RNA-protein (RNP) granules are non-membrane bound organelles that play critical roles in the stress response1,2, maternal mRNA storage3, synaptic plasticity4, tumor progression5,6 and neurodegeneration7-9. However, the dynamics of their mRNA components within and near the granule surface remain poorly characterized, particularly in the context and timing of mRNAs exiting translation. Herein, we used multicolor single molecule tracking to quantify the precise timing and kinetics of single mRNAs as they exit translation and enter RNP granules during stress. We observed single mRNAs interacting with stress granules and P-bodies, with mRNAs moving bidirectionality between them. While translating mRNAs only interact with RNP granules dynamically, non-translating mRNAs can form stable, and sometimes rigid, associations with RNP granules with stable associations increasing with both mRNA length and granule size. Live and fixed cell imaging demonstrated mRNAs can extend beyond the protein surface of a stress granule, which may facilitate interactions between RNP granules. Thus, the recruitment of mRNPs to RNP granules involves dynamic, stable, and extended interactions affected by translation status, mRNA length, and granule size that collectively regulate RNP granule dynamics.
To simultaneously visualize and quantify the exit of single mRNAs from translation and their dynamic interactions with RNP granules in living cells, we adapted the newly developed Nascent Chain Tracking technique10. The translation of individual mRNAs labeled by fluorescent MS2 coat protein (MCP-Halo with JF646 dye11) was monitored via the binding of fluorescent antibody fragments (Fab conjugated with the Cy3 dye) to epitopes at the N-terminus of the nascent peptide (Fig. 1a). Thus, translating mRNAs were labeled by both MCP and Fab, while non-translating mRNAs were only labeled by MCP. By imaging these constructs in U-2 OS cells stably expressing the stress granule (SG) marker GFP-G3BP112, we could examine the translation status of single mRNAs during arsenite stress in relation to their interactions with individual SGs.
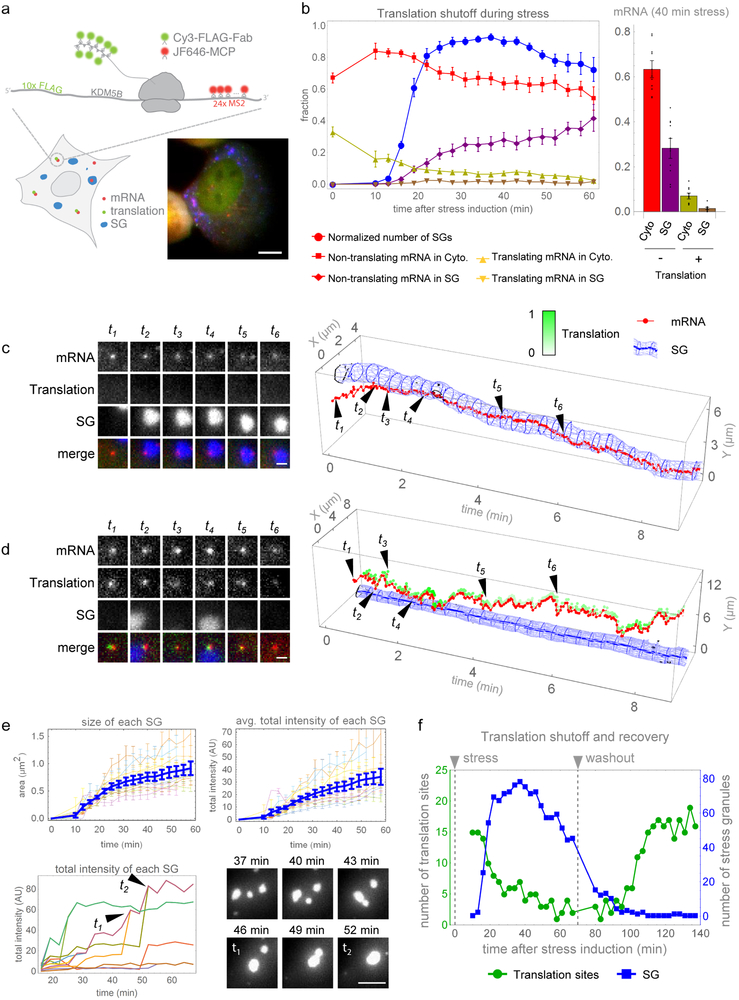
Fig. 1. mRNAs are translationally repressed prior to entry into SGs and resume translation following SG disassembly.
(a) Single mRNAs with 24×MS2 stem loops in the 3’UTR are visualized with JF646-MCP (red) and translation observed by anti-FLAG Cy3-Fab (green) binding the N-terminal ORF 10×FLAG tags in U-2 OS cells expressing the SG marker GFP-G3BP1 (blue); a representative cell (from 10 we analyzed in detail in (b)) is shown (scale bar, 10 μm). (b) Simultaneous detection of mRNA localization, translation activity, and SG formation in arsenite-stressed cells. Left, normalized number of SGs per cell (i.e. fraction of maximal number of SGs observed in each cell throughout the stress, blue); fraction of cytoplasmic non-translating (red) and translating (yellow) mRNAs; fraction of SG-localized translating (brown) and non-translating (purple) mRNAs. Right, fraction of cytoplasmic or SG-localized translating or non-translating mRNAs at 40 minutes post-stress. Avg. ± SEM shown from n=10 cells collected from 3 independent experiments. (c and d) Left, representative images of single mRNAs (red), their translation activity (green), and SG (blue) interactions (scale bars, 1 μm). Right, graphical representation of interactions with intensity of translation foci represented as white-green. (c) A representative long-term mRNA-SG interaction (>3 min) from 82 non-translating mRNAs tracked from 9 cells collected from 3 independent experiments. (d) An example of transient translating mRNA-SG interactions from 334 translating mRNAs tracked from 9 cells collected from 3 independent experiments. (e) SG growth over time, as average SG size (upper left) and average SG intensity (upper right). Growth of individual SGs (lower left) with fusion events shown graphically (t1 and t2) and as a representative time series (lower right, scale bar, 5 μm). Avg. ± SEM shown from n=10 cells collected from 3 independent experiments. (f) Representative data from a single cell (from 4 cells collected from 2 independent experiments) showing translation resumption after arsenite washout. Number of translation foci (green) and SGs (blue) during stress (0-70 min) and following washout (80-140 min). Source data are provided in Supplementary Table S2.
