Abstract
Background:
Atherosclerosis of the aorta and coronary arteries is still one of the major causes of death. We recently reported obesity paradox between body mass index (BMI) and atherosclerosis of the aortas (AA) in morbidly obese decedent patients. The cause of this obesity paradox is unknown. The aim of the present study was to carry out genomic microarray analysis in order to determine gene expression profiles in the aortas of morbidly obese decedents with either mild or severe atherosclerosis of the aorta.
Methods:
Microarray studies using Affymetrix GeneChips Clariom D Human array chips were performed on the aortas obtained from six morbidly obese decedents, three of whom had minimal AA and three who had severe disease.
Results:
Group 1 (severe AA) and group 2 (mild AA) included 3 patients each. The patients were matched by age and BMI. There were significant (p<0.005) differences in the expressions of 1,067 genes between Groups 1 and 2, including 602 upregulated and 465 downregulated genes.
Conclusions:
Our data show significantly different gene signatures between morbidly obese decedents who have mild or severe AA, suggesting that genetic factors may be important contributors to the obesity paradox as it relates to aortic atherosclerosis. Further studies are warranted to define differences in protein expression in the aortas of these two groups to further elucidate the cause of this obesity paradox.
Recently we reported that there was a significant inverse relationship between body mass index (BMI) and atherosclerosis of the abdominal aortas of morbidly obese decedents with BMI ≥40 kg/m2 (1). Degoulet et al (2) first observed a decrease in mortality of 1,453 French patients with end stage renal disease who were undergoing dialysis between 1972 and 1978. Salahudeen (3) carried out a metaanalysis of 13 different studies involving 213,616 dialysis patients and reported better survival in those with high versus normal BMI. This obesity-survival paradox first was referred to as an “obesity paradox” by Gurm et al. (4), who reported fewer complications in patients with high versus normal or low BMI who underwent percutaneous coronary arterial revascularization. The basis for this obesity paradox is unknown but we recently suggested some possible explanations, including healthy versus unhealthy adiposity, hemodynamic factors (including endothelial cell shear stress) and genetic determinants that may significantly reduce the risk of developing aortic atherosclerosis in morbidly obese individuals (5). The aim of the present study was to determine by microarray analysis if there were significant differences in the gene expression profiles of aortas obtained from morbidly obese decedents with mild and severe aortic atherosclerosis.
Materials and Methods:
Microarray analysis was performed on the aortas obtained from morbidly obese decedents, three of whom had mild atherosclerosis and three who had severe disease.
A semiquantitative scale ranging from 0 to 3 to determine the severity of aortic atherosclerosis was used, as we described earlier (1). The semiquantitative grading scale had the following gradations: 0 was no atherosclerosis; 1 was mild atherosclerotic changes, consisting of raised fibrous plaques involving <25% of the aortic endothelial surface without calcification; 2 was moderate, when the plaques involved between 25% and 50% of the arterial surface area or scattered ulcerated, calcific plaques were present; and 3 was severe, when plaques involved >50% of the aortic endothelial surface with many ulcerated and extensively calcified areas (1).
Samples of the descending thoracic aortas, taken at the level of diaphragmatic penetration, were collected during autopsies and frozen at −80°C. A PureLink™ RNA Mini Kit (Thermo Fisher Scientific, Waltham, MA) was used to extract RNAs from the aortas based on the manufacturer’s protocol. Briefly, 100 mg of aortic wall, devoid of periaortic fibroadipose tissue, was homogenized and total RNA was extracted using the kit. A GeneChip® WT PICO Reagent Kit was used to generate ss-cDNA from the RNA. The quality of the samples was confirmed by the Nanodrop test (Thermo Fisher Scientific, Waltham, MA). For microarray analysis, GeneChips Clariom D Human array chips were used. Data files were generated and processed with Affymetrix software and analyzed with Transcriptome Analysis Console Software v. 3.0 (all from Thermo Fisher Scientific, Waltham, MA). ANOVA test was used to compare differences in gene expression between the two groups and pathway analysis was performed using Ingenuity Pathway Analysis (IPA) software (Qiagen Sciences Inc U.S., Germantown, MD). Student’s two-tailed t-test was used to compare differences in demographic data. Differences were considered significant with p<0.05.
Results.
