Abstract
Methionine (Met) is an essential and multifunctional nutrient in vertebrate diets. It is a precursor of S-adenosylmethionine (SAM), the methyl donor for DNA methylation, which has an important role in the inflammatory responses. However, whether Met exerts anti-inflammatory effects by altering DNA methylation in macrophages is unclear. In this study, Met was found to diminish the activation of the mitogen-activated protein kinase signaling pathway; decrease the production of tumor necrosis factor-α, interleukin-6, and interferon-β; and enhance the levels of intracellular SAM after lipopolysaccharide (LPS) treatment in macrophages. Similarly, SAM inhibited the LPS-induced inflammatory response, consistent with the result of Met treatment. Met-treated macrophages displayed increased global DNA methylation. The DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine partially blocked the anti-inflammatory effects of Met in macrophages, suggesting a mechanism involving DNA methylation. Collectively, the results indicated that Met inhibits the LPS-induced inflammatory response by altering DNA methylation in RAW 264.7 macrophages. The findings provide new insights into the interplay between nutrition and immunology, and highlight the regulatory effects of amino acids on the host immune system.
Introduction
As an essential amino acid in animals, methionine (Met) is closely related to physiological processes including immune function, protein synthesis, growth promotion, and detoxification.1 Met is converted to S-adenosylmethionine (SAM), which is a methyl donor that has crucial roles in immune disorders owing to its activities of epigenetic regulation, especially DNA methylation.2,3
DNA methylation is an important mammalian epigenetic mechanism that is thought to be involved in inflammatory responses. Inflammation is a key biological mechanism in an organism’s defense system against harmful intruders. The methylation profiles on the cytosine-phosphate–guanine (CpG) regions are involved in inflammation-response-related genes.4,5 Macrophages are a component of the innate immune system and play important roles in its responses against bacterial and viral pathogens.6 Macrophages can be activated by several external stimuli including lipopolysaccharides (LPS), which are a component of the outer membrane of Gram-negative bacteria, lead to the activation of the Toll-like receptor-4 signaling pathway, and induce the activation of mitogen-activated protein kinases (MAPKs), resulting in the secretion of inflammatory cytokines.7 Previous studies have demonstrated that DNA methylation modulates LPS-induced macrophage inflammatory responses and maintains intestinal homeostasis by regulating mucosal inflammation in the gut.8 Moreover, Met could inhibit the LPS-induced expression of inflammation-related genes in macrophage cells.9
However, whether the mechanism by which Met exerts anti-inflammatory effects involves DNA methylation in macrophages is poorly understood. Here, we provide evidence that Met effectively inhibits LPS-induced inflammatory responses partly through DNA methylation in RAW 264.7 macrophages.
Materials and Methods
Cell Culture
RAW 264.7 cells obtained from the American Type Culture Collection (ATCC, Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in a humidified 5% CO2 atmosphere. The cells were preincubated for 12 h with Met or SAM and subsequently cultured for 1.5–24 h in the presence of 100 ng/mL LPS. For the DNA methylation inhibitor treatment, RAW 264.7 cells were pretreated with 5-aza-2′-deoxycytidine (5-aza; Sigma-Aldrich, St. Louis, MO) for 24 h and then cultured with Met (or SAM) and LPS.
Cell Viability
Cells (5000 per well) were seeded into a 96-well plate and treated with variable concentrations of Met for 24 h. A cell counting kit-8 (Dojindo, Tabaru, Japan) was used to label cells for 1 h. The absorbance at 450 nm was then measured using a plate reader, and cell viability was expressed as the percentage absorbance of treated cells relative to that of untreated cells.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and Quantitative PCR
Total cellular RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA). RT-PCR was performed using a one-step RNA-PCR kit (Takara Bio, Shiga, Japan) and SYBR Green master mix (Applied Biosystems, Foster City, CA) according to the manufacturers’ protocols. β-Actin cDNA was used as an internal control to normalize the amount of total RNA in each reaction. The results were expressed as relative abundance [log(2 – ΔΔCt)]. The RT-PCR primers used were as follows: β-actin forward 5′-GATGAGATTGGCATGGCTTT-3′, reverse 5′-CACCTTCACCGTTCCAGTTT-3′; interleukin (IL)-6 forward 5′-AGTTGCCTTCTTGGGACTGA-3′, reverse 5′-TCCACGATTTCCCAGAGAAC-3′; tumor necrosis factor-α (TNF-α) forward 5′-CTGGGACAGTGACCTGGACT-3′, reverse 5′-GCACCTCAGGGAAGAGTCTG-3′; and interferon-β (IFN-β) forward 5′-CCCTATGGAGATGACGGAGA-3′, reverse 5′-CTGTCTGCTGGTGGAGTTCA-3′.
