Graphical abstract
Keywords: CRISPR-Cas9, Carbapenem, Resistance, Beta-lactamase OXA-55, Shewanella algae
Abbreviations: Cas9, CRISPR-associated protein 9; COGs, Clusters of Orthologous Groups of proteins; CRISPR, clustered regularly interspaced short palindromic repeat; DAP, diaminopimelic acid; NCBI, National Center for Biotechnology Information; PGAAP, Prokaryotic Genomes Automatic Annotation Pipeline; SMRT, single-molecule real-time
Highlights
-
•
Emergence of carbapenem-resistant S. algae is a severe problem.
-
•
Re-sensitization of S. algae to carbapenem by CRISPR/Cas9 genome editing.
-
•
The blaOXA-55-like gene is essential for carbapenem resistance in S. algae.
-
•
One-plasmid genome editing system for CRISPR/Cas9 genome editing in S. algae.
-
•
CRISPR/Cas9 genome editing is a promising approach to validate the gene function.
Abstract
Antibiotic resistance in pathogens is a growing threat to human health. Of particular concern is resistance to carbapenem, which is an antimicrobial agent listed as critically important by the World Health Organization. With the global spread of carbapenem-resistant organisms, there is an urgent need for new treatment options. Shewanella algae is an emerging pathogen found in marine environments throughout the world that has increasing resistance to carbapenem. The organism is also a possible antibiotic resistance reservoir in humans and in its natural habitat. The development of CRISPR/Cas9-based methods has enabled precise genetic manipulation. A number of attempts have been made to knock out resistance genes in various organisms. The study used a single plasmid containing CRISPR/Cas9 and recE/recT recombinase to reverse an antibiotic-resistant phenotype in S. algae and showed blaOXA-55-like gene is essential for the carbapenem resistance. This result demonstrates a potential validation strategy for functional genome annotation in S. algae.
Introduction
Infections caused by antibiotic-resistant pathogens constitute a worldwide health crisis and have high mortality and morbidity. It is estimated that more than 20,000 deaths annually are linked to antimicrobial-resistant infections in the United States, and up to two million of such infections occur per year [1]. In the European Union, infections resulting from multidrug-resistant bacterial strains lead to 25,000 deaths and cost healthcare systems 1.5 billion euros per year [2]. The situation will almost certainly deteriorate if novel therapeutic strategies are not developed soon [3].
Resistance to carbapenem is particularly alarming, as it is a critically important class of antibiotics on the WHO List of Essential Medicines [4]. Carbapenems are often used as the last-resort treatment for many bacterial infections [5]. They are also crucial for treating life-threatening nosocomial infections and infections in compromised hosts. The emergence of carbapenem resistance first came to light in 1990 and is now recognized as a global issue that poses a significant threat to human health [6]. Therefore, new therapies for carbapenem-resistant bacteria are urgently required [7].
Shewanella algae is a gram-negative, motile, facultative anaerobic marine bacillus [8]. The organism has been proposed to be a potential source of antibiotic resistance in marine environments [9]. Known to be ubiquitous in the marine environment, the organism has become an emerging human pathogen, causing bacteremia, soft tissue and intra-abdominal infections [10], [11], [12], [13], [14], [15]. There have been increasing reports of carbapenem resistance in S. algae [16]. Previous studies have suggested that chromosome-encoded OXA-type β-lactamase is associated with carbapenem resistance in Shewanella spp. [17].
Recently, the clustered regularly interspaced short palindromic repeat (CRISPR) system and CRISPR-associated protein 9 (Cas9) were shown to provide adaptive immunity in prokaryotic defense systems, and the CRISP/Cas9 system has since been harnessed as a genome editing tool [18]. Attempts have been made to re-sensitize resistant E. coli using this system [19], [20]. In this study, we sequenced and annotated the genome of the carbapenem-resistant strain of Shewanella algae VGH117 and performed genome editing of 3 candidate genes using CRISPR/Cas9 and recE/recT recombinase to reverse the carbapenem resistance in S. algae.
Material and methods
Whole genome sequencing of S. algae VGH117
For genomic DNA preparation, the human clinical isolate S. algae VGH117 was grown on an LB agar plate (BD, Franklin Lakes, NJ, USA) at 35 °C for 12 h. Cells were harvested and suspended in nuclease-free water and OD600 was adjusted to 1 (approx. 1.0 × 109 CFU/mL). DNA was extracted from 2 mL of bacterial suspension using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). DNA quality and quantity were determined by the OD260/280 ratio using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and concentrations were read using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). DNA was sheared to the 10 kb range using g-TUBE (Covaris, Woburn, MA, USA), and the size distribution was examined by a Bioanalyzer (Agilent, Santa Clara, CA, USA). The sheared DNA was treated with DNA damage repair mix followed by end repair and ligation of SMRT adapters using the PacBio SMRTbell Template Prep Kit (Pacific Biosciences, Menlo Park, CA, USA) following the manufacturer’s instructions. The prepared libraries were then sequenced on a Pacific Biosciences RSII sequencer using three single-molecule real-time (SMRT) cells. SMRT Analysis portal version 2.1 was used for read filtering and adapter trimming with default parameters, which produced postfiltered data of approx. 1.63 Gb (approx. 332.8-fold coverage) with an N50 read length of approx. 6.3 kb (Table 1).
Table 1.
Sequencing statistic of VGH117 using three SMRT cells.
| SMRT cells | No. reads | N50 (bp) | Max. (bp) | Total bases (Mbp) | Coverage |
|---|---|---|---|---|---|
| 1 | 103,471 | 6353 | 38,351 | 537.8 | 109.76 |
| 2 | 98,213 | 6302 | 40,227 | 506.2 | 103.31 |
| 3 | 113,901 | 6316 | 40,210 | 586.7 | 119.73 |
| Total | 315,85 | 6324 | 40,227 | 1630.7 | 332.80 |
Genome assembly and gene annotation
The postfiltered reads were assembled by Canu (v1.4) [21], which produced one single large chromosomal contig (approx. 4.7 Mb) and a small plasmid contig (Table 2). Circlator was used to circularize these contigs into circular form [22]. Protein-coding and non-coding genes in the VGH117 genome and plasmid were annotated using the National Center for Biotechnology Information (NCBI) Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) (with accession numbers CP032414 and CP032415, respectively). Functional classification of these annotated genes was carried out by RPSBLAST v. 2.2.15 in conjunction with the COGs (Clusters of Orthologous Groups of proteins) databases (E-values < 0.001).