Group 1 consisted of 3 decedents with severe atherosclerosis and Group 2 consisted of 3 decedents with mild disease. The decedents in Groups 1 and 2 were matched by age (60.7 ±12 years and 66.7 ±8.5 years, respectively, p=0.2600) and BMI (59.0 ±22 kg/m2 and 44.3 ±4.3 kg/m2, respectively, p=0.1686). There were 2:1 and 1:2 females:males in Groups 1 and 2, respectively, all of whom were Caucasian.
All decedents had risk factors for atherosclerosis. Thus, all six decedents had hypertension; five decedents (two in group 1 and three in group 2) had diabetes mellitus; three decedents (one in group 1 and two in group 2) had past history of smoking (no active smoking habit at the time of death was documented for any of the decedents).
The immediate cause of death, identified on the autopsy, included myocardial infarction (1 decedent in group 1), segmental pulmonary embolism that could lead to fatal arrhythmia (1 decedent in group 1). The other decedents did not have obvious immediate anatomic cause of death; therefore, it was attributed to a fatal cardiac arrhythmia. Two of those decedents (one on each group) had clinical presentation with sepsis.
Underlying cause of death included: atherosclerosis of the coronary arteries, chronic pyelonephritis and coagulation disorder in group 1; all decedents in group 2 had a malignancy that was attributed as a significant contributing factor in the underlying cause of death.
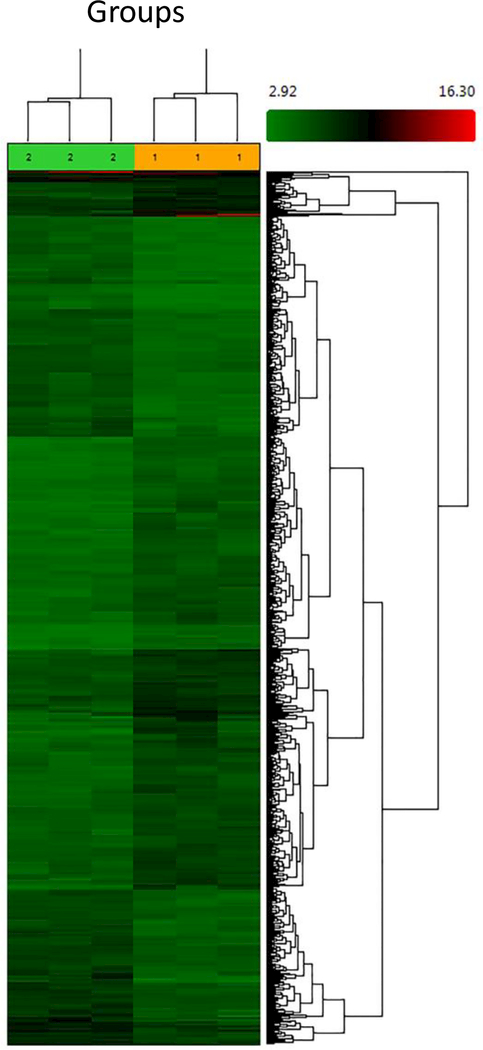
Microarray data showed significant (p<0.005) differential gene expression profiles (2X or greater) of 1067 genes between Groups 1 and 2, including 601 that were upregulated and 466 that were downregulated (Figure 1).
Figure 1. Heatmap and cluster analysis of total gene expressions in morbidly obese patients with severe and mild atherosclerosis of the aorta.
GeneChips Clariom D Human array chips were used to detect cDNA obtained from the aortas from morbidly obese decedent patients with severe (Group 1) and mild (Group 2) atherosclerosis of the aorta. The heatmap was generated by Transcriptome Analysis Console Software v. 3.0 software.
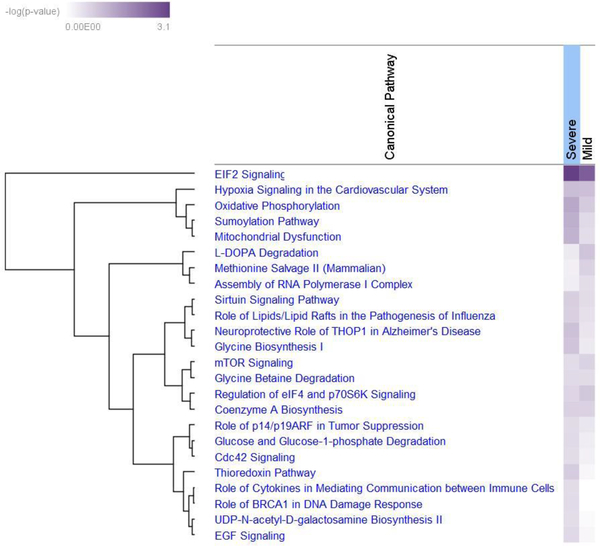
Among those genes, there were 102 were upregulated and 70 downregulated mRNAs, 11 upregulated and 10 downregulated small RNA genes, and 6 upregulated and 14 downregulated micro-RNA precursor genes. Pathway analysis showed several canonical pathways that were significantly different between Groups 1 and 2 (Figure 2). Among those were the eukaryotic initiation factor-2 (eIF2) and oxidative phosphorylation pathways, hypoxia signaling in the cardiovascular system, and the sumoylation and mitochondrial dysfunction pathways.
