Abstract
Background
The abnormal expression of leucine-rich-alpha-2-glycoprotein 1 (LRG-1) is reported to be associated with multiple malignancies, but its role in the progression of pancreatic ductal adenocarcinoma (PDAC) remains to be determined.
Methods
The expression of LRG-1 was assessed in PDAC tissues by RT-PCR, Western blot and immunohistochemistry. LRG-1-silenced or overexpressed cell lines were constructed using shRNA or LRG-1-overexpressing plasmids. EdU incorporation assay, Transwell invasion and wound-healing assays were performed to evaluate the proliferation, invasion and migration of PDAC cells. In addition, protein expression of the mitogen-activated protein kinase (MAPK) pathway was detected using Western blot. Finally, Co-immunoprecipitation assay was conducted in search of the potential interaction between LRG-1 and epidermal growth factor receptor (EGFR).
Results
The expression of LRG-1 in PDAC tissue was significantly higher than that in adjacent normal tissue, and high LRG-1 expression predicted poor survival and a late tumor stage. In addition, LRG-1 markedly promoted the viability, proliferation, migration and invasion of PDAC cells in vitro and facilitated tumor growth in vivo. More importantly, we revealed that these bioactivities of LRG-1 might result from its selective interaction with EGFR, which might further activate the p38/MAPK signaling pathways.
Conclusion
LRG-1 may prove to be a promising biomarker for predicting prognosis of PDAC patients. Inhibition of LRG-1 or its downstream pathway could be a potential therapeutic target for the treatment of PDAC.
Keywords: LRG-1, Pancreatic ductal adenocarcinoma, Epidermal growth factor receptor (EGFR), p38, Biomarker
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignancies [1] with a median survival less than 6 months and a 5-year survival rate of less than 6% [2–5]. The incidence of PDAC is increasing yearly. According to GLOBOCAN estimates, there were 458,918 newly diagnosed cases of pancreatic cancer globally in 2018 [6]. The dismal prognosis of PDAC has persisted for decades, with only minimal improvement in these years, which might be ascribed to the ambiguous mechanism underlying the development and progression of PDAC [7].
A variety of signaling pathways have been reported to be involved in the development and progression of PDAC including mitogen-activated protein kinase (MAPK) [8] and Notch [9] signaling pathways, growth factors such as epidermal growth factor (EGF) [8], fibroblast growth factor (FGF) [10] and insulin-like growth factor 1 (IGF-1) [11]. Based on these findings, scientists have uncovered various biological targets, and developed several corresponding targeted therapies [12, 13]. However, concerns regarding their low efficacy, adverse events and high risk of recurrence are frustrating challenges for clinicians. This deficiency is primarily due to the poor understanding about the exact pathophysiology and the key driven gene in the initiation and development of PDAC.
In 1977, Leucine-rich-alpha-2-glycoprotein-1 (LRG-1) was first identified as an inflammatory protein in human serum by Haupt and Baudner [14]. However, few related studies have been reported and the function of LRG-1 remained unknown until 2013, when Wang et al. reported that LRG-1 was able to promote angiogenesis by modulating endothelial transforming growth factor β (TGF-β) signaling [15]. Since then, more studies on LRG-1 have emerged gradually and LRG-1 is recognized as a new regulator of tumorigenesis and a novel oncogene-associated protein [15, 16], playing an important role in epithelial-mesenchymal transition (EMT) and angiogenesis in colon cancer [16, 17]. LRG-1 is also known as a promising tumor biomarker and an independent prognostic factor for endometrial carcinoma [18] and non-small cell lung cancer [19]. In addition, studies demonstrated that LRG-1 promoted glioma cell invasion, migration and angiogenesis in the damaged retina [20]. Some recent studies demonstrated that serum LRG-1 level was significantly increased in PDAC patients and was correlated with progressive clinical stages [21]. Another study reported that the combination of tissue inhibitors of metalloproteinase-1 (TIMP-1), and LRG-1 to carbohydrate antigen 19–9 (CA 19–9) statistically significantly improves the detection of early-stage PDAC [22]. However, the prognostic value of LRG-1 in PDAC patients has not been reported and the effect of LRG-1 on human PDAC cells and its potential molecular mechanisms remain largely unknown.
The aim of the present study was to examine LRG-1 expression in tissue samples, evaluate the prognostic value of LRG-1 in PDAC patients, clarify the effect of LRG-1 on various cellular behaviors of PDAC cell lines both in vivo and in vitro, and explore potential signaling pathways and target proteins involved in the biochemical mechanism underlying the regulatory effect of LRG-1 on the pathogenesis of PDAC.
Materials and methods
Patients
This retrospective study involved 127 consecutive PDAC patients admitted to our hospital between 2011 and 2013. Patients enrolled in this study should be (a) aged 18–75 years; (b) without distant metastasis; (c) diagnosed with resectable PDAC and confirmed by postoperative pathology. Patients with the following criteria were excluded: (a) a previous history of treatment; (b) multi-organ dysfunction; and (c) contradictions for pancreatic surgery. All patients underwent a baseline assessment within one week before pancreatic surgery.
Cell culture
Human PDAC cell lines BxPc-3 and SW1990 were purchased from the ATCC (American Type Culture Collection). Cells were cultured in DMEM (Gibco, Grand Island, New York, USA) supplemented with 10% fetal bovine serum (FBS, Gibco), 1% penicillin and 1% streptomycin, and incubated at 37 °C in a humidified atmosphere with 5% CO2.
RNA extraction and qRT-PCR
Total RNA was isolated from the PDAC and adjacent normal specimens using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The RNA was reversely transcribed into cDNA with Oligo (dT) and M-MLV Reverse Transcriptase (Thermo Fisher Scientific). GAPDH was used as a reference gene. Primers of LRG-1 were as follows: 5’-GGACACCCTGGTATTGAAAGAAA-3′ (forward) and 5’-TAGCCGTTCTAATTGCAGCGG-3′ (reverse) [23].
Immunohistochemistry and evaluation
Paraformaldehyde-fixed paraffin-embedded tissue sections (5 μm) were prepared using a rabbit monoclonal immunoglobulin IgG specific for LRG-1 or EGFR or p-p38 (Abcam, Cambridge, UK, 1:200) and incubated overnight at 4 °C. The sections were developed with diaminobenzidine and counterstained with haematoxylin after incubation with secondary antibodies [24].
Western blot
Tissues lysates were electrophoresed on SDS-PAGE gel and transferred to PVDF membranes (Millipore, Bedford, MA, USA). The membranes were blocked with 5% skim milk and the primary antibodies were used for incubation overnight at 4 °C. After washing, PVDF membranes were incubated with secondary antibodies for 1 h. Immunoreactive bands were quantitatively analyzed with ImageJ software (http://imagej.nih.gov/ij/) [24]. The primary antibodies were as follows: anti-LRG-1, 1:1000; anti-Smad1/5, 1:1000 (Abcam, Cambridge, UK); anti-GAPDH, 1:10000; anti-MMP-2, 1:1000; anti-MMP-9, 1:1000; anti-TIMP-1, 1:1000; anti-JNK, 1:1000; anti-p-JNK, 1:1000; anti-ERK, 1:1000; anti-p-ERK, 1:1000; anti-p38, 1:1000; anti-p-p38, 1:1000; anti-p-Smad1/5, 1:1000 (Cell Signaling Technology, Beverly, MA, USA).