Table 2.
Assembly statistic of VGH117. The N50, genome, and plasmid, and total sizes are shown in base pairs (bp).
| Sample | N50 size | Genome size | Plasmid size | Total size |
|---|---|---|---|---|
| VGH117 | 4,787,117 | 4,787,117 | 132,551 | 4,919,668 |
Detection of antibiotic resistance genes and target selection
Antibiotic-resistance genes were predicted using an online tool, Comprehensive Antibiotic Resistance Database (CARD, https://card.mcmaster.ca/home), that utilizes protein homology to predict gene models [23]. Two criteria were used to select S. algae VGH117 gene targets for CRISPR/Cas9 editing and functional tests in this study. The first criterion was that the minimal protein identity of the matching region must be higher than 30% and must extend over 80% of the entire protein length. The second criterion was that the E-value of BlastX must be lower than 1E−40.
Bacterial strains and growth conditions
E. coli TOP10 (Thermo Fisher Scientific, Waltham, MA, USA) and BW29427 (aka WM3064) were employed for plasmid construction and conjugation of plasmid DNA into Shewanella, respectively. All E. coli and S. algae VGH117 cells were cultivated in LB broth/agar (BD, Franklin Lakes, NJ, USA) at 28 °C with shaking at 180 rpm if broth was used. The media was supplemented with 0.3 mM of 2,6-diaminopimelic acid (DAP) (Sigma-Aldrich, St. Louis, MO, USA) when growing E. coli BW29427 and 10 mM L-arabinose (Sigma-Aldrich, St. Louis, MO, USA) was added into the media to induce the recE/recT operon expression. LB agar was supplemented with 100 µg/mL apramycin (Sigma-Aldrich, St. Louis, MO, USA) to select for cells carrying pCC1-oriT-Cas9 or with 100 µg/mL apramycin and 100 µg/mL kanamycin (Sigma-Aldrich, St. Louis, MO, USA) to select for cells carrying the pCC1-oriT-Cas9-sgRNA-donor plasmids.
Construction of the Cas9 containing plasmid pCC1-oriT-Cas9
Both the Streptococcus pyogenes Cas9 gene (SpCas9) and the E. coli Rac prophage recombinase genes recE and recT were derived from their corresponding genomic sequences, synthesized (Thermo Fisher Scientific, Waltham, MA, USA), and used in this study. The oriT sequence that is responsible for the transfer of DNA during conjugation was derived from a mariner transposon plasmid, pKMW2. All these building blocks (SpCas9, recE/recT, and oriT) were cloned into the pCC1Fos (Epicentre, Madison, WI, USA) vector backbone using Gibson assembly (SGI-DNA, La Jolla, CA, USA). In this plasmid, the expression of the Cas9 endonuclease gene is driven by its original S. pyogenes promoter. The expression of recE/recT operon is under the control of the arabinose-inducible promoter pBAD (or ParaB) and terminated by the λ tL3 terminator.
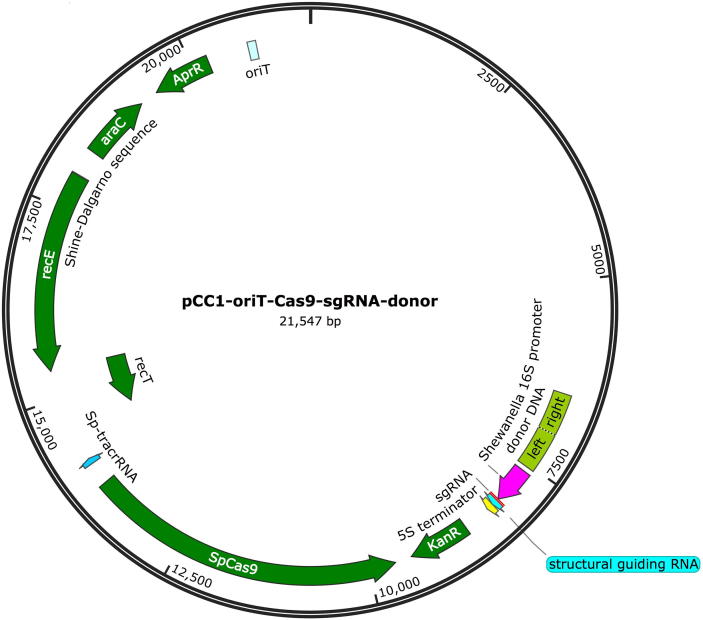
The sgRNA-scaffold DNA was synthesized (Thermo Fisher Scientific, Waltham, MA, USA), PCR fused with the 16S rRNA gene promoter amplified from Shewanella onedensis MR-1 (ATCC 700550) genomic DNA, and then inserted into the pCC1-oriT-Cas9 plasmid. Two BsaI cutting sites were designed immediately downstream of the Shewanella 16S promoter and upstream of the sgRNA scaffold, allowing insertion of a spacer sequence. A unique SacI restriction site was planned 63 bp upstream of the 16S promoter to allow cloning of the donor DNA template into the pCC1-oriT-Cas9 plasmid. The map of plasmid with genome editing function (pCC1-oriT-Cas9-sgRNA-donor) is shown in Fig. 1.
Fig. 1.
Map of pCC1-oriT-Cas9-sgRNA-donor.
