Abstract
The intestinal tract is inhabited by a tremendous number of microorganisms, termed the gut microbiota. These microorganisms live in a mutualistic relationship with their host and assist in the degradation of complex carbohydrates. Although the gut microbiota is generally considered beneficial, the vast number of microbial cells also form a permanent threat to the host. Thus, the intestinal epithelium is covered with a dense layer of mucus to prevent translocation of the gut microbiota into underlying tissues. Intestinal mucus is an organized glycoprotein network with a host-specific glycan structure. While the mucus layer has long been considered a passive, host-designed barrier, recent studies showed that maturation and function of the mucus layer are strongly influenced by the gut microbiota. In return, the glycan repertoire of mucins can select for distinct mucosa-associated bacteria that are able to bind or degrade specific mucin glycans as a nutrient source. Because the intestinal mucus layer is at the crucial interface between host and microbes, its breakdown leads to gut bacterial encroachment that can eventually cause inflammation and infection. Accordingly, a dysfunctional mucus layer has been observed in colitis in mice and humans. Moreover, the increased consumption of a low-fiber Western-style diet in our modern society has recently been demonstrated to cause bacteria-mediated defects of the intestinal mucus layer. Here, I will review current knowledge on the interaction between gut bacteria and the intestinal mucus layer in health and disease. Understanding the molecular details of this host–microbe interaction may contribute to the development of novel treatment options for diseases involving a dysfunctional mucus layer, such as ulcerative colitis.
Keywords: Dietary fiber, gut microbiota, host–microbe interaction, inflammatory bowel disease, mucus, mucin, mucosal barrier, metabolic disease, probiotics, ulcerative colitis
Introduction
The intestinal microbial community, termed the gut microbiota, lives in a mutualistic relationship with its host and produces vitamins and other metabolites that are beneficial for host physiology. However, although these microbiota-derived molecules can signal to organs distant from the intestine [1], their microbial producers need to be contained within the intestinal lumen. As the gut microbiota are separated from the host by only a single layer of enterocytes, the trillions of bacterial cells form a permanent threat. Thus, to prevent translocation of commensal and pathogenic microorganisms across the mucosal barrier, the host has developed effective defense mechanisms, including the formation of a physical mucus barrier that covers the intestinal epithelium.
Intestinal mucus is produced by goblet cells and forms a highly organized glycoprotein network, mainly consisting of mucin 2 (MUC2), but also containing a stable core proteome [2]. The mucus layer has long been considered to be a simple lubricator to facilitate passage of the fecal material through the intestinal channel, but the recent interest in the gut microbiota has also brought intestinal mucus into the focus of research. While we are still only at the beginning of understanding the function and importance of the mucus layer, recent studies have already shown that its interaction with the gut microbiota is more intense than previously thought. Moreover, the observation of mucus defects in diseases such as inflammatory bowel disease [3, 4] and hyperglycemia [5] underlines the importance of this barrier for host physiology. Here, I review recent findings on the interaction between the gut microbiota and the intestinal mucus layer in health and disease.
Structure of the intestinal mucus layer(s)
Intestinal mucus is constitutively produced by goblet cells, specialized secretory cells of the epithelial layer. In the small intestine, goblet cells are primarily localized in the crypts of Lieberkühn, but also in lower numbers on the small-intestinal villi. In the colon, the mucus-secreting cells accumulate at the opening of the colonic crypts but are also found deeper within the crypts and on the colonic surface. Recently, a specialized ‘sentinel’ goblet cell sub-type has been identified in mice at the colonic crypt opening, which orchestrates mucus secretion in response to bacterial invaders that managed to penetrate the inner colonic mucus layer [6].
The major secreted mucus protein in the intestine is MUC2, encoded by the MUC2 gene on chromosome 11 in humans. This gel-forming glycoprotein consists of more than 5100 amino acids and is highly O-glycosylated [7], generating a molecular weight of about 2.7 MDa per mucin molecule [8]. In fact, more than 80% of its molecular weight is due to oligosaccharide side chains, explaining the characteristic gel-forming properties. In addition, the mucus-specific glycan decoration is a crucial part of its interaction with the gut microbiota and will be discussed in more detail below.