LRG-1 knockdown and over-expressing in PDAC cells
The LRG-1 shRNA was constructed in the pLKO.1 shRNA expression vector. Purified plasmids were transfected into HEK-293 T cells by using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA), along with helper plasmids psPAX2 and VSV-G. The virus supernatant was added on the cell culture in the presence of 8 mg/ml polybrene. Positive clones were obtained upon puromycin selection. The interference sequence of shRNA was as follows: 5’-AGCTAAAAAGATGTTTTCCCAGAATGACTCTCTTGA AGTCATTCTGGGAAAACATCGGG-3′ (shRNA-1), 5’-GATCCCCGATGTTTTCCCAGAATGACTTCAAGAGAGTCATTCTGGGAAAACATCTTTTT-3′ (shRNA-2).
The amplified LRG-1 coding region was cloned into the pUM-T vector, and positive recombinant plasmid was sequenced. Cells were transfected and selected for stable expression clones as described above. The amplification primers for the LRG-1 coding region were as follows: forward: 5’-GGCTGAAGCTTGCAGAGCTACCATGTCCTC-3′; reverse: 5’-TGATGGATCCTGGTCTCACTGGGACTTGG-3′.
Thiazolyl blue tetrazolium bromide (MTT) assay
For the cell viability assay, cells were seeded in 96-well plates (2.5 × 103 cells per well). After 24, 48, or 72 h, cell culture medium was replaced by MTT working solution, followed by 4 h incubation at 37 °C in a 5% CO2 incubator (Sigma-Aldrich Corp., St. Louis, MO, USA). After removing the MTT working solution and adding the DMSO, the absorbance at 490 nm were detected using a microplate reader (Tecan Group AG, Männedorf, Switzerland).
EdU incorporation assay
Click-iT EdU (5-ethynyl-2′-deoxyuridine) Alexa Fluor 488 Imaging Kit (Invitrogen, Carlsbad, CA, USA) was used to detect PDAC cell line proliferation according to the manufacturer’s instructions. Fluorescence was analyzed using a Zeiss 510 laser-scanning microscope (Zeiss, Thornwood, NY, USA) [24].
Wound-healing assay
PDAC cell lines were seeded in six-well plates (5 × 105 cells per well). A scratch wound was created using a micropipette tip when cells reached confluence. The narrowing of the wound area was investigated at 0 and 48 h. The area of each wound was measured via ImageJ software.
Transwell assays
The 48-well Transwell plates (Millipore, Bedford, MA, USA) with 8 μm pore filters were used for measuring cell migration. A total of 1 × 106/well PDAC cells were seeded in the upper chambers and then incubated with medium alone at 37 °C in a 5% CO2-filled incubator. PDAC cells migrating to the lower surface were stained with 0.5% crystal violet (Sigma-Aldrich, St. Louis, MO, USA) and photographed.
Co-immunoprecipitation assays
Cell extracts was diluted in IP lysis buffer and incubated with 1.5 μg normal rabbit IgG for 2 h, followed by 2 h of incubation with 5 μl protein A magnetic beads (Millipore, Bedford, MA, USA) to precipitate proteins that interacted non-specifically with IgG and/or protein A magnetic beads. This pre-cleared lysate was then incubated with 2 μg anti-LRG-1 antibody (Abcam, Cambridge, UK), at room temperature overnight. Protein A magnetic bead (20 μl) was added and incubated at room temperature for 6 h. Beads were finally washed 3 times using lysis buffer and eluted by incubating the beads 5 min at 70 °C in 25 μl in NuPAGE LDS sample buffer. Immunoprecipitated proteins were analyzed by Western blot analysis.
Xenograft mouse model
Totally 32 BALB/c nude mice (4–5 weeks old, 18–20 g) used in this study were obtained from Shanghai Medical College of Fudan University. All nude mice were routinely bred in a specific pathogen-free (SPF) laboratory in the animal center of Shanghai Medical College of Fudan University. The Mice were randomly divided into 4 groups: normal group, LRG-1-overexpressing group, LRG-1 shRNA-1 group and LRG-1 shRNA-2 group. The subcutaneous tumor models were established by respectively injecting 1 × 107 cells suspension into the right upper flanks of BALB/c nude mice respectively. The tumor volume was calculated using the formula (width2 × length)/2. After four weeks, the mice were euthanized, and subcutaneous tumors were removed and fixed in 4% paraformaldehyde.
Statistical analysis
SPSS 21.0 (IBM, Chicago, USA) was used to perform statistical analysis, and P < 0.05 was defined as the threshold of statistical significance. Normally distributed data were expressed as mean ± standard deviation (SD), and asymmetrically distributed data were expressed as median (range). Differences in outcomes between high and low expressions of LRG-1 were assessed for significance using independent-samples t tests. ROC curve was used to determine the sensitivity and specificity of prediction of LRG-1 for PDAC diagnosis. Kaplan-Meier method was used to calculate survival curves, and the significance was analyzed by log-rank test. Multivariate survival analysis was performed by Cox proportional hazards model.
Results
High LRG-1 expression predicts poor survival and late tumor stage
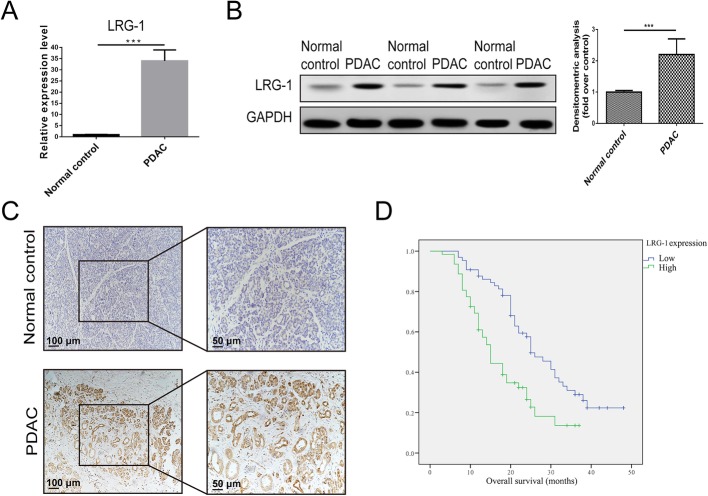
The expression level of LRG-1 was first detected in PDAC and adjacent normal tissues. It was found that mRNA and protein expression of LRG-1 in PDAC tissue was significantly higher than that in the adjacent normal tissue (Fig. 1a, b). To obtain pathological evidence, immunohistochemistry (IHC) was subsequently performed, and the results showed that the expression of LRG-1 in PDAC tissue was higher than that in the adjacent normal tissue (Fig. 1c).