Design and addition of targeted sgRNA and donor DNA into pCC1-oriT-Cas9
Plasmids with genome editing function were derived from pCC1-oriT-Cas9 by adding the targeted sgRNA sequences (spacers) and donor DNA templates. To design a spacer sequence for each target knockout, the protospacer was chosen by selecting a 20 nt sequence which was immediately flanked at the 3′ end by the NGG sequence. To ensure uniqueness of the selected spacer sequences, BLAST analyses were performed for all possible 23 nt queries (20 nt plus NGG) against the complete genome sequence of S. algae VGH117. Forward and reverse spacers were synthesized in oligonucleotides with the sticky ends GAAG (the last 4 nt of the 16S promoter) added to the beginning of the forward oligos, and AAAC (complementary to the first 4 nt of the sgRNA scaffold) added to the beginning of the reverse oligos. Paired oligonucleotides were annealed and ligated with BsaI-digested pCC1-oriT-Cas9 using T4 DNA ligase (NEB, Ipswich, MA, USA). The spacer sequence was confirmed by Sanger sequencing. Oligonucleotides employed for spacer oligo annealing and sequence verification are shown in Table 3.
Table 3.
Oligos used for spacer annealing and sequence verification.
| Oligo name | Sequence (5–3′)a |
|---|---|
| Forward | |
| sul2_gRNA.fwd | gaagCGCTGCGTTCTATCCGCAAT |
| OXA55_gRNA.fwd | gaagCTGGCCAACTGCTGATACAC |
| NmcR_gRNA.fwd | gaagTTGGGGCCGTTGAGCAGCAT |
| Reverse | |
| sul2_gRNA.rev | aaacATTGCGGATAGAACGCAGCG |
| OXA55_gRNA.rev | aaacGTGTATCAGCAGTTGGCCAG |
| NmcR_gRNA.rev | aaacATGCTGCTCAACGGCCCCAA |
| gRNA screen primer pair | |
| gRNA.screenF | CACTTAACGGCTGACATGGGAATTAGC |
| gRNA.screenR | CTGGCTTTCTACGTGTTCCGCTTCC |
Lowercase are sequences complementary to the flanking vector sequences generated by the BsaI cut.
The donor DNA is approximately 1 kb in length with sequence homology to regions upstream (500 bp) and downstream (500 bp) of the targeted genes. The donor DNA was designed to delete the complete open reading frame, including the start and stop codons. To generate the 1 kb donor DNA, two homologous arms were amplified from genomic DNA separately and were then fused together by fusion PCR. The donor DNA was then cloned into the pCC1-oriT-Cas9 plasmid carrying the corresponding spacer sequence by Gibson assembly (SGI-DNA, La Jolla, CA, USA) to yield the pCC1-oriT-Cas9-sgRNA-donor. Primers used for amplifying and PCR fusion of the donor DNA arms are shown in Table 4.
Table 4.
Primers used for donor DNA construction.
| Primer name | Sequence (5–3′)a |
|---|---|
| L arm forward | |
| sul2_Larm.fwd | ccgtattaccgcctttgagtgagctcGGCTACTTGAATGAGCAAACTGGCAG |
| OXA-55_Larm.fwd | ccgtattaccgcctttgagtgagctcTATCTCAAGTCCAAAGGAACGCCG |
| NmcR_Larm.fwd | ccgtattaccgcctttgagtgagctcCGCGATAATATATCGCTGCTGAGTCG |
| L arm reverse | |
| sul2_Larm.rev | cggtttctttcagcgGATGAGCGATTTATTCATGGGGGCT |
| OXA-55_Larm.rev | aataaaaggggaatgagGTCTGGCTGGCACGGCATAG |
| NmcR_Larm.rev | cgttacccaggccCGTTTGGTCTGTCCGTTCCAGG |
| R arm forward | |
| sul2_Rarm.fwd | atgaataaatcgctcatcCGCTGAAAGAAACCGCAAGAATTCG |
| OXA-55_Rarm.fwd | gtgccagccagacCTCATTCCCCTTTTATTCCCGGCG |
| NmcR_Rarm.fwd | acggacagaccaaacgGGCCTGGGTAACGAACAGTTCTTC |
| R arm reverse | |
| sul2_Rarm.rev | attttgttatcattcccctagagctcAAGTCGGGATTGACGGCATTGC |
| OXA-55_Rarm.rev | attttgttatcattcccctagagctcATCTGCAACCGGCTCCAGTAAAG |
| NmcR_Rarm.rev | attttgttatcattcccctagagctcAAGCTGAACCTTTGATGGACTGACTG |
| donor DNA screen primer pair | |
| Donor.screenF | CACTTAACGGCTGACATGGGAATTAGC |
| Donor.screenR | CTGGCTTTCTACGTGTTCCGCTTCC |
Lowercase are sequences complementary to the flanking vector sequences generated by the BsaI cut.
Targeted gene deletion using CRISPR/Cas9 coupled with recE/recT recombinase
The fully assembled pCC1-oriT-Cas9-sgRNA-donor was extracted from TOP10 cells and then transferred into the E. coli conjugation strain BW29427 by electroporation (0.1 cm cuvette, 1.80 kV) (Bio-Rad, Hercules, CA, USA). After electroporation, cells were immediately added into 1 mL of SOC medium (Invitrogen, Carlsbad, CA, USA) and recovered for 1 h at 28 °C prior to plating on LB agar containing kanamycin and apramycin.
Recipient S. algae VGH117 were grown overnight at 28 °C with shaking at 180 rpm in 5 mL of LB broth. Donor E. coli BW29427 (harboring pCC1-oriT-Cas9 or pCC1-oriT-Cas9-sgRNA-donor) were also grown in the same conditions as recipient but in LB broth supplemented with antibiotics and DAP. Both recipient and donor cell pellets were resuspended and diluted with LB broth to OD600 = 1. They were then mixed in a 1:3 ratio immediately by pipetting. Fifty µL of cell mixture was then placed on an LB agar plate containing DAP and L-arabinose. After incubation at 30 °C for 24 h, cells were collected and washed twice with LB broth. The bacterial pellet was resuspended in 1 mL of LB broth and then decimal dilutions from 10−2 to 10−6 of bacterial suspension were prepared by transferring 100 µL of the previous dilution to 900 µL of LB broth. One hundred µL of each dilution was pipetted onto separate LB agar plates containing L-arabinose and antibiotics, and then spread across the surface of each plate and allowed to dry. Plates were promptly incubated for 24 h at 30 °C. Sixteen colonies were randomly picked for further target knockout screening.