Within the endoplasmic reticulum of goblet cells, MUC2 monomers dimerize via their C-terminal disulphide bridges [9] and subsequently trimerize via intermolecular disulphide bridges through the characteristic N-terminal von Willebrand D-domains in the Golgi [10]. These mucin oligomers are then densely packaged in secretory vesicles, where low pH, high Ca2+ concentration and the absence of water facilitate organized storage conditions. The subsequent mucus secretion is best characterized in the small intestine: hydrogen carbonate (HCO3–) secretion by the cystic fibrosis transmembrane conductance regulator (CFTR) channel leads to alkalizing conditions and a decrease in Ca2+ concentration compared to the environment in the mucus granules. These environmental changes allow massive expansion of the mucus oligomers, generating 2D, highly organized sheets that interact with the previously formed ones to build up the 3D mucus layer [11, 12]. At the same time, hydration leads to swelling of the mucus, causing a 100- to 1000-fold expansion in the volume of the packaged mucin [13]. However, recent findings suggest that mucin assembly is likely even more complicated and may also involve non-mucin molecules: for example, the presence of trefoil factor-3 (TFF3), keratins and calcium-binding proteins in native mucin preparation may indicate their relevance for mucin assembly, even though their individual roles and contributions need to be tested experimentally [14].
Upon secretion into the small intestine, mucus forms a single layer that extends over the tips of the small-intestinal villi [15]. Whereas only a few studies have investigated the small-intestinal mucus in detail, it appears that, in mice, the mucus is not completely impenetrable to bacteria and released from the epithelium by the microbially induced protease meprin-β [15, 16]. Given that the small intestine is a metabolically active organ that is specialized in nutrient uptake, a loose mucin network may facilitate the uptake of dietary molecules across the epithelial border. However, to prevent opportunistic bacteria escaping from their luminal environment, small-intestinal mucus is charged with defense molecules, comparable to an electric fence (Figure 1) [17]. These antibacterial proteins and peptides include, among others, Paneth-cell-produced defensins, lysozyme, regenerating islet-derived protein 3α (REG3α; REG3γ in mice) and phospholipase A2-IIA [17–20]. In addition, aggregation of bacteria is induced by molecules such as immunoglobulin A or alpha-defensin 6, which thereby prevent mucosal barrier crossing by size exclusion [21, 22]. Of note, despite a sealed physical barrier being required to efficiently protect the host against the gut microbiota, intended loopholes within the mucus layer are found. They are localized at the areas above the Peyer’s patches, which are focal structures characterized by the presence of microfold (M) cells within the follicle-associated epithelium [23]. As M cells are specialized in sampling of microbial and dietary antigens from the intestinal lumen, the physical mucus barrier on top of these structures is penetrable to bacteria or may even be absent [15, 23].
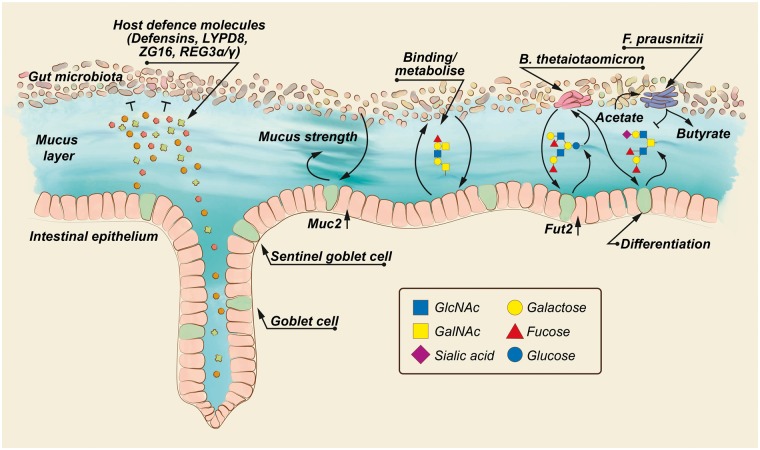
Figure 1.
The structure of the mucus layer is affected by the gut microbiota. Gut bacteria are separated from the host epithelium by the intestinal mucus layer, which is fortified with host defense molecules, such as defensins, Ly6/PLAUR domain containing protein 8 (LYPD8), zymogen granulae protein 16 (ZG16), Regenerating islet-derived proteins 3 (REG3α/γ) and others. The presence of gut microbiota induces expression of the genes encoding mucin 2 (Muc2) and galactoside 2-alpha-L-fucosyltransferase 2 (Fut2), thereby affecting mucus strength and mucin glycan structure. This can in return affect gut-microbiota composition, as specific gut bacteria, such as Bacteroides thetaiotaomicron, can bind and metabolize mucin glycans as an energy source. Degradation of glycans leads to production of microbial metabolites, which not only affect goblet cell differentiation, but also via cross-feeding the abundance of other microbial species, for example Faecalibacterium prausnitzii. Accordingly, the composition of the gut microbiota affects mucus function through the availability of microbial metabolites.