Fig. 1.
LRG-1 is overexpressed in PDAC tissues. Overexpression of LRG-1 (a) mRNA and (b) protein in PDAC tissues than normal tissues. c Representative images of immunohistochemical staining of LRG-1 in normal tissues and PDAC tissues. d Survival analysis of PDAC patients with different LRG-1 expressions. Data are presented as the mean ± SD; ***p < 0.001
We further analyzed the expression of LRG-1 by IHC and other clinical factors in 127 PDAC patients. Our data showed that high LRG-1 expression (P = 0.001), late tumor stage (P < 0.001), and CA 19–9 levels (P = 0.020) were significant independent prognostic factors (Table 1). Patients with high LRG-1 levels had significantly poor survival than those with low LRG-1 levels (mean survival: 18.13 months vs. 28.66 months, P < 0.001) (Fig. 1d, Table 1). In addition, the logistic regression model showed that a higher LRG-1 expression was correlated with a later tumor stage (HR = 1.95, P = 0.034), a higher CA 125 (HR = 2.92, P = 0.023) and CA 19–9 level (HR = 1.70, P = 0.028) (Table 2).
Table 1.
Prognostic value of different risk factors
| Risk factors | Univariate regression analysis | Multivariate regression analysis | Overall survival | |||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | Mean ± SD, months | P value | |
| LRG-1 expression | ||||||||
| Group 1: Low | 2.29 | 1.47–3.57 | <0.001 | 2.30 | 1.43–3.71 | 0.001 | 28.66 ± 1.73 | <0.001 |
| Group 2: High | 18.13 ± 1.40 | |||||||
| Tumor stage | ||||||||
| Group 1: Stage I | 3.02 | 1.97–4.62 | <0.001 | 2.48 | 1.59–3.87 | <0.001 | 32.08 ± 2.53 | <0.001 |
| Group 2: Stage II | 22.41 ± 1.37 | |||||||
| Group 3: Stage III | 10.08 ± 1.24 | |||||||
| R1 resection | ||||||||
| Group 1: Yes | 2.87 | 1.23–6.66 | 0.015 | 2.16 | 0.89–5.25 | 0.089 | 12.50 ± 2.48 | 0.009 |
| Group 2: No | 24.88 ± 1.36 | |||||||
| Carbohydrate antigen 125 | ||||||||
| Group 1: <35 | 2.04 | 1.25–3.32 | 0.004 | 1.39 | 0.82–2.34 | 0.219 | 26.06 ± 1.51 | 0.003 |
| Group 2: ≥35 | 16.91 ± 2.06 | |||||||
| Carbohydrate antigen 19–9 | ||||||||
| Group 1: <37 | 1.68 | 1.26–2.24 | <0.001 | 1.42 | 1.06–1.90 | 0.020 | 31.01 ± 2.67 | 0.001 |
| Group 2: 37–200 | 23.78 ± 1.55 | |||||||
| Group 3: >200 | 18.00 ± 1.89 | |||||||
Table 2.
Risk factors correlated with LRG-1 expression using Logistic regression model
| Risk factors | HR | 95% Confidence Interval | P value |
|---|---|---|---|
| Tumor stage | 1.95 | 1.05–3.61 | 0.034 |
| R1 resection | 8.15 | 0.97–68.27 | 0.053 |
| Carbohydrate antigen 125 | 2.92 | 1.16–7.32 | 0.023 |
| Carbohydrate antigen 19–9 | 1.70 | 1.06–2.72 | 0.028 |
LRG-1 promotes PDAC cell viability, proliferation, invasion and migration
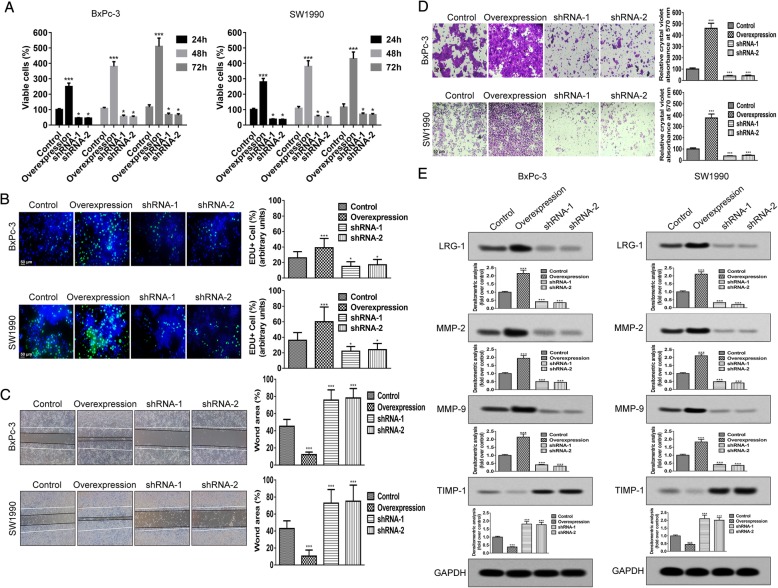
To determine the effect of LRG-1 in PDAC cells, LRG-1 knockdown or overexpressing experiments were performed in BxPc-3 and SW1990 cells. Once LRG-1 was attenuated by specific shRNA, the viability of BxPc-3 and SW1990 cells was significantly inhibited (Fig. 2a). Additionally, our EdU incorporation assay demonstrated that the proportion of EdU-positive cells decreased markedly after LRG-1 downregulation (Fig. 2b). Further wound healing assays revealed a profound migratory defect in LRG-1-depleted cells (Fig. 2c) and transwell assays observed a consistent reduction (Fig. 2d). Moreover, the downregulation of LRG-1 also reduced the expression of critical factors participating in tumor metastasis, including MMP-2 and MMP-9 (Fig. 2e). The expression of TIMP-1 was correspondingly up-regulated (Fig. 2E). In contrast, over-expression of LRG-1 could significantly promote cell viability (Fig. 2a), proliferation (Fig. 2b), migration (Fig. 2c) and invasion (Fig. 2d). In addition, enhanced LRG-1 expressions also upregulated the expression of MMP-2 and MMP-9, and reduced TIMP-1 expression simultaneously (Fig. 2e).
Fig. 2.