Measurement of Cytokines
The protein concentrations of interleukin IL-6, TNF-α, and IFN-β in cell culture supernatants were measured using murine cytokine-specific Quantikine enzyme-linked immunosorbent assay (ELISA) kits (eBioscience, San Diego, CA).
Western Blot
Protein extracts were separated on 10% sodium dodecyl sulfate-polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked with 5% fat-free milk at room temperature for 1 h and then incubated overnight at 4 °C with a primary antibody (Ab), followed by incubation with the appropriate horseradish-peroxidase-conjugated reporter Ab (eBioscience, San Diego, CA) for 1 h at room temperature. The enhanced chemiluminescence FluorChem E system (Bio-Techne, Minneapolis, MN) was used for developing the reaction. Abs specific for the total and phosphorylated forms of ERK1/2 (Thr202/Tyr204), JNK1/2 (Thr183/Tyr185), and p38 (Thr180/Tyr182) were obtained from Cell Signaling Technology (Danvers, MA). An Ab against β-actin was purchased from Santa Cruz Biotechnology (Dallas, TX).
High-Performance Liquid Chromatography (HPLC) Analysis
The intracellular concentration of SAM was measured by HPLC. RAW 264.7 cells were incubated with Met for 12 and 24 h. Then, the culture medium was aspirated and the cell monolayer was washed with cold phosphate-buffered saline. The cells were treated with 20% trichloroacetic acid at 4 °C for 4 h and then immediately frozen at −80 °C until measurement. HPLC was performed on a model 1260 Infinity II LC system (Agilent Technologies, Santa Clara, CA) using a ZORBAX Eclipse Plus C18 column (Agilent Technologies) at 30 °C. The mobile phase consisted of 0.01 mol/L ammonium formate/methanol (97:3, v/v, pH 3.0). The flow rate was 0.8 mL/min, and the detection was performed at 260 nm.
Global DNA Methylation
DNA isolated from RAW 264.7 cells was used for global DNA methylation analyses.
Global DNA methylation was determined by DNA quantification using a monoclonal Ab against 5-methylcytosine (5-mC) in an ELISA-like reaction using the MethylFlash methylated DNA quantification kit (Epigentek, Farmingdale, NY), according to the manufacturer’s instructions. The 5-mC amount was calculated using a standard curve created using defined dilutions of methylated genomic DNA. Methylation levels were calculated relative to the methylated control DNA included in the kit and were expressed as a percentage of total methylated DNA.
Statistical Analysis
All data were expressed as mean ± standard deviation (SD). Comparisons between two groups were performed using the Student’s t-test. Comparisons among more than two groups were performed using one-way analysis of variance with post hoc comparisons using Dunnett’s test. All of the analyses were performed using GraphPad Prism version 4.0 software (GraphPad Software, LaJolla, CA). Differences were considered significant at p < 0.05.
Results
Met Reduces the Production of LPS-Induced Proinflammatory Mediators in Macrophages
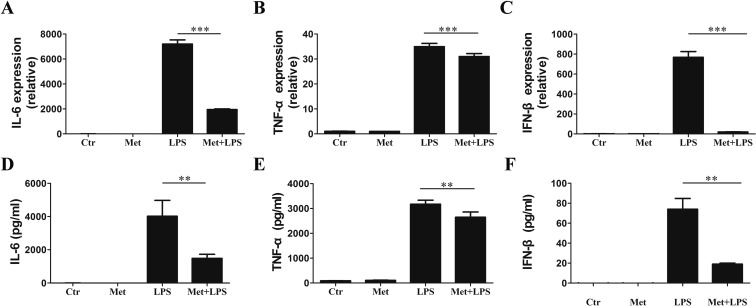
The viability of RAW 264.7 cells did not change following Met treatment (Supporting Information Figure S1). RAW 264.7 cells were incubated with the various concentrations of Met (0.01–10 mM) for 12 h, followed by exposure to LPS (100 ng/mL) for 3–12 h to evaluate the anti-inflammatory effects of Met. The expression of TNF-α, IL-6, and IFN-β mRNA was dramatically lower in the Met-treated cells as compared to the control cells, and 10 mM Met was chosen as the optimal concentration for use in subsequent experiments (Supporting Information Figure S2). The relative expression of genes encoding TNF-α, IL-6, and IFN-β was considerably lower with 10 mM Met for 12 h (Figure 1A–C). Meanwhile, the results of ELISA analysis to determine the concentrations of TNF-α, IL-6, and IFN-β in the culture supernatant at 24 h were consistent with the enhanced gene expression (Figure 1D–F). Collectively, the data indicated that Met inhibited the LPS-induced inflammatory response in RAW 264.7 macrophages.