In the colon, mucus is characterized by a distinct stratified inner layer that is virtually free of bacteria and a less defined outer layer, which is composed of mucus, gut bacteria and dietary material [24]. Due to its blending with intestinal content, the outer layer is often difficult to observe in colonic tissue sections or by other imaging techniques. In contrast, the inner colonic mucus layer is clearly visible, revealing that its thickness is maintained at a rather constant level: ∼50–80 µm in mice and ∼100 µm in humans [4, 15, 24, 25]. Since mucus is continuously secreted at a basal level, this indicates that the transition from the inner to the outer layer must be tightly regulated. Indeed, the calcium-activated chloride channel regulator 1 (CLCA1), which is—despite its name—a metalloprotease with high abundance in colonic mucus, has recently been shown to contribute to this transition [26]. However, deletion of Clca1 in mice revealed that further, yet unidentified cysteine proteases can compensate for the lack of this enzyme, suggesting a level of redundancy in this process. Whether the transition between the inner and outer mucus layer is only mediated by host proteases, or whether bacterial enzymes may add an additional layer of complexity, will be an interesting question that needs to be investigated in future research.
In contrast to the ileum, in which antimicrobial peptides kill microorganisms that reach proximity to the epithelium, the colonic bacteria form an active internal bioreactor that produces a tremendous number of metabolites, including vitamins and short-chain fatty acids (SCFAs), which influence host physiology mostly in a beneficial way [1, 27–31]. Therefore, the internal bioreactor needs to be maintained in a regulated manner—hence with sufficient distance from the epithelium. Accordingly, the colonic epithelium produces fewer antibacterial peptides that stick to the mucus, but rather secretes peptides that bind and aggregate bacteria and consequently inhibit bacterial trespassing by physical-size exclusion and inhibition of motility. Remarkably, despite the relatively macroscopic mechanism, these peptides are rather specific. Recently, Ly6/PLAUR domain containing protein 8 (Lypd8) has been shown to specifically bind flagellated bacteria and thus blocks their motility in the mouse colon [32]. In addition, the lectin-like protein zymogen granulae protein 16 (ZG16) specifically binds the bacterial cell-wall compound peptidoglycan of Gram-positive bacteria and thereby inhibits their penetration into the inner colonic mucus layer [33]. However, this binding/aggregation defense system is complemented by additional antimicrobial peptides, such as REG3γ, resistin-like molecule β (RELMβ) and others [19, 34]. Consequently, mucus in the small and large intestine protects the host against the gut microbiota by different, yet similarly effective mechanisms.
Specific interaction between gut microbiota and the mucus layer
Despite being a barrier against the gut microbiota, intestinal mucus requires the presence of bacteria to develop its full functionality, as has been shown in germ-free (GF) animals. Early histochemical analyses already described a thinner or locally even absent mucus layer in the colon of GF rats [35]. Moreover, by Periodic acid–Schiff (PAS) staining—a technique used to stain polysaccharides and glycoproteins—the same study revealed structural differences within the layer, since the colonic mucus layer appeared less compact in GF rats when compared to conventionally raised rats (Figure 1) [35]. These findings were complemented by more specific lectin-stainings, which identified less intense reactivity upon UEA-1 (Ulex europaeus agglutinin I; α-L-fucose) and DBA (Dolichis biflorus agglutinin; α-N-Acetyl-D-galactosamine) incubation of goblet cells in the cecum, but not the small or large intestine of GF mice [36]. While this indicates that the terminal glycan residues in the cecum of GF mice differ from their conventional counterparts, secreted mucus was not analysed in this study. Importantly, however, the observation that mucin glycosylation differs already intracellularly before mucus secretion suggests that not only bacterial degradation of specific mucus glycans (see below) but also bacteria-mediated host processes contribute to mucus fucosylation. This hypothesis is supported by data comparing fucosylated glycoconjugates between GF and conventionally raised mice: in the small intestine, colonized mice displayed an increasing gradient of fucosylated epitopes in enterocytes, Paneth cells and goblet cells, whereas small-intestinal fucosylation in GF mice could only be detected in Paneth cells in the bottom of the crypts of Lieberkühn [37]. Further experiments revealed that the microbiota specifically induced expression of host fucosyltransferases that add L-fucose in the α-1,2 position [37].