LRG-1 promotes PDAC cell viability, proliferation, invasion and migration in vitro. a MTT assays at 24, 48, or 72 h in LRG-1 knockdown and overexpression PDAC cells. b EdU incorporation assay at 48 h in LRG-1 knockdown and overexpression PDAC cells. EdU (green) was used to label proliferating cells and the nucleus was stained with DAPI (blue). Wound-healing assay (c) and Transwell cell invasion assay (d) at 48 h in LRG-1 knockdown and overexpression PDAC cells. e The protein level of MMP-2, MMP-9 and TIMP-1 at day 3 in LRG-1 knockdown and overexpression PDAC cells. Data are presented as the mean ± SD, *p < 0.05; **p < 0.01; ***p < 0.001
LRG-1 promotes PDAC growth in vivo
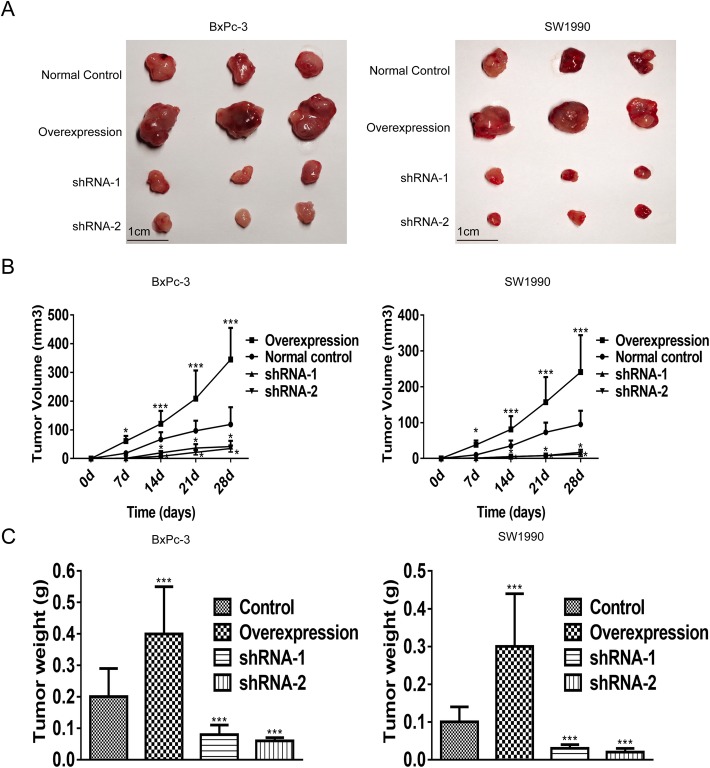
To assess the function of LRG-1 in promoting the development of PDAC in vivo, a nude mice xenograft model was established and the results showed that both the tumor weight and size were reduced significantly at all designated time points in the nude mice injected with LRG-1 knock-down cells as compared with the nude mice injected with scramble plasmids (Fig. 3a, b, c). On the other hand, xenograft assays showed that the tumor burden in the nude mice injected with LRG-1 over-expressing PDAC cells was heavier than that in the nude mice injected with control cells (Fig. 3a, b, c). These data suggested that LRG-1 played an important role in the development and progression of PDAC in vivo.
Fig. 3.
LRG-1 promotes PDAC growth in vivo. a Images of tumors at day 28 of the xenograft mouse model. b Tumor volume was measured using a caliper at indicated time points. c The tumor weight was measured at the end of the experiments. Data are presented as the mean ± SD, *p < 0.05; **p < 0.01; ***p < 0.001
LRG-1 promotes PDAC cell proliferation and migration via p38/MAPK signaling pathways
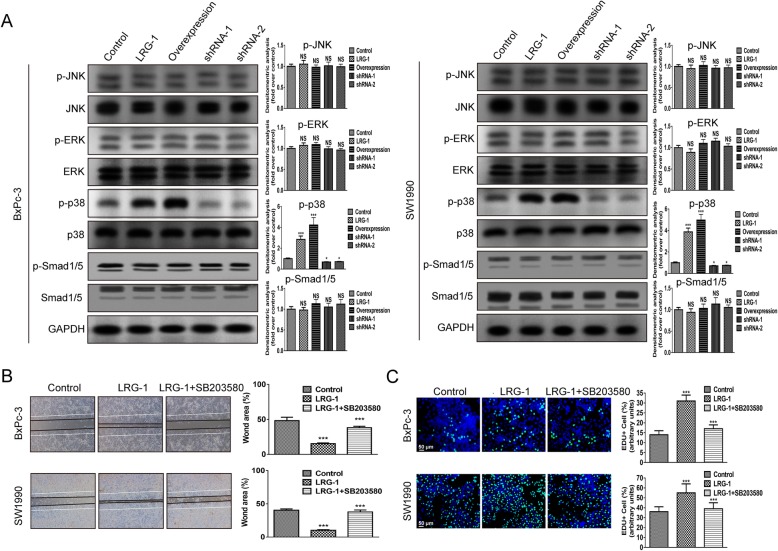
MAPK pathway is known as one of the key signaling pathways that regulate the pathological process of pancreatic cancer [25, 26], and LRG-1 was reported to modulate the activation of MAPK signaling downstream protein [27]. To further explore the underlying mechanism of how LRG-1 promoted the malignant behaviors of PDAC cells, we analyzed the activation of the downstream mediators of the MAPK signaling pathway using Western blot assay, and found that LRG-1 incubation and LRG-1 overexpression significantly upregulated p38 phosphorylation in PDAC cells. The phosphorylation of p38 was remarkably decreased in LRG-1 knockdown PDAC cells (Fig. 4a). However, JNK and ERK phosphorylation were not obviously affected by LRG-1 (Fig. 4a). Previous study has reported that the addition of LRG-1 could induce Smad1/5 phosphorylation in endothelial cells in the presence of TGF-β1. Our Western blot assay demonstrated that exogenous LRG-1 could not significantly upregulate the expression level of p-Smad1/5 in human PDAC cells in the presence of TGF-β1 suggesting that LRG-1 activation in Smads signaling was not available in PDAC cells (Fig. 4a).
Fig. 4.
LRG-1 promotes PDAC cells proliferation and migration via p38/MAPK signaling. a Levels of phosphorylated and total JNK, ERK, p38 and Smad 1/5 were examined by Western blot after 30-min incubation of BxPc-3 and SW1990 cells with LRG-1 (500 ng/ml) for. Wound-healing assay (b) and EdU incorporation assay (c) in cultured PDAC cells after 48 h incubation with SB203580 (0.5 μM) and LRG-1 (500 ng/ml). Data are presented as the mean ± SD, NS = not significant; ***p < 0.001
To confirm whether the enhancement of the malignant behavior of PDAC cells by LRG-1 was dependent on the activation of p38/MAPK signaling pathway, SB203580 (a specific inhibitor of this pathway) was used for the subsequent experiment. It was found that the promoting effect of LRG-1 on cell proliferation and migration was blocked after treatment of PDAC cells with LRG-1 and SB203580 (Fig. 4b, c) indicating that LRG-1 might promote the malignant behavior of PDAC cells via the p38/MAPK signaling pathway.
