Abstract
Metagenomic sequencing of bacterial samples has become the gold standard for profiling microbial populations, but 16S rRNA profiling remains widely used due to advantages in sample throughput, cost, and sensitivity even though the approach is hampered by primer bias and lack of specificity. We hypothesized that a hybrid approach, that combined targeted PCR amplification with high-throughput sequencing of multiple regions of the genome, would capture many of the advantages of both approaches. We developed a method that identifies and quantifies members of bacterial communities through simultaneous analysis of multiple variable regions of the bacterial 16S rRNA gene. The method combines high-throughput microfluidics for PCR amplification, short read DNA sequencing, and a custom algorithm named MVRSION (Multiple 16S Variable Region Species-Level IdentificatiON) for optimizing taxonomic assignment. MVRSION performance was compared to single variable region analyses (V3 or V4) of five synthetic mixtures of human gut bacterial strains using existing software (QIIME), and the results of community profiling by shotgun sequencing (COPRO-Seq) of fecal DNA samples collected from gnotobiotic mice colonized with a defined, phylogenetically diverse consortium of human gut bacterial strains. Positive predictive values for MVSION ranged from 65%−91% versus 44%−61% for single region QIIME analyses (p<0.01, p<0.001), while the abundance estimate r2 for MVRSION compared to COPRO-Seq was 0.77 vs. 0.46 and 0.45 for V3-QIIME and V4-QIIME, respectively. MVRSION represents a generally applicable tool for taxonomic classification that is superior to singleregion 16S rRNA methods, resource efficient, highly scalable for assessing the microbial composition of up to thousands of samples concurrently, with multiple applications ranging from whole community profiling to targeted tracking of organisms of interest in diverse habitats as a function of specified variables/perturbations.
Keywords: Microbial community analysis, Microbial diversity, Next generation sequencing, 16S rRNA gene
1. Introduction
Shotgun sequencing of total community DNA and sequencing of amplicons generated from SSU rRNA genes (notably bacterial 16S rRNA), are widely used to profile microbial populations; the former method captures gene content and allows species and strain-level resolution while the latter method remains popular due to low cost, high sample throughput, sensitivity, and fewer sample limitations (e.g. cases of host DNA contamination). However, amplicon sequencing is confounded by primer bias and challenges related to the specificity/resolution that can be achieved for taxonomic assignments (Liu, DeSantis, Andersen, & Knight, 2008; Vetrovsky & Baldrian, 2013; Yang, Wang, & Qian, 2016)
Current 16S rRNA profiling methods commonly target only one of the gene’s nine variable regions; amplicons produced by PCR from a given region or a portion of that region are then sequenced. One solution to the problems of primer biases and taxonomic resolution is to amplify and sequence multiple amplicons generated from multiple variable regions of a given taxon’s 16S rRNA gene(s).
Producing sequence data from multiple variable regions is technically trivial; however, analysis of such data represents a significant challenge. Existing taxonomic classifiers such as the UCLUST (Edgar, 2010) consensus taxonomy assigner in QIIME (Caporaso et al., 2010), the RDP Classifier (Wang, Garrity, Tiedje, & Cole, 2007), the k-Nearest Neighbor algorithm in mothur (Schloss et al., 2009), and SPINGO (Allard, Ryan, Jeffery, & Claesson, 2015) are designed to assign taxonomy to each variable region individually rather than integrating information from multiple amplicons.
2. Results
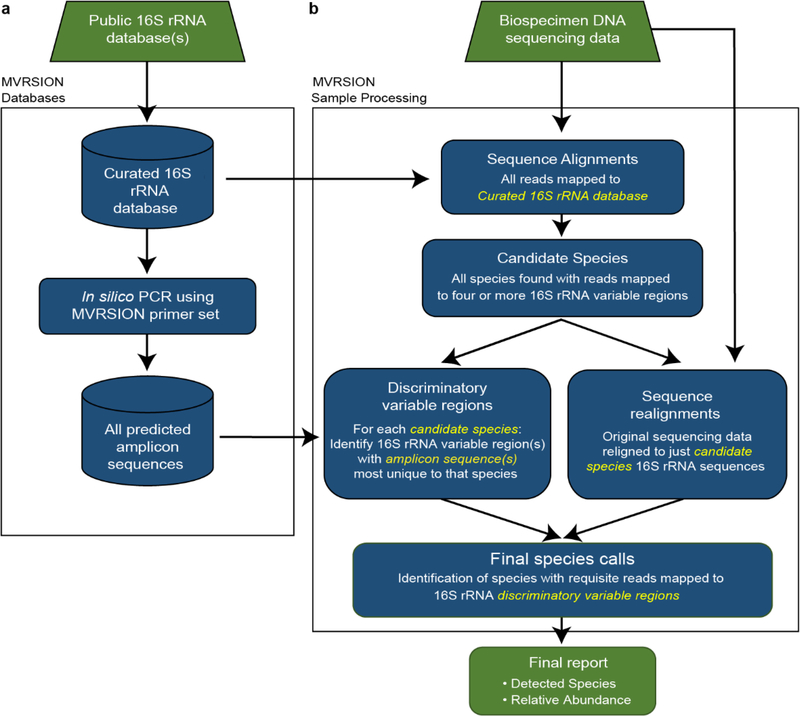
To achieve an improvement in specificity while maintaining the efficiency of an amplicon-based sequencing approach, MVRSION was designed with the ability to utilize, in parallel, multiple 16S rRNA variable regions in the absence of information linking amplicons from different regions together. A multistep filtering strategy is employed to first reduce the complexity of the analysis followed by dynamic “discriminatory variable region” selection, a process that utilizes information from the initial alignment regarding which bacterial species may be present, combined with an assessment of which variable regions contain the necessary information to correctly distinguish species with high 16S rRNA sequence identity. Figure 1 provides an overview of the process; a complete description of the analytical method is presented in Section 5.5.
Figure 1. Overview of the MVRSION method for identifying microbial species.