Whereas the comparison between adult GF and conventionally raised rodents shows a clear difference in mucus thickness and composition [35, 36, 38, 39], the GF mouse model is an extreme case of the natural situation, in which the newborn intestine becomes colonized with its individual microbial population after birth. Interestingly, while expression of the mouse mucin genes Muc1–4 increased between postnatal Days 1 and 6 even in the absence of a microbiota [40], the presence of a complex microbiota resulted in higher baseline expression of the gene encoding the secreted MUC2, but not of the membrane-bound mucins 1, 3 and 4. In contrast, mice mono-colonized with the probiotic bacterial strains Lactobacillus acidophilus NCFM or Escherichia coli Nissle 1917 displayed similar Muc2 expression as compared with the level of GF mice [40]. This indicates that other members in the complex microbial community are required to stimulate Muc2 expression or that a potential metabolic interaction between microbial species is required to produce the Muc2-inducing signal.
Since Muc2 mRNA expression does not fully match MUC2 production, and mucus function is largely regulated on a post-translational level, analysis of Muc2 expression is insufficient to fully evaluate the function of the intestinal mucus layer [12]. Thus, MUC2 protein content and, by using an ex vivo technique to investigate mucus function on living tissue [25], mucus thickness and mucus penetrability were investigated in GF mice after being colonized with a complex microbial community [41]. GF mice had a penetrable inner colonic mucus layer that contained lower relative abundance of MUC2 protein when compared with conventionally raised mice. Remarkably, the thickness of the inner colonic mucus layer did not differ between the two mouse groups by using this ex vivo technique but, since a penetrable mucus that contains a lower concentration of MUC2 protein possibly shrinks more than an intact layer during the fixation procedure [42], these findings likely describe the same biologic defect as has been observed in fixed tissue sections [38, 39, 43]. After GF mice were colonized with a complex microbial community, the colonic mucus layer required about 5 weeks to become impenetrable—the same time as was needed to allow normal detachment of the small-intestinal mucus from the intestinal epithelium [41].
Despite a core gut-microbiota community that is observed in most individuals, there are considerable differences in intestinal microbial composition between individual hosts [44–48]. Considering the crucial effect of the gut microbiota on mucus function [41], it is not surprising that differences in microbiota composition affect mucus properties. Accordingly, in two mouse colonies that were housed in different rooms of the same animal facility, one colony displayed a normal, impenetrable inner colonic mucus layer while the second colony had a penetrable mucus. This difference could be linked to the specific gut-microbiota composition, as the mucus phenotype was transferrable to GF mice by cecal microbiota transplantation [49]. However, although differences in microbial composition between the two mouse colonies were observed, strong inter-individual variations within the mouse colonies did not allow causally linking specific bacterial taxa to mucus function. Consequently, although it is obvious that the microbial community composition affects mucus function, the effects of specific members of the gut microbial community are largely unknown and the effects on mucus function may depend on the community structure rather than on individual taxa. However, increasing evidence suggests that certain bacterial strains have stronger potential than others to affect mucus function (see below).
Of note, gut-microbiota analyses and microbiota transplantations often focus on bacterial community members, neglecting the potential influence of viruses and fungi that are likewise transplanted in complex communities. Especially bacteriophages—bacteria-specific viruses that populate the gut in high diversity [50]—have been shown to localize to mucosal surfaces and are specifically enriched in mucus when compared to the surrounding environment [51]. According to the proposed ‘bacteriophage adherence to mucus’ (BAM) model, phages can adhere to glycan residues of the mucin glycoproteins via interaction with Ig-like domains of their virus capsid proteins, thereby forming an additional host-independent antimicrobial shield against the gut microbiota [51]. The observation that the viral composition in the intestine of patients with inflammatory bowel disease (IBD) differs from healthy individuals, mainly by an expansion of Caudovirales bacteriophages, further supports the importance of a complex phage–bacterial interaction in the intestine that is relevant for health and disease [52, 53].