We first imaged cells at lower temporal resolution (1 volume every 3 minutes for 1 hour) to determine the timing of translation repression during stress. To avoid background from single mature proteins and limit photobleaching, we lowered laser powers such that only mRNAs translating in polysomes were detectable. As the majority of translation output is thought to occur in polysomes, we considered mRNAs without a detectable associated nascent peptide to be in a translationally repressed state. The translation of single KDM5B reporter mRNAs declined upon stress, with SGs forming after 10 minutes of stress (Fig. 1b, Movie S1). The interaction of KDM5B mRNAs with SGs slightly lagged behind SG assembly, with ~30-40% of the mRNAs associating with SGs 40-60 minutes after arsenite addition, which is similar to what is predicted of endogenous KDM5B mRNAs from the SG transcriptome (~39%)13. 98% of the KDM5B SG-associated mRNAs were translationally repressed, implying translation repression is a general prerequisite for mRNA recruitment to SGs. Surprisingly, 1-2% of SG-associated mRNAs retained nascent chains, suggesting that mRNAs can interact with SGs while still associated with polysomes. To verify these observations, we imaged samples at a higher temporal resolution (1 volume every 2 seconds for 10 minutes) between 10 and 30 minutes post-stress, a period when both translating mRNAs and SGs are observed (Fig. 1b). This confirmed that most of the mRNAs entered SGs in a translationally repressed state (Fig. 1c, Movie S2). 1-2% of the mRNAs interacting with SGs were associated with nascent chains, but these mRNAs only interacted with SGs transiently for a few seconds (Fig. 1d; Movie S3). Out of a total of 336 interactions detected, only two transcripts associated with nascent chains interacted with stress granules for longer than two minutes (Fig. 1d, Fig. S1, Movie S3).
Translation repression positively correlates with the growth of SGs, which increases throughout the stress response14-16. SGs grew by both incremental accumulation of material and SG fusion (Fig. 1e). Incremental growth dominated during the early phases of stress before ~40 minutes post-stress, when the number and size of SGs continued to increase, whereas fusion events dominated at later times after ~40 minutes post-stress when the number of SGs ceased increasing but SG size continued to increase. SG growth also correlated with increased KDM5B mRNAs in SGs, consistent with mRNA recruitment contributing to SG growth.
During the recovery from arsenite stress, SG disassembly (quantified as number of individual SGs) occurred a few minutes prior to translation resumption in single cells (Fig. 1f, Fig. S2, Movie S4). We verified that these translating spots in the recovery phase were bona-fide translating mRNAs by observing their disappearance following puromycin treatment (Movie S5). KDM5B mRNAs did not resume translation until SGs had dissolved completely, arguing SGs disassemble prior to translation resumption.
To determine how mRNAs interact with RNP granules, we measured the duration of individual SG interactions with mRNAs encoding either KDM5B, H2B or p300 (Fig. 2a&b). In all three cases, the probability a SG-mRNA interaction lasted a set duration of time (i.e. the interaction time survival probability) could be fit by a two-state model with fast interactions on the order of seconds and slow interactions on the order of many minutes (Fig. 2c&d). These results suggest mRNAs frequently ‘sample’ SGs and occasionally enter a stable association.
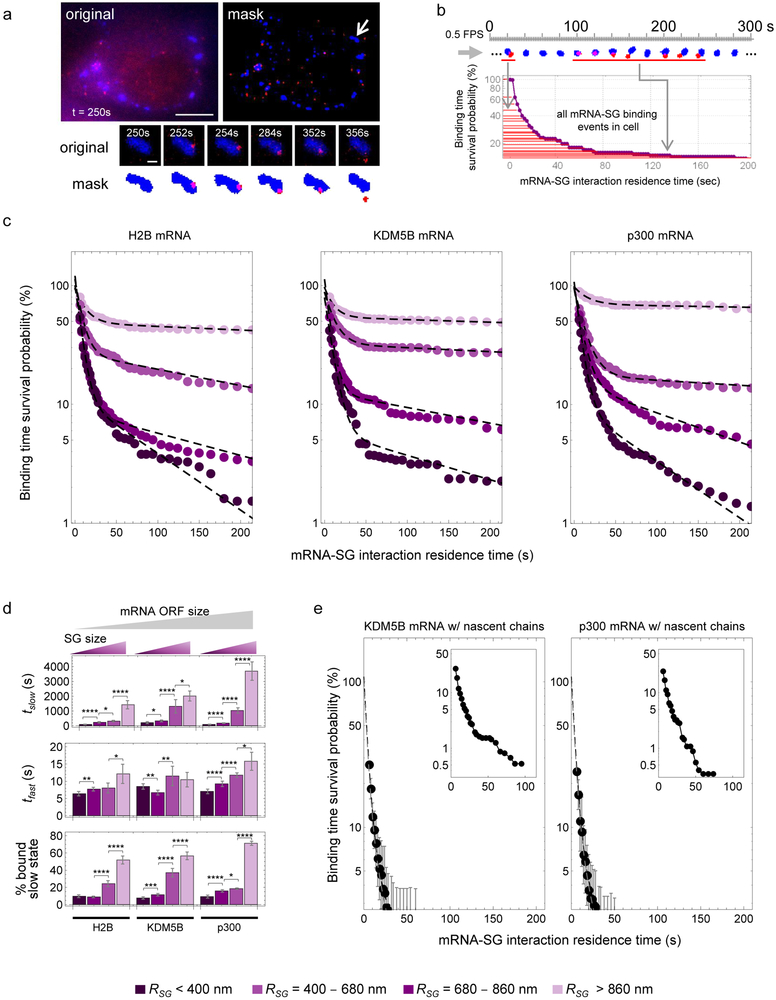
Fig. 2. mRNAs interact transiently and stably with SGs.