There are two major components to the MVRSION algorithm. (a) Two MVRSION databases are compiled for use with all sequencing datasets. A curated database contains accurately annotated (bacterial species-level), full length 16S rRNA sequences from public sources (SILVA). Using this curated 16S rRNA database and known amplicon primer sequences, in silico PCR predicts a list of all amplicon sequences across the nine 16S rRNA variable regions of all bacterial species in the curated database. These databases are subsequently used for processing all input datasets. (b) For sample processing, amplicon sequencing reads are mapped to the 16S rRNA sequences in the curated database. A list of candidate species is generated from all species with reads mapping to four or more variable regions. For each candidate species, the predicted amplicons are compared to all other candidate species. From the predicted amplicon comparison, variable region(s) with sequences most unique to that candidate species versus all other candidates are selected as its “discriminatory variable regions”. In parallel, the original input reads are re-aligned to just the candidate species, as all other species from the curated 16S rRNA database have been eliminated from consideration. If a requisite number of reads have been mapped to a candidate species discriminatory variable region(s) from this realignment, the species is called present. For all species called present, the abundance is estimated as described in Section 5.5.5.
The initial MVRSION amplicon panel consisted of 14 primer pairs targeting the nine variable regions in 16S rRNA (Table 1). In silico analysis with PrimerProspector (Quast et al., 2013) indicated that seven of the primer pairs (V1, V3_1, V5_1, V5_2, V6_2, V6_3, V9) would amplify very few species present in the SILVA rRNA database (Table 2).
Table 1. Primer Sequences and Sources.
Primer sequences associated with the 14 PCR amplicons covering 9 variable regions tested for the MVRSION method.
| Bacterial 16S rRNA Variable Region Amplicon Primers | ||||||
|---|---|---|---|---|---|---|
| Region | Amplicon Size (bp) | Name | Primer Sequence | Name | Sequence | Reference PMID |
| V1-V2 | 300 | V1-V2 F | AGAGTTTGATCCTGGCTCAG | V1-V2 R | TGCTGCCTCCCGTAGGAGT | 19892944, 18264105, 22179717 |
| V1 | 113 | V1 F | AGAGTTTGATCMTGGCTCAG | V1 R | TTACTCACCCGTICGCCRCT | 18047683 |
| V2 | 261 | V2 F | AGYGGCGIACGGGTGAGTAA | V2 R | CYIACTGCTGCCTCCCGTAG | 18047683 |
| V3 | 200 | V3_1 F | ACTYCTACGGRAGGCWGC | V3_1 R | GTGCCAGCMGCCGCGGTAA | 23579286, 18047683 |
| V3 | 170 | V3_2 F | CCTACGGGAGGCAGCAG | V3_2 R | GTATTACCGCGGCTGCTGG | 22853944, 21460107 |
| V4 | 250 | V4 F | GTGCCAGCMGCCGCGGTAA | V4 R | GGACTACHVGGGTWTCTAAT | 22179717, 20534432 |
| V5 | 140 | V5_1 F | ATTAGATACCYTGTAGTCC | V5_1 R | CCGTCAATTCMTTTGAGTTT | 18047683 |
| V5 | 100 | V5_2 F | AGGATTAGATACCCT | V5_2 R | CRTACTHCHCAGGYG | 22853944 |
| V5-V6 | 280 | V5-V6 F | AGGATTAGATACCCTGGTA | V5-V6 R | CRRCACGAGCTGACGAC | 23791918, 18665274 |
| V6 | 167 | V6_1 F | AAACTCAAAKGAATTGACGG | V6_1 R | ACGAGCTGACGACARCCATG | 18047683 |
| V6 | 100 | V6_2 F | CNACGCGAAGAACCTTANC | V6_2 R | CGACAGCCATGCANCACCT | 20962877 |
| V6 | 76 | V6_3 F | CAACGCGARGAACCTTACC | V6_3 R | ACAACACGAGCTGACGAC | 20711427 |
| V7-V8 | 300 | V7-V8 F | GYAACGAGCGCAACCC | V7-V8 R | GACGGGCGGTGWGTRC | 20880993 |
| V9 | 116 | V9F | GTACACACCGCCCGT | V9 R | TACCTTGTTACGACTT | 18047683 |
Table 2. Amplicon Coverage and Model Utilization.
Amplicon sequencing data were evaluated using the 20 taxa HM-782D and 48 taxa 48G-Eq samples. Several of the amplicons where predicted, by in silico PCR, to have poor amplification for the species present in the SILVA 16S rRNA database. A species is ‘Predicted to amplify’ by a primer pair if the primers amplify 80% or more of the 16S rRNA sequences belonging to the species in the database. Predictions of poor amplification were empirically confirmed by the negligible number of species with a significant number of mapped reads (> 1% of total) when these two mock community samples were sequenced.
| Variable Region Amplicon | % SILVA Species Predicted to Amplify | HM-782D (20-taxa consortium) |
48G-Eq (48 taxa consortium; even) |
||
|---|---|---|---|---|---|
| % Total Reads | % Species >1% Total Reads | % Total Reads | % Species >1% Total Reads | ||
| V1 | 0.2% | 5.9% | 0 | 3.3% | 0 |
| V1-V2 | 37.9% | 4.6% | 24 | 2.4% | 21 |
| V2 | 91.3% | 7.6% | 28 | 9.7% | 21 |
| V3_1 | 0.01% | 9.7% | 1 | 0.5% | 2 |
| V3_2 | 98.3% | 10.6% | 31 | 14.6% | 23 |
| V4 | 97.7% | 7.9% | 32 | 7.1% | 23 |
| V5-V6 | 96.6% | 7.1% | 27 | 6.3% | 21 |
| V5_1 | 73.8% | 5.6% | 0 | 1.7% | 0 |
| V5_2 | 0.1% | 5.5% | 1 | 4.6% | 0 |
| V6_1 | 98.6% | 6.4% | 27 | 14.2% | 21 |
| V6_2 | 0.04% | 5.30% | 0 | 7.6% | 0 |
| V6_3 | 0.04% | 5.8% | 0 | 8.3% | 0 |
| V7-V8 | 94.0% | 9.0% | 30 | 9.1% | 22 |
| V9 | 0.1% | 7.1% | 0 | 5.8% | 0 |
Follow-up in vitro testing of this set of primers was performed using DNA isolated from two mixtures of human-associated bacterial taxa: HM-782D, which contains equivalent amounts of 20 species (BEI Resources, ATCC, Manassas, VA) and 48G-Eq, which contains equivalent amounts of 48 strains representing 45 species (Faith et al., 2013) (Table 3). These samples were each sequenced to a depth of ~400,000 reads (2×150 nt paired-end; Table 4). Reads associated with all 14 amplicons were observed (range, 0.5% to 14.6% of total reads/sample). The results verified the in silico prediction as very few species received a significant number (>1%) of the total reads with the seven primer pairs described above (Table 2). Consequently, amplicons produced with these primers were removed leaving amplicons corresponding to eight variable regions (V1, V2, V3, V4, V5, V6, V7, and V8) for the MVRSION method.