Mucus glycans as an energy source for the gut microbiota
O-linked oligosaccharides contribute by about 80% to the total molecular weight of intestinal mucus [7, 54]. As selective intestinal bacteria can degrade mucus glycans as an energy source [39, 55, 56], it is likely that the species-specific gut-microbiota composition is influenced by the glycosylation profile of the mucus layer, which also differs between species [54, 57–60]. In support of this theory, mice that are lacking expression of core 1-derived O-glycans have subtle microbiota differences in the cecal lumen [61]. However, as the mucus glycosylation pattern rather affects mucosa-associated bacteria, it is conceivable that the microbial population at the intestinal mucosa would be affected to a larger extent. Indeed, in humans, expression of the galactoside 2-α-L-fucosyltransferase 2 (FUT2), encoded by the FUT2 (secretor) gene, has been shown to affect the colonic mucosa-associated gut-microbiota composition [62]. FUT2 transfers a terminal fucose residue to glycoproteins of the mucus layer and other body secretions and is responsible for the expression of ABO histo-blood group antigens [63]. A nonsense-mutation in the FUT2 gene that inactivates the enzyme and leads to a ‘non-secretor’ phenotype has been associated with Crohn’s disease [64, 65]. Moreover, the interaction between Crohn’s disease and secretor genotype has been associated with alterations in bacterial communities: on a species level (≥97% identity), one operational taxonomic unit (OTU) linked to Lactobacillus and two OTUs linked to Stenotrophomonas were more abundant among healthy secretor genotypes while five OTUs belonging to Prevotella, Brevundimonas, Sutterella, Faecalibacterium and an unclassified Lachnospiraceae were associated with heathy non-secretors. In contrast, three OTUs belonging to Alistipes, Coprococcus and an unclassified Lachnospiraceae were more abundant in non-secretor individuals with Crohn’s disease [62]. This suggests that a combination of genotype-determined glycan structure and disease status may affect the gut microbial community at the intestinal mucosa. However, the reduced abundance of bifidobacteria that has been reported for stool samples of non-secretors in a cohort of 79 Finnish individuals [66] could not be confirmed when larger cohorts were tested [67, 68]. Yet, in agreement with the previously mentioned mouse study [61], it is clear that FUT2 genotype only affects the mucosa-associated microbial community and does not modulate fecal microbiota composition that has been analysed in the large cohorts.
The interaction between mucus glycan structure and gut-microbiota composition has been demonstrated to be more systematic than previously thought: in a landmark study, Bry and colleagues [37] found that Bacteroides thetaiotaomicron (B. theta), a common member of the normal mouse and human intestinal microbiota, is able to induce fucosylation in the mouse ileum (Figure 1). Remarkably, the induction of host fucosyltransferases by B. theta was regulated by the actual host-derived fucose concentration [69] and absent in an isogenic bacterial mutant that was unable to metabolize L-fucose as an energy source [37]. This indicates that B. theta induces mucosal fucosylation in order to establish its own nutrient-providing niche. While this phenomenon illustrates a bona fide host–microbe interaction, the molecular signal that stimulates epithelial fucosyltransferases remains unknown, but involves signaling through extracellular signal-regulated kinases (ERK) and c-Jun N-terminal kinases (JNK) signal transduction pathways in intestinal mucosa [70].
Besides affecting mucus fucosylation, B. theta has also been shown to promote goblet cell differentiation by stimulating the secretory lineage in the colonic epithelium of rats—an effect mediated by the SCFA acetate [71]. In addition, the presence of B. theta led to increased expression of glycans carrying sialic acid residues of mucus glycans, whereas expression of sulfated and neutral oligosaccharides decreased. Interestingly, when B. theta-mono-colonized rats were supplemented with Faecalibacterium prausnitzii, an acetate consumer and butyrate producer, the increase in goblet cell differentiation and the alteration in mucus glycan profile was abrogated [71]. Consequently, this study clearly showed that not all mucus is created equally, but that instead the presence of bacteria, and more specifically the presence and relative abundance of specific bacteria, shapes the glycan profile of the mucus layer (Figure 1).
The gut microbiota degrades dietary substrates that are not metabolized and taken up by the small intestine and thus reach the colonic lumen. These dietary substrates are mainly plant-derived polysaccharides, for which the host has only a limited enzymatic repertoire of approximately 17 carbohydrate-active enzymes (CAZymes) [72]. In contrast, the genetic repertoire of the human gut microbiota (the gut microbiome) encodes for at least 89 CAZyme families, which suggests the capability of degrading a broad range of carbohydrates [72, 73]. In fact, the proteome of B. theta alone includes at least 172 glycosylhydrolases, thereby exceeding the capacity of many other sequenced gut microbiota [74]. Remarkably, the prevalence of CAZymes in the gut microbiota has been found to be geographically and age-specific, suggesting that a diet-dependent adaptation is possible [72].
