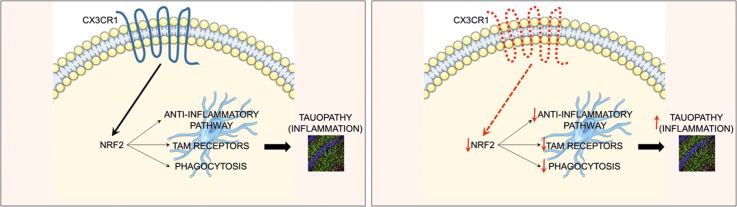
Abstract
TAU protein aggregation is the main characteristic of neurodegenerative diseases known as tauopathies. Low-grade chronic inflammation is also another hallmark that indicates crosstalk between damaged neurons and glial cells. Previously, we have demonstrated that neurons overexpressing TAUP301L release CX3CL1, which activates the transcription factor NRF2 signalling to limit over-activation in microglial cells in vitro and in vivo. However, the connection between CX3CL1/CX3CR1 and NRF2 system and its functional implications in microglia are poorly described. We evaluated CX3CR1/NRF2 axis in the context of tauopathies and its implication in neuroinflammation. Regarding the molecular mechanisms that connect CX3CL1/CX3CR1 and NRF2 systems, we observed that in primary microglia from Cx3cr1-/- mice the mRNA levels of Nrf2 and its related genes were significantly decreased, establishing a direct linking between both systems. To determine functional relevance of CX3CR1, migration and phagocytosis assays were evaluated. CX3CR1-deficient microglia showed impaired cell migration and deficiency of phagocytosis, as previously described for NRF2-deficient microglia, reinforcing the idea of the relevance of the CX3CL1/CX3CR1 axis in these events. The importance of these findings was evident in a tauopathy mouse model where the effects of sulforaphane (SFN), an NRF2 inducer, were examined on neuroinflammation in Cx3cr1+/+ and Cx3cr1-/- mice. Interestingly, the treatment with SFN was able to modulate astrogliosis but failed to reduce microgliosis in Cx3cr1-/- mice. These findings suggest an essential role of the CX3CR1/NRF2 axis in microglial function and in tauopathies. Therefore, polymorphisms with loss of function in CX3CR1 or NRF2 have to be taken into account for the development of therapeutic strategies.
Abbreviations: AD, Alzheimer's disease; ARE, Antioxidant response element; NRF2, Nuclear Factor (erythroid-derived 2)-like 2; SFN, sulforaphane; TAM receptors, Tyro3, Axl and Mer
Keywords: Inflammation, Neurodegeneration, TAU, Migration, TAM receptors, AXL, Microgliosis, Sulforaphane
Graphical abstract
Highlights
-
•
CX3CR1-deficient primary microglial cells present impaired expression of the transcription factor NRF2 signature.
-
•
TAM receptors expression is decreased in CX3CR1-deficient microglia.
-
•
AXL receptor is a NRF2-dependent gene.
-
•
Loss of CX3CR1 expression led to impaired phagocytosis and migration of microglia.
-
•
Sulforaphane treatment did not reverse rAAV-TAUP301L induced microgliosis in CX3CR1-deficient mice.
1. Introduction
Neurodegenerative diseases such as Alzheimer's disease (AD), progressive supranuclear palsy among others, are characterized by the deposition of microtubule-associated protein TAU and are known collectively as tauopathies. These diseases share important clinical, pathological, biochemical and genetic characteristics, although the molecular events that lead from conformation changes in normal TAU protein to neuronal dysfunction and cell death are essentially unknown and are probably diverse [1]. Besides neuronal degeneration, it is also known that the neuron environment contributes to this event, where glial cells play a crucial role [2]. In this context, neuroinflammation with a reactive morphology of astrocytes and microglia, together with low to moderate levels of proinflammatory markers is a key factor in tauopathies. The implication of inflammation in neurodegeneration is supported by several evidence. First, neurodegenerative diseases have genetic hallmarks of autoinflammatory diseases [3]. Second, immune memory in the brain is an important modifier of neuropathology [4], [5], [6]. Third, the existence of genetic variants only expressed by microglia in the central nervous system (CNS) such as TREM2, CD33 and CR1, which have been associated with AD and other neurodegenerative diseases [7], [8], [9], [10], [11], [12]. For instance, polymorphisms in CX3CR1 influence disease progression but not risk in Alzheimer's disease and amyotrophic lateral sclerosis, two diseases characterized by neuroinflammation [13], [14]. Moreover, our group has described that CX3CR1-deficiency exacerbates α-synuclein-A53T induced neuroinflammation and neurodegeneration in a mouse model of Parkinson's disease [15]. CX3CR1 is the receptor for the chemokine fractalkine (CX3CL1) and is a critical signalling pathway for microglia-neuron crosstalk [16], [17]. Interestingly, the CX3CL1/CX3CR1 axis is implicated in the regulation of cognitive functions and synaptic plasticity, particularly in the hippocampus. Disruption in this pathway has been associated with impaired neurogenesis. In aged rats, there are decreased levels of hippocampal CX3CL1 protein [18], [19]. These data indicate that the CX3CL1/CX3CR1 axis declines with age, which is an essential key factor for neurodegeneration. As to tauopathies, we previously described that hippocampal neurons expressing human TAUP301L mutant protein produce CX3CL1 and the hippocampus of patients with AD also exhibited increased expression of CX3CL1 in TAU-injured neurons that recruit microglia [20] as a help-me signal. One of the mechanisms triggered by CX3CL1 is the activation of the transcription factor Nuclear Factor (erythroid-derived 2)-like 2 (herein referred as NRF2). NRF2 recognizes an enhancer sequence termed antioxidant response element (ARE) that is present in the regulatory regions of over 250 genes (ARE genes) such as antioxidant enzymes and biotransformation reactions, lipid and iron catabolism and mitochondrial bioenergetics [21]. Furthermore, evidence showed that NRF2 is also implicated in the modulation of inflammatory processes through crosstalk with the transcription factor NF-κB, the principal regulator of inflammation [22]. Additionally, it has been described that NRF2 is essential in proteostasis, which modulates the proteasome and autophagy processes [23], [24]. Therefore, modulation of NRF2 activity has the potential to alter neurodegenerative disease course [25], [26], [27], [28]. Interestingly, previous work showed that NRF2- and CX3CR1-knockout mice did not express heme oxygenase 1 (HO1), a NRF2-dependent enzyme, in microglial cells, which led to increased microgliosis and astrogliosis in response to neuronal TAUP301L expression [20]. Related to inflammation, in toxin A-induced enteritis it has been observed that HO1 expression was detected mainly in F4/80-positive bone marrow cells expressing CX3CR1, and Cx3cr1-/- mice failed to increase HO1 expression after toxin A treatment [29]. Taken together, all these evidence indicate a connection between CX3CR1 and NRF2 in inflammatory processes.
Therefore, in this work, we analysed in depth the molecular mechanisms implicated in the CX3CR1/NRF2 axis in microglial cells and the consequences for tauopathies. For this purpose, we evaluated the role of CX3CR1 receptor expression in the modulation of NRF2 signature and its relevance in microglia phagocytosis and migration. Finally, to evaluate the role of CX3CR1/NRF2 in neurodegeneration, we determined whether the treatment with sulforaphane, an NRF2 activator, could modulate neuroinflammation in a tauopathy mouse model in absence of CX3CR1, which would indicate the relevance of CX3CR1 and NRF2 loss of function polymorphisms in developing therapeutic strategies for humans.
2. Methods
2.1. Cell culture
Primary astrocytes and microglia were prepared from neonatal (P0-P2) mouse cortex from Cx3cr12+/+ and Cx3cr1-/- and grown and isolated as described in [20]. Briefly, neonatal (P0-P2) mouse cortex were mechanically dissociated and the cells were seeded onto 75 cm2 flasks in DMEM:F12 supplemented with 10% FCS and penicillin/streptomycin. After 2 weeks in culture, flasks were trypsinized and separated using CD11b MicroBeads for magnetic cell sorting (MACS Miltenyi Biotec, Germany). Microglial and astroglial cultures were at least 99% pure, as judged by immunocytochemical criteria. Medium was changed to Dulbecco's Modified Eagle Medium:F12 (DMEM:F12) serum-free without antibiotics 16 h before treatment. Immortalized microglial cell line (IMG) isolated from the brains of adult mice, were purchased from Kerafast Inc., and were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin and 2 mM L-glutamine, in 5% CO2 at 37 °C, 50% relative humidity. Medium was changed to serum-free DMEM without antibiotics 16 h before treatments. CX3CL1 was obtained from PreproTech (Catalog# 400-26) and solubilized in water at 46 μM and used at 100 nM. Sulforaphane was obtained from LKT Labs (Catalog# S8044) and used at 15 μM for short-time treatment (6 h) or 5 μM for long-time treatment (16 h).
2.2. Analysis of mRNA levels by quantitative real-time PCR
Total RNA extraction, reverse transcription, and quantitative polymerase chain reaction (PCR) were done as detailed in previous articles [20]. Primer sequences are shown in Supplementary Table S2. Data analysis was based on the ΔΔCT method with normalization of the raw data to housekeeping genes (Applied Biosystems). All PCRs were performed in triplicates.
2.3. Migration assay
Cell migration was assayed using the CytoSelect 96-well Cell Migration Assay according to the manufacturer's instruction (Cell Biolabs, Cambridge, UK). In brief, primary microglia from Cx3cr1+/+ and Cx3cr1-/- mice were collected as described above and were resuspended in serum-free DMEM containing 0.1% bovine serum albumin (BSA). As positive control medium containing 15% fetal bovine serum was added to the bottom chamber (100 μl per well). Primary microglial cells were added to the top insert and cells were incubated at 37 °C for 16 h before lysis of migratory cells and quantified using CyQuant® GR Fluorescent Dye, using a fluorescence plate reader with excitation at 480 nm and emission at 520 nm.
2.4. Luciferase assays
Transient transfections of HEK293T cells were performed with the expression vectors pGL3_AxlP1-LUC (gift of Prof. Dr H. Allgayer, Department of Experimental Surgery-Cancer Metastasis, Medical Faculty, Ruprecht-Karls-University Heidelberg, Germany). pTK-Renilla was used as an internal control vector (Promega). Luciferase assays were performed as described in [22].