Figure 1.
Met inhibits the LPS-induced inflammatory stress in RAW 264.7 macrophages. RAW 264.7 cells were pretreated with 10 mM Met for 12 h prior to stimulation with 100 μg/mL LPS for 3 h. The gene expression of (A) IL-6, (B) TNF-α, and (C) IFN-β was analyzed by RT-qPCR. (D–F) Effects of Met on LPS-stimulated IL-6, TNF-α, and IFN-β secretion from RAW 264.7 cells were analyzed by ELISA. Cells were cultured for 12 h with Met (10 mM) and then treated with LPS (100 ng/mL) for 12 h. Data represent the mean ± SD of three independent experiments, each performed in five samples. Comparisons among means used t-tests (*p < 0.05, **p < 0.01, ***p < 0.001).
Met Suppresses the LPS-Induced Activation of MAPK Signaling in Macrophages
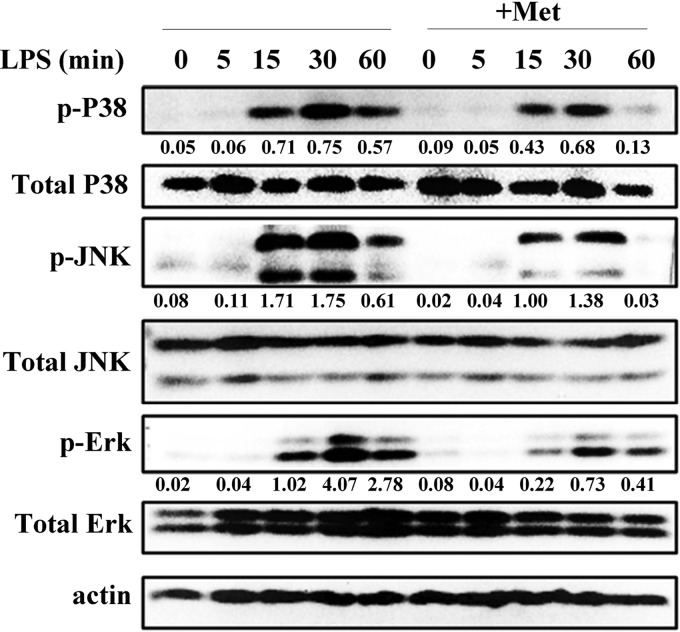
To evaluate the inhibitory effect of Met on LPS-induced proinflammatory responses in RAW 264.7 cells, the protein phosphorylation levels of three MAPK proteins (ERK, p38, and JNK) were determined. As shown in Figure 2, LPS treatment induced the phosphorylation of ERK, p38, and JNK, and their phosphorylation levels peaked at 30 min. The phosphorylation levels of p38, ERK1/2, and JNK in the Met-treated group were lower than those in the LPS-treated group. These data indicated that Met suppressed the LPS-induced activation of MAPK signaling in RAW 264.7 cells.
Figure 2.
Met inhibits the LPS-induced phosphorylation of mitogen-activated protein kinases (MAPK) in RAW 264.7 macrophages. RAW 264.7 cells were cultured for 12 h with Met (10 mM) and then treated with LPS (100 ng/mL) for 0, 5, 15, 30, and 60 min. Western blotting assays for phospho-ERK1/2 (p-ERK1/2), ERK1/2, p-JNK, JNK, p-p38, and p38 were performed. β-Actin was used as the loading control. Levels for target phosphorylated proteins were normalized to total proteins. All data shown are representative of three independent experiments.