Figure 2. Heatmap and cluster analysis of canonical pathways in morbidly obese patients with severe and mild atherosclerosis of the aorta.
Ingenuity Pathway Analysis software was used to analyze different signaling pathways that are statistically associated with differentially expressed genes in the aortas from morbidly obese decedent patients with severe and mild atherosclerosis of the aorta.
eIF2 is a GTP-binding protein that escorts the initiation-specific form of met-tRNA onto the ribosome. Important functions of eIF2 include delivery of charged initiator methionyl-tRNA to the ribosome, as well as a role in identifying the translational start site. There were 40 and 38 upregulated genes associated with the eIF2 pathway in Groups 1 and 2, respectively.
The oxidative phosphorylation pathway consists of five protein-lipid enzyme complexes located in the mitochondrial inner membrane. In Groups 1 and 2 there were, respectively, 18 and 15 upregulated genes associated with the oxidative phosphorylation pathway.
The hypoxia pathway induces a group of physiologically important genes such as erythropoietin and vascular endothelial growth factor. These genes are transcriptionally up-regulated by hypoxia-inducible factor 1 (HIF-1) which is the primary effector in oxygen regulated gene expression. There were 13 upregulated genes associated with the hypoxia pathway in each of Groups 1 and 2.
The small ubiquitin like modifier (SUMO) conjugation pathway modifies numerous proteins that participate in diverse cellular processes, most commonly in the nucleus. Sumoylation is highly analogous to ubiquitination, using a sequence of E1, E2, and E3 enzymes. In Groups 1 and 2 there were, respectively, 17 and 13 upregulated genes associated with the sumoylation pathway.
Mitochondria are the primary consumers of cellular oxygen and contain a multitude of redox carriers that are capable of transferring single electrons to oxygen. This results in the formation of the reactive oxygen species (ROS) superoxide. Mitochondrial dysfunction occurs when the ROS-mediated oxidative stress overpowers the antioxidant defense system. There were 24 and 19 upregulated genes associated with the mitochondrial dysfunction pathway in Groups 1 and 2, respectively.
Discussion:
To the best of our knowledge, this is the first attempt to detect differences in gene expression that may be associated with the obesity paradox, in this case, as it specifically related to aortic atherosclerosis (1, 5). We performed microarray analysis of samples of thoracic aorta obtained from morbidly obese decedents with severe or mild atherosclerosis of the aorta. Our data revealed significantly different gene signatures between these two groups of individuals. There were 1,067 genes with greater than a twofold difference in expression between the two groups. Although the sample size was small, these differences were statistically significant.
Cardiovascular diseases, and especially atherosclerosis, are still one of the main causes of death worldwide (6). Atherosclerosis of the aorta can be a causative factor in the development of abdominal aortic aneurysms (AAA), which can lead to either rupture or dissection. As reported by Tzani et al., the 30-day all-cause mortality rate in patients who had a repair of AAA was 1.7% for patients with non-ruptured AAA and as high as 33.8% for those with ruptured AAA (7). Recently Gäbel et al reported that the “molecular fingerprint” of the aortas of patients with AAA had two gene sets, one containing 5 genes linked to terminal progression and a second set containing 5 genes exclusively upregulated in ruptured AAA. Several pathways were discovered, including HIF-1α signaling (8), ALOX5 (9), MSN, PSMB10, and STIM1 (10), cytokine-cytokine receptor interaction pathway, chemokine signaling pathway, and antigen processing and presentation pathway (11), calcium signaling, development and differentiation, and cell adhesion (12). These studies primarily were carried out in two regions, one of the aneurysm itself and the other in an uninvolved part of the aorta. This could result in a bias because different pathways might be activated in the area of active expanding aneurysm, including inflammation and tissue repair. Our data were obtained in the same region of the aorta of all of the decedents, thus minimizing the sampling error. Furthermore, we evaluated aortas from different decedents so our data might detect common pathways that participate in the pathogenesis of atherosclerosis rather than changes associated with the location. The decedents in our study were of similar age and BMI, which also could reduce age- and BMI-associated errors.