Screening of targeted gene deletion
The screening of S. algae VGH117 colonies for targeted gene deletion was performed by PCR using primers designed to flank the target and donor DNA sequence. Colonies carrying the targeted deletion would have a different size PCR product compared to the wild-type strain. The primer sequences for screening targeted deletion are listed in Table 5. The control strain (harboring pCC1-oriT-Cas9) and PCR-confirmed knockout strains (harboring pCC1-oriT-Cas9-sgRNA-donor plasmid) were subjected to the antimicrobial susceptibility tests described below.
Table 5.
Primers used for target gene deletion PCR screen.
| Primer name | Sequence (5–3′) |
|---|---|
| Forward | |
| sul2_del_screen.fwd | GGCCATGAAGGCCGCTTATTGA |
| OXA-55_del_screen.fwd | AACTGGATGTTCAGTTTACCGATGCC |
| NmcR_del_screen.fwd | CTGAAGATGTGTCTGCGGTTCATGG |
| Reverse | |
| sul2_del_screen.rev | CCTAAAACTCTTCAATGCACGGGTCT |
| OXA-55_del_screen.rev | GGTCGAATCCGGTCAGCAGTATC |
| NmcR_del_screen.rev | AGACCTATCTGCTCTATTGCGATCGC |
Antimicrobial susceptibility testing
Minimal inhibitory concentration (MIC) were determined by the E-test (bioMérieux, Marcy-l'Étoile, France) following the manufacturer's instructions. Ampicillin, ciprofloxacin, levofloxacin, imipenem, and trimethoprim/sulfamethoxazole were used in this study. EUCAST Clinical Breakpoint Tables v 8.0 (online available at http://www.eucast.org/clinical_breakpoints/) were used for interpreting the MIC values.
Results
Antibiotics resistance profile of S. algae VGH117
S. algae VGH117 was resistant to both ampicillin and imipenem on the E-test strips in the antimicrobial susceptibility tests. According to the manufacturer's instructions, the MIC values of ampicillin and imipenem were reported as ≥256 and ≥32 µg/mL, respectively. The MICs of ciprofloxacin, levofloxacin and trimethoprim/sulfamethoxazole were 0.19, 0.19 and 1.5 µg/mL respectively. Based on the EUCAST Clinical Breakpoint v 8.0, S. algae VGH117 showed a high level of resistance to ampicillin and imipenem, but was reported as susceptible to ciprofloxacin and levofloxacin. For trimethoprim/sulfamethoxazole, EUCAST had no breakpoint data that we could reference at the time of this writing.
Detection of potential antibiotics resistance genes
With the available genome sequence, the CARD Resistance Gene Identifier (RGI) was used to detect a potential sulfonamide resistance gene in S. algae VGH117 with 100% identity in protein sequence with dihydropteroate synthase (sul2) (E-value = 4.0E−148). RGI also detected a total of 18 genes that may be responsible for the resistance of ampicillin and imipenem, including 1 ambler class D β-lactamase, 1 SRT β-lactamase, 1 subclass B3 LRA β-lactamase, 1 class C/D LRA β-lactamase, and 14 NmcA β-lactamase genes. There were 2 candidates that reached the target selection criteria we set. One had 97.92% identity (E-value = 1.3E−160) in protein sequence with OXA-55 β-lactamase (blaOXA-55), and the other had 33.22% identity (E-value = 6.9E−40) with NmcA β-lactamase (NmcR). In this study, three genes including sul2, blaOXA-55-like and NmcR-like were chosen for the CRISPR/Cas9-mediated genome editing experiment (Table 6).
Table 6.
Statistics of selected targets for CRISPR/Cas9 genome editing.
CRISPR/Cas9-mediated deletion of sul2, blaOXA-55-like, and NmcR-like and antibiotic resistance tests
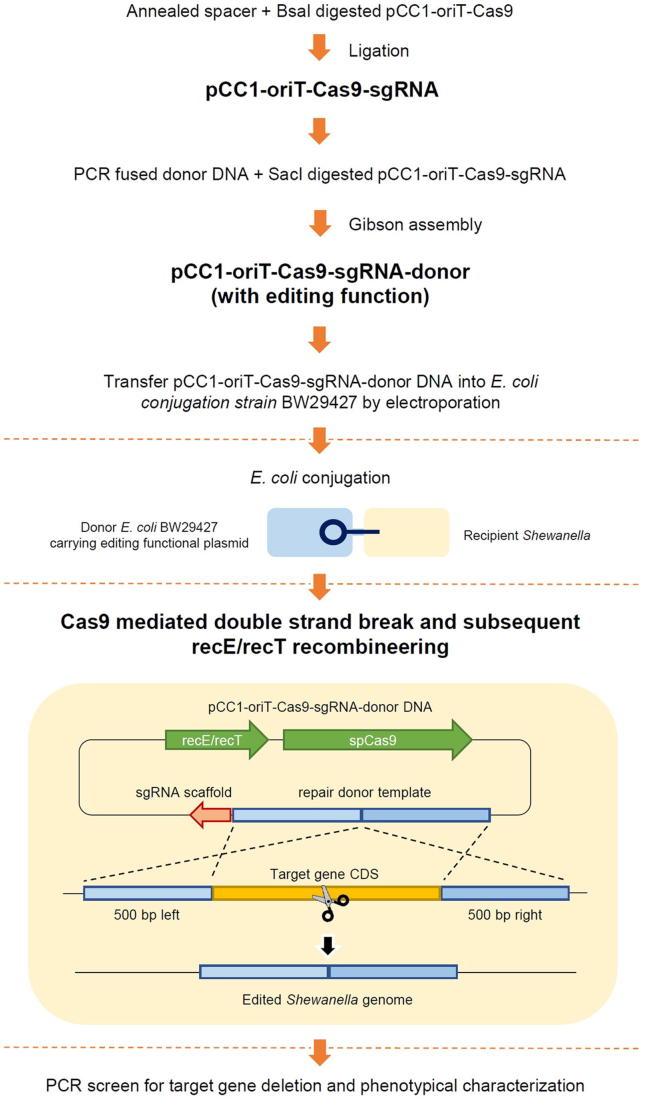
The plasmid with genome editing function (pCC1-Cas9-oriT-sgRNA-donor) was derived from pCC1-oriT-Cas9 by adding the targeted spacer and donor DNA. By delivering the pCC1-Cas9-oriT-sgRNA-donor from E. coli BW29427 into S. algae VGH117, a single gene deletion could be achieved. The editing approach flow chart is shown in Fig. 2. This one-plasmid genome editing system appears to work well with S. algae VGH117 cells. After screening 16 colonies for each editing experiment, we observed 68.75%, 31.25%, and 75% gene knockout efficiency in sul2, blaOXA-55-like, and NmcR-like, respectively (Table 7).