EGF receptor (EGFR) mediates LRG-1 induced phosphorylation of p38 signaling
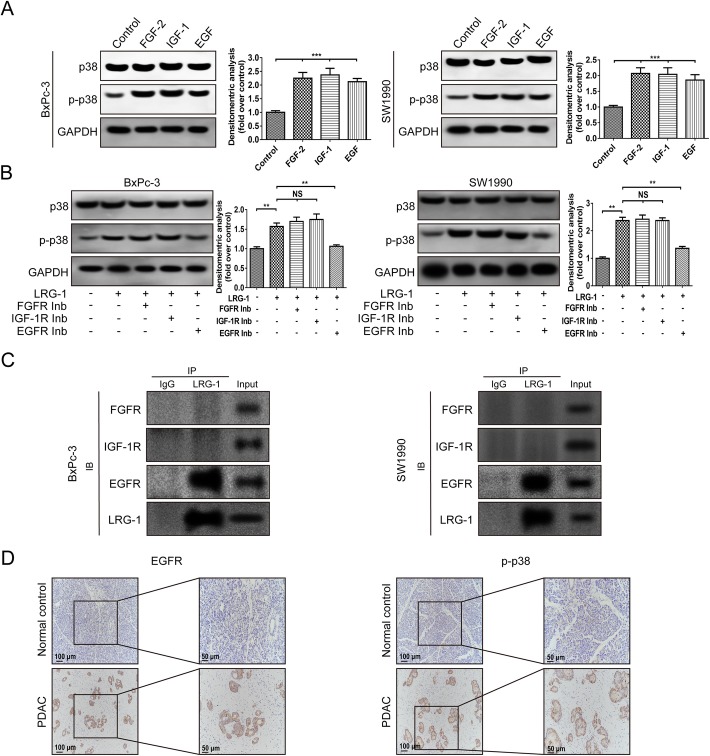
During the pathological development and progression of PDAC, many growth factors play an important role, and the exaggerated activation of their downstream signaling has been found to be associated with the malignant behavior of PDAC cells [8, 28–31]. Among them, FGF [32], IGF [33] and EGF [8, 34] were the most important cytokines in PDAC formation and have proved to interact with p38 pathway. Consistent with other studies [35–37], our Western blot analysis indicated that FGF-2, IGF-1 and EGF significantly induced p38 phosphorylation in PDAC cells (Fig. 5a). Next, we investigated whether these cytokines took part in LRG-1 mediated p38 phosphorylation, and found that LRG-1 induced upregulation of p38 phosphorylation was almost completely abolished by the EGFR inhibitor Erlotinib, while the FGFR and IGFR-1 inhibitors had little effect on LRG-1 induced upregulation of p38 phosphorylation (Fig. 5b).
Fig. 5.
The role of EGFR in LRG-1-induced p38 phosphorylation. a Levels of phosphorylated and total p38 were examined by Western blot after 30-min incubation of cells with FGF-2 (20 ng/ml) or IGF-1 (100 ng/ml) or EGF (100 ng/ml). b The expression of phosphorylated and total p38 in both cell lines after 30-min incubation with LRG-1 (500 ng/ml) and FGFR inhibitor (PD173074, 0.2 μM) or IGF-1R inhibitor (OSI-906, 5 μM) or EGFR inhibitor (erlotinib, 5 μM). c Co-immunoprecipitation (Co-IP) between LRG-1 and FGFR or IGF-1R or EGFR. d Immunohistochemistry images of EGFR and p-p38 in human PDAC and normal pancreas tissues. Data are presented as the mean ± SD, NS = not significant; **p < 0.01; ***p < 0.001
EGFR is critical for LRG-1-mediated malignant behaviors of PDAC cells
Recently, researchers found that LRG-1 could bind directly to endoglin, promoting the pro-angiogenic Smad1/5 signaling pathway (PMID: 23868260). We were therefore curious to know whether LRG-1 bound directly to the EGFR to enhance its activation and further induce p38 phosphorylation. Therefore, we performed Co-immunoprecipitation (Co-IP) assays to determine whether LRG-1 might bind directly to EGFR. Co-IP assay showed that EGFR and LRG-1 could form a complex, but no obvious interaction between FGFR and IGF-1R with LRG-1 was observed (Fig. 5c). We further tested the expression of EGFR and p-p38 in both PDAC tissues and normal pancreas tissues and found the expression of EGFR and p-p38 was significantly higher in PDAC tissues than that in normal pancreas tissues (Fig. 5d). The expressions of LRG-1, EGFR, p-p38 were evaluated in all PDAC tissues, the correlation analysis showed that EGFR (r = 0.224, P = 0.012) and p-p38 (r = 0.507, P < 0.001) expression was positively correlated with LRG-1 expression.
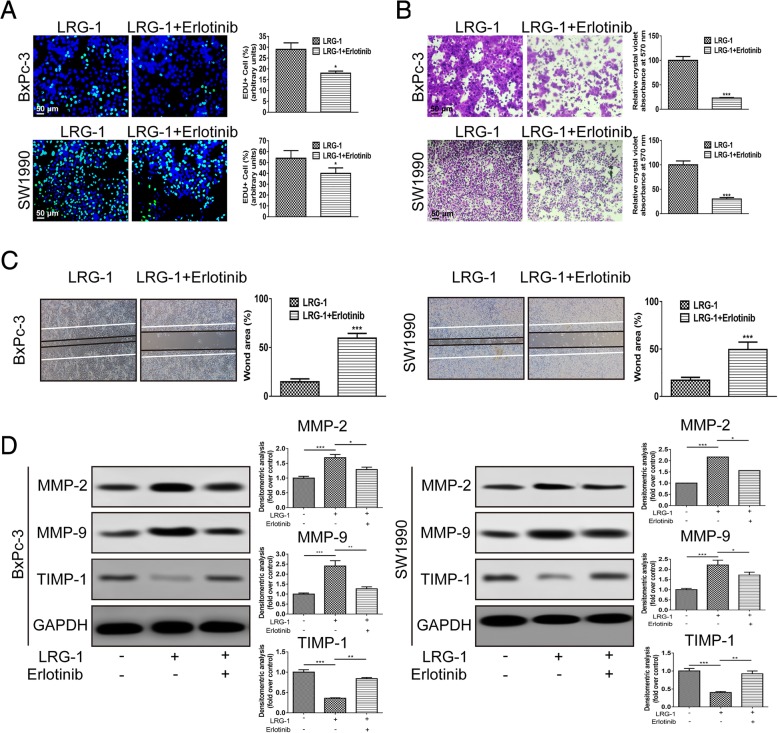
To determine whether the LRG-1-induced malignant behavior of PDAC cells was mediated by EGFR. EdU, Transwell assays and wound healing assays were performed. The results demonstrated that the promoting effect induced by LRG-1 was restored when Erlotinib was added in the BxPc-3 cell and SW1990 cell cultures (Fig. 6a, b and c). Erlotinib also reversed the upregulation of the protein levels of MMP-2 and MMP-9 induced by LRG-1 (Fig. 6d). These results indicate that EGFR played a crucial role in LRG-1-mediated malignant behavior of PDAC cells.
Fig. 6.