(a) Determination of mRNA-SG interaction times. Representative image (from 9 cells expressing KDM5B mRNA we analyzed in detail in (c)) showing mRNAs (red) and SGs (blue) (upper left, scale bar, 10 μm) and the masked image (upper right) with a representative mRNA-SG interaction (arrow) shown below (scale bar, 1 μm). (b) Survival probability distribution of mRNA-SG interaction times (red) from one representative cell from 9 cells expressing KDM5B mRNA we analyzed in detail in (c). (c) The binding time survival probability (%) of H2B (left; data were calculated from 492 tracked SGs from 11 cells collected from 3 independent experiments), KDM5B (middle; data were collected from 409 tracked SGs from 9 cells collected from 4 independent experiments) and p300 (right; data were calculated from 824 tracked SGs from 16 cells from 4 independent experiments) mRNA-SG interactions are shown partitioned by SG size (legend at bottom). (d) Data in (c) was fit, resulting in average (±90% CI) fitted slow and fast mRNA-SG interaction times, tslow (top), tfast (middle), and % mRNA bound to SGs in the slow interaction mode (“slow state”). Fitted results are shown for H2B, KDM5B and p300 mRNAs for a given effective SG radius (“RSG”, legend at bottom). Two-sided t-tests were performed with * indicating p ≤ 0.05, ** p ≤ 0.01, ** p ≤ 0.005, **** p ≤ 0.001 and df=44. Statistics source data for Fig 2d can be found in Tables S2 and S3. Fitting was performed once to the collective data set shown in (c). (e) The average binding time survival probability (%) (±90% CI) of KDM5B (left; data were calculated from 326 tracked SGs from 9 cells collected from 3 independent experiments) and p300 (right; data were calculated from 336 tracked SGs from 10 cells collected from 4 independent experiments) mRNAs associated with nascent chains is shown with graphical insets showing the full distribution (adjusted xy-axes). Source data is provided in Tables S2.
More generally, three parameters influenced the nature of SG-mRNA interactions. First, we compared the dynamic interactions of reporter mRNAs containing the KDM5B ORF (4632 nt), the shorter H2B ORF (375 nt) or the longer p300 ORF (8265 nt) with SGs by determining the dwell time of mRNAs in SGs and found mRNA ORF length correlated with duration of interactions with large SGs (Fig. 2c&d, tslow(H2B) = 1400 ± 300 s, tslow(KDM5B) = 2000 ± 300 s, and tslow(p300) = 3700 ± 600 s). We also observed that the duration of the slow interactions and the fraction of mRNAs engaged in slow interactions with SGs for the H2B, KDM5B and p300 mRNAs varied depending on the size of the SG. Larger SGs showed more and longer stable interactions with mRNAs than smaller SGs (Fig. 2c&d, Movie S6 & S7). The observation that longer mRNAs interact more stably with SGs explains why longer mRNAs were more likely to be highly enriched in the steady-state SG transcriptome13,17. Finally, we observed that KDM5B and p300 mRNAs still engaged with polysomes, as assessed by the detection of associated nascent peptides, could interact with SGs transiently but did not engage in the stable interaction mode (Fig. 2e). We did not analyze translating H2B transcripts because they were more difficult to track due to their dimness and low number per cell, even before stress10. Translating mRNAs in the cytoplasm moved slower than translationally repressed mRNAs (Fig 4a), ruling out the possibility that the transient association of translating mRNAs with SGs was due to faster overall mobility. Taken together, these observations suggest that mRNA length, SG size and mRNA translation status are strongly correlated with mRNA partitioning into SGs, although there are likely other mRNP factors that influence the dynamics of mRNA-SG interactions.
Fig. 4. mRNAs can be rigidly positioned within SGs and/or tethered to them.
(a) Average mean squared displacement (MSD) (±90%CI) of cytoplasmic KDM5B mRNAs (red; fitted D=0.016±0.004 μm2/sec), mRNAs with nascent chains (green; fitted D=0.011±0.002 μm2/sec), SGs (blue; fitted D=0.003±0.0006 μm2/sec), SGs containing mRNAs (purple; fitted D=0.004±0.0004 μm2/sec), and mRNAs in SGs (gray; fitted D=10±2 nm2/sec) from n=7 cells from 2 independent experiments; 1243 mRNAs, 108 mRNAs with nascent chains, 1049 SGs, 92 SGs containing mRNAs and 3 mRNAs tracked in 3D within one SG). (b) Representative time series (from 2 cells collected from 2 independent experiments) showing three KDM5B mRNAs (red foci marked by red, blue and green arrows, top, scale bar, 1 μm) and their 3D positions (as red, green or blue dots, bottom) in a SG. (c) Projected 2D positions (top) and relative 3D positions (bottom) of the three mRNAs in (b) plotted over time (green mRNA position fixed and blue mRNA oriented relative to green mRNA). Frames in (b) indicated as t1, t2 and t3 (black arrows). Yellow arrows indicate exit and entrance of the red mRNA from the SG (from 2 cells collected from 2 independent experiments). (d) Representative immunofluorescence-smFISH images for AHNAK (n=4 cells) and NORAD (n=4 cells) RNAs (red) and SGs (G3BP1, green) at 60 min. post-stress. Left, representative images of RNAs clustered near SGs (white circles). Images shown represent data from 4 cells collected from one experiment. Scale bar, 0.5μm. Right, relative frequency of distances of 50 AHNAK or NORAD RNAs to the nearest SG. (e) Schematic of AHNAK smFISH probe positions (top). (Bottom) Representative smFISH images where one end of the AHNAK mRNA was outside the SG while the other end was inside the SG (5’ probes in red, 3’ probes in green; SG in grey) (n=14 cells collected from one experiment). Scale bars, 1μm. Source data in Tables S2. (f) Model. Translation repression causes ribosomes to run off transcripts, which interact transiently with SGs via docking and undocking. Some transcripts then enter a stable association (‘lock’) with SGs and may engage in multivalent interactions with other RNAs and proteins. Transcripts may be tethered to the SG surface, perhaps facilitating SG growth or docking of P-bodies. Those transcripts remaining in translation complexes can only transiently interact with SGs.