Fig. 2.
The flow chart of editing approach used in this study.
Table 7.
Selected target spacer design and editing efficiency.
| Target | Del size (bp) | Editing efficiency | Spacer statistics |
||
|---|---|---|---|---|---|
| % of editing (n = 16) | PAM | Tm (°C) | %GC | ||
| sul2 | 769 | 68.75% | TGG | 59 | 55 |
| blaOXA-55-like | 870 | 31.25% | AGG | 57 | 55 |
| NmcR-like | 648 | 75.00% | TGG | 64 | 60 |
In antibiotics resistance testing, the MIC value of the NmcR-like knockout strain (SH042) showed no obvious difference with the wild-type strain. This result suggested NmcR-like gene is not related to the resistance. However, the sul2 and blaOXA-55-like knockout strains (SH041 and SH036 respectively) showed diminished resistance to sulfonamides, ampicillin and imipenem (Table 8, Fig. 3C, 3F, 3I). The sulfonamide MIC of SH041 was 0.38 µg/mL, which was 4 times lower than that of wild-type (1.5 µg/mL). SH036 showed a significantly decreased MIC to ampicillin and imipenem (Table 8). The inhibition ellipse clearly appeared around the ampicillin and imipenem strips, and the E-test results indicated that the MIC was 0.38 µg/mL in both antibiotics (Fig. 3F, 3I). Based on the EUCAST clinical breakpoints and the resulting MIC values, SH036 had become susceptible to ampicillin and imipenem. This finding demonstrates the feasibility of using the one-plasmid CRISPR/Cas9 system to perform genome editing in S. algae VGH117 cells and to verify predicted antibiotic resistance genes experimentally. Taken together, these results also suggest this system may have a potentially broader functional applicability in various other Shewanella species.
Table 8.
MIC values of S. algae strains and the breakpoint of EUCAST.
| S. algae strains | MIC (µg/mL) |
||||
|---|---|---|---|---|---|
| AMc | IPd | CIe | LEf | TSg | |
| VGH117a | ≥256 | ≥32 | 0.19 | 0.19 | 1.5 |
| SH024b | ≥256 | ≥32 | 0.19 | 0.19 | 1.5 |
| SH041 (sul2 KO) | ≥256 | ≥32 | 0.125 | 0.25 | 0.38 |
| SH036 (blaOXA-55-like KO) | 0.38 | 0.38 | 0.125 | 0.19 | 1.5 |
| SH042 (NmcR-like KO) | ≥256 | ≥32 | 0.125 | 0.19 | 1.0 |
| EUCAST MIC breakpoint | |||||
| Susceptible ≤ | 2 | 2 | 0.25 | 0.5 | – |
| Resistance > | 8 | 8 | 0.5 | 1 | – |
Wild type strain.
VGH117 carrying control plasmid pCC1-oriT-Cas9.
Ampicillin.
Imipenem.
Ciprofloxacin.
Levofloxacin.
Trimethoprim/sulfamethoxazole.
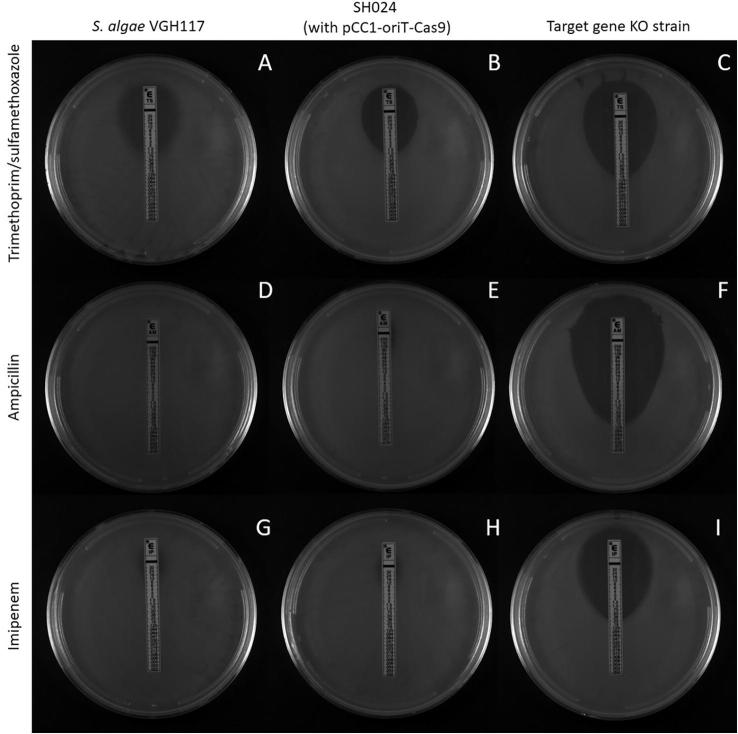
Fig. 3.