Table 3. Sample Species Compositions.
Species information for each of the tested samples. HM-782D (20 species), and all 48G (45 species), samples have known DNA quantities added for each of the species. The species composition of fecal microbiota samples recovered from gnotobiotic mice was determined by short read shotgun sequencing of community DNA and mapping the results reads to the genomes of members of the consortium of cultured human gut bacterial strains that had been introduced into the animals. The COPRO-Seq percent relative abundance of each species is given as the mean for all 92 gnotobiotic samples plus or minus the standard deviation.
| HM-782D (20-taxa consortium) | 48G 45-taxa consortium | Gnotobiotic Mice 68-taxa consortium | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Known Species | Known Species | Stg1 Rel. % | Stg2 Rel. % | Stg3 Rel. % | Known Species | COPRO-Seq Rel. % | |||||||
| Acinetobacter baumannii | Akkermansia muciniphila | 4.2 | 0.3 | 4.2 | Akkermansia muciniphila | 3.5E+01 ± 6.8E+00 | |||||||
| Actinomyces odontolyticus | Alistipes indistinctus | 0.1 | 0.5 | 4.2 | Anaerotruncus colihominis | 2.7E-01 ± 1.3E-01 | |||||||
| Bacillus cereus | Anaerococcus hydrogenalis | 0.3 | 4.2 | 8.4 | Bacteroides WH2 | 7.9E+00 ± 2.4E+00 | |||||||
| Bacteroides vulgatus | Anaerotruncus colihominis | 8.4 | 1.1 | 0.1 | Bacteroides caccae | 1.6E+01 ± 2.7E+00 | |||||||
| Clostridium beijerinckii | Bacteroides cellulosilyticus | 0.3 | 0.5 | 0.1 | Bacteroides cellulosilyticus | 3.0E-01 ± 6.4E-01 | |||||||
| Deinococcus radiodurans | Bacteroides dorei | 8.4 | 2.1 | 1.1 | Bacteroides coprophilus | 4.2E-04 ± 6.0E-04 | |||||||
| Enterococcus faecalis | Bacteroides eggerthii | 1.1 | 8.4 | 0.3 | Bacteroides dorei | 5.3E+00 ± 4.9E+00 | |||||||
| Escherichia coli | Bacteroides finegoldii | 0.1 | 0.1 | 0.5 | Bacteroides eggerthii | 1.8E+00 ± 8.3E-01 | |||||||
| Helicobacter pylori | Bacteroides intestinalis | 1.1 | 4.2 | 1.1 | Bacteroides finegoldii | 1.5E-01 ± 1.0E-01 | |||||||
| Lactobacillus gasseri | Bacteroides ovatus | 8.4 | 1.1 | 0.1 | Bacteroides intestinalis | 1.2E-01 ± 1.4E-01 | |||||||
| Listeria monocytogenes | Bact. thetaiotaomicron | 1.1 | 17.3 | 5.4 | Bacteroides ovatus | 1.5E-01 ± 1.7E-01 | |||||||
| Neisseria meningitidis | Bacteroides uniformis | 2.1 | 0.3 | 4.2 | Bacteroides plebeius | 6.1E-04 ± 6.6E-04 | |||||||
| Propionibacterium acnes | Bacteroides vulgatus | 1.1 | 0.1 | 0.3 | Bact, thetaiotaomicron | 6.9E+00 ± 1.3E+00 | |||||||
| Pseudomonas aeruginosa | Bacteroides xylanisolvens | 0.1 | 2.1 | 0.5 | Bacteroides uniformis | 8.3E-01 ± 2.3E-01 | |||||||
| Rhodobacter sphaeroides | Bifidobacterium bifidum | 0.3 | 0.3 | 2.1 | Bacteroides vulgatus | 4.3E+00 ± 2.6E+00 | |||||||
| Staphylococcus aureus | Bifid. pseudocatenulatum | 0.1 | 4.2 | 8.4 | Bacteroides xylanisolvens | 1.2E-01 ± 5.8E-02 | |||||||
| Staph. epidermidis | Blautia hansenii | 8.4 | 0.3 | 0.1 | Bifidobacterium adolescentis | 1.7E-02 ± 7.9E-03 | |||||||
| Streptococcus agalactiae | Blautia luti | 2.1 | 1.1 | 2.1 | Bifidobacterium bifidum | 1.5E-05 ± 1.0E-04 | |||||||
| Streptococcus mutans | Clostridium asparagiforme | 0.1 | 0.5 | 8.4 | Bifid. pseudocatenulatum | 3.6E-04 ± 8.2E-04 | |||||||
| Strep. pneumoniae | Clostridium hathewayi | 0.3 | 0.3 | 0.1 | Blautia hansenii | 6.7E-01 ± 2.3E-01 | |||||||
| Clostridium leptum | 0.5 | 0.1 | 2.1 | Blautia hydrogenotrophica | 9.8E-06 ± 9.6E-05 | ||||||||
| Clostridium nexile | 4.7 | 2.2 | 0.5 | Blautia luti | 2.5E-04 ± 5.2E-04 | ||||||||