2.5. Bioinformatics analysis
A putative antioxidant response element (ARE) in Axl gene promoters was identified in The Encyclopedia of DNA Elements at UCSC (ENCODE)25 for the human genome (Feb. 2009) taking as reference the available information from chromatin immunoprecipitation (ChIP) of ARE binding factors MAFK and BACH1. The putative MAFK and BACH1 binding regions were localized in 200- to 400-base pair-long DNase-sensitive and H3K27Ac-rich regions, i.e., most likely regulatory promoter regions. In addition, a frequency matrix of the consensus ARE sequence based on the JASPAR database26 was converted to a position-specific scoring matrix (PSSM) by turning the frequencies into scores through the log(2) [odd-ratio (odd ratio: observed frequency/expected frequency)]. One unit was added to each frequency to avoid log(0). Then a script was generated with the Python 3.4 program to scan the promoter sequences with candidate AREs retrieved from ENCODE with the PSSM. The max score was calculated by adding the independent scores for each of the 11 base pairs of the consensus ARE sequence with the PSSM. The relative score (score relative) was calculated from this max score (score of the sequence max) as: score relative = (score of the sequence max − score min possible)/(score max possible − score min possible). The min possible score (score min possible) is calculated as the lowest possible number obtained for a sequence from the PSSM and the max possible score (score max possible) is the highest possible score that can be obtained. We considered putative ARE sequences those with a score relative over 80%, which is a commonly used threshold for the computational framework for transcription factor binding site/TFBS analyses using PSSM.
2.6. Phagocytosis assay
Primary microglia from Cx3cr1+/+ and Cx3cr1-/- mice were collected as described above and 150.000 cells were plated on coverslips for 16 h. Then, the medium was replaced with serum-free DMEM:F12 without antibiotics for 24 h before adding fluorescent microspheres (150 microspheres per cell) (FluoSpheres polystyrene microspheres, Invitrogen) and CX3CL1 (100 nM) and incubating for 2 h. Then, cells were washed with PBS, fixed with 4% paraformaldehyde, and stained with DAPI. The images were captured using 90i Nikon microscope (Nikon, Montreal, QC, Canada) at 40×.
2.7. Animals and treatments
Colonies of Cx3cr1-/- (B6.129P-Cx3cr1tm1Litt/J) mice and Cx3cr1+/+ littermates were obtained from Jackson Laboratory, Bar Harbor, ME [30]. Each experimental group comprised 5–8 animals. Recombinant AAV vectors of hybrid serotype 1/2 express mutant hTAUP301L under control of the human synapsin 1 gene promoter and were used as described [31]. Surgical procedures and unilateral intracerebral injection of viral particles into the right hemisphere were performed as described before [20]. In brief, 2 μl of viral suspension containing 10E8 T.U. were injected at the stereotaxic coordinates −1.94 mm posterior, −1.4 mm lateral, and −1.8 mm ventral relative to bregma. All experiments were performed by certified researchers according to regional, national, and European regulations concerning animal welfare and animal experimentation, and were authorized by the Ethics Committee for Research of the Universidad Autónoma de Madrid and the Comunidad Autónoma de Madrid, Spain, with Ref PROEX 279/14, following institutional, Spanish and European guidelines (Boletín Oficial del Estado (BOE) of 18 March 1988 and 86/609/EEC, 2003/65/EC European Council Directives). SFN (50 mg/kg) (LKT Laboratories, St. Paul, MN) was prepared in saline solution just before use and i.p. injected. We did not detect significant weight loss, hair loss or other gross alterations in the SFN-treated mice either in the 3-weeks administration every day.
2.8. Immunofluorescence on mouse tissues
The protocol was previously described [23]. Primary antibodies are described in Supplementary Table S1. Secondary antibodies were: Alexa Fluor 546 goat anti-mouse, Alexa 546 goat anti-rabbit and Alexa Fluor 488 goat anti-mouse (1:500, Life Technologies, Madrid, Spain). Control sections were treated following identical protocols but omitting the primary antibody.
2.9. Stereological analysis of microgliosis and astrogliosis
Cell counts were performed every four sections (30 μm-thick) using Fiji Software (http://fiji.sc/Fiji) in 5 sections of the hippocampus separated [32]. The error coefficient attributable to the sampling was calculated according to Gundersen and Jensen (1987), and values ≤ 0.10 were accepted. (n = 4–5 animals per experimental group).
2.10. Statistical analyses
Data are presented as mean ± SEM. To determine the statistical test to be used, we employed GraphPad Instat 3, which includes the analysis of the data to normal distribution via the Kolmogorov-Smirnov test. In addition, statistical assessments of differences between groups were analysed (GraphPad Prism 5, San Diego, CA) by unpaired Student's t-tests when normal distribution and equal variances were fulfilled, or by the non-parametric Mann–Whitney test. One and two-way ANOVA with post hoc Newman-Keuls test or Bonferroni's test were used, as appropriate.
3. Results
3.1. CX3CR1-deficient primary microglial cells present impaired levels of the transcription factor NRF2 signalling
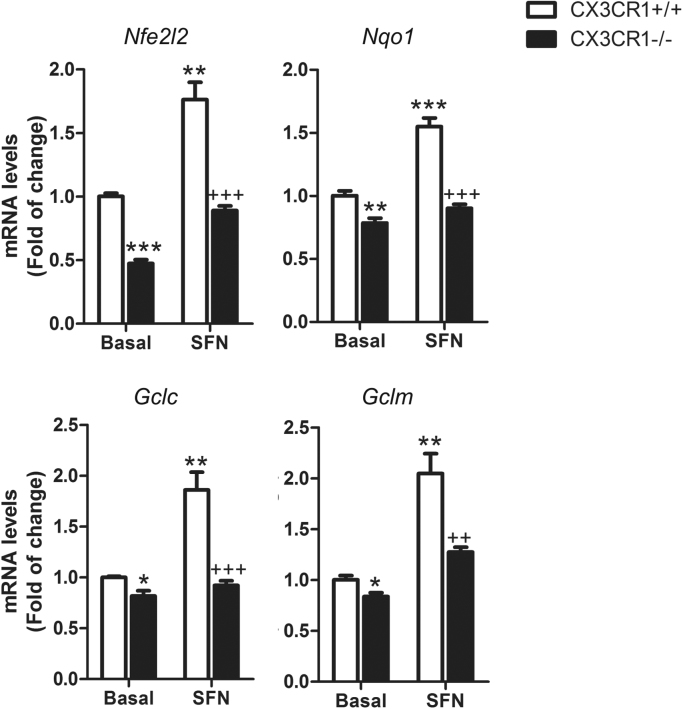
Previous results showed that CX3CR1-deficient bone marrow cells [29], macrophages [33] and microglial cells [20] displayed lack of HO1 expression, suggesting an alteration in NRF2 signalling. To gain more insight into the role of CX3CR1 axis on NRF2 signalling, we analysed the expression pattern of NRF2 pathway in Cx3cr1-deficient primary microglia compared to wild-type. Nfe2l2 mRNA expression levels were decreased in the absence of CX3CR1 as well as NRF2-dependent genes like Nqo1, Gclc and Gclm (Fig. 1). Moreover, to determine whether NRF2 activation could improve this impairment, Cx3cr1+/+ and Cx3cr1-/- primary microglia were treated with sulforaphane (SFN) (15 μM, 6 h), a NRF2 inducer [34]. Although Cx3cr1+/+ microglia showed significant induction of Nfe2l2, Nqo1, Gclc and Gclm expression levels, Cx3cr1-/- failed to replicate this effect to a greater extent. These results are specific for CX3CR1-expressing microglia given that astrocytes obtained in the same purification setting did not show those effects (Suppl. Fig. 1) and exhibited SFN dependent induction.
Fig. 1.
CX3CR1 receptor implications in the transcription factor NRF2 signalling in microglia. Primary cultures of microglia from control wild-type mice (Cx3cr1+/+) and Cx3cr1-knockout mice (Cx3cr1−/−) were incubated with vehicle or SFN (15 μM, 6 h). Quantitative real-time PCR determination of messenger RNA levels of NRF2-regulated genes coding Nfe2l2, Nqo1, Gclc and Gclm, respectively, normalized by Actb (β-Actin) messenger RNA levels. Two-way ANOVA followed by Bonferroni post-test was used to assess significant differences among groups. Asterisks denote significant differences *p < 0.05, **p < 0.01 respect to the basal Cx3cr1+/+ group and #p < 0.05 and ##p < 0.01 respect to the SFN-treated Cx3cr1+/+ group.