SAM Reduces the Production of LPS-Induced Proinflammatory Mediators in Macrophages
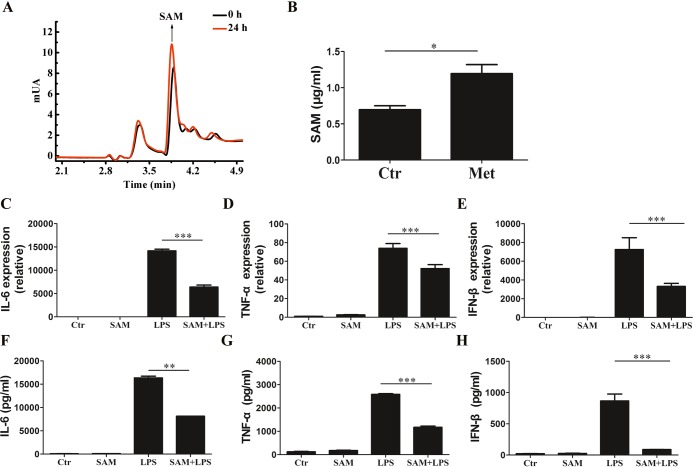
HPLC analysis revealed that the concentration of SAM in cell culture supernatants significantly increased following Met treatment (Figure 3A,B). We explored whether the Met-derivative SAM was involved in the regulation of the LPS-induced inflammatory response in macrophages. The production of TNF-α, IL-6, and IFN-β in RAW 264.7 cells upon LPS stimulation was significantly inhibited at the mRNA and protein levels after SAM treatment (Figure 3C–H).
Figure 3.
Met-derivative SAM inhibits the LPS-induced inflammatory stress in RAW 264.7 macrophages. (A, B) Intracellular concentration of SAM was determined by high-performance liquid chromatography. RAW 264.7 cells were pretreated with 0.5 mM SAM for 12 h prior to stimulation with 100 μg/mL LPS for 3 h. The gene expression levels of (C) IL-6, (D) TNF-α, and (E) IFN-β were analyzed by RT-qPCR. (F–H) Effects of Met on LPS-stimulated IL-6, TNF-α, and IFN-β in RAW 264.7 cells were analyzed by ELISA. The cells were cultured for 12 h with Met (10 mM) and then treated with LPS (100 ng/mL) for 12 h. Data represent the mean ± SD of three independent experiments, each performed in five samples. Comparisons among means used t-tests (*p < 0.05, **p < 0.01, ***p < 0.001).
SAM Inhibits LPS-Induced MAPK Signaling in Macrophages
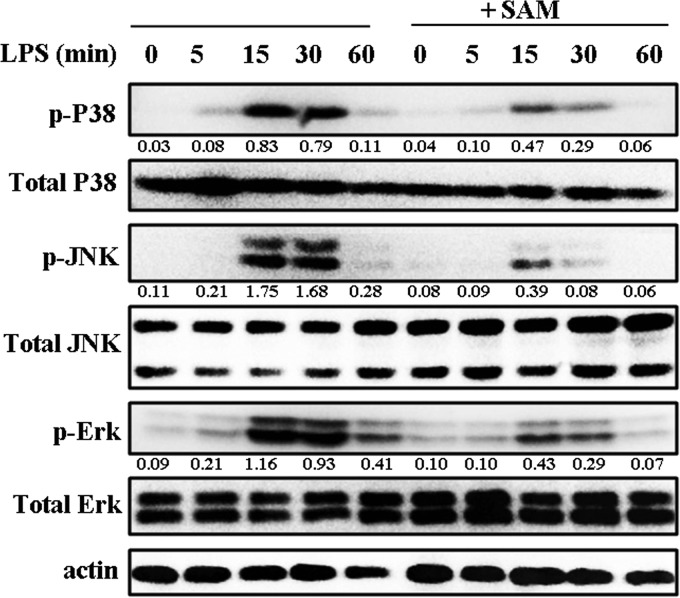
To confirm the effects of the Met-derivative SAM on the LPS-initiated activation of MAPK, we examined phosphorylation levels of ERK1/2, JNK1/2, and p38 in RAW 264.7 cells by western blotting. SAM inhibited the LPS-induced activation of all three MAPKs (Figure 4), consistent with the results of Met treatment.
Figure 4.