While dietary carbohydrates are the main source of nutrients for the gut microbiota, the mucus layer is an alternative source of host-derived glycans and contributes to bacterial colonization and persistence in the human gut [55, 75, 76]. Such metabolic flexibility is especially evident and beneficial for the microbe when complex carbohydrates—or microbiota-accessible carbohydrates (MACs [77])—are lacking in the diet, as exemplified by studies investigating the glycan metabolism of B. theta in newborn mice [78] or in mice fed a diet that is devoid of dietary fiber [79]. The ability of switching from dietary glycans to host-derived glycans accordingly determines which gut microbiota can persist in the gut when dietary MACs are scarce.
Among a synthetic community consisting of 13 human-derived gut microbial species that cover members from the dominant phyla in the human gut, Akkermansia muciniphila and Barnesiella intestinihominis have been found to be mucus specialist and exclusively feed on mucin O-glycans. In contrast, B. theta and Bacteroides caccae demonstrated the metabolic flexibility mentioned above, and were thus termed mucus generalists that can feed on a variety of polysaccharides [39]. Remarkably, when mice that were colonized with this synthetic community were switched from a fiber-rich to a fiber-free diet, expression and activity of CAZymes responded by an increase in mucus-targeting enzymes and a decrease in fiber-targeting enzymes. As a consequence, mice fed a fiber-free diet had decreased thickness of the colonic mucus layer, which increased susceptibility towards infection by the enteric mucosal mouse pathogen Citrobacter rodentium (Figure 2) [39]. Of note, mice defective in the major mucus protein MUC2 (Muc2–/–) were similarly susceptible to C. rodentium infection [80], which resembles infection with enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli (EHEC) in humans [81]. Thus, the parallel between diet-dependent destabilization of the mucus layer and the absence of MUC2 indicates the crucial role of the gut microbiota in forming the protective mucus shield against infection by enteric pathogens.
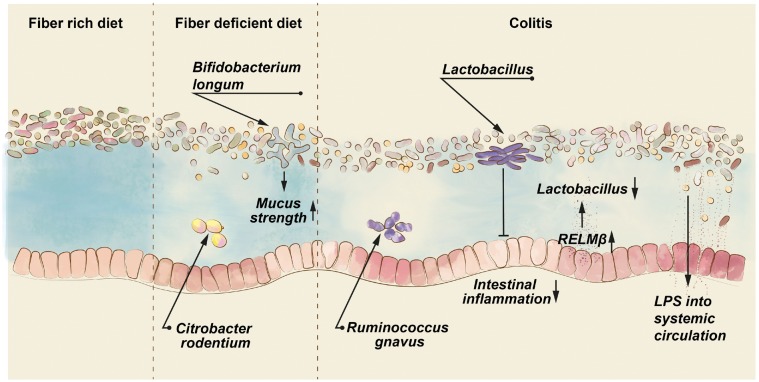
Figure 2.
Defects of the intestinal mucus layer exacerbate intestinal infection and inflammation. Diets lacking microbiota-accessible carbohydrates, as contained in dietary fiber, direct gut microbial species to degrade host glycans of the intestinal mucus layer (depicted by lighter mucus color), thereby deteriorating mucus strength. A defective mucus layer increases the risk for intestinal infections, for example by the mouse pathogen Citrobacter rodentium. The diet-mediated mucus defect can be prevented by specific gut bacteria, such as Bifidobacterium longum. During colitis (depicted by intense red epithelial cells), increased expression of resistin-like molecule β (Relmβ) induces production of the antimicrobial protein regenerating islet-derived protein 3β (REG3β), which reduces beneficial Lactobacillus species. Correspondingly, application of Lactobacillus can ameliorate colitis. In the mucus of inflammatory bowel disease patients, an increased number of the mucus-degrading bacterium Ruminococcus gnavus has been observed, while a defective mucus layer may lead to increased translocation of bacterial lipopolysaccharide, thereby contributing to metabolic diseases.