3.2. CX3CR1-deficient microglia exhibit decreased expression of TAM receptors
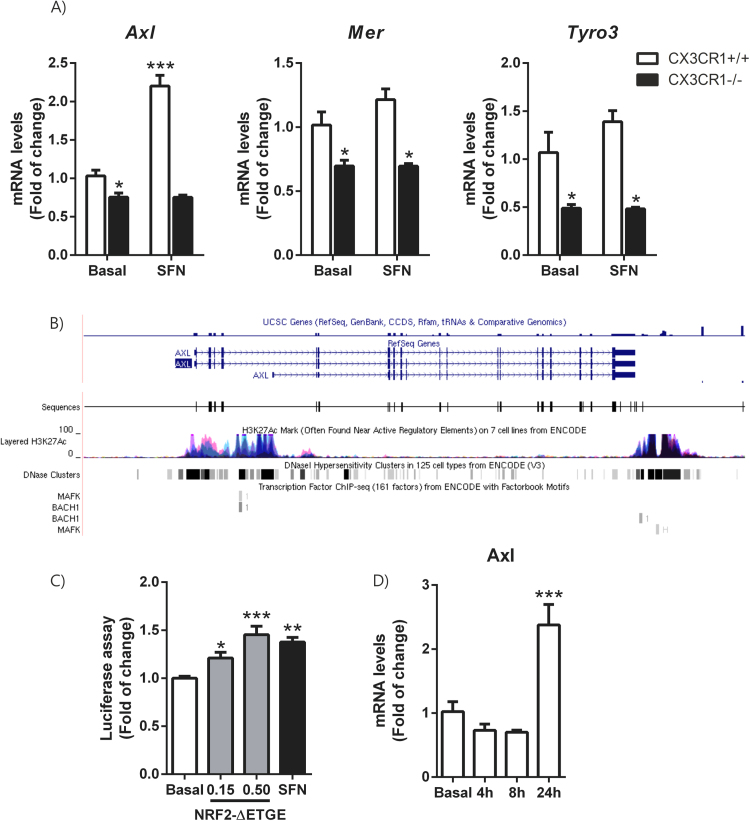
Microglia is mobilized in response to nearly any brain modification, and can act to both resolve and exacerbate central nervous system diseases [35]. It has been described that TAM receptor tyrosine kinases Mer, Axl and Tyro3 regulate these microglial functions [36]. These receptors are essential for the phagocytosis of apoptotic cells. In the immune system, they act as pleiotropic inhibitors of the innate inflammatory response to pathogens [37]. Deficiencies in TAM signalling are implicated in chronic inflammatory and autoimmune disease in humans [23], [38]. In primary culture of microglial cells from Cx3cr1+/+ and Cx3cr1-/- mice, we observed a reduction of the expression of Axl, Mer and Tyro3 mRNA levels (Fig. 2A) in Cx3cr1-deficient microglia. These results are very similar to the reduction of the same set of genes observed in primary microglia from Nrf2-/- mice [23], suggesting that CX3CR1/NRF2 axis modulates TAM receptor expression. Then, we analysed whether induction of NRF2 by SFN could restore TAM expression levels. SFN treatment (15 μM, 6 h) only induced the expression of Axl in those primary microglial cells, suggesting that Axl promoter could possess an antioxidant response element (ARE) and be an NRF2-dependent gene. In order to test this hypothesis, we first searched the Encyclopedia of DNA Elements at UCSC (ENCODE) of the human genome (Feb. 2009) to look whether TAM receptors possess putative AREs. This database contains the experimental data from chromatin immunoprecipitation (ChIP) studies of several transcription factors, like MAFK and BACH1 which are ARE binding factors, because NFE2L2 is not included in ENCODE (Fig. 2B). We found evidence of MAFK and BACH1 consensus binding sites only in Axl. Then, we developed a Python-based bioinformatics analysis script [39], to compare the human consensus ARE sites from the JASPAR database26 with putative AREs in the promoter regions of AXL. We detected one ARE (relative score over 80%) in this gene. To corroborate this finding we analysed the induction of a firefly luciferase reporter containing − 2376 to + 7 Axl promoter (full promoter of Axl) [40] by a stable mutant of NRF2, NRF2ΔETGE-V5, that lacks four residues (ETGE) essential for recognition by the E3 ligase complex Cul3/Keap1 or treatment with SFN (5 μM, 16 h). We found a dose-dependent activation of Axl reporter (Fig. 2C) by NRF2ΔETGE-V5 and at the same level as SFN. These data indicate that AXL is a NRF2-dependent gene. Additionally, CX3CL1 (100 nM) was able to induce Axl expression after 24 h of treatment (Fig. 2D).
Fig. 2.
TAM receptors expression are decreased in Cx3cr1-deficient microglia. Axl could be modulated by the transcription factor NRF2. (A) Primary cultures of microglia from Cx3cr1+/+ and Cx3cr1−/− mice were incubated with vehicle or SFN (15 μM, 6 h). Quantitative real-time PCR determination of messenger RNA levels of Axl, Mer and Tyro3 respectively, normalized by Actb (β-Actin) messenger RNA levels. Values are mean±SEM (n = 4). (B) To analyze the role of NFE2L2 in the transcriptional regulation of TAM receptors, we searched the Encyclopedia of DNA Elements at UCSC (ENCODE)25 of the human genome (Feb. 2009) for putative AREs. This database contains the experimental data from chromatin immunoprecipitation (ChIP) studies of several transcription factors. Although NFE2L2 is not included, we analysed 2 other ARE binding factors, MAFK and BACH1, for which information is available. We found evidence of MAFK or BACH1 binding in the Axl gene sequence. (C) HEK293T cells were co-transfected with NRF2ΔETGE-V5 expression vector, AXL-LUC reporter, Renilla control vector and empty vector, or treated with vehicle or SFN (5 μM for 16 h). Luciferase experiments were performed at least three times using three samples per group. (D) IMG cells were incubated in the presence of recombinant CX3CL1 (100 nM) for 4, 8 and 24 h and quantitative real-time PCR determination of messenger RNA for Axl was analysed and normalized by Actb (β-Actin) messenger RNA levels. Values are mean±SEM. Statistical analyses were performed with one-way ANOVA followed by Newman–Keuls multiple-comparison test: *p < 0.05, **p < 0.01, and ***p < 0.001, comparing the indicated groups.
3.3. CX3CR1-deficient primary microglial cells show impaired phagocytosis and migration
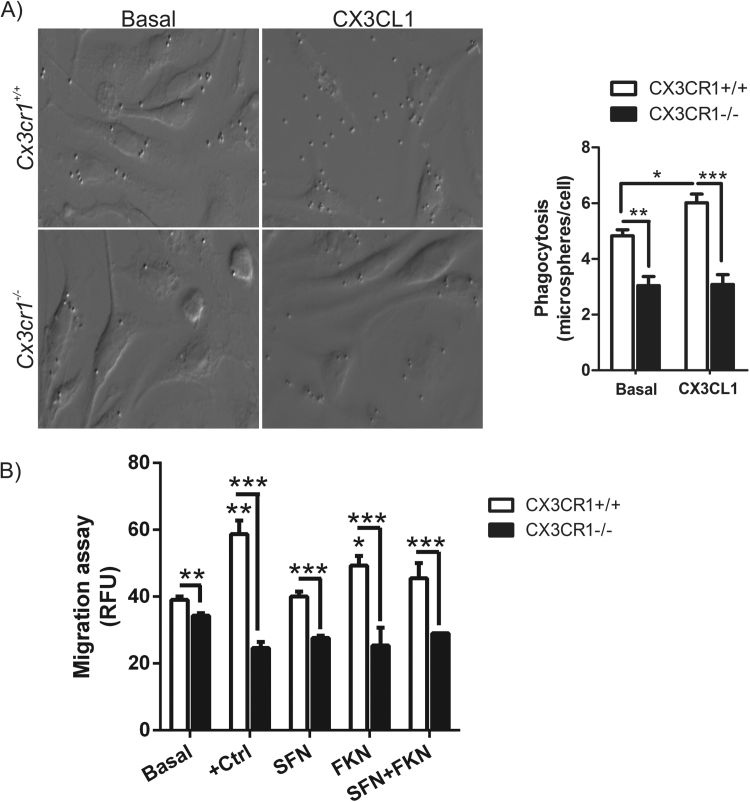
To get insight in the functional implications of CX3CR1 deficiency in microglia, we analysed the phagocytic capacity and migration of Cx3cr1+/+ and Cx3cr1-/- primary microglial murine cells at basal levels and after stimulation with CX3CL1. Lack of CX3CR1 reduces significantly the phagocytic capacity of microglial cells (Fig. 3). Besides, CX3CL1 is able to increase phagocytosis in Cx3cr1+/+ microglia but not in Cx3cr1-/- cells, indicating the relevance of CX3CR1 in one of the main function of microglia. Interestingly, these results are very similar to those obtained in microglia deficient for NRF2, where we observed that treatment with dimethyl fumarate (DMF), an NRF2 inducer, had no effect on phagocytosis [23], [28].
Fig. 3.
Impaired phagocytic response and migration of Cx3cr1-deficient microglia. (A) Microglia from Cx3cr1+/+ or Cx3cr1−/− mice were incubated with fluorescent microspheres in the absence or presence of 100 nM of CX3CL1 for 2 h. Phagocytic efficiency was calculated as a number of microspheres per cell. One-way ANOVA followed by Newman–Keuls test was used to assess differences among groups. Asterisks denote significant differences: *p < 0.05, **p < 0.01, ***p < 0.001 compared with the indicated groups. (B) Motility was determined by using CytoSelect 96-well cell migration assay from primary cultures of microglia from Cx3cr1+/+ and Cx3cr1−/− mice in accordance with the manufacturer's instructions, described in “Materials and Methods”. Positive control: 15% FBS; SFN (5 μM); CX3CL1 (100 nM) for 16 h. Values are mean±SEM (n = 3, performed two times).
CX3CL1 has been implicated in microglial migration. In its soluble form, it acts as an extracellular chemoattractant promoting cellular migration [35], but there is no evidence about the role of CX3CR1 deficiency in microglial migration. We observed that Cx3cr1-/- primary microglia showed impaired migration properties compared to Cx3cr1+/+ microglia at basal level. This effect was more evident using a control of positive migration with serum (15% of FCS) (Fig. 3B). Moreover, the presence of CX3CL1 (100 nM, 16 h) in the bottom chamber increased microglia migration of Cx3cr1+/+ cells but not of cells with the Cx3cr1-/- genotype. The addition of the NRF2 inducer SFN (5 μM, 16 h) did not show any significant effect in microglial migration in both genotypes. These data suggest that CX3CR1 has a significant role in microglial migration and can be induced by its ligand.