EGFR is critical for LRG-1-mediated malignant behavior of PDAC cells. a EdU incorporation assay, (b) Transwell assay and (c) Wound-healing assay in PDAC cells after 48 h incubation with LRG-1 (500 ng/ml) or LRG-1 + erlotinib (5 μM). d The protein level of MMP-2, MMP-9 and TIMP-1 in PDAC cells after 3-day incubation with LRG-1 (500 ng/ml) or LRG-1 + erlotinib (5 μM). Data are presented as the mean ± SD, *p < 0.05; **p < 0.01; ***p < 0.001
Discussion
The incidence of PDAC is increasing rapidly during the past five years, accounting for about 90% of all pancreatic malignancies [38]. Current therapeutic approaches for PDAC mainly include pancreatic surgery, cytotoxic medication and radiation therapy [39]. However, survival of PDAC patients remains unsatisfied. Although some molecular-targeting therapies have been developed in recent years, survival benefits are still very limited. The unfavorable outcomes of these approaches could be ascribed to the poor understanding about the mechanisms underlying the pathogenesis of PDAC, knowing that the key oncogene causing PDAC remains undiscovered. Thus, it is urgent to find the relevant signaling pathways or target proteins, and clarify the exact mechanism about how PDAC develops and progresses.
In the present study, we found that LRG-1, a kind of secretive glycoprotein, could serve as an efficient biomarker for predicting the prognosis of PDAC patients. LRG-1 has been proven to be associated with inflammation and autoimmune disease in the past few years [40, 41]. Shinzaki et al. demonstrated that LRG-1 was a serum biomarker of mucosal healing in ulcerative colitis (UC) and serum LRG-1 concentrations in active UC patients was significantly higher than that in patients who had complete mucosal healing and deep remission [40, 42]. In osteoarthritis, tumor necrosis factor-α (TNF-α) induced LRG-1 expression in the subchondral bone and articular cartilage, and LRG-1 contributed to angiogenesis-coupled de novo bone formation in the subchondral bone of osteoarthritis joints [27]. Persistent inflammation could be an overture for malignant transformation of the normal tissue. Besides LRG-1 overexpression of in patients with inflammatory disease, LRG-1 expression of was also significantly higher in patients with malignancy. LRG-1 expression (serum or immunohistochemical staining) in patients with gastric cancer was higher than that in healthy controls, and LRG-1 expression increased with the progression of the pathological stage [43]. What’s more, LRG-1 expression was high in the malignant tissues of patients with colorectal cancer [17] and endometrial carcinoma [18], and it was correlated with tumor stage and lymph node metastasis. In patients with non-small cell lung cancer, serum LRG-1 expression was significantly higher than that in healthy volunteers, and LRG-1 was found as an outstanding tool in predicting prognosis and relapse [19]. It was found that in our study that LRG-1 expression in PDAC tissues was significantly higher than that in adjacent normal tissues. Survival analysis suggested that LRG-1 was an independent prognostic factor, and subsequent regression analysis indicated that LRG-1 level was correlated with more advanced tumor stage, higher CA 125 and CA 19–9 levels.
The malignant behavior of PDAC cells, including over proliferation, invasion and migration, is the pathophysiological characteristics in the development and progression of PDAC. Here, we demonstrated that LRG-1 could promote the viability, proliferation, migration and invasion of PDAC cells. Our in vivo experiment also showed that the tumor weight and size in mice injected with LRG-1 over-expressing PDAC cells exhibited obviously increased than that in mice injected with control PDAC cells. On the contrary, the tumor burden in nude mice injected with LRG-1 knockdown cells was significantly lower than that in control mice. This appeared to be consistent with previous publications demonstrating a potential tumor-promoting effect of LRG-1 on other malignancies, such as hepatocellular carcinoma [44], glioblastoma [20, 45] and colon cancer [46].
Furthermore, we found that the positive action in carcinogenesis was owing to the enhancing effect of LRG-1 on p38/MAPK signaling pathway in PDAC cells. MAPK signaling played important roles in regulating tumorigenesis, metastasis and chemoresistance of PDAC [47, 48]. Among all the downstream proteins of MAPK signaling tested in our experiments, the level of p38 phosphorylated was significantly increased after LRG-1 treatment, while the level of JNK and ERK phosphorylated remained unaffected. In addition, inhibition of the p38 pathway was sufficient to block LRG-1-induced malignant behavior of PDAC cells. This finding was also in line with a previous study reporting that LRG-1 significantly induced p38 phosphorylation in human bone marrow mesenchymal stem cells, thus promoting their migration and aberrant bone formation [27].
EGFR is a transmembrane glycoprotein that is conserved and overexpressed in pancreatic cancer [49]. EGFR over-expression has been confirmed to confer a poor survival, correlating with a more advanced stage and the presence of metastasis in pancreatic cancer [50]. EGFR phosphorylation initiates downstream signaling cascades such as MAPK, PI3K/Akt and Src pathways, which have been implicated in tumorigenesis by affecting cell proliferation, invasion and metastasis [51]. Interestingly, we found that EGFR inhibitor reduced LRG-1-induced p38 phosphorylation remarkably. Consistently, EdU incorporation assay and transwell assays showed that LRG-1 induced cell proliferation and invasion were almost abolished by the EGFR inhibitor. Further Co-IP assay revealed that LRG-1 could bind directly to EGFR to form a complex. Similarly, a previous study found that LRG-1 could bind to endoglin, a TGF-β accessory receptor, promoting the pro-angiogenic Smad1/5 signaling pathway [15].
Conclusion
The expression of LRG-1 in PDAC tissue was significantly higher than that in adjacent normal tissue, and high LRG-1 expression predicted poor survival and aan dvanced tumor stage. LRG-1 remarkably promoted the viability, proliferation, migration and invasion of PDAC cells and facilitated tumor growth in vivo. This kind of bioactivity of LRG-1 may be ascribed to its selective interaction with EGFR and subsequent activation of the p38/MAPK signaling pathway. LRG-1 could serve as a promising biomarker for predicting prognosis in PDAC patients. Targeting LRG-1 or its downstream pathway may provide a novel and efficient strategy for the treatment of PDAC.
Acknowledgments
We thank Pro Shun-Xing Zhang for language editing.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81472221) (D.L.F.), and Clinical key projects of the National Health and Family Planning (Oncology 2013–2015) (D.L.F.)