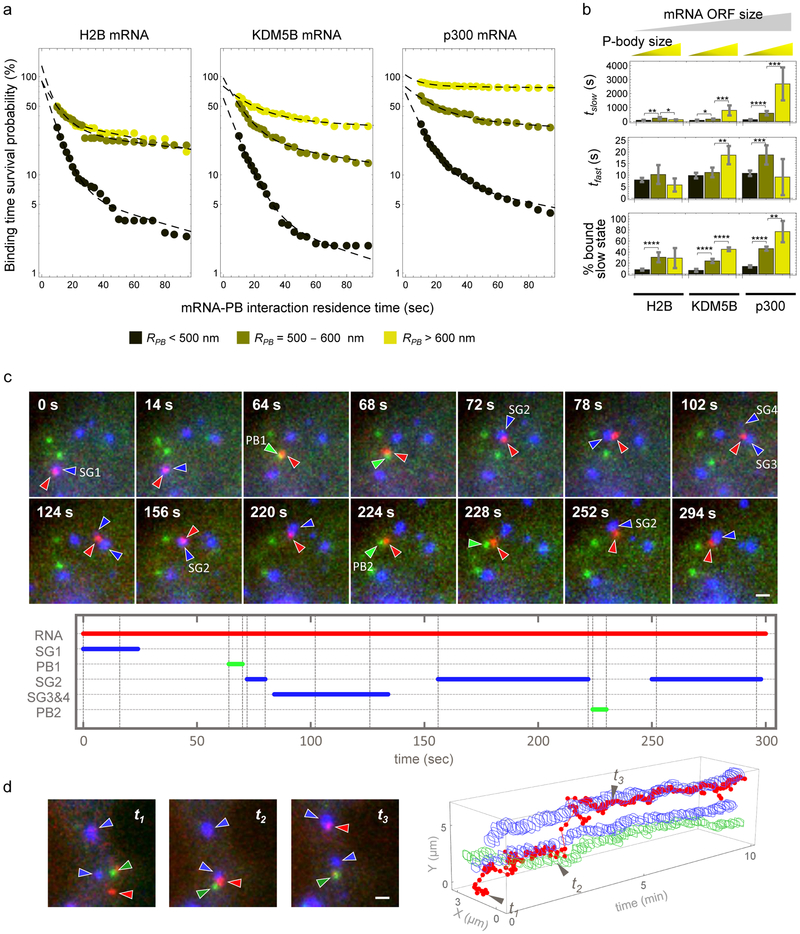
To determine if other RNP granules also exhibit bimodal interactions with mRNAs, we examined the dynamics of mRNA interactions with P-bodies (PBs), marked with mRFP-DCP1a in stressed U-2 OS cells also expressing the SG marker GFP-G3BP1. We observed that H2B, KDM5B and p300 mRNAs all showed rapid and stable interactions with PBs (Fig. 3a), that mRNA ORF length was correlated with duration of mRNA interaction with PBs, as p300 interacted most stably with PBs and H2B interacted most unstably with PBs (Fig. 3a&b), and that the degree of stable interactions for all mRNAs analyzed increased with the size of the PB (Fig. 3a&b; Movie S8). These observations indicate that mRNAs can dock and undock to SGs and PBs through transient interactions, and in some cases form a stable set of interactions that ‘locks’ the mRNA into the granule for prolonged periods. Stable association could be more prevalent on longer mRNAs and in larger SGs and PBs due to increased sites for additional interactions. This model implies that the association of an mRNA with a granule can be influenced by cis or trans inputs that modify either the docking/undocking step, or the rate of entry into a stable, locked interaction state, which is supported by the observation that knockdown of the RNA binding protein LARP1 can reduce the association of some mRNAs with PBs and SGs18.
Fig. 3. mRNAs interact transiently and stably with PBs in stressed cells and traffic bidirectionally between PBs and SGs.
(a) Average binding time survival probability distributions of H2B mRNA-PB (left; data were calculated from 106 tracked PBs of 4 cells from one experiment), KDM5B mRNA-PB (middle; data were calculated from 137 tracked PBs of 7 cells collected from 3 independent experiments), and p300 mRNA-PB (right; data were calculated from 244 tracked PBs of 16 cells collected from 4 independent experiments) for a given effective PB radius (“RPB”, legend at bottom). (b) Data in (a) was fitted, resulting in average fitted fast and slow interaction times and % mRNA bound to PBs in the slow interaction mode (“slow state”). Fitted results are shown (±90% CI) for H2B, KDM5B or p300 mRNAs for each effective PB radius (legend in (a)). Two-sided t-tests were performed with * indicating p ≤ 0.05, ** p ≤ 0.01, ** p ≤ 0.005, **** p ≤ 0.001, df = 31. Statistics source data for Fig 3b can be found in Tables S2 and S3. Fitting was performed once to the collective data set shown in (a). (c) Representative images (top, scale bar, 1 μm) (from 6 cells collected from 4 independent experiments) of a single KDM5B mRNA (red, red arrows) that interacted with four SGs (blue, blue arrows) and two PBs (green, green arrows). The duration of each mRNA-RNP granule interaction was plotted (bottom). (d) A representative mRNA trajectory (from 3 cells collected from 2 independent experiments) between two SGs and a PB visualized by plotting the position of the mRNA (red), SG (blue), and PB (green) over time. Example cropped images corresponding to time points t1 to t3 are shown (left, scale bar, 1 μm). Source data is provided in Supplementary Table S2.
Non-translating mRNAs can accumulate in both SGs and PBs. A general model is that following translation repression mRNAs first associate with SGs, and then can be sorted for targeting to PBs19. In contrast, it has been proposed that mRNAs move from PBs to SGs upon glucose deprivation in yeast20. To determine if there is a preferred movement of mRNAs between SGs and PBs, we simultaneously imaged SGs, PBs and KDM5B mRNAs during stress. We observed mRNAs exchange between SGs and PBs in a bidirectional manner (Fig. 3c&d). These interactions included cases of mRNAs transitioning from a SG to a PB and back to a different SG within as little as ~30 seconds (Fig. 3c, Movie S9) and instances where the mRNA was localized between a PB and a SG for a few minutes (Fig. 3d & Movie S10). Transcripts were considered to be associated with a SG or PB when they co-moved with these granules over several frames (or at least 4 seconds). These rapid bidirectional movements between SGs and PBs indicate that there is no obligate path for mRNA movement between these granules, which also demonstrates that an individual mRNA with its associated proteins is capable of interacting with either SGs or PBs, or exchanges proteins that enable SG or PB interaction within seconds.
SGs, and other RNP granules, have been proposed to be liquid-like compartments with components showing rapid diffusion within the compartment21-24 and/or contain liquid-like areas surrounding densely compacted ‘cores’14,25. Those mRNAs that were stably associated with RNP granules for many minutes adopted the motility of SGs (Fig. 4a) and could be observed at the surface or embedded within SGs (Fig. 4b). To examine the liquid-like nature of SGs, we examined the movement of mRNAs relative to one another within SGs. We observed two cases where the relative localization of mRNAs, as assessed by quantifying the distance and angle between three individual KDM5B mRNAs in one SG, remained relatively constant over time (Fig. 4b&c, Fig. S3, Fig. S4, Movie S11, Movie S12), consistent with limited movement of the mRNA within the larger assembly. Quantification revealed the intra-SG diffusion of mRNAs to be 280 times slower than the SG as a whole and over 1600 times slower than free mRNAs within the cytoplasm (Fig. 4a). These observations demonstrate that SGs contain solid or gel-like components, and mRNAs within large SGs can be rigidly positioned, with relatively limited movements of mRNAs.