3.4. SFN treatment reverses astrogliosis but not microgliosis in the Cx3cr1-/- mice in the rAAV-TAUP301L mouse model
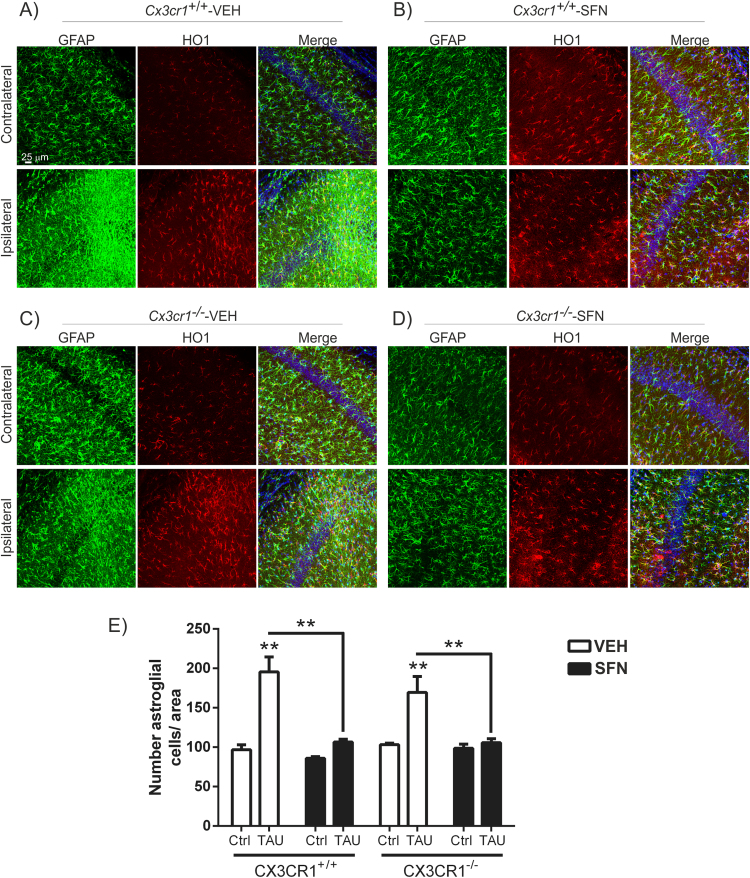
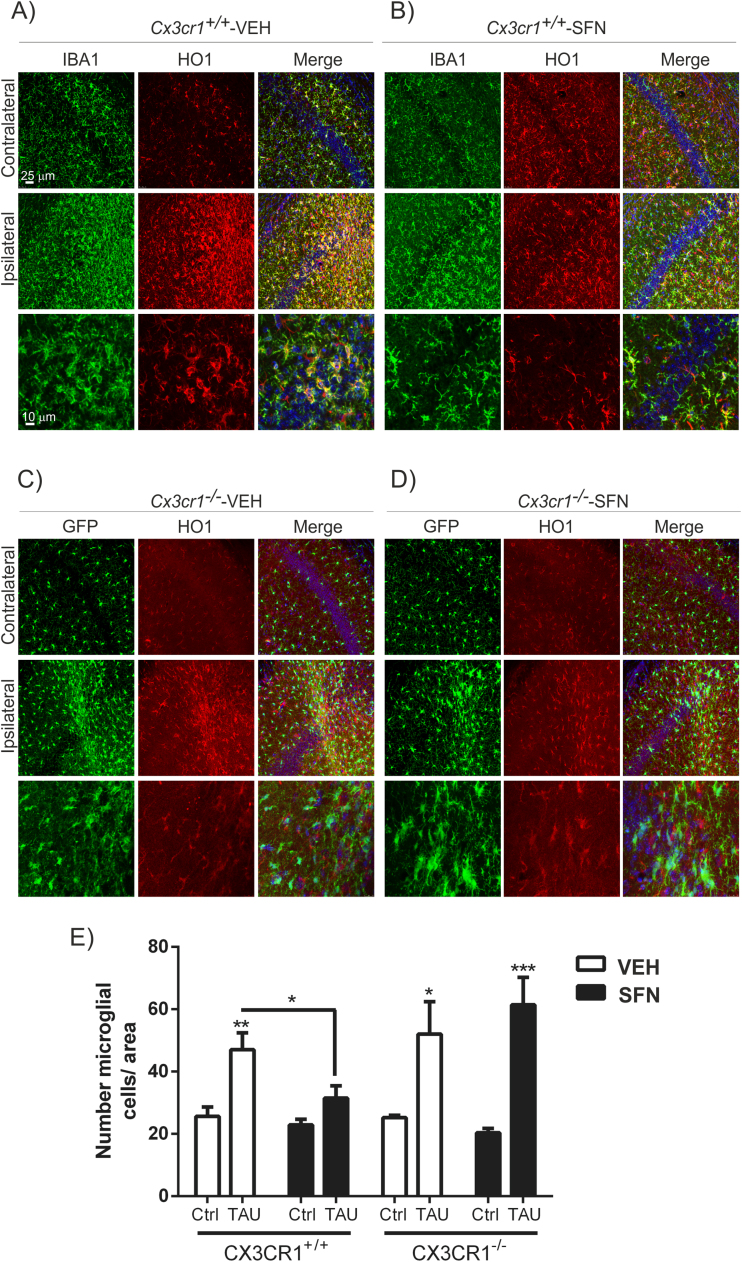
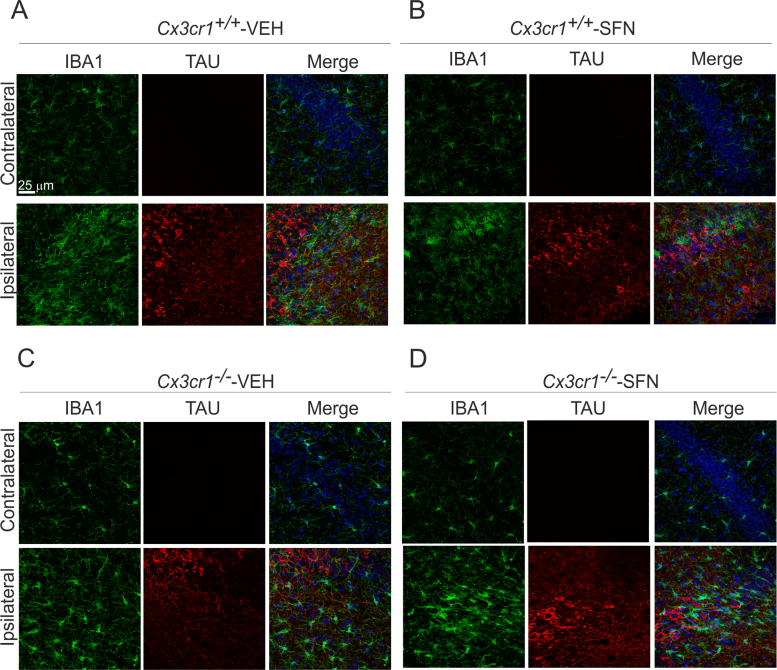
Previous work from our group showed the relevance of the CX3CR1/NRF2 axis in a tauopathy mouse model [20]. This model is based on stereotaxic delivery in hippocampus of an adeno-associated viral vector for expression of TAUP301L, under the control of the human synapsin 1 gene promoter (AAV-TAUP301L) [41], [42]. Our previous results confirmed that TAU-injured neurons express CX3CL1, and that NRF2- and CX3CR1-knockout mice not only do not express heme oxygenase 1 (HO1) in microglia but also exhibited exacerbated microgliosis and astrogliosis [20], [31], [42]. Considering that in CX3CR1-deficient mice there is a reduction of the NRF2 axis in microglial cells, we evaluated whether an inducer of NRF2 could modulate this effect with putative implications in the neuroinflammation associated to tauopathies. Moreover, it has been reported that NRF2 induction has neuroprotective effects in several neurodegenerative mouse models [28], [34], [43], [44] as well as in tauopathies [26]. Thus, the effects of SFN on neuroinflammation were examined in Cx3cr1+/+ and Cx3cr1-/- mice stereotaxically injected in the right hippocampus with AAV-TAUP301L and the left hippocampus was used as control (contralateral side), and treated daily with SFN (50 mg/kg, i.p) during three weeks. A control adeno-associated virus vector expressing green fluorescence protein did not elicit significant changes in inflammation or gliosis (data not shown). Three weeks after, we observed hTAU protein expression in the hippocampi of both genotypes, indicating that SFN did not influence AAV-TAUP301L expression (data not shown). Moreover, in this model TAUP301L-expression did not induce noticeable hippocampal neuronal cell death as measured by Nissl-staining, FluoroJade or Bielschowsky-staining [20], [26]. Regarding astrogliosis, TAUP301L toxicity associated with a very significant increase in GFAP+ astrocytes at the ipsilateral hippocampal side of Cx3cr1+/+-VEH and Cx3cr1−/−-VEH mice (Figs. 4A and 4C), which was confirmed by stereological quantification (Fig. 4E). This reactive astrogliosis was significantly reduced by the treatment with SFN both genotypes, indicating that SFN has anti-inflammatory effect related to astrocytes (Figs. 4B, 4D and 4E). Looking at the morphology, astrocytes displayed enlarged bodies and ramifications, consistent with a reactive state after TAUP301L expression in vehicle treated mice (Figs. 4A and 4C), which was reversed by SFN to a resting morphology (Figs. 4B and 4D). Concerning microglia, TAUP301L expression induced a very significant increase in IBA1 + microglia at the ipsilateral hippocampal side of Cx3cr1+/+-VEH and Cx3cr1−/−-VEH mice (Figs. 5A, 5C and Suppl. Fig. 2), which was confirmed by stereological quantification (Fig. 5E). SFN treatment reduced significantly this microgliosis in Cx3cr1+/+ but not in Cx3cr1−/− mice, reinforcing the idea of CX3CR1/NRF2-dependent anti-inflammatory effect (Figs. 5B, 5D and 5E) in Cx3cr1+/+ mice. Regarding morphology, we observed that SFN treatment preserved microglia in a quiescent state in comparison to a more phagocytic-activation state induced by TAUP301L expression in Cx3cr1+/+ mice (Figs. 5A-5B, Suppl. Fig. 2). As SFN treatment had no effect in Cx3cr1−/− mice, there was no changes in microglia morphology between Cx3cr1−/−-VEH and Cx3cr1−/−-SFN (Figs. 5C-5D, Suppl. Fig. 2), observing in both cases increased phagocytic-activation microglial state induced by TAUP301L expression.
Fig. 4.
SFN treatment attenuates TAUP301L-induced astrogliosis. Photographs show the astrocyte marker GFAP (green) and HO1 (red) as a NRF2 reporter gene, in 30 µm-thick sections of hippocampus from mice with the genotypes (A) Cx3cr1+/+-VEH, (B) Cx3cr1+/+-SFN, (C) Cx3cr1-/--VEH and (D) Cx3cr1-/--SFN. (E) Stereological quantification of the number of astrocytes in the control side and the TAUP301L expressing side of all the experimental groups. Differences among groups were assessed by two-way ANOVA followed by Bonferroni's test. Asterisks denote significant differences **p < 0.01, comparing the indicated groups.
Fig. 5.
CX3CR1 receptor is required to induce HO1 in microglia in response to TAUP301Lexpression and SFN treatment. Photographs show the microglial marker IBA1 (green) and HO1 (red) as a NRF2 reporter gene, in 30 µm-thick sections of hippocampus from mice with the genotypes (A) Cx3cr1+/+-VEH, (B) Cx3cr1+/+-SFN, (C) Cx3cr1-/--VEH and (D) Cx3cr1-/--SFN. Microglia from Cx3cr1−/− mice, stained with anti-GFP antibody (green), do not express HO1 (red) in response to TAUP301L. (E) Stereological quantification of the number of microglial cells in the control side and the TAUP301L expressing side of all experimental groups. Two-way ANOVA followed by Bonferroni post-test was used to assess significant differences among groups. Asterisks denote significant differences *p < 0.05, **p < 0.01 and ***p < 0.001, comparing the indicated groups.