| Clost. saccharolyticum | 8.4 | 0.1 | 4.2 | Citrobacter youngae | 4.9E-02 ± 5.8E-02 | ||||||||
| Clostridium sporogenes | 2.1 | 2.1 | 0.5 | Clostridium asparagiforme | 1.2E-01 ± 6.0E-02 | ||||||||
| Collinsella intestinalis | 1.1 | 2.1 | 1.1 | Clostridium bartlettii | 5.7E-06 ± 5.6E-05 | ||||||||
| Coprococcus comes | 4.2 | 1.1 | 0.5 | Clostridium bolteae | 1.1E+00 ± 2.6E-01 | ||||||||
| Dorea formicigenerans | 4.2 | 1.1 | 0.1 | Clostridium hathewayi | 3.6E+00 ± 1.3E+00 | ||||||||
| Dorea longicatena | 0.1 | 0.5 | 8.4 | Clostridium hylemonae | 5.2E-02 ± 7.1E-02 | ||||||||
| Edwardsiella tarda | 0.1 | 8.4 | 0.1 | Clostridium leptum | 3.0E-02 ± 1.3E-02 | ||||||||
| Enterobacter cancerogenus | 1.1 | 0.1 | 0.5 | Clostridium nexile | 2.0E-01 ± 1.6E-01 | ||||||||
| Escherichia coli | 0.1 | 0.1 | 8.4 | Clostridium ramosum | 5.6E-01 ± 2.6E-01 | ||||||||
| Escherichia fergusonii | 1.1 | 0.5 | 0.1 | Clostridium scindens | 4.2E-01 ± 1.4E-01 | ||||||||
| Eubacterium biforme | 2.1 | 8.4 | 2.1 | Clostridium spM62 1 | 4.5E-05 ± 2.0E-04 | ||||||||
| Eubacterium eligens | 0.3 | 4.2 | 0.5 | Clostridium sporogenes | 1.1E-02 ± 9.6E-03 | ||||||||
| Eubacterium ventriosum | 0.3 | 0.1 | 2.1 | Clostridium symbiosum | 5.1E-01 ± 1.6E-01 | ||||||||
| Faecalibacterium prausnitzii | 4.2 | 0.1 | 0.3 | Collinsella aerofaciens | 9.6E-01 ± 6.6E-01 | ||||||||
| Parabacteroides johnsonii | 0.1 | 0.1 | 8.4 | Collinsella intestinalis | 4.3E-01 ± 2.7E-01 | ||||||||
| Proteus penneri | 4.2 | 0.1 | 0.1 | Collinsella stercoris | 8.6E-01 ± 6.2E-01 | ||||||||
| Providencia alcalifaciens | 2.1 | 0.1 | 4.2 | Coprococcus comes | 2.9E-02 ± 2.4E-02 | ||||||||
| Roseburia intestinalis | 8.4 | 1.1 | 0.3 | Desulfovibrio piger | 3.1E+00 ± 7.6E-01 | ||||||||
| Ruminococcus gnavus | 0.5 | 4.2 | 1.1 | Dorea formicigenerans | 2.5E-03 ± 5.7E-03 | ||||||||
| Ruminococcus lactaris | 0.5 | 4.2 | 0.1 | Dorea longicatena | 2.4E-04 ± 5.2E-04 | ||||||||
| Ruminococcus torques | 2.1 | 2.1 | 1.1 | Edwardsiella tarda | 2.5E-01 ± 2.0E-01 | ||||||||
| Streptococcus infantarius | 0.1 | 0.3 | 0.1 | Enterobacter cancerogenus | 3.8E-02 ± 9.0E-02 | ||||||||
| Subdoligranulum variabile | 0.1 | 8.4 | 2.1 | Escherichia coli | 1.6E-03 ± 5.8E-03 | ||||||||
| Escherichia fergusonii | 2.7E-01 ± 2.5E-01 | ||||||||||||
| Eubacterium biforme | 7.0E-04 ± 9.7E-04 | ||||||||||||
| Eubacterium cylindroides | 2.1E-03 ± 1.8E-03 | ||||||||||||
| Eubacterium dolichum | 3.5E-05 ± 1.6E-04 | ||||||||||||
| Eubacterium hallii | 2.0E-04 ± 4.6E-04 | ||||||||||||
| Eubacterium rectale | 2.2E-05 ± 1.3E-04 | ||||||||||||
| Faecalibacterium prausnitzii | 4.1E-05 ± 1.9E-04 | ||||||||||||
| Flavonifractor plautii | 1.1E-03 ± 1.2E-03 | ||||||||||||
| Fusobacterium varium | 1.7E+00 ± 6.4E-01 | ||||||||||||
| Holdemania filiformis | 3.6E-01 ± 1.0E-01 | ||||||||||||
| Lactobacillus ruminis | 4.6E-04 ± 1.2E-03 | ||||||||||||
| Marvin, formatexigens | 9.1E-06 ± 8.9E-05 | ||||||||||||
| Megamonas funiformis | 9.2E-02 ± 7.3E-02 | ||||||||||||
| Mitsuokella multacida | 1.3E-04 ± 6.6E-04 | ||||||||||||
| Parabacteroides distasonis | 2.2E-02 ± 5.2E-03 | ||||||||||||
| Parabacteroides johnsonii | 3.8E+00 ± 1.2E+00 | ||||||||||||
| Parabacteroides merdae | 3.4E-01 ± 4.2E-01 | ||||||||||||
| Proteus penneri | 2.7E-02 ± 4.9E-02 | ||||||||||||
| Providencia stuartii | 1.9E-04 ± 4.3E-04 | ||||||||||||
| Ruminococcus gnavus | 4.7E-01 ± 2.1E-01 | ||||||||||||
| Ruminococcus lactaris | 7.5E-05 ± 2.4E-04 | ||||||||||||
| Ruminococcus torques | 7.2E-01 ± 8.5E-01 | ||||||||||||
| Subdoligranulum variabile | 4.0E-01 ± 1.2E-01 | ||||||||||||
Table 4.