Unexpectedly, we observed two cases where the MCP signal, which represents the 3’ end of a KDM5B mRNA, exited the SG and then returned to approximately the same position within the SG (Fig. 4c, yellow arrows, Movie S11, Movie S12, Fig. S3, Fig. S4). This observation suggested that the 3’ end of this mRNA might exit the GFP-G3BP1-marked portion of the SG while the remainder of the mRNA is still within the SG. Consistent with the body of an mRNA extending beyond the G3BP1-marked SG, we observed that endogenous RNAs that localize to SGs (AHNAK and NORAD) are often detected near, but not in SGs in U-2 OS cells by smFISH (Fig. 4d), with the shorter NORAD RNA (~5 kb) being closer to the periphery of a SG than the longer AHNAK RNA (18kb). Furthermore, using smFISH probes to the 5’ or 3’ ends of the AHNAK mRNA, we observed ~15% of ~1000 AHNAK mRNAs were at the SG periphery with one end of the mRNA outside the SG while the other end co-localized with GFP-G3BP1, with a ~two-fold bias for the 3’ end to be within the SG (Fig. 4e). These observations suggest that the surfaces of these membraneless organelles are not uniform and can have RNP extensions into the cytosol beyond boundaries as defined by protein components. Such surface extensions thus could provide a set of interactions to promote fusion of smaller SG and docking of SG and PBs, which is consistent with our observations of individual mRNAs localizing between a PB and a SG for several minutes (Fig. 3d & Movie S10).
These experiments demonstrate that mRNAs interact in both stable and transient manners with SG and PBs. In model assemblies, exchange rates of components are related to both the strength of individual interactions, and the valency, with higher valency having more stable interactions26. This suggests that the transient interactions with SGs or PBs represent a low valency interaction mode but provide a “docked” state whereby mRNPs can form additional interactions with SG components to perhaps enter a stable “locked” state with higher valency (Fig. 4f). Higher valency would be expected on longer mRNAs, and larger SGs or PBs, providing an explanation for why longer mRNAs and larger RNP granules have more stable RNA-granule interactions. Our experiments provide unambiguous evidence that mRNAs stably associated with SGs are translationally repressed, although mRNAs associated with polysomes can transiently interact with SGs. This may be because the 3’UTR and/or partially exposed coding regions may be sufficient to form transient protein-protein or RNA-RNA interactions, but a fully exposed coding region is required to make sufficient interactions for stable SG association. This work identifies the docking and locking steps as two distinct steps in RNP association with any RNP granule that can be modulated to affect RNA-granule association. We propose this bimodal nature of RNA-granule interaction will be a general principle of any self-organized RNP granule where RNP recruitment requires entry into a multivalent state.
Methods
Cell culture
U-2 OS cells stably expressing the SG marker GFP-G3BP1 and the P-body marker mRFP-DCP1a (a generous gift from the Kedersha lab12) were maintained in DMEM supplemented with 10% fetal bovine serum and 1% streptomycin/penicillin. Cells harboring the GFP-G3BP1 SG marker alone were isolated by cell sorting of the GFP-G3BP1 and mRFP-DCP1 mixed cell pool (BioFrontiers Institute Flow Cytometry Core). U-2 OS cells were validated by STR profiling and morphological assessments.
Fab Generation and Dye-Conjugation
Fab generation and dye-conjugation was done essentially as described in Morisaki et al., (2016)10. Briefly, Pierce mouse IgG1 preparation kit (Thermo Scientific) was used to generate Fab according to the manufacturer’s instruction. Immobilized ficin in the presence of 25 mM cysteine was used to digest FLAG (Wako, 012-22384 Anti DYKDDDDK mouse IgG2b monoclonal) antibodies to create Fab. Fab were separated from the Fc region using NAb Protein A columns. After elution Fab were concentrated to 1 mg/ml and conjugated to Cy3. Cy3 N-hydroxysuccinimide ester (Invitrogen) were dissolved in DMSO and stored at −20°C. 100 μg of Fab were diluted into 100 μl of 100 mM NaHCO3 (pH 8.5). 1.33 μl of Cy3 was added to this solution and incubated with end-over-end rotation for 1-2 hours at room temperature. The conjugated Fab were then eluted from a PBS pre-equilibrated PD-mini G-25 desalting column (GE Healthcare) that removed unconjugated dye. Conjugated Fabs were then concentrated using an Ultrafree 0.5 filter (10k-cut off; Millipore) to 1 mg/ml. The Fab:dye ratio was calculated using the absorbance at 280 and 550 nm, and using the extinction coefficient of Fab with the dye correction factor at 280 nm provided by the manufacturers (0.08 for Cy3). The degree of labeling was calculated using the following formula:
| (1) |
Only Fab with a DOL of ~1 were used.
MCP Purification
MCP was purified as described in Morisaki et al., (2016)10. His-tagged MCP was purified with Ni-NTA-agarose (Qiagen) following the manufacturer’s instructions with minor modifications. Bacteria were lysed in a PBS-based buffer containing a complete set of protease inhibitors (Roche), and binding to the Ni-NTA resin was carried out in the presence of 10 mM imidazole. After washing with 20 and 50 mM imidazole in PBS, the protein was eluted with 300 mM imidazole in PBS, and directly used for experiments. The rest was dialyzed against a HEPES-based buffer (10% glycerol, 25 mM HEPES pH 7.9, 12.5 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 0.01 % NP-40 detergent, and 1 mM DTT) and stored at −80 °C after snap-freezing in liquid nitrogen.
Nascent Chain Tracking
Nascent Chain Tracking was done essentially as described in Morisaki et al., (2016)10. Briefly, a plasmid expressing a reporter mRNA encoding the FLAG spaghetti monster tag N-terminal to H2B (Addgene plasmid #81082), KDM5B (Addgene plasmid #81084) or p300 open reading frames, followed by the 3’ UTR of beta-actin with 24x MS2 stem loops was either transfected into cells two hours prior to bead-loading or bead-loaded into cells together with purified Fab and/or MCP. 100 μg/ml of fluorescently labeled Fab and/or 33 μg/ml of purified MCPHaloTag protein in 4 μl of PBS were bead loaded. Cells were incubated for 2-3 hours prior to imaging in DMEM lacking phenol red with 10% fetal bovine serum, 1% streptomycin/penicillin and 1% glutamine. Cells were incubated for 30 minutes in the presence of JF646-HaloTag ligand to label MCP-HaloTag protein and washed thrice immediately before imaging.