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Abbreviations
- CA
Carbohydrate antigen
- Co-IP
Co-immunoprecipitation
- EGFR
Epidermal growth factor receptor
- EMT
Epithelial-mesenchymal transition
- FGF
Fibroblast growth factor
- IGF-1
Insulin-like growth factor1
- IHC
Immunohistochemistry
- LRG-1
Leucine-rich-alpha-2-glycoprotein-1
- MAPK
Mitogen-activated protein kinase
- MMP
Matrix metalloproteinase
- PDAC
Pancreatic ductal adenocarcinoma
- SD
Standard deviation
- SPF
Specific pathogen-free
- TGF-β
Transforming growth factor β
- TIMP-1
Tissue inhibitors of metalloproteinase-1
Author’s contributions
Conception and design: ZBX, and DLF; administrative support: ZBX, YFZ and DLF; provision of study materials or patients: CJ, YSM and DLF; collection and assembly of data: ZBX, YFZ and DLF; data analysis and interpretation: ZBX, YFZ and DLF; manuscript writing: ZBX, YFZ and DLF; final approval of manuscript: All authors.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of Shanghai Medical College of Fudan University (Shanghai, China), and conducted in accordance with the Declaration of Helsinki and internationally accepted ethical guidelines. The use of human tissue samples and clinical data was approved by the Clinical Research Ethics Committee of Huashan Hospital affiliated to Fudan University. All donors provided written informed consent to donate their samples. All animal experiments were carried out according to the standards of animal care as outlined in the NIH guide for the Care and Use of Laboratory Animals. All methods were taken in accordance with the approved guidelines of Shanghai Medical College of Fudan University.
Consent for publication
Written informed consent was obtained from all patients.
Competing interests
The authors disclose no conflicts of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhi-Bo Xie, Email: 15111220054@fudan.edu.cn.
Yi-Fan Zhang, Phone: +(86)-21-23271699-5615, Email: zhangyifan82@126.com.
Chen Jin, Email: jinchen_sh@163.com.
Yi-Shen Mao, Email: maoyishen_sh@163.com.
De-Liang Fu, Phone: +(86)-21-52887164, Email: surgeonfu@163.com.
References
- 1.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371(22):2140–2141. doi: 10.1056/NEJMc1412266. [DOI] [PubMed] [Google Scholar]
- 3.Zell JA, Rhee JM, Ziogas A, et al. Race, socioeconomic status, treatment, and survival time among pancreatic cancer cases in California. Cancer Epidemiol Biomark Prev. 2007;16(3):546–552. doi: 10.1158/1055-9965.EPI-06-0893. [DOI] [PubMed] [Google Scholar]
- 4.Lau MK, Davila JA, Shaib YH. Incidence and survival of pancreatic head and body and tail cancers: a population-based study in the United States. Pancreas. 2010;39(4):458–462. doi: 10.1097/MPA.0b013e3181bd6489. [DOI] [PubMed] [Google Scholar]
- 5.Quaresma M, Coleman MP, Rachet B. 40-year trends in an index of survival for all cancers combined and survival adjusted for age and sex for each cancer in England and Wales, 1971-2011: a population-based study. Lancet. 2015;385(9974):1206–1218. doi: 10.1016/S0140-6736(14)61396-9. [DOI] [PubMed] [Google Scholar]
- 6.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 7.Vincent A, Herman J, Schulick R, et al. Pancreatic cancer. Lancet. 2011;378(9791):607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu H, Han T, Zhuo M, et al. Elevated COX-2 expression promotes angiogenesis through EGFR/p38-MAPK/Sp1-dependent Signalling in pancreatic Cancer. Sci Rep. 2017;7(1):470. doi: 10.1038/s41598-017-00288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y, Su H, Li X, et al. The NOTCH ligand JAGGED2 promotes pancreatic cancer metastasis independent of NOTCH signaling activation. Mol Cancer Ther. 2015;14(1):289–297. doi: 10.1158/1535-7163.MCT-14-0501. [DOI] [PubMed] [Google Scholar]
- 10.Matsuda Y, Yoshimura H, Suzuki T, et al. Inhibition of fibroblast growth factor receptor 2 attenuates proliferation and invasion of pancreatic cancer. Cancer Sci. 2014;105(9):1212–1219. doi: 10.1111/cas.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H, Fang W, Liu M, et al. Complement component 1, q subcomponent binding protein (C1QBP) in lipid rafts mediates hepatic metastasis of pancreatic cancer by regulating IGF-1/IGF-1R signaling. Int J Cancer. 2017;141(7):1389–1401. doi: 10.1002/ijc.30831. [DOI] [PubMed] [Google Scholar]
- 12.Rozengurt E, Sinnett-Smith J, Eibl G. Yes-associated protein (YAP) in pancreatic cancer: at the epicenter of a targetable signaling network associated with patient survival. Signal Transduct Target Ther. 2018;3:11. doi: 10.1038/s41392-017-0005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanteti R, Mirzapoiazova T, Riehm JJ, et al. Focal adhesion kinase a potential therapeutic target for pancreatic cancer and malignant pleural mesothelioma. Cancer Biol Ther. 2018;19(4):316–327. doi: 10.1080/15384047.2017.1416937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haupt H, Baudner S. Isolation and characterization of an unknown, leucine-rich 3.1-S-alpha2-glycoprotein from human serum (author's transl) Hoppe Seylers Z Physiol Chem. 1977;358(6):639–646. doi: 10.1515/bchm2.1977.358.1.639. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Abraham S, McKenzie JAG, et al. LRG1 promotes angiogenesis by modulating endothelial TGF-beta signalling. Nature. 2013;499(7458):306–311. doi: 10.1038/nature12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Zhu L, Fang J, et al. LRG1 modulates epithelial-mesenchymal transition and angiogenesis in colorectal cancer via HIF-1alpha activation. J Exp Clin Cancer Res. 2016;35:29. doi: 10.1186/s13046-016-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ladd JJ, Busald T, Johnson MM, et al. Increased plasma levels of the APC-interacting protein MAPRE1, LRG1, and IGFBP2 preceding a diagnosis of colorectal cancer in women. Cancer Prev Res. 2012;5(4):655–664. doi: 10.1158/1940-6207.CAPR-11-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen SY, Zhang LN, Yang XM, et al. LRG1 is an independent prognostic factor for endometrial carcinoma. Tumor Biol. 2014;35(7):7125–7133. doi: 10.1007/s13277-014-1953-6. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Zhang Y, Qiu F, et al. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. 2011;32(15):1976–1983. doi: 10.1002/elps.201000598. [DOI] [PubMed] [Google Scholar]
- 20.Zhong D, He G, Zhao S, et al. LRG1 modulates invasion and migration of glioma cell lines through TGF-beta signaling pathway. Acta Histoch. 2015;117(6):551–558. doi: 10.1016/j.acthis.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Furukawa K, Kawamoto K, Eguchi H, et al. Clinicopathological significance of leucine-rich alpha2-Glycoprotein-1 in sera of patients with pancreatic Cancer. Pancreas. 2015;44(1):93–98. doi: 10.1097/MPA.0000000000000205. [DOI] [PubMed] [Google Scholar]