Sequencing read information
| Sequencing Run | Samples | Total Reads (2×150 nts) | Ave. Reads/Sample |
|---|---|---|---|
| MiSeq Run1 | HM-782D, 48G, 44 fecal samples from gnotobiotic mice | 20,174,270 | 411,720 |
| MiSeq Run2 | 48 fecal samples from gnotobiotic mice | 16,415,978 | 342,000 |
| HiSeq Run1 | 48 fecal samples from gnotobiotic mice (same samples characterized in MiSeq Run2) | 200,411,162 | 4,175,233 |
For the purposes of performance evaluation, MVRSION was compared to single variable region (V3 or V4) QIIME analyses of the test samples. This evaluation used comparative measures of calculated True Positives (TP), False Positives (FP), False Negatives (FN), Positive Predictive Value (PPV), and Sensitivity. Detailed descriptions of the comparative measures are presented in Section 5.7. MVRSION thresholds used for the analyses are presented Section 5.5 and the QIIME commands are in Section 5.6.
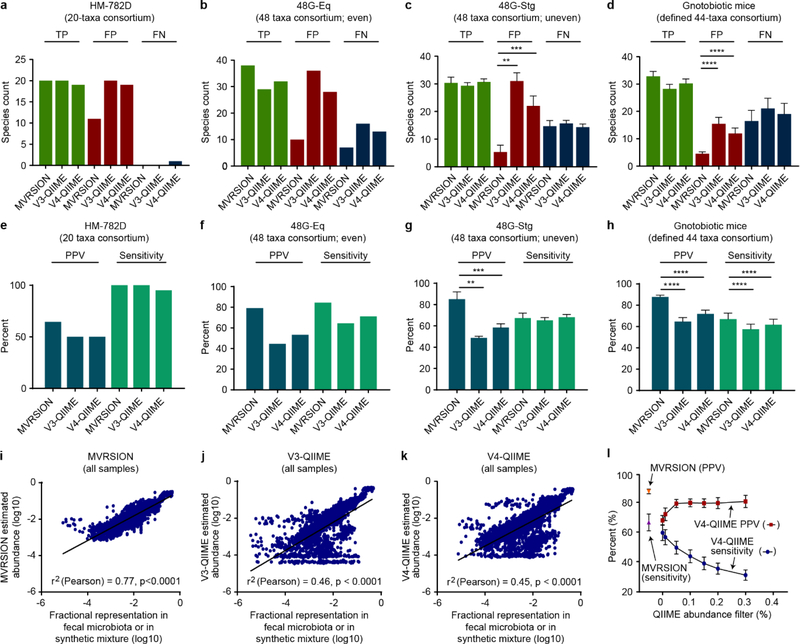
In the initial comparison utilizing HM-782D, MVRSION and the V3-QIIME analyses identified all 20 bacterial species in the sample (100% sensitivity), while V4-QIIME identified 19 (95% sensitivity) (Figure 2a). However, both the V3-QIIME and V4-QIIME analyses produced nearly double the number of false positives (20 and 19, versus 11 for MVRSION), resulting in Positive Predictive Values (PPVs) for V3-QIIME and V4-QIIME of 50% versus 65% for MVRSION (Figures 2e). Similarly, for the 48GEq sample, MVRSION demonstrated superiority in both PPV (79%) and sensitivity (84%) compared to V3-QIIME (45%, 64%) and V4-QIIME (53%, 71%) (Figures 2b,f).
Figure 2. MVRSION and single 16S rRNA variable region QIIME comparative analyses.
(a-d) Species-level assessments of the comparative measures True Positives (TP), False Positives (FP), and False Negatives (FN) as computed for the three analytical methods (MVRSION, V3-QIIME, and V4- QIIME) utilizing the synthetic mixtures (HM-782D, 48G-Eq, combined 48G-Stg1–3), and 92 fecal samples from gnotobiotic mice, respectively. (e-h) Calculated Positive Predictive Values (PPV) and Sensitivity (Sens) for the three analytical methods and samples. Statistical comparisons are significantly improved for MVRSION compared to single variable region analyses with QIIME, **, P<0.01; ***, P<0.001; ****, P<0.0001 (two-tailed unpaired t-test, equal variance). (i-k) MVRSION, V3-QIIME, and V4-QIIME estimated relative abundance for all taxa identified in all samples, compared to their relative abundance based on known DNA concentrations in synthetic mixtures or COPRO-Seq analysis of DNAs prepared from fecal samples obtained from gnotobiotic mice harboring a defined model human gut microbiota. While all correlations are significant (P<0.0001), MVRSION demonstrates a markedly higher r2 value (0.77) compared to V3-QIIME (0.46) and V4-QIIME (0.45). (l) V4-QIIME analysis was run at multiple levels of abundance filtering, as described in Section 5.6, to illustrate the optimization of MVRSION for both sensitivity and specificity.
Microbial community samples rarely contain equivalent mixtures of all members. Therefore, three synthetic DNA mixtures of known uneven composition were tested; 48G-Stg1, 48G-Stg2, and 48G-Stg3 contain genomic DNA from the same strains as 48G-Eq but at relative abundances varying over a 1000-fold range (Table 3). Analysis of these uneven mixtures indicated equivalent sensitivity for MRVSION and the two QIIME single-region analyses, while MVRSION was significantly better in its ability to correctly identify component species [PPV of 85±6.9% (mean ± SD) compared to 49±1.6% for V3-QIIME (p<0.001, two-tailed unpaired t-test, equal variance) and 58±3.5% for V4-QIIME (p<0.01); see Figure 2c,g for results combined from all three uneven communities].
To further benchmark MVRSION performance, 92 fecal DNA samples were profiled from gnotobiotic mice harboring a consortium of 68 sequenced members of the human gut microbiota. These mice had been subjected to a series of diet oscillations where the representation of various micronutrients was intentionally manipulated (Hibberd et al., 2017). Consistent with our findings with the synthetic communities that had been assembled in vitro, a substantial reduction in false positives was noted for MVRSION compared to QIIME single variable region analyses (Figure 2d). Consequentially, MVRSION demonstrated a significant PPV advantage (88±1.6%; mean ± SD) over V3-QIIME (65%±3.7%) and V4-QIIME (72±3.5%) (p<0.0001, two-tailed unpaired t-test, equal variance; Figure 2h). Though modest in magnitude, a significant improvement in mean sensitivity was also observed for MVRSION (67±5.7%; mean ± SD) versus V3-QIIME (58±4.7%) and V4-QIIME (62±5.2%) (p<0.0001, two-tailed unpaired t-test, equal variance; Figure 2h).