Imaging conditions
To track single mRNAs and their translation status with RNP granules, we used a custom-built widefield fluorescence microscope with a highly inclined illumination scheme10,27. Briefly, the excitation beams, 488, 561 and 637 nm solid-state lasers (Vortran), were coupled and focused on the back focal plane of the objective (60X, NA 1.49 oil immersion objective, Olympus). The emission signals were split by an imaging grade, ultra-flat dichroic mirror (T660lpxr, Chroma), and detected by two aligned EM-CCD (iXon Ultra 888, Andor) cameras by focusing with 300 mm tube lenses (this lens combination produces 100X images with 130 nm/pixel).
Live cells were placed into an incubation chamber at 37 °C and 5% CO2 (Okolab) on a piezoelectric stage (PZU-2150, Applied Scientific Instrumentation). The focus was maintained with the CRISP Autofocus System (CRISP-890, Applied Scientific Instrumentation). The lasers, the cameras, and the piezoelectric stage were synchronized by an Arduino Mega board (Arduino). Image acquisition was performed using open source Micro-Manager software (1.4.22)28,29. Imaging size was set to 512 × 512 pixels2 (66.6 × 66.6 μm2), and exposure time was selected as 53.64 msec. The readout time of the cameras from the combination of our imaging size and the vertical shift speed we selected was 23.36 msec, resulting in our imaging rate of 13 Hz (70 msec per image). The excitation laser lines were digitally synched such that they illuminate the cells only when the camera is exposing to avoid any excess observational photobleaching. The far-red signal of mRNA visualized by JF646-MCP was imaged on one camera, and the red signal of translation (or P-bodies) visualized by Cy3-FLAG-Fab (or mRFP-DCP1) and the green signal of SGs visualized by GFP-G3BP1 were imaged on the other camera. Cy3/mRFP and GFP signals were imaged alternately. To minimize the bleed-through, the JF646 signal was simultaneously imaged with the GFP signal. We also employed a filter wheel (HS-625 HSFW TTL, Finger Lakes Instrumentation) equipped with a filter for Cy3 and mRFP (593/46 nm BrightLine, Semrock), and for GFP (510/42 nm BrightLine, Semrock). The filter position was changed during the camera read out time (23.36 msec) by the Arduino Mega board.
To capture the whole thickness of the cytoplasm of U2OS cells, 13 z-stacks of step size 500 nm (6 μm in total) were imaged using the piezoelectric stage such that the z-position changed every 2 images (Cy3 and GFP). This resulted in our total cellular imaging rate of 0.5 Hz (2 sec per volume). For Fig. 1b&e, the cells were first imaged at a single time point with 13 z-stacks prior to arsenite addition (time point ‘0’). Arsenite was then added (0.5 mM) and after ten minutes cells were imaged every 3 minutes with 13 z-stacks per timepoint for up to 60 minutes. For Fig. 1f, image acquisitions were performed with the same conditions described above, except that the cells were washed with fresh media 10 times at 70 minutes post-stress. Ten minutes after washout, cells were imaged again every 3 min with 13 z-stacks per timepoint for another 60 minutes. For all other figures, the cells were imaged continuously at 0.5 Hz with 13 z-stacks per timepoint for 10 minutes beginning between 10 and 30 minutes post-stress (when SGs were visible). Note that all colors described in the text and figures are based on the color of excitation lasers as mentioned above, namely mRNA in red (JF646), translation or PB in green (Cy3 or mRFP), and SG in blue (GFP). Following this color scheme, purple corresponds to mRNA (red) in SGs (blue), while yellow corresponds to mRNA in PBs and brown corresponds to translating mRNAs in SGs.
Analysis of stress granule formation, mRNA localization, and translation
Single particle detection was performed on XY maximum projections of the stacks to detect mRNA, translation, and SGs, using custom Mathematica code (version 11.2.0.0). Briefly, for each image channel a bandpass filter was used to highlight particles within a given size range and the resulting image was binarized using a reasonable threshold to create a mask of the cell in which the vast majority of mRNA, translation, or SGs were detectable. The Mathematica command ComponentMeasurements was then used to measure the intensity-centroid position, area, total intensity, and convex vertices of each masked object, either mRNA, translation, or SGs. The precise coordinates (super-resolved locations) of mRNAs and nascent peptides were determined by fitting (using the built-in Mathematica routine NonlinearModelFit) the original, pre-processed sub-images of detected particles to 2D Gaussians of the following form:
where IBG is the background fluorescence, I the particle intensity, (σx, σy) spreads of the particle, and (x0, y0) the particle location. The offset between the two cameras was registered using the built-in Mathematica routine FindGeometricTransform to find the transform function that best aligned the fitted positions of 200 nm diameter Tetraspeck beads evenly spread out across the image field-of-view. Translating mRNAs were found by selecting mRNAs which colocalize with a translation signal location within 390 nm of the mRNA. mRNAs within the SGs were found by selecting mRNAs within detected SG masked regions. The number of translating mRNAs in the cytosol or SGs, and the number of non-translating mRNAs in the cytosol or SGs were counted and divided by the total number of mRNAs at each time point to determine the fraction of each population throughout the time course during stress. The number of SGs were renormalized to the fraction of the maximum number of SGs detected in the cell at a single timepoint. Since the timing of the beginning of SG formation varies from cell to cell, all the curves were aligned first before averaging data from different cells. This alignment was done as follows. First, the SG formation curve was fit with the following equation:
where NSG is the number of SGs, A, B, and C are normalization factors, and t is the frame number. Then, from the fitted results, all the single cells curves were shifted such that their values at 10% of their maxima were aligned. These aligned curves were then averaged.