- 22.Capello M, Bantis LE, Scelo G, et al. Sequential Validation of Blood-Based Protein Biomarker Candidates for Early-Stage Pancreatic Cancer. J Nation Cancer Ins. 2017;109(4). [DOI] [PMC free article] [PubMed]
- 23.Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang W, Zhang H, Hao Y, et al. A non-synonymous single nucleotide polymorphism in the HJURP gene associated with susceptibility to hepatocellular carcinoma among Chinese. PLoS One. 2016;11(2):e0148618. doi: 10.1371/journal.pone.0148618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins MA, Yan W, Sebolt-Leopold JS, et al. MAPK signaling is required for dedifferentiation of acinar cells and development of pancreatic intraepithelial neoplasia in mice. Gastroenterology. 2014;146(3):822–834. doi: 10.1053/j.gastro.2013.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jun S, Lee S, Kim HC, et al. PAF-mediated MAPK signaling hyperactivation via LAMTOR3 induces pancreatic tumorigenesis. Cell Rep. 2013;5(2):314–322. doi: 10.1016/j.celrep.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Xu J, Zhang X, et al. TNF-alpha-induced LRG1 promotes angiogenesis and mesenchymal stem cell migration in the subchondral bone during osteoarthritis. Cell Death Dis. 2017;8(3):e2715. doi: 10.1038/cddis.2017.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun TY, Xie HJ, Li Z, et al. miR-34a regulates HDAC1 expression to affect the proliferation and apoptosis of hepatocellular carcinoma. Am J Transl Res. 2017;9(1):103–114. [PMC free article] [PubMed] [Google Scholar]
- 29.O'Sullivan H, Kelleher FC, Lavelle M, et al. Therapeutic potential for FGFR inhibitors in SOX9-FGFR2 Coexpressing pancreatic Cancer. Pancreas. 2017;46(8):e67–e69. doi: 10.1097/MPA.0000000000000870. [DOI] [PubMed] [Google Scholar]
- 30.Yin H, Chen N, Guo R, et al. Antitumor potential of a synthetic interferon-alpha/PLGF-2 positive charge peptide hybrid molecule in pancreatic cancer cells. Sci Rep. 2015;5:16975. doi: 10.1038/srep16975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balogh J, Victor D, 3rd, Asham EH, et al. Hepatocellular carcinoma: a review. J Hepat Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuyama D, Kawahara K. Proliferation of neonatal cardiomyocytes by connexin43 knockdown via synergistic inactivation of p38 MAPK and increased expression of FGF1. Basic Res Cardiol. 2009;104(6):631–642. doi: 10.1007/s00395-009-0029-z. [DOI] [PubMed] [Google Scholar]
- 33.Reckenbeil J, Kraus D, Stark H, et al. Insulin-like growth factor 1 (IGF1) affects proliferation and differentiation and wound healing processes in an inflammatory environment with p38 controlling early osteoblast differentiation in periodontal ligament cells. Arch Oral Biol. 2017;73:142–150. doi: 10.1016/j.archoralbio.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Jiang W, Tian W, Ijaz M, et al. Inhibition of EGF-induced migration and invasion by sulfated polysaccharide of Sepiella maindroni ink via the suppression of EGFR/Akt/p38 MAPK/MMP-2 signaling pathway in KB cells. Biomed Pharmacother. 2017;95:95–102. doi: 10.1016/j.biopha.2017.08.050. [DOI] [PubMed] [Google Scholar]
- 35.Kim BS, Park JY, Kang HJ, et al. Fucoidan/FGF-2 induces angiogenesis through JNK- and p38-mediated activation of AKT/MMP-2 signalling. Biochem Biophys Res Commun. 2014;450(4):1333–1338. doi: 10.1016/j.bbrc.2014.06.137. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Deng W, Zhang Y, et al. MICAL2 promotes breast cancer cell migration by maintaining epidermal growth factor receptor (EGFR) stability and EGFR/P38 signalling activation. Acta Physiol. 2018;222(2). [DOI] [PubMed]
- 37.Ma Y, Fu S, Lu L, et al. Role of androgen receptor on cyclic mechanical stretch-regulated proliferation of C2C12 myoblasts and its upstream signals: IGF-1-mediated PI3K/Akt and MAPKs pathways. Mol Cell Endocrinol. 2017;450:83–93. doi: 10.1016/j.mce.2017.04.021. [DOI] [PubMed] [Google Scholar]
- 38.Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleeff J, Korc M, Apte M, et al. Pancreatic cancer. Nat Review Dis Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 40.Shinzaki S, Matsuoka K, Iijima H, et al. Leucine-rich Alpha-2 glycoprotein is a serum biomarker of mucosal healing in ulcerative colitis. J Crohns Colitis. 2017;11(1):84–91. doi: 10.1093/ecco-jcc/jjw132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Y, He Z, Li Z, et al. Irgm1 is required for the inflammatory function of M1 macrophage in early experimental autoimmune encephalomyelitis. J Leukoc Biol. 2017;101(2):507–517. doi: 10.1189/jlb.3A0116-028RR. [DOI] [PubMed] [Google Scholar]
- 42.Serada S, Fujimoto M, Terabe F, et al. Serum leucine-rich alpha-2 glycoprotein is a disease activity biomarker in ulcerative colitis. Inflamm Bowel Dis. 2012;18(11):2169–2179. doi: 10.1002/ibd.22936. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto M, Takahashi T, Serada S, et al. Overexpression of leucine-rich alpha2-glycoprotein-1 is a prognostic marker and enhances tumor migration in gastric cancer. Cancer Sci. 2017;108(10):2052–2060. doi: 10.1111/cas.13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang CH, Li M, Liu LL, et al. LRG1 expression indicates unfavorable clinical outcome in hepatocellular carcinoma. Oncotarget. 2015;6(39):42118–42129. doi: 10.18632/oncotarget.5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong D, Zhao S, He G, et al. Stable knockdown of LRG1 by RNA interference inhibits growth and promotes apoptosis of glioblastoma cells in vitro and in vivo. Tumor Biol. 2015;36(6):4271–4278. doi: 10.1007/s13277-015-3065-3. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y, Zhang X, Zhang J, et al. LRG1 promotes proliferation and inhibits apoptosis in colorectal cancer cells via RUNX1 activation. PLoS One. 2017;12(4):e0175122. doi: 10.1371/journal.pone.0175122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang K, Li Y, Lian G, et al. KRAS promotes tumor metastasis and chemoresistance by repressing RKIP via the MAPK-ERK pathway in pancreatic cancer. Int J Cancer. 2018;142(11):2323–2334. doi: 10.1002/ijc.31248. [DOI] [PubMed] [Google Scholar]
- 48.Sheng W, Chen C, Dong M, et al. Calreticulin promotes EGF-induced EMT in pancreatic cancer cells via integrin/EGFR-ERK/MAPK signaling pathway. Cell Death Dis. 2017;8(10):e3147. doi: 10.1038/cddis.2017.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tzeng CW, Frolov A, Frolova N, et al. EGFR genomic gain and aberrant pathway signaling in pancreatic cancer patients. J Surg Res. 2007;143(1):20–26. doi: 10.1016/j.jss.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 50.Tzeng CW, Frolov A, Frolova N, et al. Epidermal growth factor receptor (EGFR) is highly conserved in pancreatic cancer. Surgery. 2007;141(4):464–469. doi: 10.1016/j.surg.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 51.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Reviews Mol Cell Biol. 2001;2(2):127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.