Since variation in sequence depth could affect sensitivity and PPV, 48 of the 92 mouse fecal DNA samples were re-sequenced to approximately 4,000,000 reads per sample (2×150 nt paired-end reads; Table 4). Comparison to the original dataset disclosed that this 10-fold increase in sequencing depth had no significant effect on sensitivity or PPV for the MVRSION or QIIME analyses (p>0.05, two-tailed unpaired t-test, equal variance).
In addition to the sensitivity and specificity analyses, abundance estimates were computed for all samples with MVRSION and QIIME (n=97). These estimates were correlated with the known relative abundances of members of the synthetic mixtures or, in the case of the mouse fecal samples, to abundances determined by short read shotgun sequencing of the fecal DNAs (Community PROfiling by Sequencing; COPRO-Seq). MVRSION demonstrated superior correlation with known composition (r2 = 0.77 versus 0.46 for V3-QIIME and 0.45 for V4-QIIME; Figure 2i-k).
3. Discussion
3.1. QIIME abundance filtering
In both the mixture and gnotobiotic mice samples, MVRSION has a marked advantage in specificity compared to QIIME. It is possible to reduce false positives and increase specificity in QIIME by filtering low abundance taxonomic calls. To evaluate the effect of abundance filtering on QIIME sensitivity and specificity (PPV), multiple abundance filter levels were tested for the 92 fecal samples obtained from gnotobiotic mice colonized with the defined model human gut microbial community. Adjusting the minimum abundance filter above 0% for the QIIME analyses did introduced improvements in PPV. However, this improvement came at substantial cost to sensitivity. At the commonly used filter level of 0.1%, V4-QIIME sensitivity was reduced from 60±5.3% (mean ± SD) to 44±4.3% to achieve a PPV of 80±2.6%, which is still significantly lower than that of MVRSION (88±1.6%, P < 0.0001; two-tailed unpaired t-test, equal variance) (Figure 2l). Thus, the single variable region QIIME analyses could be optimized to approach MVRSION on either axis of sensitivity or specificity (PPV) but not both.
3.2. Fluidigm Platform
The Fluidigm platform used to generate amplicons for sequencing provides additional benefits. The practical lower limit of input DNA mass with the Fluidigm Access Array platform is not currently known. Sample concentrations below the limit of detection of the Qubit dsDNA High-Sensitivity kit (<500 pg/microliter) have been tested; these low concentration samples have produced results comparable on all levels (mapped reads, species called, etc.) to their higher concentration counterparts. Furthermore, recent improvements in Fluidigm microfluidic circuits allow up to 192 samples to be processed on the Access Array system per run to generate multiplexed amplicon libraries. As ever more sequencing indexes become available, the number of amplicon libraries that can be processed by short read sequencing platforms (Illumina) scales, allowing for very high through-put sample handling.
3.3. MVRSION Software
A python implementation of the MVRSION algorithm is available at https://bitbucket.org/WUGTAC/mvrsion. Instructions for downloading the MVRSION formatted SILVA database used in this paper can also be found on the website. The software requires a NovoAlign license to run although BWA support may be added in the future. On a machine with eight AMD Opteron 2435 (2009) cores and 8GB of memory, MVRSION processed the validation samples at an average wall time of 382 seconds per 100,000 150nt paired-end reads (±229 seconds STD) and a maximum of 1307 seconds (i.e., an average wall time of 25 minutes per sample and a worst case of 1 hour 30 minutes.) For processing samples in parallel, the MVRSION implementation supports submitting jobs via PBS/TORQUE, SLURM, and SGE.
4. Conclusions
In summary, selection of multiple variable regions of the bacterial 16S rRNA gene provides clear advantages compared to traditional single variable region approaches, particularly in regard to detection specificity as measured by PPV. Although this study focused on bacterial 16S rRNA variable regions, in principle any collection of informative sequences, including additional SSU rRNAs, could be employed to classify closely related species or strains of other organisms. As such, MVRSION has multiple applications ranging from whole community profiling to targeted tracking of the representation of a series of organisms of interest in diverse environmental and animal host habitats as a function of specified variables/perturbations.
5. Methods
5.1. Selection of Primers
Published literature was searched for bacterial 16S rRNA primer pairs that satisfied the following criteria: (i) specificity validated in published literature; (ii) product ㍤300bp in length, and (iii) detect bacterial taxa represented across a variety of animal and environmental habitats (Table 1). 14 primer pairs were selected that amplify all nine variable regions in the bacterial 16S rRNA gene.
5.2. Synthetic Community Mixtures
Five DNA samples consisting of synthetic mixtures of bacteria for validation of the method were obtained; Human Microbiome Project sample, HM-782D (Microbial Mock Community B; Even, Low concentration, v5.1L; BEI Resources, Manassas, VA), which is composed of DNA from 20 diverse bacterial species represented in equivalent abundances, and four samples containing DNA from 48 human gut bacterial isolates, in equal concentration (48G-Eq) or various staggered concentrations (48G-Stg1, 48G-Stg2, 48G-Stg3) spanning roughly four orders of magnitude (Faith et al., 2013). This collection of 48 isolates included different strains of the same species with the result that a total of 45 species were represented (see Table 3).
5.3. Fecal Samples from Gnotobiotic Mice
Genomic DNA was prepared from fecal pellets collected from gnotobiotic mice. Animals were initially fed a nutritionally-sufficient defined diet and then gavaged with a mixture of 92 cultured sequenced human gut bacterial strains. Animals were subsequently exposed to a series of manipulations of dietary micronutrient content (Hibberd et al, 2017). DNA was isolated from fecal pellets that were collected over the course of the experiment. The relative abundances of each strain in these samples were determined by short-read shotgun sequencing (Hibberd et al., 2017); the 68-species identified across these samples are listed in Table 3.