SAM inhibits the LPS-induced phosphorylation of MAPKs in RAW 264.7 macrophages. RAW 264.7 cells were cultured for 12 h with SAM (0.5 mM) and then treated with LPS (100 ng/mL) for 0, 5, 15, 30, and 60 min. Western blotting assays for phospho-ERK1/2 (p-ERK1/2), ERK1/2, p-JNK, JNK, p-p38, and p38 were performed. β-Actin was used as the loading control. Levels for target phosphorylated proteins were normalized to total proteins. All data shown are representative of three independent experiments.
Global Methylation Level Following Met Treatment
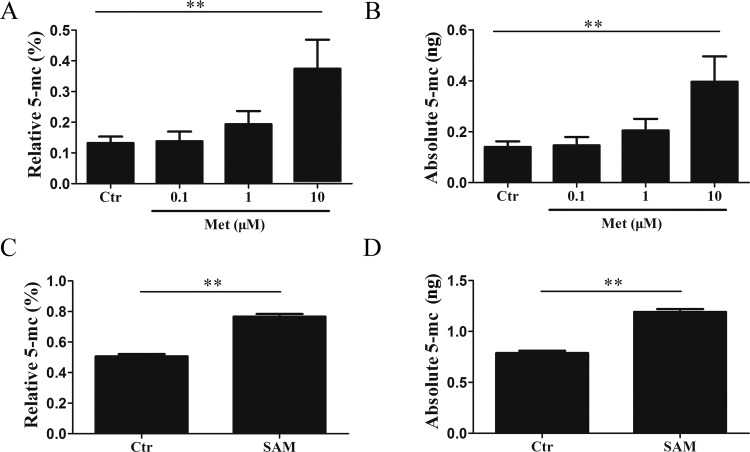
To analyze the influence of Met on DNA methylation, a monoclonal Ab against 5-mC was utilized to quantify the global methylation level using the MethylFlash methylated DNA quantification kit. The relative genome-wide methylated DNA methylation level of the control was 0.13 ± 0.02% (Figure 5A). After treatment with 10 μM Met, the methylation level significantly increased to 0.37 ± 0.09% (p < 0.05; Figure 5A). The absolute amount of 5-mC in the control cells was 0.14 ± 0.02 ng, whereas it increased to 0.40 ± 0.09 ng in the Met-treated cells (Figure 5B). To check whether Met-derivative SAM was involved in DNA methylation, the DNA methylation of SAM-treated macrophages was further analyzed. We also found that SAM effectively enhanced the level of DNA methylation in macrophages (Figure 5C,D). These data suggested that Met treatment significantly induced genome-wide methylation in macrophages.
Figure 5.
Met increases the global methylation level of RAW 264.7 macrophages. RAW 264.7 cells were cultured for 12 h with Met (0.1, 1, and 10 mM) and SAM (0.5 mM). (A) Relative and (B) absolute amounts of 5-mC were analyzed with Met treatment. (C) Relative and (D) absolute amounts of 5-mC were analyzed with SAM treatment. A statistically significant difference in the genome methylation was seen between the control and Met-treated macrophages. Comparisons among means were performed using t-tests (*p < 0.05, **p < 0.01, ***p < 0.001). All data shown are representative of three independent experiments. aThe absolute amount of 5-mC in the control cells was 0.14 ± 0.02 ng, whereas it increased to 0.40 ± 0.09 ng in the Met-treated cells (Figure 5B). To check whether Met-derivative SAM was involved in DNA methylation, the DNA methylation of SAM-treated macrophages was further analyzed. We also found that SAM effectively enhanced the level of DNA methylation in macrophages (Figure 5C,D).
DNA Methylation is Required for the Anti-Inflammatory Effects of Met in Macrophages
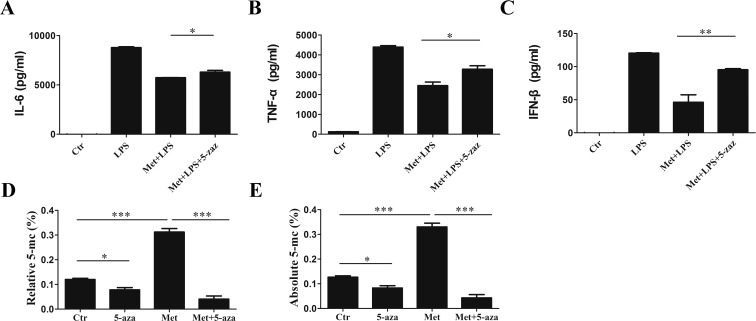
The DNA methyltransferase inhibitor 5-aza influences immune regulation.10 The secretion of TNF-α, IL-6, and IFN-β from RAW 264.7 cells significantly increased following the treatment with 2 μM 5-aza (Figure 6A–C). Thus, 5-aza treatment reduced the anti-inflammatory effects of Met in macrophages. With 5-aza pretreatment, a reduction in global DNA methylation was also detected in Met-treated macrophages (Figure 6D,E). Together, these data demonstrated that DNA methylation mediated the anti-inflammatory effects of Met in macrophages.