Other reports have described differences in gene expression associated with atherosclerosis of the aorta in rodents, primarily in mice. Microarray data showed 22 dysregulated pathways that were common to both adipose tissue and aorta in obese LDL-receptor knockout mice (13). There were 22 dysregulated pathways common to both tissues, with p values <0.05 and selected inflammatory response and oxidative phosphorylation pathways. Analysis revealed great overlap in gene expression alterations in obese adipose tissue and atherosclerosis, provide evidence for common pathogenic pathways for obesity-driven insulin resistance and atherogenesis. Using laser microdissection, Yin et al (14) have generated transcript atlases of gene expression in atherosclerotic plaques, aortic media, adventitia, and adjacent aorta-draining lymph nodes (LN) in ApoE knockout and wild-type mice. Similar studies were performed by Beer et al. (15). Our data were generated from humans and therefore provide information that is more relevant to the clinical practice than animal data.
All decedents had a comorbidity that could affect the progression of atherosclerosis. However, such comorbidities were present in both groups equally. Thus, history of hypertension was noted in all six decedents, diabetes mellitus was documented in two decedents with mild atherosclerosis (group 2) and in one patient with severe atherosclerosis (group 1). Of note, all these decedents had metabolic syndrome associated with morbid obesity; therefore, some may have had undiagnosed diabetes mellitus. None of the decedents had smoking habit at the time of death, but 2 decedents from group 2 (mild atherosclerosis) and one decedent from group 1 (severe atherosclerosis) were smokers in the past. Therefore, we believe that these factors did not affect the microarray data. Similarly, in all decendents the immediate cause of death was related to cardiovascular disease (either acute myocardial infarction or fatal cardiac arrhythmia, because no other anatomic causes of death were identified during the autopsy). Therefore, we believe that our microarray data were not affected by these acute processes as well.
There are several limitations in the present study. First, we have only three decedents in each group. Second, we only have microarray data and this will require further confirmation by PCR and proteomic analysis. Nevertheless, we already have identified a number of genes that show significant changes even in this small sample of the aortas. It is possible that by increasing the sample size other genes and associated pathways might be identified that also show significant changes. Third, additional studies are warranted to detect gender- and race-associated changes in gene expression. Fourth, our data were generated from morbidly obese decedents and there may be different mechanisms of atherogenesis than those in normal weight decedents. Fifth, we used autopsy tissue with varying intervals of time between death and sample procurement, with varying degrees of autolysis and degradation of RNA. Therefore, further studies should be carried out from tissues obtained from surgical specimens.
These caveats notwithstanding, our data provide a roadmap for future studies to define genomic differences between obese and non-obese decedents. Although we focused on morbidly obese decedents, similar mechanisms could be important in the pathogenesis of aortic atherosclerosis in general, and more specifically to define genomic differences associated with athoergenesis in obese and non-obese individuals.