5.4. Generation of Amplicon Libraries
DNA samples were processed using the Fluidigm Access Array System according to the manufacturer’s protocol (Fluidigm Access Array Users Guide). Up to 48 samples were loaded onto the Access Array using an integrated fluidic circuit (model LP 48.48 IFC). For each sample, 1μl of DNA (5 ng) was mixed with 4μl of PCR reaction buffer containing High Fidelity FastStart Reaction Buffer without MgCl2 (Roche), 4.5 mM MgCl2 (Roche), 5% DMSO (Roche), 200 μM PCR Grade Nucleotide Mix (Roche), 0.05 U/μl FastStart High Fidelity Enzyme Blend (Roche), and 1X Access Array Loading Reagent (Fluidigm). Four microliters of the reaction mixture were loaded into the Sample Inlets of the fluidic circuit. Sequencing primers for the 14 amplicons were loaded into the Assay Inlets at a concentration of 200 nM in 1X Access Array Loading Reagent (Fluidigm). PCR amplification was performed on the BioMark HD system from Fluidigm. Each sample (consisting of the 14 amplicons) was harvested and Illumina sequencing adapters were attached using 14 rounds of PCR where the 3’ ends of the primer anneal to Fluidigm specific sequences attached to the 5’ end of the 16S rRNA primers. Samples were pooled, subjected to a bead-based purification (AMPure XP, Beckman) and then sequenced on a single lane of a flow cell using an Illumina MiSeq or HiSeq 3000 instrument (see Table 4 for sequencing depths). Reads from the individual samples were de-multiplexed based on their unique index tags.
5.5. MVRSION Algorithm
An overview of the analysis method is provided in Figure 1. A detailed technical description of each component of the process is provided below.
5.5.1. Curated 16S rRNA database preparation, in silico PCR, and predicted amplicon sequences
MVRSION relies on a curated 16S rRNA database to identify known bacterial sequences within a sample. For this work, we started with the non-redundant, Silva SSU database (Ref NR 99 release 123) (Quast et al., 2013). The annotated species were compared to accepted species names from the List of Prokaryotic Names with Standing in Nomenclature (LPSN) (Euzeby, 1997). Sequences in SILVA that did not contain a species identifier within the LPSN list were removed. All information beyond the species name was subsequently discarded. The final form of the database included the following files; (i) A fasta file of full length 16S rRNA sequences annotated with unique sequence ids and a matching NovoAlign index file, and (ii) a table matching each sequence id with a species-level taxonomic string
The predicted amplicon sequence dataset was prepared by performing in silico PCR on the curated database with the primer set using PrimerProspector (Walters et al., 2011) commands: “analyze_primers.py” (default parameters)
“get_amplicons_and_reads.py –min_seq_len 100 –read_len 151”
This dataset consists of (i) Fasta files for each primer pair containing amplicon sequences from running PrimerProspector on the full length 16S rRNA sequences, and (ii) a table detailing what fraction of a species’ full-length 16S rRNA sequences have in silico amplification for each amplicon.
5.5.2. Preprocessing of sequencing data
Raw fastq files are demultiplexed by sample and then processed with the adapter trimming program scythe (Buffalo, 2011). The following procedure is applied to each read pair to identify the origin amplicon and to remove primer sequences:
Let 𝐿(𝑁, 𝑠1,𝑠2) be the Levenshtein distance between the first N letters of two strings
Let 𝑅1 and 𝑅2 be a read pair
Let be the forward Fluidigm primer and the reverse Fluidigm primer from amplicon A
Annotate read pair as originating from amplicon A if the following condition is met:
where N is set to the length of the longest primer in the amplicon panel. M was set to 4 in our validation tests. Any reads with multiple or zero amplicon annotations are removed from the data. For each read remove the 3’ subsequence 𝑅′ such that
5.5.3. Candidate Species Identification
Annotated reads are aligned to the database with NovoAlign (version 3.07.00) with the options:
“novoalign –t 60 –Hk –I PE 50–400 –r Random –d database”
Alignments are filtered to consider only read pairs that are both aligned to sequences from the same species. For each species, we define a set of 𝐷 = {𝛼1, 𝛼2,…, 𝛼𝑁} where 𝛼𝑖 is the set of all reads originating from amplicon i that were mapped to any reference sequence belonging to the species. We define a filter function f (D, δ) = {α ∈ D: |α\≥ δ} which returns the α sets from D that have greater than or equal to δ reads. The decision function used to determine the presence of a species is
F takes as input the data D, a baseline number of reads θ, a scaling factor β, and a maximum number of amplicons ρ. k is the number of α sets that the filter function must return for F to return a positive result. The scaling factor β exponentially increases the number of reads required by the filter function when k is fewer than ρ amplicons are required. The decision function F is applied to all species that have at least one alignment. A species is defined as present if F does not return the null set.
For the initial filter stage β =5, ρ = 3 and θ = 10−4∑i|αi|
For the second filter stage β =5, ρ = 4 and θ = 10−4∑i|αi|
Species passing the initial filter stage are regarded as candidate species because the final calls of the algorithm will come from this group. A sub-database is created that contains only 16S rRNA sequences from the candidate species. The purpose of this sub-database is to eliminate erroneous mapping of reads to species that are likely not present in the sample.
5.5.4. Dynamic Discriminatory Variable Region Selection
For final species calls, the decision function is applied to a group of species-specific amplicon sets instead of the entire amplicon panel. These amplicon sets are chosen by their ability to distinguish a species from the other candidate species. To generate these amplicon sets, fasta files created by PrimerProspector for each amplicon are aligned by NovoAlign to the sub-database:
“novoalign –t 60 –Hk –I PE 50–400 –r All 400”
From this alignment, a list for each sequence is created of possible multi-mappings to sequences from other species. For each species, the list of possible multi-mapping to other species is fed into the procedure MakeModel and its component subroutines.
Intersection:
Find the smallest set of amplicons such that the intersection of their multi-mapping lists is the empty set.
Backup:
Sort the amplicons by increasing length of multi-mapping lists. Return all sets of amplicons from the first r+1 sorted amplicons.
MakeModel:
Remove from consideration any amplicons with predicted amplification of less than 80% of a species’ sequences. Run Intersection recursively, removing the selected amplicons from consideration in the following Intersection call. If no valid intersections can be found, run Backup and return.
MakeModel returns one or more amplicon sets, any of which can be used to call an organism. Multiple models are generated to allow for the unexpected failure of one of the amplicons.