It has been described that expression of TAU and HO1 may be regulated by oxidative stress in a coordinated manner and play a pivotal role in the cytoprotection of neuronal cells [45]. On the other hand, NRF2 activation induced HO1 overexpression in astrocytes and microglia [20], [23], [26], [27] as an anti-inflammatory mechanism. Our results shown that TAUP301L expression induced HO1 expression in astrocytes (Fig. 4) as well as in microglia (Figs. 5A and 5B) in Cx3cr1+/+-VEH mice. In Cx3cr1−/−-VEH mice there is no microglial HO1 expression (Figs. 5C and 5D), results that support previous experiments obtained with these mice in the tauopathy model [20]. SFN treatment induced HO1 expression in both microglia and astrocytes of the contralateral hippocampus in Cx3cr1+/+-SFN mice (Figs. 4B and 5B). On the other hand, SFN treatment did not increase HO1 expression in the ipsilateral hippocampus where TAUP301L was overexpressed. As to CX3CR1-deficient mice, SFN increased astrocytic HO1 expression in both, contralateral and ipsilateral sides (Figs. 4C and 4D). Interestingly, SFN was not able to induce HO1 expression in microglia of Cx3cr1−/−-SFN mice (Fig. 5D), indicating that SFN is not activating the signalling pathway of NRF2 in microglia deficient in CX3CR1. These results corroborate those obtained in vitro (Fig. 1). Altogether, these data suggest that SFN treatment cannot induce NRF2/HO1 signalling in Cx3cr1-/- microglia and therefore it cannot modulate the microgliosis induced by TAUP301L overexpression.
3.5. SFN treatment slightly protects from hippocampal neuronal damage in the AAV-TAUP301L mouse model
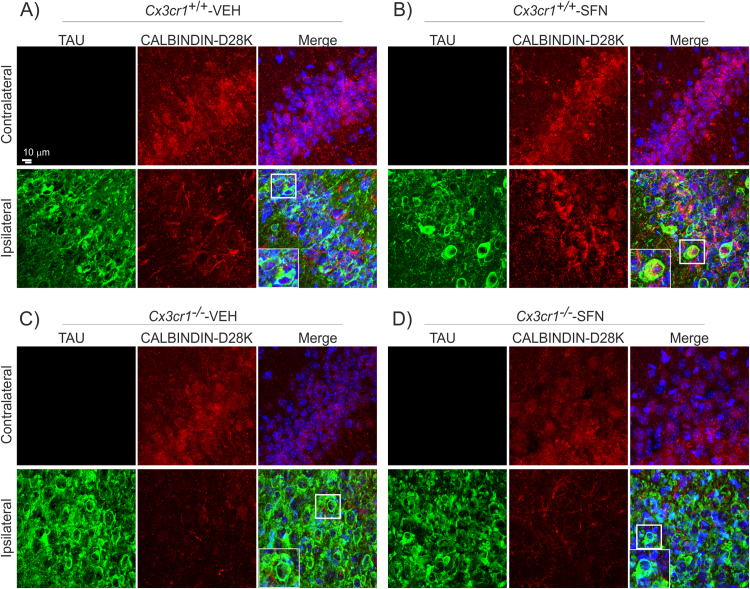
As we have previously described in this model of tauopathy, twenty-one days of TAUP301L-expression did not induce noticeable hippocampal neuronal cell death [26], but we demonstrated that TAUP301L-expressing neurons do not express calbindin-D28K, a major calcium-binding and buffering protein, that has a critical role in preventing neuronal death as well as maintaining calcium homeostasis and synaptic plasticity. Here, immunofluorescence analysis with anti- calbindin-D28K antibody corroborated that TAUP301L-expressing neurons do not express calbindin-D28K, in Cx3cr1+/+ mice but also in Cx3cr1-/- mice (Figs. 6A and 6C), indicating that TAUP301L- participates in dysregulation of synaptic plasticity. Very relevant, SFN treatment had a slight protective effect against calbindin-D28K loss in TAUP301L-expressing neurons of Cx3cr1+/+ mice (Fig. 6B), but not in Cx3cr1-/- mice (Fig. 6D). Taken together, this data indicates that the treatment with SFN is able to have a mild neuroprotective effect for which the modulation of the microgliosis in relation with CX3CR1 has a key role.
Fig. 6.
TAUP301Lexpression induces hippocampus degeneration which is slightly prevented by SFN in Cx3cr1+/+mouse. Immunohistochemical staining with anti-human-TAU antibody (green) and CALBINDIN D-28K (red) in 30 µm-thick sections of hippocampus of (A) Cx3cr1+/+-VEH, (B) Cx3cr1+/+-SFN, (C) Cx3cr1-/--VEH and (D) Cx3cr1-/--SFN. White square indicates TAU+/calbindin--neurons in Cx3cr1+/+ animals treated with vehicle (VEH), while in Cx3cr1+/+-SFN animals, square indicates TAU+/calbindin+-neurons. In Cx3cr1-/- animals, white squares indicate TAU+/calbindin--neurons.
4. Discussion
Neurodegenerative diseases such as Alzheimer's disease and other tauopathies are among the most burdensome health concerns in societies with pronounced aging tendencies. Although neurons are essential targets in neurodegeneration, there are other environmental factors that contribute to neurodegeneration such as neuroinflammation, which involves other cell types of the brain. In this context, the study of non-cell-autonomous neurodegenerative process would open new windows to understand how neurodegeneration could be promoted by local neuroinflammation. Microglia play an essential role in development, homeostasis, synapse modulation, migration, phagocytosis and neurogenesis, indicating that impairment in the crosstalk between neurons and microglia are determinant in neurodegenerative processes [46]. Fractalkine (CX3CL1) and its cognate receptor (CX3CR1) are key factors in this crosstalk, and are implicated in many processes like synaptic integration of adult-born hippocampal granule neurons [47], synaptic refinement and transmission in the developing hippocampus [48] as well as the progression of neurodegenerative disorders [15], [20], [49]. Previously, our laboratory described that CX3CL1 activates the transcription factor NRF2 and its target genes including HO1 [20]. Related to neurodegenerative diseases, in a mouse model of tauopathy, we confirmed that TAU-injured neurons express CX3CL1. Moreover, NRF2- and CX3CR1-knockout mice did not express HO1 in microglia and exhibited increased microgliosis and astrogliosis in response to neuronal TAUP301L expression, demonstrating a crucial role of the CX3CL1/NRF2/HO1 pathway in attenuating the pro-inflammatory phenotype. Therefore, we went further to get insight in the CX3CR1/NRF2 axis and its implications in tauopathies. Our results show for the first time that CX3CR1-deficient primary microglial cells present impaired levels of the transcription factor NRF2, affecting it signalling and that SFN treatment could not circumvent this effect (Fig. 1). As a consequence, microglia deficient in CX3CR1 behave like microglia deficient in NRF2. As microglia deficient in NRF2 [23], [28], the absence of CX3CR1 triggers a significant decrease in TAM receptors (Fig. 2A). These receptors are essential in the regulation of immune responses and phagocytosis [50]. It has been described that TAM-deficient microglia display reduced processes motility and delayed convergence to sites of injury [36]. Our results are in agreement with these findings because in the absence of CX3CR1 we observed a decrease in the expression of TAM receptors as well as a decrease in phagocytic capacity and migration of microglia (Fig. 2A and Fig. 3). These observations are supported by the results from Dr. Jesus Avila's laboratory, which demonstrated that the CX3CL1/CX3CR1 axis played a key role in the phagocytosis of TAU by microglia in vitro and in vivo and that this was also affected as AD progressed. Moreover, they found a novel mechanism of TAU internalization by microglia through direct binding to CX3CR1 [51]. Taken together, these data suggest that CX3CR1 could be a critical key factor in microglial function and tauopathies progression.
The involvement of the CX3CL1/CX3CR1 axis in the microglial phenotype has always been a very controversial issue. Consequently, CX3CL1 appears to have anti-inflammatory/neuroprotective activity in some settings, whereas it contributes to neurotoxicity in others [16], [52]. Our results show that in primary microglia obtained from CX3CR1-deficient mice, there is a significant decrease in the anti-inflammatory phenotype (M2), evidenced by a decrease in the expression of the anti-inflammatory NRF2 transcription factor (Fig. 1). Furthermore, CX3CR1-deficient primary microglia also showed impaired phagocytosis and migration (Fig. 3), functions that are associated with an M2 phenotype. These data indicate that deficiency in CX3CR1 leads to impaired M2 phenotype. The consequence of the loss of this M2 phenotype can be clearly seen in the tauopathy model, in vivo. Overexpression of TAUP301L induces neuroinflammation, increasing astrogliosis (Fig. 4) and microgliosis (Fig. 5). Interestingly, Cx3cr1-/- mice showed overactivation of microglial cells, what is in agreement with the in vitro data. Faced with the damage caused by overexpression of TAUP301L, the microglia deficient in CX3CR1 is not able to activate the signalling pathway of NRF2, so we showed no expression of HO1 in the microglia (Fig. 5C) and they are not able to migrate to the focus of the lesion and phagocytose neurons damaged by the overexpression of TAUP301L. This could lead to enhanced the neurodegenerative process, as has been demonstrated in the same tauopathy mouse models [42].
Our results point out that in CX3CR1 deficient microglia, there is about 50% downregulation of Nfe2l2 transcription (NRF2 gene). These data indicate that the CX3CR1 axis modulates transcriptionally to NRF2. In the case of the treatment with SFN, a modulation of the stability of the protein occurs, by destabilization of the interaction of NRF2 with the chaperone KEAP1. Under physiological conditions, NRF2 is regulated by cytoplasmic KEAP1, an adopter protein for CULLIN3-based ubiquitin E3 ligase that continuously ubiquitinates NRF2 for proteasomal degradation. Upon exposure to electrophiles (in our case SFN treatment) or oxidative stress, KEAP1 is inactivated due to electrophile binding. The inactivation leads to dislodging of NRF2 from KEAP1, and allows NRF2 to escape proteasomal degradation [53], [54]. Moreover, SFN may also mediate the phosphorylation of NRF2 by activating various kinases (MAP, PKC and AKT), which alter nuclear and cytoplasmic trafficking and NRF2 integrity and stability [55], [56], [57], [58]. So it seems that CX3CR1 and SFN exert very different effects on the NRF2 pathway.
The transcriptional modulation of NRF2 by the CX3CR1 pathway may also explain why NRF2-deficient and CX3CR1-deficient mice have a phenotypic similarity. We still do not know how modulation of the NRF2 pathway is produced by CX3CR1, but we cannot rule out any possibility. Unpublished data from the group indicate the possible involvement of the transcription factor NF-kB, which is induced by CX3CL1/CX3CR1 signalling. NRF2 has a kB site in their promoter region [59] and therefore could also be modulated by this other way in a transcriptional level.