Analysis of mRNA interactions with stress granules and P-bodies
The location and the area of mRNA, SGs, and PBs were detected and tracked as described above. The effective radius was then calculated from the area to be the radius of an equivalent-area circle. Although the effective radius is a good approximation of the size of the object, it can be influenced by the brightness of the object since it is calculated from a binarized mask. Thus, small objects that are extremely bright (in particular PBs), may have effective radii that are a bit larger than they are in reality. The option “CornerNeighbors → False” was used to prevent the detection of any SGs or mRNAs that may have been clipped near the edge of the image frame. From these tracks and the binarized masks, the presence or absence of mRNA inside tracked SGs or PBs could be easily determined by the overlap of mRNA masks with the tracked SG or PB mask. The length of consecutive frames a tracked SG or PB mask had an overlapping mRNA mask was then set equal to the length of the interaction time. Interactions lasting fewer than 2 frames (1 second) were ignored, as they could be influenced by imaging noise. If more than one mRNA mask overlapped with a SG or PB mask, it was counted as a single interaction (since most of the time these mRNA masks themselves overlapped, making it difficult to know how many mRNAs were actually present). This tends to slightly underestimate the number of long interactions. Finally, to validate our image analysis pipeline, we manually assessed the interaction times of individual KDM5B mRNAs in SGs and PBs using Fiji30. In all cases, manual measurements agreed well with the image analysis results in Mathematica.
To calculate the probability of an interaction lasting a certain duration of time (i.e. the survival probability of the interaction), we counted the fraction of tracks lasting longer than N seconds, with N ranging from 6 to 300. A curve of the fraction of tracks versus time was then fit to a double exponential of the form A e−t/t1 + B e−t/t2. Data was then renormalized according to the fit such that at t = 0 sec the fraction was 100% (i.e. A + B = 100). The renormalized data represents the survival probability versus time and is displayed in the plots in the main text, along with the fitted fast interaction time t1, slow interaction time t2, and the slow fraction B/(A+B). The 90% confidence interval was calculated for the fitted values, from which the standard deviation was estimated. From the standard deviation and fitted mean values from each fit, effect size and p-values between fits were calculated. All source data and information used to derive statistics are provided in Table S2.
Analysis of mean squared displacement and 3D movement of mRNAs within SGs
The mean squared displacement (MSD) was calculated using the tracked mRNA super-resolved locations for mRNA in the cytoplasm or in SGs. The MSD of mRNA inside SGs was calculated by dividing by a factor of two the MSD of the position of one mRNA with respect to the other. The MSD of SGs was calculated using the intensity-centroid of the SG.
The 3D coordinates of mRNAs were determined by fitting (using the built-in Mathematica routine NonlinearModelFit) the original, pre-processed sub-image stacks of the detected particle to 3D Gaussians of the following form:
where IBG is the background fluorescence, I the particle intensity, (σx, σy, σz) spreads of the particle, and (x0, y0, z0) the particle location. The convex vertices of SGs and the locations of mRNAs were plotted. For these analyses, mRNA spots were linked to the nearest neighbor mRNA in the consecutive frame. With the sparse mRNA density in the analyzed SGs and the high temporal imaging frequency, it is unlikely different mRNAs will be linked frame to frame. Additionally, the quality of automated tracking was validated by eye to ensure proper tracking.
Sequential immunofluorescence and single-molecule fluorescence in situ hybridization (smFISH)
The protocol was performed as described previously13. Briefly, U-2 OS cells were seeded on sterilized coverslips in 6-well tissue culture plates. At ~80% confluency, media was exchanged 60 min. before stress with fresh media. Cells were then stressed by addition of sodium arsenite (0.5 mM) for 60 min. Media was then removed and cells were washed with pre-warmed PBS. Cells were fixed with 500 μL 4% paraformaldehyde for 10 min. at room temperature. After fixation, cells were washed twice with PBS, permeabilized in 0.1% Triton X-100 in PBS for 5 min. and washed once with PBS. Coverslips were transferred to a humidifying chamber and cells were incubated in 5 μg/mL mouse α-G3BP1 antibody (ab56574, Abcam) in PBS for 60 min. at room temperature. Afterward, the coverslips were transferred to a 6-well plate and washed 3 times with PBS. Coverslips were then transferred back to the humidifying chamber and incubated in goat α-Mouse FITC-conjugated antibody in PBS (1:1000 dilution, ab6785, Abcam) for 60 min. at room temperature. The coverslips were transferred to 6-well plate and washed 3 times with PBS. Antibodies binding to cells were fixed on cells by incubating coverslips with 500 μL 4% paraformaldehyde for 10 min. at room temperature.
After immunofluorescence, smFISH was performed as described previously13 which was adapted from protocol provided by Biosearch Technologies website (https://biosearchassets.blob.core.windows.net/assets/bti_custom_stellaris_immunofluorescence_seq_protocol.pdf). Biosearch Technologies Stellaris Buffers were also used (SMF-HB1-10, SMF-WA1-60, and SMF-WB1-20). Specific smFISH probes were created by Biosearch Technologies. Probes to the 5’ end of the AHNAK mRNA and for NORAD were described in (14). Probes to the 3’ end of the AHNAK mRNA are described in Table S1. Imaging was done using a widefield DeltaVision Elite microscope with a 100x objective and a PCO Edge sCMOS camera and softWoRx. Images shown in Figure 4d were deconvolved and max intensity projections made from at least 30 Z-stacks to span the entire cell using Fiji30. Figure 4e shows images from a single z-plane.
Image analysis with Bitplane Imaris analysis software
Image analysis of SGs and smFISH spots was done using Bitplane Imaris analysis software (8.4.1) as described previously13. To measure the distances between smFISH spots to the nearest SG, smFISH diffraction limited spots and SGs were first identified by Imaris imaging software using the spot and cell component respectively. Recognition of the smFISH diffraction limited spot by the spot component was determined using the following parameters: diameter (0.2 μm) and thresholding (manually determined for each image). Recognition of SGs (G3BP1 staining) by the cell component was determined using the following parameters: minimum size (≥1 voxel) and thresholding (manually determined for each image). A measuring tool (measuring points) was then manually applied to measure the distance between the center of the spot and the surface of the SG. This was applied to 50 individual NORAD or AHNAK RNAs as described in Fig. 4d.
Code availability
Custom Mathematica (version 11.2.0.0) code was deposited on GitHub and can be accessed at https://raw.githubusercontent.com/TatsuyaMorisaki/Translation-Stress/master/Translation-Stress.nb.