Figure 6.
DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine attenuates the anti-inflammatory effects of Met in macrophages. RAW 264.7 cells were pretreated with 2 μM 5-aza for 24 h prior to stimulation with 100 ng/mL LPS for 3 h. (A–C) Effects of Met (10 mM) on the LPS-induced production of IL-6, TNF-α, and IFN-β in RAW 264.7 cells were analyzed by ELISA. Cells were cultured for 24 h with 5-aza-2′-deoxycytidine (2 μM) and then treated with Met (10 mM) for 12 h. (D, E) Percentage of cytosine bases modified with a 5-methyl group was determined. Data represent the mean ± SD of three independent experiments, each performed in five samples. Comparisons among means used t-tests (*p < 0.05, **p < 0.01, ***p < 0.001).
Discussion
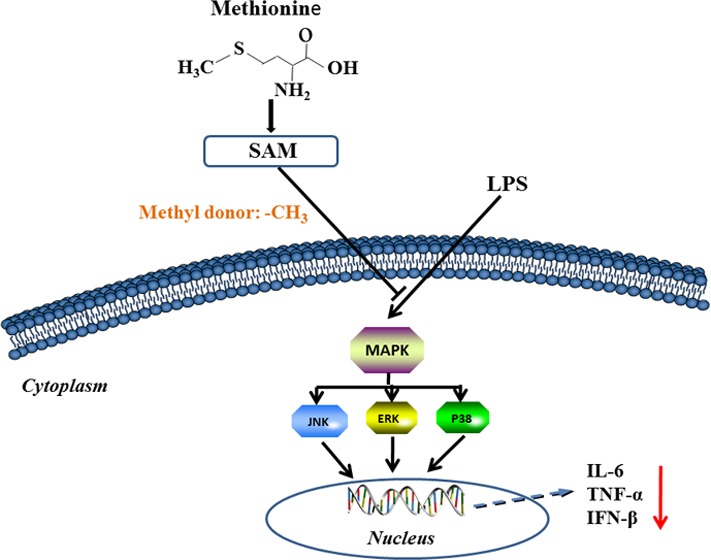
In this study, we demonstrated that Met exerted anti-inflammatory effects in LPS-stimulated RAW 264.7 macrophages by inhibiting the LPS-induced activation of p38, ERK, and JNK MARKs. The Met-derivative SAM also inhibited the inflammatory response via DNA methylation in RAW 264.7 cells.
Met is an essential and multifunctional nutrient in vertebrate diets. Met is a precursor of SAM, which is the methyl donor for DNA methylation.11 LPS is expressed in the outer membrane of Gram-negative bacteria and plays a key role in the initiation of inflammation by binding to Toll-like receptor-4; activation of downstream inflammatory signaling pathways, including MAPK; and induction of the production of inflammatory mediators.12 We found that the production of TNF-α, IL-6, and IFN-β in RAW 264.7 cells after LPS stimulation was significantly inhibited at the mRNA and protein levels with Met treatment (Figure 1). Furthermore, Met inhibited the LPS-induced phosphorylation and activation of the JNK, ERK, and p38 MAPKs (Figure 2). MAPK signaling can upregulate the production of proinflammatory cytokines, including TNF-α and IL-6, in LPS-stimulated macrophages.13,14 Our findings suggest that the anti-inflammatory properties of Met in LPS-treated RAW 264.7 macrophages are probably attributable to its inhibition of the LPS-stimulated activation of MAPKs.
