. Detrimental effects for colonocytes of an increased exposure to luminal hydrogen sulfide: the adaptive response. Free Radical Biology & Medicine. 2016;93:155–164. doi: 10.1016/j.freeradbiomed.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 73.Matallo J., Vogt J., McCook O., et al. Sulfide-inhibition of mitochondrial respiration at very low oxygen concentrations. Nitric Oxide. 2014;41:79–84. doi: 10.1016/j.niox.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abou-Hamdan A., Ransy C., Roger T., Guedouari-Bounihi H., Galardon E., Bouillaud F. Positive feedback during sulfide oxidation fine-tunes cellular affinity for oxygen. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2016;1857(9):1464–1472. doi: 10.1016/j.bbabio.2016.04.282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding authors upon request.