E-test results of S. algae strains. A, S. algae VGH117 trimethoprim/sulfamethoxazole MIC 1.5; B, SH024 (carrying pCC1-oriT-Cas9) trimethoprim/sulfamethoxazole MIC 1.5; C, SH041 (sul2 KO strain) trimethoprim/sulfamethoxazole MIC 0.38; D, S. algae VGH117 ampicillin MIC > 256; E, SH024 (carrying pCC1-oriT-Cas9) ampicillin MIC > 256; F, SH036 (blaOXA-55-like KO strain) ampicillin MIC 0.38; G, S. algae VGH117; imipenem MIC > 32; H, SH024 (carrying pCC1-oriT-Cas9) imipenem MIC > 32; and I, SH036 (blaOXA-55-like KO strain) imipenem MIC 0.38.
Discussion
In the study, the carbapenem-resistant S. algae was successfully re-sensitized using the CRISPR/Cas9 system. To the best of our knowledge, this is the first report of re-sensitization of resistant nonfermentative, gram-negative bacilli. The results potentially open up a new strategy for reversing carbapenem resistance using the CRISPR/Cas9 system. Further studies are needed to optimize the efficacy of this method and its delivery system [24].
Conventional methods of studying antibiotic resistance genes in Shewanella were usually carried out by large-scale PCR screening of candidate genes [25] or genomic expression library cloning [17], [26], [27], [28]. These approaches have their own limitations because PCR can only be used to screen a set of known genes, and the use of genomic clone libraries requires a large volume of phenotypic screenings. The CRISPR/Cas9 system with or without recombineering has provided an efficient strategy in several bacteria such as E. coli [29], [30], [31], Streptococcus [31], Streptomyces [32], Lactobacillus [33] and Clostridium [34]. CRISPR/Cas9 is an adaptive immune system in bacteria and archaea that confers resistance to foreign genetic elements [35]. The class 2 type II CRISPR/Cas9 system consists of a Cas nuclease (Cas9), a trans-activating CRISPR RNA (tracrRNA) and a programmable CRISPR-targeting RNA (crRNA), which generates a Cas9-mediated double-strand break (DSB) at almost any target locus [36], [37]. Since the DSB is deadly, bacterial cells possessing specific modification generated via recombination can be easily selected. If a repair template with homology arms flanking the target is supplied, the break could be repaired according to this template, allowing for precise gene deletions by either the Rac prophage RecE/RecT system or the bacteriophage lambda Red system [38], [39]. In this study, a plasmid which contains all of the above elements to perform CRISPR/Cas9-mediated gene deletion was constructed. By comparing the resistance levels of the wild-type strain and target gene knockout strains, we were able to functionally assess whether the predicted target genes were responsible for the antibiotic resistance.
In recent years, the CRISPR/Cas9 method has rapidly advanced, allowing precise genome editing for the purposes of understanding the function of a given gene and linkages between genetic variations and biological phenotypes [19], [31]. Compared with previous genome engineering technologies, such as zinc finger nucleases (ZENs) [40] and transcription activator-like effector nucleases (TALENs) [41], the CRISPR/Cas9 system is simpler and more efficient in modifying genomic targets [42]. Several attempts have been made to deliver the CRISPR-Cas–encoding cassette into pathogen by phage and demonstrated the approach is capable to eliminate specific pathogen [43], [44]. Yosef et al. further used lytic phages to transfer CRISPR-Cas system to sensitize antibiotic-resistant bacteria and showed the potential applicability in the antibiotic resistance problem [45]. In this study, the customizable modular design of pCC1-Cas9-oriT enabled construction of 3 different functional pCC1-Cas9-oriT-sgRNA-donors in vitro that targeted different genomic regions for editing. Interestingly, the editing efficiency of blaOXA-55-like gene was two-fold lower compared with other targets without any significant difference in spacer sequence design or target gene size (Table 7). To further understand the factors that may affect efficiency, different spacer sequences designed for the same target need to be evaluated, such as Tm(°C) and %GC of spacer sequence, and the chosen PAM sequence should be considered and tested.
Using whole genome sequencing and consecutive antibiotic resistance gene prediction, a sulfonamide resistance gene (sul2) was found in the genome of S. algae VGH117. This in silico analysis result was confirmed by antibiotic sensibility test using the EUCAST guidelines. A previous study indicated that some Shewanella clinical isolates carry the sul2 gene [46]; however, the results did not demonstrate whether the presence of sul2 was related to sulfonamide resistance. The sul2 gene expresses dihydropteroate synthase, which facilitates the production of para-aminobenzoate (PABA), an intermediate of folate synthesis, capable of reducing the growth inhibition effect caused by sulfonamide competing for the PABA [47]. According to the result of this study, the sul2 knockout strain (SH041) showed a reduction of trimethoprim/sulfamethoxazole MIC from 1.5 µg/mL to 0.38 µg/mL (Table 8). It suggests that the presence of sul2 gene might confer sulfonamide resistance in Shewanella.
Shewanella spp. are able to resist many β-lactams including cephalosporins and penams. We identified an OXA-55-like β-lactamase in the genome of S. algae VGH117. This 289-amino-acid OXA-55-like protein shared 97.92% identity with the first reported carbapenem-hydrolyzing ambler class D β-lactamase OXA-55 (blaOXA-55) from Shewanella algae KB-1 (GenBank ID: AAR03105.1) [17]. There are also several variants of class D β-lactamase-encoding genes harbored by various Shewanella strains, such as blaOXA-54 in S. oneidensis MR-1 [48] and many other blaOXA-48-like β-lactamases including blaOXA-48a, blaOXA-48b, blaOXA-151, blaOXA-181, blaOXA-199, blaOXA-252, blaOXA-514 and blaOXA-515 in other Shewanella species [25], [49], [50]. However, they were mostly reported as possessing a narrow spectrum of hydrolysis activity for penicillins, cephalosporins and imipenem [17], [49], [50]. S. algae KB-1 was the first Shewanella strain to be descried as having the blaOXA-55 gene and only showed an MIC of 4 µg/mL for imipenem [17]. The imipenem MIC of S. algae VGH117 was higher than 32 µg/mL, indicating that VGH117 is more resistant to imipenem than KB-1. The blaOXA-55-like gene found in VGH117 had 17 nucleotide differences from blaOXA-55 and encoded the OXA-55-like protein with 6 amino acid substitutions (C41G, E43G, L98I, V128A, E167D, and V261I) (Fig. 4). Knockout of the blaOXA-55-like gene resulted in an imipenem MIC of S. algae SH036 that showed more susceptibility to imipenem than wild-type (Table 8). This result suggests that the substitution of amino acids in OXA-55-like β-lactamase may play an important role in imipenem resistance. To address this question, further research is recommended ncluding gene cloning and protein expression experiments, enzyme kinetic analysis, and CRISPR/Cas9 mediated genome editing.