The fact that microglia deficient in CX3CR1 behave like microglia deficient in NRF2 has its correlation in murine models, which could explain the phenotypic similarity between mice deficient for CX3CR1 and NRF2. Related to tauopathies, it has been shown that CX3CR1 deficiency enhanced TAU pathology and exacerbated degeneration [42], [60], [61] as observed in NRF2 deficient mice [20], [62], [63]. Moreover, lipopolysaccharide (LPS) sensitizes mice microglial activation in CX3CR1-deficient mice [64] as well as in NRF2-deficient mice [65]. In general, deficiency in those factors showed an exacerbated inflammatory condition after pro-inflammatory stimuli [20], [42] and cognitive decline [52], [66], [67] in the central nervous system (CNS). This fact is of high relevance when designing new therapeutic strategies that modulate neuroinflammation/neurodegeneration. Regarding to CX3CR1 receptor, 2 polymorphisms named V249I and T280M have been described. These polymorphisms induce a low availability of receptors on the cell surface or have reduced receptor affinity for CX3CL1, respectively. It has been shown that CX3CR1 is a gene involved in the modification of survival and progression in amyotrophic lateral sclerosis [14], and that there is an association of the variant CX3CR1-V249I with a progression of neurofibrillary pathology in late-onset Alzheimer's disease [13]. These studies indicate that polymorphisms in CX3CR1 could modulate neurodegenerative progression (Suppl. Fig. 3). As in the case of CX3CR1, it has been observed that haplotypes for NRF2 gene (NFE2L2) only influence the progression of the Alzheimer's disease patients, but do not increase the risk of suffering the disorder [68], suggesting the relevance of both genes in the evolution of the disease. In addition, our data indicate that the modulation of microgliosis by treatment with SFN is key in neuroprotection processes. Microglia is key in neuroinflammatory processes, where it has been described that inflammatory factors produced by microglia and astrocytes can damage local tissue and, together with released damage-associated molecular patterns (DAMPs), can further increase inflammation and glial activation, leading to a vicious inflammatory cycle [69]. Therefore, our study pioneers the description and analysis of the CX3CR1/NRF2 axis in tauopathies and its implications in the treatment of these neurodegenerative disorders.
Regarding TAM receptors, we found for the first time that CX3CR1 deficiency decreased TAM mRNA expression levels, and that only Axl could be modulated by the activation of NRF2. Interestingly, these results are comparable to those obtained in primary microglia of NRF2-deficient mice [20], [23], indicating a crosstalk between CX3CR1/NRF2/TAM receptors and also its implication in phagocytosis. It has been shown that Gas6-Axl signalling plays an important role in maintaining axonal integrity in addition to regulating and reducing the CNS inflammation that cannot be compensated for by ProS1/Tyro3/MerTK signalling [70]. The function of activated Axl in normal tissues includes the efficient clearance of apoptotic material and the dampening of TLR-dependent inflammatory responses and natural killer cell activity [71]. Hence, modulation of Axl by NRF2-inducers could become a new potential target for therapeutic intervention in CNS diseases.
In recent years, NRF2 has shown promise as a novel therapeutic target in neurodegenerative diseases, due to the oxidative and inflammatory stress components associated with these disorders [72]. Among the NRF2 inducers, sulforaphane (SFN) has demonstrated neuroprotective effects in several in vitro and in vivo studies [73]. For example, it has been suggested that SFN can inhibit Aβ oligomer production in AD [74]. Furthermore, SFN was able to reverse iron-induced decrease in mitochondrial fission protein, DNM1L, as well as synaptophysin levels in the hippocampus, leading to a recovery of recognition memory impairment induced by iron [75]. Animals treated with SFN displayed a reduction in the number of microglial cells in the hippocampus and an attenuated production of inflammation markers in response to LPS [65]. Therefore, we examined the effects of SFN on neuroinflammation in Cx3cr1+/+ and Cx3cr1-/- mice stereotaxically injected in the right hippocampus with AAV-TAUP301L and treated daily with SFN (50 mg/kg, i.p) during three weeks. Whereas SFN treatment was able to reverse astrogliosis induced by TAUP301L expression in both genotypes (Fig. 4), we did not see any improvement in the Cx3cr1-/- mice at the microglia level (Fig. 5). As in vitro, SFN could not induce NRF2 signalling in microglia, and none HO1 expression was observed (Fig. 5D). Our data indicate that deficiency in CX3CR1 in microglia develops an NRF2 signalling impairment (Fig. 1 and Fig. 5) associated with decreased phagocytosis and migration ability (Fig. 3). The transcriptional reduction of NRF2 in the CX3CR1-deficient microglia cannot be restored by treatment with SFN (Fig. 1), so that SFN is not able to reverse the microgliosis induced by overexpression of TAUP301L (Fig. 5). These results showed very similar effects as in NRF2-deficient mice [26]. Altogether, these data reinforce the idea that one must take into account the relevance of polymorphisms of CX3CR1 and NRF2 when using NRF2 inducers as a therapeutic strategy, for example in AD.
Interestingly, it seems that NRF2 has different roles in different cell types, depending on the neurodegenerative process. Indeed, astroglia and microglia are also relevant in the response of the whole brain parenchyma. It has been reported that depending on the neurodegenerative process NRF2 can be induced in neurons, astrocytes and / or microglia [76]. For example, in Parkinson's disease (PD) it has been reported that analysis of substantia nigra (SN) neurons of post-mortem PD brain reveals robust accumulation of nuclear NRF2 as compared to normal brain, and aberrant localisation of KEAP1 to Lewy bodies. The increase in nuclear NRF2 evident in the SN of human post-mortem PD brain (as compared to normal brain) appears to be restricted to neurons. In contrast, NQO1, HO1 and peroxiredoxin 6 are strongly expressed in astrocytes and or microglia in the SN of PD brain, with more infrequent expression in neurons [23], [28]. In our experience, related to tauopathies, TAU expression in neurons do not induced NRF2 signalling in HT22 neuronal cell cultures, in vivo rAAV-TAUP301L mouse model and human AD samples [20]. In contrast, the overexpression of TAU in neurons induces the activation of the NRF2 pathway in astrocytes and microglia, in vivo rAAV-TAUP301L mouse model and human AD samples [20] and as we described in the present manuscript. Therefore, we believe that the role of NRF2 in relation to tauopathies is essential at the level of gliosis, although its possible neuroprotective involvement should not be ruled out. This is supported by previous data of our group, where the treatment with DMF modulates TAU phosphorylation, neuronal impairment measured by calbindin-D28K and BDNF expression, and inflammatory processes involved in astrogliosis, microgliosis and pro-inflammatory cytokines production [26].
5. Conclusions
We demonstrated that CX3CR1/NRF2 plays multifaceted roles in microglia-mediated functions. These novel findings suggest that the activation of the CX3CR1-NRF2 axis is essential for the modulation of microglial activation associated with tauopathies. In addition, associated polymorphisms of CX3CR1 and NRF2 (Suppl. Fig. 3, [77]) must be taken into account in the design of pharmacological strategies aimed to the treatment of these diseases.
Acknowledgements
The authors are grateful to Marta Pajares-Cabetas for her help with the bioinformatics analysis and to David Nonis for his helpful comments and constructive feedback.
Acknowledgments
Declarations
Ethics approval and consent to participate
All experiments were performed by certified researchers according to regional, national, and European regulations concerning animal welfare and animal experimentation, and were authorized by the Ethics Committee for Research of the Universidad Autónoma de Madrid and the Comunidad Autónoma de Madrid, Spain, with Ref PROEX 279/14, following institutional, Spanish and European guidelines (Boletín Oficial del Estado (BOE) of 18 March 1988 and 86/609/EEC, 2003/65/EC European Council Directives).
Consent for publication
Not applicable.
Availability of supporting data
Not applicable.
Competing interests
All authors declare no competing interests.
Funding
This work was supported by a Spanish Ministry of Economy and Competitiveness (Grants refs. SAF2016-76520-R).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2019.101118
Contributor Information
Sara Castro-Sánchez, Email: scastro@iib.uam.es.
Ángel J. García-Yagüe, Email: ajgarcia@iib.uam.es.
Sebastian Kügler, Email: sebastian.kuegler@med.uni-goettingen.de.
Isabel Lastres-Becker, Email: ilbecker@iib.uam.es.
Appendix A. Supplementary material
Supplementary material
Fig. S1.
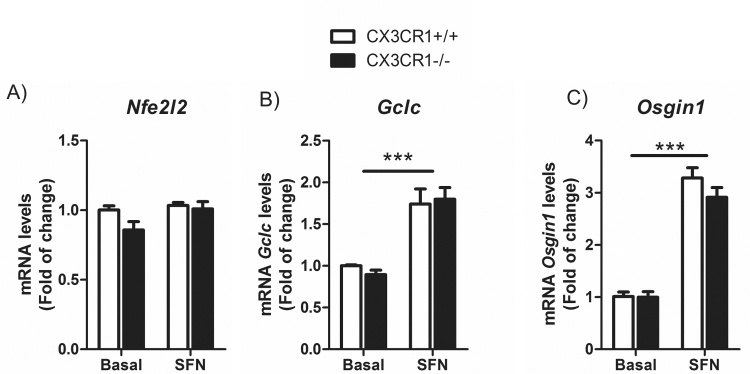
Astrocytes from Cx3cr1-/-mice do not shown any alterations in the transcription factor NRF2 signalling. Primary cultures of astrocytes from Cx3cr1+/+ and Cx3cr1−/− mice were incubated with SFN (15 μM, 6 h). Quantitative real-time PCR determination of messenger RNA levels of NRF2-regulated genes coding Nfe2l2, Nqo1, Gclc and Osgin1, respectively, normalized by Actb (β-Actin) messenger RNA levels. Two-way ANOVA followed by Bonferroni post-test was used to assess significant differences among groups. Asterisks denote significant differences ***p < 0.001 respect to the basal Cx3cr1+/+ group.
Fig. S2.
TAUP301 Loverexpression induced microglial activation of hippocampus from Cx3cr1+/+and Cx3r1-/-mice. Immunohistochemical staining with anti-human-TAU antibody (red) and IBA1 (green) in 30 µm-thick sections of hippocampus of (A) Cx3cr1+/+-VEH, (B) Cx3cr1+/+-SFN, (C) Cx3cr1-/--VEH and (D) Cx3cr1-/--SFN.
Fig. S3.