Acknowledgements:
This study was supported by the OSU Center of Clinical and Translation Science voucher #5094 (supported by CTSA Grant UL1TR001070) and the Department of Pathology Start-Up Fund to SVB. We thank the Genomics Shared Resource at The Ohio State University Comprehensive Cancer Center (OSU CCC), Columbus, OH for conducting the Affymetrix gene expression analyses. This works was supported in part by the OSU CCC and the National Institutes of Health grant P30 CA16058. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest: None
Literature cited:
- 1.Brodsky SV, Barth RF, Mo X, Yildiz V, Allenby P, Ivanov I, et al. An obesity paradox: an inverse correlation between body mass index and atherosclerosis of the aorta. Cardiovascular pathology : the official journal of the Society for Cardiovascular Pathology. 2016;25(6):515–20. Epub 2016/09/30. doi: 10.1016/j.carpath.2016.09.002. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 2.Degoulet P, Legrain M, Reach I, Aime F, Devries C, Rojas P, et al. Mortality risk factors in patients treated by chronic hemodialysis. Report of the Diaphane collaborative study. Nephron. 1982;31(2):103–10. Epub 1982/01/01. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 3.Salahudeen AK. Obesity and survival on dialysis. American journal of kidney diseases : the official journal of the National Kidney Foundation. 2003;41(5):925–32. Epub 2003/05/02. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 4.Gurm HS, Brennan DM, Booth J, Tcheng JE, Lincoff AM, Topol EJ. Impact of body mass index on outcome after percutaneous coronary intervention (the obesity paradox). Am J Cardiol. 2002;90(1):42–5. Epub 2002/06/29. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 5.Barth RF, Maximilian Buja L, Cao L, Brodsky SV. An Obesity Paradox: Increased Body Mass Index Is Associated with Decreased Aortic Atherosclerosis. Current hypertension reports. 2017;19(7):55 Epub 2017/06/09. doi: 10.1007/s11906-017-0753-y. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 6.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–128. doi: 10.1016/S0140-6736(12)61728-0. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzani A, Doulamis IP, Katsaros I, Martinou E, Schizas D, Economopoulos KP. Mortality after endovascular treatment of infrarenal abdominal aortic aneurysms - the newer the better? VASA Zeitschrift fur Gefasskrankheiten. 2018:1–10. Epub 2018/01/16. doi: 10.1024/0301-1526/a000685. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 8.Gabel G, Northoff BH, Weinzierl I, Ludwig S, Hinterseher I, Wilfert W, et al. Molecular Fingerprint for Terminal Abdominal Aortic Aneurysm Disease. Journal of the American Heart Association. 2017;6(12). Epub 2017/12/02. doi: 10.1161/JAHA.117.006798. PubMed PMID: ; PubMed Central PMCID: PMC5779007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang YG, Li MX, Kou L, Zhou Y, Qin YW, Liu XJ, et al. Long Noncoding RNA Expression Signatures of Abdominal Aortic Aneurysm Revealed by Microarray. Biomedical and environmental sciences : BES. 2016;29(10):713–23. Epub 2016/12/09. doi: 10.3967/bes2016.096. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 10.Butt HZ, Sylvius N, Salem MK, Wild JB, Dattani N, Sayers RD, et al. Microarray-based Gene Expression Profiling of Abdominal Aortic Aneurysm. European journal of vascular and endovascular surgery : the official journal of the European Society for Vascular Surgery. 2016;52(1):47–55. Epub 2016/05/10. doi: 10.1016/j.ejvs.2016.03.016. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 11.Biros E, Moran CS, Rush CM, Gabel G, Schreurs C, Lindeman JH, et al. Differential gene expression in the proximal neck of human abdominal aortic aneurysm. Atherosclerosis. 2014;233(1):211–8. Epub 2014/02/18. doi: 10.1016/j.atherosclerosis.2013.12.017. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 12.Hinterseher I, Erdman R, Elmore JR, Stahl E, Pahl MC, Derr K, et al. Novel pathways in the pathobiology of human abdominal aortic aneurysms. Pathobiology : journal of immunopathology, molecular and cellular biology. 2013;80(1):1–10. Epub 2012/07/17. doi: 10.1159/000339303. PubMed PMID: ; PubMed Central PMCID: PMC3782105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreno-Viedma V, Amor M, Sarabi A, Bilban M, Staffler G, Zeyda M, et al. Common dysregulated pathways in obese adipose tissue and atherosclerosis. Cardiovascular diabetology. 2016;15(1):120 Epub 2016/08/27. doi: 10.1186/s12933-016-0441-2. PubMed PMID: ; PubMed Central PMCID: PMC5000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin C, Mohanta S, Ma Z, Weber C, Hu D, Weih F, et al. Generation of Aorta Transcript Atlases of Wild-Type and Apolipoprotein E-null Mice by Laser Capture Microdissection-Based mRNA Expression Microarrays. Methods in molecular biology. 2015;1339:297–308. Epub 2015/10/09. doi: 10.1007/978-1-4939-2929-0_20. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 15.Beer M, Doepping S, Hildner M, Weber G, Grabner R, Hu D, et al. Laser-capture microdissection of hyperlipidemic/ApoE(−)/(−) mouse aorta atherosclerosis. Methods in molecular biology. 2011;755:417–28. Epub 2011/07/16. doi: 10.1007/978-1-61779-163-5_35. PubMed PMID: . [DOI] [PubMed] [Google Scholar]