Fig. 4.
Comparison of amino acid sequences of OXA-55 to the OXA-55-like β-lactamases from S. algae VGH117. The asterisk indicates the positions of amino acid substitution.
Conclusions
This study presents a one-plasmid CRISPR/Cas9 strategy that is flexible and involves simple assembly of crRNA (spacer) from the type II CRISPR/Cas9 system. In this system, the repair template (homology donors) for HDR can also be easily added using well-developed DNA assembly techniques. Simple conjugation of pCC1-Cas9-oriT-sgRNA-donor into the Shewanella host results in highly efficient gene editing. This method reduces the time, cost and labor needed to perform precise genome manipulation compared with previous genome engineering technologies. Combining whole-genome sequencing data and subsequent genome annotation or gene prediction provides an alternative to previous large-scale PCR screening or genomic expression library cloning for gene function research. This result demonstrates a potential validation strategy for clinically relevant gene function in S. algae.
Funding
This work was supported in part by the Taichung Veterans General Hospital (TCVGH-1083901B, TCVGH-PU1088101) and the National Chung Hsing University (GP-01565-YOU).
Conflict of interest
The authors have declared no conflict of interest.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Acknowledgements
I would like to take this opportunity to thank Dr. Wayne Huey-Herng Sheu. This work would not have been possible without his gratitude, encouragement and support. The authors would like to acknowledge Yasuo Yoshikuni, Ze Peng, Dave Robinson and Rita C. Kuo from DOE Joint Genome Institute for sharing the experience of molecular biological techniques and the fruitful discussions on genome engineering. The authors would also like to thank Hirofumi Sawa, Michihito Sasaki, Manabu Igarashi, and Junya Yamagishi from Hokkaido University Research Center (Zoonosis Control) for sharing the experience of next-generation sequencing.
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
Jan-Fang Cheng, Email: jfcheng@lbl.gov.
Po-Yu Liu, Email: pyliu@vghtc.gov.tw.
References
- 1.Prestinaci F., Pezzotti P., Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109:309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hojgard S. Antibiotic resistance – why is the problem so difficult to solve? Infect Ecol Epidemiol. 2012;2 doi: 10.3402/iee.v2i0.18165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shallcross L.J., Howard S.J., Fowler T., Davies S.C. Tackling the threat of antimicrobial resistance: from policy to sustainable action. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140082. doi: 10.1098/rstb.2014.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen O., Denning D. World health organization ranking of antimicrobials according to their importance in human medicine. Clin Infect Dis. 2017;64:986–987. doi: 10.1093/cid/cix059. [DOI] [PubMed] [Google Scholar]
- 5.Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 2016;3:15–21. doi: 10.1177/2049936115621709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papp-Wallace K.M., Endimiani A., Taracila M.A., Bonomo R.A. Carbapenems: past, present, and future. Antimicrob Agents Chemother. 2011;55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theuretzbacher U., Gottwalt S., Beyer P., Butler M., Czaplewski L., Lienhardt C. Analysis of the clinical antibacterial and antituberculosis pipeline. Lancet Infect Dis. 2018 doi: 10.1016/S1473-3099(18)30513-9. [DOI] [PubMed] [Google Scholar]
- 8.Gram L., Bundvad A., Melchiorsen J., Johansen C., Fonnesbech Vogel B. Occurrence of Shewanella algae in Danish coastal water and effects of water temperature and culture conditions on its survival. Appl Environ Microbiol. 1999;65:3896–3900. doi: 10.1128/aem.65.9.3896-3900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tacao M., Araujo S., Vendas M., Alves A., Henriques I. Shewanella species as the origin of blaOXA-48 genes: insights into gene diversity, associated phenotypes and possible transfer mechanisms. Int J Antimicrob Agents. 2018;51:340–348. doi: 10.1016/j.ijantimicag.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Poovorawan K., Chatsuwan T., Lakananurak N., Chansaenroj J., Komolmit P., Poovorawan Y. Shewanella haliotis associated with severe soft tissue infection, Thailand, 2012. Emerg Infect Dis. 2013;19:1019–1021. doi: 10.3201/eid1906.121607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janda J.M. Shewanella: a marine pathogen as an emerging cause of human disease. Clin Microbiol Newsl. 2014;36:25–29. [Google Scholar]
- 12.Byun J.H., Park H., Kim S. The phantom menace for patients with hepatobiliary diseases: shewanella haliotis, often misidentified as Shewanella algae in biochemical tests and MALDI-TOF analysis. Jpn J Infect Dis. 2017;70:177–180. doi: 10.7883/yoken.JJID.2015.658. [DOI] [PubMed] [Google Scholar]
- 13.Liu P.Y., Lin C.F., Tung K.C., Shyu C.L., Wu M.J., Liu J.W. Clinical and microbiological features of shewanella bacteremia in patients with hepatobiliary disease. Intern Med. 2013;52:431–438. doi: 10.2169/internalmedicine.52.8152. [DOI] [PubMed] [Google Scholar]
- 14.Liu P.Y., Lin C.F., Wu M.J., Kao C.C., Shi Z.Y. Shewanella bloodstream infection. Hemodial Int. 2012;16:578–579. doi: 10.1111/j.1542-4758.2012.00664.x. [DOI] [PubMed] [Google Scholar]
- 15.Tadera K., Shimonaka A., Ohkusu K., Morii D., Shimohana J., Michinaka T. A case report of Shewanella haliotis showing a phlegmonous inflammation of right lower leg with sepsis. JSCM. 2010;20:239–244. [Google Scholar]
- 16.Yousfi K., Bekal S., Usongo V., Touati A. Current trends of human infections and antibiotic resistance of the genus Shewanella. Eur J Clin Microbiol Infect Dis. 2017;36:1353–1362. doi: 10.1007/s10096-017-2962-3. [DOI] [PubMed] [Google Scholar]