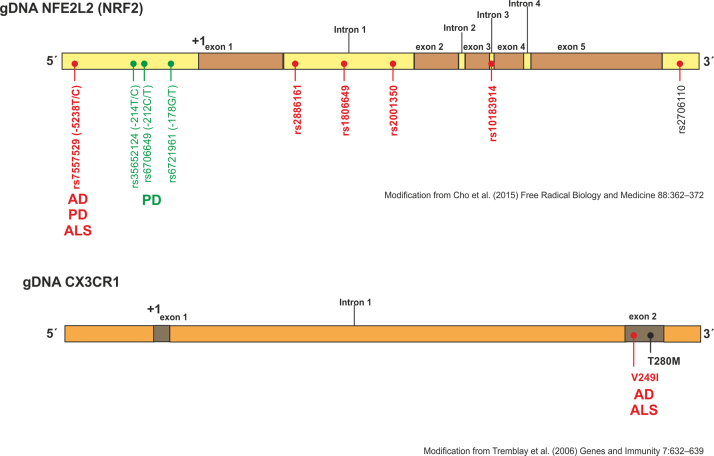
Polymorphisms in NRF2 and CX3CR1 genes associated with neurodegenerative disorders. Scheme of human NRF2 and CX3CR1 genes (modified from (68) and (77) respectively). In this figure, it is depicted the risk of genetic variations in the promoter, exon and introns of the Nfe2l2 and Cx3cr1 genes (red dots denote haplotypes that confer increased susceptibility in Alzheimer’s disease (AD), Parkinson’s disease (PD) and Amyloid lateral sclerosis (ALS) whereas green dots show haplotypes that are protective in PD).
References
- 1.Williams D.R. Tauopathies: classification and clinical update on neurodegenerative diseases associated with microtubule-associated protein tau. Intern. Med. J. 2006;36(10):652–660. doi: 10.1111/j.1445-5994.2006.01153.x. [DOI] [PubMed] [Google Scholar]
- 2.Ransohoff R.M. How neuroinflammation contributes to neurodegeneration. Science. 2016;353(6301):777–783. doi: 10.1126/science.aag2590. [DOI] [PubMed] [Google Scholar]
- 3.Richards R.I., Robertson S.A., Kastner D.L. Neurodegenerative diseases have genetic hallmarks of autoinflammatory disease. Hum. Mol. Genet. 2018 doi: 10.1093/hmg/ddy139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pascual G., Wadia J.S., Zhu X., Keogh E., Kukrer B., van Ameijde J. Immunological memory to hyperphosphorylated tau in asymptomatic individuals. Acta Neuropathol. 2017;133(5):767–783. doi: 10.1007/s00401-017-1705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wendeln A.C., Degenhardt K., Kaurani L., Gertig M., Ulas T., Jain G. Innate immune memory in the brain shapes neurological disease hallmarks. Nature. 2018;556(7701):332–338. doi: 10.1038/s41586-018-0023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laurent C., Buée L., Blum D. Tau and neuroinflammation: What impact for Alzheimer's disease and Tauopathies? Biomed. J. 2018;41(1):21–33. doi: 10.1016/j.bj.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J.C., Carrasquillo M.M. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 2011;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert J.C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 9.Sims R., van der Lee S.J., Naj A.C., Bellenguez C., Badarinarayan N., Jakobsdottir J. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer's disease. Nat. Genet. 2017;49(9):1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J.T., Zhang Y. TREM2 regulates innate immunity in Alzheimer's disease. J. Neuroinflamm. 2018;15(1):107. doi: 10.1186/s12974-018-1148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wes P.D., Sayed F.A., Bard F., Gan L. Targeting microglia for the treatment of Alzheimer's disease. Glia. 2016;64(10):1710–1732. doi: 10.1002/glia.22988. [DOI] [PubMed] [Google Scholar]
- 12.Villegas-Llerena C., Phillips A., Garcia-Reitboeck P., Hardy J., Pocock J.M. Microglial genes regulating neuroinflammation in the progression of Alzheimer's disease. Curr. Opin. Neurobiol. 2016;36:74–81. doi: 10.1016/j.conb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Lopez A., Gelpi E., Lopategui D.M., Vidal-Taboada J.M. Association of the CX3CR1-V249I variant with neurofibrillary pathology progression in late-onset alzheimer's disease. Mol. Neurobiol. 2017 doi: 10.1007/s12035-017-0489-3. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Lopez A., Gamez J., Syriani E., Morales M., Salvado M., Rodriguez M.J. CX3CR1 is a modifying gene of survival and progression in amyotrophic lateral sclerosis. PLoS One. 2014;9(5):e96528. doi: 10.1371/journal.pone.0096528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castro-Sanchez S., Garcia-Yague A.J., Lopez-Royo T., Casarejos M., Lanciego J.L., Lastres-Becker I. Cx3cr1-deficiency exacerbates alpha-synuclein-A53T induced neuroinflammation and neurodegeneration in a mouse model of Parkinson's disease. Glia. 2018 doi: 10.1002/glia.23338. [DOI] [PubMed] [Google Scholar]
- 16.Lauro C., Catalano M., Trettel F., Limatola C. Fractalkine in the nervous system: neuroprotective or neurotoxic molecule? Ann. N.Y. Acad. Sci. 2015;1351:141–148. doi: 10.1111/nyas.12805. [DOI] [PubMed] [Google Scholar]
- 17.Mecca C., Giambanco I., Donato R., Arcuri C. Microglia and aging: the role of the TREM2–DAP12 and CX3CL1-CX3CR1 axes. Int. J. Mol. Sci. 2018;19(1):318. doi: 10.3390/ijms19010318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachstetter A.D., Morganti J.M., Jernberg J., Schlunk A., Mitchell S.H., Brewster K.W. Fractalkine and CX 3 CR1 regulate hippocampal neurogenesis in adult and aged rats. Neurobiol. Aging. 2011;32(11):2030–2044. doi: 10.1016/j.neurobiolaging.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Miranda A.S., Zhang C.-J., Katsumoto A., Teixeira A.L. Hippocampal adult neurogenesis: does the immune system matter? J. Neurol. Sci. 2017;372:482–495. doi: 10.1016/j.jns.2016.10.052. [DOI] [PubMed] [Google Scholar]
- 20.Lastres-Becker I., Innamorato N.G., Jaworski T., Rabano A., Kugler S., Van Leuven F. Fractalkine activates NRF2/NFE2L2 and heme oxygenase 1 to restrain tauopathy-induced microgliosis. Brain: J. Neurol. 2014;137(Pt 1):78–91. doi: 10.1093/brain/awt323. [DOI] [PubMed] [Google Scholar]
- 21.Dinkova-Kostova A.T., Kostov R.V., Kazantsev A.G. The role of Nrf2 signaling in counteracting neurodegenerative diseases. FEBS J. 2018 doi: 10.1111/febs.14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuadrado A., Martin-Moldes Z., Ye J., Lastres-Becker I. Transcription factors NRF2 and NF-kappaB are coordinated effectors of the Rho family, GTP-binding protein RAC1 during inflammation. J. Biol. Chem. 2014;289(22):15244–15258. doi: 10.1074/jbc.M113.540633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lastres-Becker I., Ulusoy A., Innamorato N.G., Sahin G., Rabano A., Kirik D. alpha-Synuclein expression and Nrf2 deficiency cooperate to aggravate protein aggregation, neuronal death and inflammation in early-stage Parkinson's disease. Hum. Mol. Genet. 2012;21(14):3173–3192. doi: 10.1093/hmg/dds143. [DOI] [PubMed] [Google Scholar]
- 24.Pajares M., Jimenez-Moreno N., Garcia-Yague A.J., Escoll M., de Ceballos M.L., Van Leuven F. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12(10):1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burnside S.W., Hardingham G.E. Transcriptional regulators of redox balance and other homeostatic processes with the potential to alter neurodegenerative disease trajectory. Biochem. Soc. Trans. 2017;45(6):1295–1303. doi: 10.1042/BST20170013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuadrado A., Kugler S., Lastres-Becker I. Pharmacological targeting of GSK-3 and NRF2 provides neuroprotection in a preclinical model of tauopathy. Redox Biol. 2018;14:522–534. doi: 10.1016/j.redox.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lastres-Becker I. Role of the transcription factor Nrf2 in Parkinson's disease: new insights. J. Alzheimers Dis. Park. 2017;7(4):9. [Google Scholar]
- 28.Lastres-Becker I., Garcia-Yague A.J., Scannevin R.H., Casarejos M.J., Kugler S., Rabano A. Repurposing the NRF2 activator dimethyl fumarate as therapy against synucleinopathy in parkinson's disease. Antioxid. Redox Signal. 2016;25(2):61–77. doi: 10.1089/ars.2015.6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inui M., Ishida Y., Kimura A., Kuninaka Y., Mukaida N., Kondo T. Protective roles of CX3CR1-mediated signals in toxin A-induced enteritis through the induction of heme oxygenase-1 expression. J. Immunol. 2011;186(1):423–431. doi: 10.4049/jimmunol.1000043. [DOI] [PubMed] [Google Scholar]
- 30.Jung S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell Biol. 2000;20(11):4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaworski T., Dewachter I., Lechat B., Croes S., Termont A., Demedts D. AAV-tau mediates pyramidal neurodegeneration by cell-cycle re-entry without neurofibrillary tangle formation in wild-type mice. PLoS One. 2009;4(10):e7280. doi: 10.1371/journal.pone.0007280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marelli G., Erreni M., Anselmo A., Taverniti V., Guglielmetti S., Mantovani A. Heme-oxygenase-1 production by intestinal CX3CR1(+) macrophages helps to resolve inflammation and prevents carcinogenesis. Cancer Res. 2017;77(16):4472–4485. doi: 10.1158/0008-5472.CAN-16-2501. [DOI] [PubMed] [Google Scholar]
- 34.Jazwa A., Rojo A.I., Innamorato N.G., Hesse M., Fernandez-Ruiz J., Cuadrado A. Pharmacological targeting of the transcription factor Nrf2 at the basal ganglia provides disease modifying therapy for experimental parkinsonism. Antioxid. Redox Signal. 2011;14(12):2347–2360. doi: 10.1089/ars.2010.3731. [DOI] [PubMed] [Google Scholar]
- 35.Fourgeaud L., Través P.G., Tufail Y., Leal-Bailey H., Lew E.D., Burrola P.G. TAM receptors regulate multiple features of microglial physiology. Nature. 2016;532:240. doi: 10.1038/nature17630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fourgeaud L., Traves P.G., Tufail Y., Leal-Bailey H., Lew E.D., Burrola P.G. TAM receptors regulate multiple features of microglial physiology. Nature. 2016;532(7598):240–244. doi: 10.1038/nature17630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemke G. Biology of the TAM receptors. Cold Spring Harb. Perspect. Biol. 2013;5(11):a009076. doi: 10.1101/cshperspect.a009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Q., Lemke G. Homeostatic regulation of the immune system by receptor tyrosine kinases of the Tyro 3 family. Science. 2001;293(5528):306–311. doi: 10.1126/science.1061663. [DOI] [PubMed] [Google Scholar]