5.5.5. Final Species Calls – Assignment and Abundance Estimation
The processed fastq files are realigned to the sub-database sequences using NovoAlign.
“novoalign –t 60 –Hk –I PE 50–400 –r Random”
For each species, apply the procedure FinalCall.
Call:
Apply the decision function F (defined in the Initial Filter section) to data containing only reads originating from amplicons in the model amplicon set.
FinalCall:
Run Call on each of the amplicon sets returned by MakeModel and call species as present if Call returns true for any of the sets. Re-run MakeModel removing any species not called as present from the multi-mapping lists. Apply Call again to each of the amplicon sets returned by the second MakeModel call and return the final set of called species.
The final output of MVRSION is a list of species that were called ‘present ‘in the sample along with a relative abundance p based on the number of reads that aligned to the species.
For a species S, relative abundance is calculated as:
Let N be the number of amplicons.
Let 𝛼𝑖 be the set of reads aligning to S from amplicon i and
Let 𝐴𝑖 be the set of reads aligning to any species from amplicon i.
5.6. QIIME Analysis
QIIME comparator data were constructed with both V3_2 and V4 amplicons (Table 1) to provide insights about variation between regions. For both amplicons, the paired-end amplicon-annotated reads were joined using fastq-join with default parameters (Aronesty, 2011). Reads were then processed with QIIME version 1.9.0
“pick_open_reference_otus –m uclust –p qiime_parameters.txt”
The qiime_parameters.txt file is available upon request. OTUs at 97% sequence identity were generated with UCLUST using the MVRSION database full-length 16S rRNA fasta file as the --reference_fp value. Taxonomy was assigned by QIIME’s UCLUST consensus taxonomy assigner at the default similarity parameter of 0.9, and using the MVRSION database sequence id to taxonomy table as the --id_to_taxonomy_fp value.
The final set of QIIME relative abundance values was generated with
“summarize_taxa.py –i otu_table_mc2_w_tax_no_pynast_failures.biom”
The species level taxonomic summary file generated by this command is processed by the procedure NonSpeciesFilter and then the procedure AbundanceFilter with filter value λ.
NonSpeciesFilter:
Remove any taxonomic call that has an empty value for the species name from all samples.
AbundanceFilter
For each sample, remove any taxonomic calls that have relative abundance less than λ. Calculate PPV, sensitivity, and specificity using all remaining taxonomic calls.
Figure 2l was generated by performing multiple QIIME analyses on the gnotobiotic mice samples with different values of λ. The QIIME results presented in Figures 2a-k were run with λ = 0.
5.7. Assessment of Species Calls
For each sample and both MVRSION and QIIME, the True Positives (TP), False Positives (FP) and False Negatives (FN) are defined in the following way. Let 𝑅𝑆 be the relative abundance of species S called by a method and let 𝐾𝑆 be the known abundance or the COPRO-Seq determined relative abundance value for species S. Each species call is classified accordingly:
| TP is 𝑅𝑠𝑆 > 0 and 𝐾𝑆 > 0 |
| FP is 𝑅𝑆 > 0 and 𝐾𝑆 = 0 |
| FN is 𝑅𝑆 = 0 and 𝐾𝑆 > 0 |
MVRSION performs microbial taxonomic profiling utilizing multiple 16S rRNA amplicons
The method was validated utilizing samples containing well-characterized microbial DNA
Improved specificity and sensitivity as compared to single-region QIIME analysis
Improved relative abundance estimates as compared to single-region QIIME analysis
Acknowledgments
We thank Mary Preuss (Webster University) for sharing additional testing samples. This work was supported by grants from the NIH [NCI Cancer Center Support Grant P30 CA91842, ICTS/CTSA Grant UL1TR000448 (NCRR), DK30292, DK70977, and DK 78669].
Footnotes
DECLARATIONS
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Availability of data and materials
Sequencing data from all the validation samples discussed are available for download here:
Schriefer, Andrew (2018), “MVRSION validation data”, Mendeley Data, v1 http://dx.doi.org/10.17632/c3yzpm2428.1
Competing interests
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allard G, Ryan FJ, Jeffery IB, & Claesson MJ (2015). SPINGO: a rapid species-classifier for microbial amplicon sequences. BMC Bioinformatics, 16, 324. doi: 10.1186/s12859-015-0747-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty E (2011). ea-utils : Command-line tools for processing biological sequencing data. Github repository. Retrieved from https://github.com/ExpressionAnalysis/ea-utils
- Buffalo V (2011). Scythe - A Bayesian adapter trimmer (Version 0.994). Github repository Retrieved from https://github.com/vsbuffalo/scythe
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, . . . Knight R (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods, 7(5), 335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26(19), 2460–2461. doi: 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Euzeby JP (1997). List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet. Int J Syst Bacteriol, 47(2), 590–592. doi: 10.1099/00207713-47-2-590 [DOI] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, . . . Gordon JI (2013). The long-term stability of the human gut microbiota. Science, 341(6141), 1237439. doi: 10.1126/science.1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd MC, Wu M, Rodionov DA, Li X, Cheng J, Griffin NW, . . . Gordon JI (2017). The effects of micronutrient deficiencies on bacterial species from the human gut microbiota. Sci Transl Med, 9(390). doi: 10.1126/scitranslmed.aal4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, DeSantis TZ, Andersen GL, & Knight R (2008). Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res, 36(18), e120. doi: 10.1093/nar/gkn491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, . . . Glockner FO (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res, 41(Database issue), D590–596. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, . . . Weber CF (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol, 75(23), 7537–7541. doi: 10.1128/aem.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetrovsky T, & Baldrian P (2013). The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PloS one, 8(2), e57923. doi: 10.1371/journal.pone.0057923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters WA, Caporaso JG, Lauber CL, Berg-Lyons D, Fierer N, & Knight R (2011). PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics, 27(8), 1159–1161. doi: 10.1093/bioinformatics/btr087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, & Cole JR (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol, 73(16), 5261–5267. doi: 10.1128/aem.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Wang Y, & Qian PY (2016). Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinformatics, 17, 135. doi: 10.1186/s12859-016-0992-y [DOI] [PMC free article] [PubMed] [Google Scholar]