- 17.Heritier C., Poirel L., Nordmann P. Genetic and biochemical characterization of a chromosome-encoded carbapenem-hydrolyzing ambler class D beta-lactamase from Shewanella algae. Antimicrob Agents Chemother. 2004;48:1670–1675. doi: 10.1128/AAC.48.5.1670-1675.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo M.L., Leenay R.T., Beisel C.L. Current and future prospects for CRISPR-based tools in bacteria. Biotechnol Bioeng. 2016;113:930–943. doi: 10.1002/bit.25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu H., Gong J., Butaye P., Lu G., Huang K., Zhu G. CRISPR/Cas9/sgRNA-mediated targeted gene modification confirms the cause-effect relationship between gyrA mutation and quinolone resistance in Escherichia coli. FEMS Microbiol Lett. 2018;365 doi: 10.1093/femsle/fny127. [DOI] [PubMed] [Google Scholar]
- 20.Kim J.S., Cho D.H., Park M., Chung W.J., Shin D., Ko K.S. CRISPR/Cas9-mediated re-sensitization of antibiotic-resistant Escherichia coli harboring extended-spectrum beta-lactamases. J Microbiol Biotechnol. 2016;26:394–401. doi: 10.4014/jmb.1508.08080. [DOI] [PubMed] [Google Scholar]
- 21.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt M., Silva N.D., Otto T.D., Parkhill J., Keane J.A., Harris S.R. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015;16:294. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McArthur A.G., Waglechner N., Nizam F., Yan A., Azad M.A., Baylay A.J. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goren M., Yosef I., Qimron U. Sensitizing pathogens to antibiotics using the CRISPR-Cas system. Drug Resist Updat. 2017;30:1–6. doi: 10.1016/j.drup.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Zong Z. Discovery of bla(OXA-199), a chromosome-based bla(OXA-48)-like variant, in Shewanella xiamenensis. PLoS One. 2012;7:e48280. doi: 10.1371/journal.pone.0048280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel L., Heritier C., Nordmann P. Genetic and biochemical characterization of the chromosome-encoded class B beta-lactamases from Shewanella livingstonensis (SLB-1) and Shewanella frigidimarina (SFB-1) J Antimicrob Chemother. 2005;55:680–685. doi: 10.1093/jac/dki065. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J.Y., Zhao S.M., Mu X.D., Xiao Z. Genetic characterization of plasmid-mediated quinolone resistance gene qnrS2 in Pseudoalteromonas and Shewanella isolates from seawater. FEMS Microbiol Lett. 2017;364 doi: 10.1093/femsle/fnw295. [DOI] [PubMed] [Google Scholar]
- 28.Zhao J.Y., Mu X.D., Zhu Y.Q., Xi L., Xiao Z. Identification of an integron containing the quinolone resistance gene qnrA1 in Shewanella xiamenensis. FEMS Microbiol Lett. 2015;362:fnv146. doi: 10.1093/femsle/fnv146. [DOI] [PubMed] [Google Scholar]
- 29.Pyne M.E., Moo-Young M., Chung D.A., Chou C.P. Coupling the CRISPR/Cas9 system with lambda red recombineering enables simplified chromosomal gene replacement in Escherichia coli. Appl Environ Microbiol. 2015;81:5103–5114. doi: 10.1128/AEM.01248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H., Zheng G., Jiang W., Hu H., Lu Y. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin (Shanghai) 2015;47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 33.Oh J.H., van Pijkeren J.P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014;42:e131. doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Zhang Z.T., Seo S.O., Choi K., Lu T., Jin Y.S. Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system. J Biotechnol. 2015;200:1–5. doi: 10.1016/j.jbiotec.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 36.Wang H., La Russa M., Qi L.S. CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 37.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy K.C. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Buchholz F., Muyrers J.P., Stewart A.F. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 40.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y., Cobb R.E., Zhao H. High-efficiency genome editing of streptomyces species by an engineered CRISPR/Cas system. Methods Enzymol. 2016;575:271–284. doi: 10.1016/bs.mie.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Bikard D., Euler C.W., Jiang W., Nussenzweig P.M., Goldberg G.W., Duportet X. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Citorik R.J., Mimee M., Lu T.K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol. 2014;32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yosef I., Manor M., Kiro R., Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci USA. 2015;112:7267–7272. doi: 10.1073/pnas.1500107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soledad Ramirez M., Merkier A.K., Almuzara M., Vay C., Centron D. Reservoir of antimicrobial resistance determinants associated with horizontal gene transfer in clinical isolates of the genus Shewanella. Antimicrob Agents Chemother. 2010;54:4516–4517. doi: 10.1128/AAC.00570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sköld O. Resistance to trimethoprim and sulfonamides. Vet Res. 2001;32:261–273. doi: 10.1051/vetres:2001123. [DOI] [PubMed] [Google Scholar]
- 48.Yin J., Sun L., Dong Y., Chi X., Zhu W., Qi S.H. Expression of blaA underlies unexpected ampicillin-induced cell lysis of Shewanella oneidensis. PLoS One. 2013;8:e60460. doi: 10.1371/journal.pone.0060460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ceccarelli D., van Essen-Zandbergen A., Veldman K.T., Tafro N., Haenen O., Mevius D.J. Chromosome-based blaOXA-48-like variants in Shewanella species isolates from food-producing animals, fish, and the aquatic environment. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.01013-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cimmino T., Olaitan A.O., Rolain J.M. Whole genome sequence to decipher the resistome of Shewanella algae, a multidrug-resistant bacterium responsible for pneumonia, Marseille, France. Expert Rev Anti Infect Ther. 2016;14:269–275. doi: 10.1586/14787210.2016.1106936. [DOI] [PubMed] [Google Scholar]