- 39.Pajares M., Jiménez-Moreno N., García-Yagüe Á.J., Escoll M., de Ceballos M.L., Van Leuven F. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12(10):1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mudduluru G., Allgayer H. The human receptor tyrosine kinase Axl gene--promoter characterization and regulation of constitutive expression by Sp1, Sp3 and CpG methylation. Biosci. Rep. 2008;28(3):161–176. doi: 10.1042/BSR20080046. [DOI] [PubMed] [Google Scholar]
- 41.Jaworski T., Dewachter I., Lechat B., Croes S., Termont A., Demedts D. AAV-Tau mediates pyramidal neurodegeneration by cell-cycle re-entry without neurofibrillary tangle formation in wild-type mice. PLoS One. 2009;4(10) doi: 10.1371/journal.pone.0007280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaworski T., Lechat B., Demedts D., Gielis L., Devijver H., Borghgraef P. Dendritic degeneration, neurovascular defects, and inflammation precede neuronal loss in a mouse model for tau-mediated neurodegeneration. Am. J. Pathol. 2011;179(4):2001–2015. doi: 10.1016/j.ajpath.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrillo S., Piermarini E., Pastore A., Vasco G., Schirinzi T., Carrozzo R. Nrf2-inducers counteract neurodegeneration in frataxin-silenced motor neurons: disclosing new therapeutic targets for friedreich's ataxia. Int. J. Mol. Sci. 2017;18(10) doi: 10.3390/ijms18102173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips J.T., Fox R.J. BG-12 in multiple sclerosis. Semin. Neurol. 2013;33(1):56–65. doi: 10.1055/s-0033-1343796. [DOI] [PubMed] [Google Scholar]
- 45.Takeda A., Perry G., Abraham N.G., Dwyer B.E., Kutty R.K., Laitinen J.T. Overexpression of heme oxygenase in neuronal cells, the possible interaction with Tau. J. Biol. Chem. 2000;275(8):5395–5399. doi: 10.1074/jbc.275.8.5395. [DOI] [PubMed] [Google Scholar]
- 46.Lannes N., Eppler E., Etemad S., Yotovski P., Filgueira L. Microglia at center stage: a comprehensive review about the versatile and unique residential macrophages of the central nervous system. Oncotarget. 2017;8(69):114393–114413. doi: 10.18632/oncotarget.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolós M., Perea J.R., Terreros-Roncal J., Pallas-Bazarra N., Jurado-Arjona J., Ávila J. Absence of microglial CX3CR1 impairs the synaptic integration of adult-born hippocampal granule neurons. Brain Behav. Immun. 2018;68:76–89. doi: 10.1016/j.bbi.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 48.Paolicelli R.C., Bolasco G., Pagani F., Maggi L., Scianni M., Panzanelli P. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333(6048):1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 49.Hickman S., Izzy S., Sen P., Morsett L., El Khoury J. Microglia in neurodegeneration. Nat. Neurosci. 2018;21(10):1359–1369. doi: 10.1038/s41593-018-0242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ji R., Meng L., Li Q., Lu Q. TAM receptor deficiency affects adult hippocampal neurogenesis. Metab. Brain Dis. 2015;30(3):633–644. doi: 10.1007/s11011-014-9636-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolos M., Llorens-Martin M., Perea J.R., Jurado-Arjona J., Rabano A., Hernandez F. Absence of CX3CR1 impairs the internalization of Tau by microglia. Mol. Neurodegener. 2017;12(1):59. doi: 10.1186/s13024-017-0200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheridan G.K., Murphy K.J. Neuron-glia crosstalk in health and disease: fractalkine and CX3CR1 take centre stage. Open Biol. 2013;3(12):130181. doi: 10.1098/rsob.130181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kubo E., Chhunchha B., Singh P., Sasaki H., Singh D.P. Sulforaphane reactivates cellular antioxidant defense by inducing Nrf2/ARE/Prdx6 activity during aging and oxidative stress. Sci. Rep. 2017;7(1):14130. doi: 10.1038/s41598-017-14520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki T., Yamamoto M. Molecular basis of the Keap1–Nrf2 system. Free Radic. Biol. Med. 2015;88:93–100. doi: 10.1016/j.freeradbiomed.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 55.Huang H.C., Nguyen T., Pickett C.B. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002;277(45):42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 56.Huang H.C., Nguyen T., Pickett C.B. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc. Natl. Acad. Sci. USA. 2000;97(23):12475–12480. doi: 10.1073/pnas.220418997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Z., Huang Z., Zhang D.D. Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS One. 2009;4(8):e6588. doi: 10.1371/journal.pone.0006588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magesh S., Chen Y., Hu L. Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Med. Res. Rev. 2012;32(4):687–726. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39(4):199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 60.Bhaskar K., Konerth M., Kokiko-Cochran O.N., Cardona A., Ransohoff R.M., Lamb B.T. Regulation of tau pathology by the microglial fractalkine receptor. Neuron. 2010;68(1):19–31. doi: 10.1016/j.neuron.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho S.H., Sun B., Zhou Y., Kauppinen T.M., Halabisky B., Wes P. CX3CR1 protein signaling modulates microglial activation and protects against plaque-independent cognitive deficits in a mouse model of Alzheimer disease. J. Biol. Chem. 2011;286(37):32713–32722. doi: 10.1074/jbc.M111.254268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rojo A.I., Pajares M., Rada P., Nunez A., Nevado-Holgado A.J., Killik R. NRF2 deficiency replicates transcriptomic changes in Alzheimer's patients and worsens APP and TAU pathology. Redox Biol. 2017;13:444–451. doi: 10.1016/j.redox.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rojo A.I., Pajares M., Garcia-Yague A.J., Buendia I., Van Leuven F., Yamamoto M. Deficiency in the transcription factor NRF2 worsens inflammatory parameters in a mouse model with combined tauopathy and amyloidopathy. Redox Biol. 2018;18:173–180. doi: 10.1016/j.redox.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corona A.W., Huang Y., O'Connor J.C., Dantzer R., Kelley K.W., Popovich P.G. Fractalkine receptor (CX3CR1) deficiency sensitizes mice to the behavioral changes induced by lipopolysaccharide. J. Neuroinflamm. 2010;7:93. doi: 10.1186/1742-2094-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Innamorato N.G., Rojo A.I., Garcia-Yague A.J., Yamamoto M., de Ceballos M.L., Cuadrado A. The transcription factor Nrf2 is a therapeutic target against brain inflammation. J. Immunol. 2008;181(1):680–689. doi: 10.4049/jimmunol.181.1.680. [DOI] [PubMed] [Google Scholar]
- 66.Robledinos-Anton N., Rojo A.I., Ferreiro E., Nunez A., Krause K.H., Jaquet V. Transcription factor NRF2 controls the fate of neural stem cells in the subgranular zone of the hippocampus. Redox Biol. 2017;13:393–401. doi: 10.1016/j.redox.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rogers J.T., Morganti J.M., Bachstetter A.D., Hudson C.E., Peters M.M., Grimmig B.A. CX3CR1 deficiency leads to impairment of hippocampal cognitive function and synaptic plasticity. J. Neurosci.: Off. J. Soc. Neurosci. 2011;31(45):16241–16250. doi: 10.1523/JNEUROSCI.3667-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cho H.Y., Marzec J., Kleeberger S.R. Functional polymorphisms in Nrf2: implications for human disease. Free Radic. Biol. Med. 2015;88(Pt B):362–372. doi: 10.1016/j.freeradbiomed.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cherry J.D., Olschowka J.A., O'Banion M.K. Neuroinflammation and M2 microglia: the good, the bad, and the inflamed. J. Neuroinflamm. 2014;11:98-. doi: 10.1186/1742-2094-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ray A.K., DuBois J.C., Gruber R.C., Guzik H.M., Gulinello M.E., Perumal G. Loss of Gas6 and Axl signaling results in extensive axonal damage, motor deficits, prolonged neuroinflammation, and less remyelination following cuprizone exposure. Glia. 2017;65(12):2051–2069. doi: 10.1002/glia.23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rothlin C.V., Ghosh S., Zuniga E.I., Oldstone M.B., Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131(6):1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 72.Copple I.M., Dinkova-Kostova A.T., Kensler T.W., Liby K.T., Wigley W.C. NRF2 as an emerging therapeutic target. Oxid. Med. Cell. Longev. 2017;2017:8165458. doi: 10.1155/2017/8165458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tarozzi A., Angeloni C., Malaguti M., Morroni F., Hrelia S., Hrelia P. Sulforaphane as a potential protective phytochemical against neurodegenerative diseases. Oxid. Med. Cell. Longev. 2013;2013:415078. doi: 10.1155/2013/415078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hou T.T., Yang H.Y., Wang W., Wu Q.Q., Tian Y.R., Jia J.P. Sulforaphane inhibits the generation of amyloid-beta oligomer and promotes spatial learning and memory in Alzheimer's disease (PS1V97L) transgenic mice. J. Alzheimer's Dis.: JAD. 2018;62(4):1803–1813. doi: 10.3233/JAD-171110. [DOI] [PubMed] [Google Scholar]
- 75.Lavich I.C., de Freitas B.S., Kist L.W., Falavigna L., Dargel V.A., Kobe L.M. Sulforaphane rescues memory dysfunction and synaptic and mitochondrial alterations induced by brain iron accumulation. Neuroscience. 2015;301:542–552. doi: 10.1016/j.neuroscience.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 76.Liddell R.J. Are astrocytes the predominant cell type for activation of Nrf2 in aging and neurodegeneration? Antioxidants. 2017;6(3) doi: 10.3390/antiox6030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tremblay K., Lemire M., Provost V., Pastinen T., Renaud Y., Sandford A.J. Association study between the CX3CR1 gene and asthma. Genes Immun. 2006;7(8):632–639. doi: 10.1038/sj.gene.6364340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Not applicable.