Met is converted to SAM by Met adenosyltransferase. SAM is a cofactor that serves mainly as the principal methyl donor for DNA methylation.11 To further determine whether SAM has an anti-inflammatory activity, we first checked concentration of SAM in RAW 267.4 macrophages following Met treatment. Treatment with Met increased the cellular concentration of SAM 2-fold compared with control cells (Figure 3). Moreover, SAM inhibited the LPS-induced activation of the p38, JNK, and ERK1/2 MARKs and reduced LPS-induced production of proinflammatory mediators (Figures 3 and 4), consistent with the results of Met treatment. SAM is the principal methyl donor in biological reactions including DNA methylation, which plays a critical role in immune function, and is associated with several immune deficiencies and autoimmune disorders.15
Our observation that Met increased the global methylation level compared with the control indicates that the anti-inflammatory effects of Met may operate through changes in DNA methylation (Figure 5). Therefore, we next investigated whether Met modulates DNA methylation, to regulate the LPS-induced inflammatory response in macrophages. Previous studies showed that 5-aza is an epigenetic modifier and inhibits DNA methyltransferase activity, which results in DNA demethylation.16−18 We treated macrophages with the nucleoside-based DNA methyltransferase inhibitor 5-aza and evaluated whether it affects the anti-inflammatory effects of Met in macrophages. The 5-aza treatment attenuated the anti-inflammatory effects of Met and augmented the production of proinflammatory mediators by inhibiting DNA methylation in macrophages (Figure 6). Changes in the methylation status of promoter regions and especially CpG sites can regulate the expression and activation of various cytokines.19 Similarly, the IL-1β promoter of promyeloid cells is demethylated during their differentiation into monocytic cells to facilitate the accessibility of the transcriptional machinery to the promoter, which affects the subsequent production of IL-1β.20 Our understanding of the roles played by epigenetic modifications in inflammation remains rudimentary. Further research will be needed to study the associations between Met and global DNA methylation, gene-level methylation, and the silencing of gene-associated inflammation responses.
In summary, the results indicate that Met attenuates the LPS-induced inflammation associated with the Met-derivative SAM-mediated DNA methylation in RAW 264.7 macrophages. The findings provide new insights into the regulation of the actions of amino acids on the host immune system.
Acknowledgments
We thank the Key Laboratory of Poultry Genetics and Breeding, Ministry of Agriculture, and Guangdong Public Laboratory of Animal Breeding and Nutrition for providing services.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b03571.
Different concentrations of Met have no effect on the viability of RAW 264.7 cells (Figure S1); effects of Met at different concentrations on LPS-induced inflammatory cytokines in RAW 264.7 cells (Figure S2) (PDF)
Author Contributions
§ J.J. and Y.X. are co-first authors. J.J. and D.S. conceived and designed the experiments. J.J., Y.X., and H.L. performed the experiments. J.J., M.Z., C.L., D.S., and H.Q. analyzed the data. J.J. wrote the article. All authors discussed the results and approved the manuscript. The authors declare that they have no competing interest.
Financial support was provided by the National Natural Science Foundation of China (Grant No. 31601959), the Pearl River S&T Nova Program of Guangzhou (Grant No. 201806010101), the Science and Technology Program of Guangzhou (Grant Nos. 201504010017 and 201803060016), and the Earmarked Fund for Modern Agro-industry Technology Research System (Grant No. CARS-41).
The authors declare no competing financial interest.
Supplementary Material
References
- Dong Z.; Sinha R.; Richie J. P. Disease prevention and delayed aging by dietary sulfur amino acid restriction: translational implications. Ann. N. Y. Acad. Sci. 2018, 1418, 44–55. 10.1111/nyas.13584. [DOI] [PubMed] [Google Scholar]