Statistics and Reproducibility
For live cell imaging, all experiments were performed with at least 10 cells collected from 3-4 independent experiments to account for cell-to-cell variability. Images were discarded if they were of insufficient quality to accurately analyze. The numbers of images deemed of sufficient quality to be included in each analysis are described below. For the mRNA-SG interaction experiments: H2B, data were calculated from 492 tracked SGs from 11 cells collected from 3 independent experiments; KDM5B, data were calculated from 409 tracked SGs from 9 cells collected from 4 independent experiments; p300, data were calculated from 824 tracked SGs from 16 cells from 4 independent experiments. For mRNA-PB interaction experiments: H2B, data were calculated from 106 tracked PBs of 4 cells from 1 experiment; KDM5B, data were calculated from 137 tracked PBs of 7 cells collected from 3 independent experiments; p300, data were calculated from 244 tracked PBs of 16 cells collected from 4 independent experiments. For fixed cell experiments, each cell was considered as an independent biological replicate. The smFISH experiments were performed once to detect AHNAK and once to detect NORAD, and the distance of 50 AHNAK or 50 NORAD RNAs from the nearest SG was determined in 4 cells each. The analysis of AHNAK 5’ and 3’ ends within SGs and outside SGs was done using data from one experiment, with 153 mRNAs analyzed in 14 cells.
For live cell imaging experiments, the standard errors of the mean representing variation between individual cells were calculated, except for Fig. 2d and 3b. Figs. 2d and 3b were created using the fitted results from plots in Figs. 2c and 3a, respectively. Each bar represents a fitted value and each error bar represents a 90% confidence interval of the fit, calculated using Mathematica’s built-in function “NonlinearModelFit”. The degree of freedom (df) of the fits was 44 and 31 for Figs. 2d and 3b, respectively. The standard error of the mean was calculated from the 90% confidence intervals (i.e. the 90% confidence intervals were divided by 1.645), from which the two-sided T-test was performed and the p-values were calculated.
Data availability
Source data for Fig 1b-1f, Fig 2c-2e, Fig 3a-3d, Fig 4a and 4c-4e, and Supplementary Fig 1, 2 and 4 are provided in Supplementary Table S2. Raw images were deposited to figshare and can be accessed at: https://figshare.com/articles/Dataset_associated_with_Multicolor_single-molecule_tracking_of_mRNA_interactions_with_RNP_granules_−1_2/7427816 and https://figshare.com/articles/Dataset_associated_with_Multicolor_single-molecule_tracking_of_mRNA_interactions_with_RNP_granules_-2_2/7427918. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgements:
We thank Nancy Kedersha for providing GFP-G3BP1/mRFP-DCP1a U-2 OS cells and Esther Braselmann and Theresa Nahreini for isolating GFP-G3BP1 U-2 OS cells at the BioFrontiers Institute Flow Cytometry Core facility. We thank Benjamin Dodd and lab members for assistance and helpful suggestions.
Funding: S.L.M. was funded by the Anna and John J. Sie Foundation; R.P. by HHMI; T.J.S. by the NIH (R35GM119728) and the Boettcher Foundation’s Webb-Waring Biomedical Research Program.
Footnotes
Competing interests: The authors have no financial or non-financial competing interests.
References:
- 1.Buchan JR & Parker R Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kedersha N, Ivanov P & Anderson P Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494–506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schisa JA Effects of stress and aging on ribonucleoprotein assembly and function in the germ line. Wiley Interdiscip. Rev. RNA 5, 231–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudhakaran IP et al. FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control. Proc. Natl. Acad. Sci. U. S. A 111, E99–E108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Naggar AM & Sorensen PH Translational control of aberrant stress responses as a hallmark of cancer. J. Pathol 244, 650–666 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Grabocka E & Bar-Sagi D Mutant KRAS Enhances Tumor Cell Fitness by Upregulating Stress Granules. Cell 167, 1803–1813.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang K et al. Stress Granule Assembly Disrupts Nucleocytoplasmic Transport. Cell 173, 958–971.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumann M et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Li YR, King OD, Shorter J & Gitler AD Stress granules as crucibles of ALS pathogenesis. J. Cell Biol 201, 361–372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morisaki T et al. Real-time quantification of single RNA translation dynamics in living cells. Science 352, 1425–1429 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Grimm JB et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kedersha N, Tisdale S, Hickman T & Anderson P Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 448, 521–552 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Khong A et al. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol. Cell 68, 808–820.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler JR, Matheny T, Jain S, Abrisch R & Parker R Distinct stages in stress granule assembly and disassembly. eLife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kedersha N et al. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol 151, 1257–1268 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohshima D, Arimoto-Matsuzaki K, Tomida T, Takekawa M & Ichikawa K Spatio-temporal Dynamics and Mechanisms of Stress Granule Assembly. PLoS Comput. Biol 11, e1004326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Namkoong S, Ho A, Woo YM, Kwakm H, & Lee JH Systematic characterization of stress-induced RNA granulation. Mol Cell. 70, 175–187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilbertz JH et al. Single-molecule imaging of mRNA localization and regulation during the integrated stress response. bioRxiv 332502 (2018). doi: 10.1101/332502 [DOI] [PubMed] [Google Scholar]
- 19.Kedersha N et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol 169, 871–884 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson CE, Lui J, Kershaw CJ, Sims PFG & Ashe MP mRNA localization to Pbodies in yeast is biphasic with many mRNAs captured in a late Bfr1pdependent wave. J Cell Sci 127, 1254–1262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei M-T et al. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem 9, 1118–1125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feric M et al. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molliex A et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sfakianos AP, Whitmarsh AJ & Ashe MP Ribonucleoprotein bodies are phased in. Biochem. Soc. Trans 44, 1411–1416 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Niewidok B et al. Single-molecule imaging reveals dynamic biphasic partition of RNA-binding proteins in stress granules. J. Cell Biol (2018). doi: 10.1083/jcb.201709007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banani SF et al. Compositional Control of Phase-Separated Cellular Bodies. Cell 166, 651–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokunaga M, Imamoto N & Sakata-Sogawa K Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Edelstein AD et al. Advanced methods of microscope control using μManager software. J. Biol. Methods 1, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edelstein A, Amodaj N, Hoover K, Vale R & Stuurman N Computer Control of Microscopes Using μManager in Current Protocols in Molecular Biology (eds. Ausubel FM et al. ) (John Wiley & Sons, Inc., 2010). doi: 10.1002/0471142727.mb1420s92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Fig 1b-1f, Fig 2c-2e, Fig 3a-3d, Fig 4a and 4c-4e, and Supplementary Fig 1, 2 and 4 are provided in Supplementary Table S2. Raw images were deposited to figshare and can be accessed at: https://figshare.com/articles/Dataset_associated_with_Multicolor_single-molecule_tracking_of_mRNA_interactions_with_RNP_granules_−1_2/7427816 and https://figshare.com/articles/Dataset_associated_with_Multicolor_single-molecule_tracking_of_mRNA_interactions_with_RNP_granules_-2_2/7427918. All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.