- Hote P. T.; Sahoo R.; Jani T. S.; Ghare S. S.; Chen T.; Joshi-Barve S.; McClain C. J.; Barve S. S. Ethanol inhibits methionine adenosyltransferase II activity and S-adenosylmethionine biosynthesis and enhances caspase-3-dependent cell death in T lymphocytes: relevance to alcohol-induced immunosuppression. J. Nutr. Biochem. 2008, 19, 384–391. 10.1016/j.jnutbio.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M. L.; Gee A. J.; Gee R. J.; Zurita-Lopez C. I.; Khare S.; Clarke S. G.; Mamula M. J. Lupus autoimmunity altered by cellular methylation metabolism. Autoimmunity 2013, 46, 21–31. 10.3109/08916934.2012.732133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Novo C. A.; Bachert C. DNA methylation, bacteria and airway inflammation: latest insights. Curr. Opin. Allergy Clin. Immunol. 2015, 15, 27–32. 10.1097/ACI.0000000000000130. [DOI] [PubMed] [Google Scholar]
- Wichnieski C.; Maheshwari K.; Souza L. C.; Nieves F.; Tartari T.; Garlet G. P.; Carneiro E.; Letra A.; Silva R. M. DNA methylation profiles of immune response-related genes in apical periodontitis. Int. Endod. J. 2019, 5–12. 10.1111/iej.12966. [DOI] [PubMed] [Google Scholar]
- Pollard J. W. Trophic macrophages in development and disease. Nat. Rev. Immunol. 2009, 9, 259–270. 10.1038/nri2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Su B.; Karin M. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr. Opin. Immunol. 1996, 8, 402–411. 10.1016/S0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]; b Wu G. Functional amino acids in nutrition and health. Amino Acids 2013, 45, 407–411. 10.1007/s00726-013-1500-6. [DOI] [PubMed] [Google Scholar]
- Takahashi K.; Sugi Y.; Hosono A.; Kaminogawa S. Epigenetic regulation of TLR4 gene expression in intestinal epithelial cells for the maintenance of intestinal homeostasis. J. Immunol. 2009, 183, 6522–6529. 10.4049/jimmunol.0901271. [DOI] [PubMed] [Google Scholar]
- Shen Y.; Yang S.; Shi Z.; Lin T.; Zhu H.; Bi F.; Liu A.; Ying X.; Liu H.; Yu K.; Yan S. Mediates Anti-inflammation in LPS -Induced U937 Cells Targeting NF-κB Signaling Pathway. Inflammation 2015, 38, 736–744. 10.1007/s10753-014-9984-0. [DOI] [PubMed] [Google Scholar]
- Hu Y.; Cui Q.; Gu Y.; Sheng L.; Wu K.; Shi J.; Tan Y.; Fu H.; Liu L.; Fu S.; Yu X.; Huang H. Decitabine facilitates the generation and immunosuppressive function of regulatory γδT cells derived from human peripheral blood mononuclear cells. Leukemia 2013, 27, 1580–1585. 10.1038/leu.2012.345. [DOI] [PubMed] [Google Scholar]
- Lu S. C.; Mato J. M. S-Adenosylmethionine in cell growth, apoptosis and liver cancer. J. Gastroenterol. Hepatol. 2008, 23, S73–S77. 10.1111/j.1440-1746.2007.05289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T.; Akira S. TLR signaling. Cell Death Differ. 2006, 13, 816–825. 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- Ajizian S. J.; English B. K.; Meals E. A. Specific inhibitors of p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways block inducible nitric oxide synthase and tumor necrosis factor accumulation in murine macrophages stimulated with lipopolysaccharide and interferon-γ. J. Infect. Dis. 1999, 179, 939–944. 10.1086/314659. [DOI] [PubMed] [Google Scholar]
- Chan E. D.; Riches D. W. IFN-γ + LPS induction of iNOS is modulated by ERK, JNK/SAPK, and p38 (mapk) in a mouse macrophage cell line. Am. J. Physiol.: Cell Physiol. 2001, 280, C441–C450. 10.1152/ajpcell.2001.280.3.C441. [DOI] [PubMed] [Google Scholar]
- Suarez-Alvarez B. I.; Rodriguez R. M.; Fraga M. F.; López-Larrea C. DNA methylation: a promising landscape for immune system-related diseases. Trends Genet. 2012, 28, 506–514. 10.1016/j.tig.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Christman J. K. 5-Azacytidine and 5-aza-2-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 2002, 21, 5483–5495. 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Patra S. K.; Bettuzzi S. Epigenetic DNA-(cytosine-5-carbon) modifications: 5-aza-2′-deoxycytidine and DNA-demethylation. Biochemistry 2009, 74, 613–619. 10.1134/S0006297909060042. [DOI] [PubMed] [Google Scholar]
- Stresemann C.; Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer 2008, 123, 8–13. 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- Fu L. H.; Ma C. L.; Cong B.; Li S. J.; Chen H. Y.; Zhang J. G. Hypomethylation of proximal CpG motif of interleukin-10 promoter regulates its expression in human rheumatoid arthritis. Acta Pharmacol. Sin. 2011, 32, 1373–1380. 10.1038/aps.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels I.; Fleischer D.; Rink L.; Uciechowski P. Changes in chromatin structure and methylation of the human interleukin-1beta gene during monopoiesis. Immunology 2010, 130, 410–417. 10.1111/j.1365-2567.2009.03243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.