In addition to the protection against enteric pathogens, defense against the trillions of commensal gut bacteria is an equally important task for the intestinal mucus. Indeed, feeding a diet devoid of MACs to gnotobiotic mice led to a thinning of the colonic mucus layer and consequently increased the proximity of commensal gut bacteria to the mucosa [38]. This was accompanied by increased production of the antimicrobial defense molecule REG3β in the ileum. Remarkably, a similar up-regulation of Reg3β and Reg3γ has been observed in the small and large intestines of Muc2–/– mice, which lack the protective mucus barrier and eventually develop colitis [4, 82]. Thus, these studies further demonstrate that the lack of dietary MACs may lead to detrimental changes of the mucus layer, which eventually may cause intestinal diseases.
In a recent study, we could show that feeding a Western-style diet (WSD)—a diet with high content of saturated fatty acids and sucrose but only little dietary fiber—led to increased penetrability of the colonic mucus layer [83]. While this can in part be explained by gut microbial degradation of host glycans [38, 39, 55], we also detected less MUC2 protein in the colonic mucus of WSD-fed mice when compared with the mucus of control-diet-fed mice [83]. This indicates that the penetration of gut bacteria into the mucus layer may be due to increased bacterial glycan degradation, but may also be caused by a host defect that leads to decreased production, secretion or assembly of the MUC2 mucin.
The healthy inner colonic mucus layer expands towards the intestinal lumen at a speed of about 2 µm/min (‘mucus growth rate’), thereby actively pushing the gut microbiota away from the epithelial surface [25]. However, upon WSD feeding, this growth rate decreased to about 0.5 µm/min—a defect that could be prevented by supplementing the WSD with a microbiota transplant from mice fed a fiber-rich control diet, or even with a single probiotic Bifidobacterium longum strain [83]. Consequently, diet-mediated alterations of the gut microbiota led to defects of the mucus layer that are likely caused by a combination of host and microbial factors (Figure 2).
Microbiota–mucus interaction and the relevance for disease
MUC2-deficient mice are incapable of preventing contact of the intestinal microbiota with the host epithelium. Accordingly, these mice develop spontaneous colitis and augmented dissemination of colonic C. rodentium infection, highlighting the contribution of the mucus layer to intestinal disease [4, 80, 84]. Similarly, in humans, the mucus of patients with ulcerative colitis, self-limiting colitis and acute appendicitis was penetrable to bacteria and revealed reduced mucus thickness that correlated with disease severity [3, 4, 85]. Moreover, increased numbers of mucosa-adherent bacteria were detected in patients with IBD when compared to healthy controls or patients with irritable bowel syndrome (IBS) [86]. Accordingly, characterization of the mucosa-associated bacteria is a superior predictor of disease phenotype than the analysis of fecal microbiota composition [87].
While changes in gut-microbiota composition in IBD patients have been observed in several larger and small cohorts, methodological differences, cohort heterogeneity and increased fluctuations of the microbiome in IBD patients failed to identify specific common microbial taxa that were associated with either form of IBD [88–91]. Still, a recent longitudinal study investigating stool microbiota in patients with IBD for up to 12 months identified a novel Ruminococcus gnavus clade that was transiently enriched in IBD patients [91]. Of note, when investigating specific genomic adaptations of the R. gnavus clades, the authors identified genes involved in mucus and sugar utilization exclusively in the IBD-specific R. gnavus clade (Figure 2). This specificity of mucus degradation pathways in R. gnavus is in line with a previous study that analysed mucus degradation and utilization of two human-derived R. gnavus strains: Only strain ATCC 29149, but not E1, was able to grow on mucin as a single carbohydrate source—a feature that could be linked to the presence of a specific sialidase and fucosidases [92].
Bacterial mucin degradation that exceeds mucus renewal by the host is an evident factor that leads to barrier dysfunction and likely contributes to disease. However, recent studies suggest that a penetrable or absent mucus layer itself may not be sufficient to cause disease. For example, in the absence of MUC2, increased expression of the Retnlb gene (encoding RELMβ) induced production of the antimicrobial protein REG3β, which reduced the number of beneficial Lactobacillus species [93]. Correspondingly, application of two murine-derived lactobacilli, but not of a commercial probiotic mixture containing three human-derived Lactobacillus strains, could ameliorate colitis in Muc2–/– mice (Figure 2). Although Lactobacillus-treated mice had increased SCFAs in the colon, it has to be determined whether these organic acids indeed mediate amelioration of colitis, especially since the comparison with human-derived Lactobacillus strains demonstrates that the modulation of mucosal barrier function is strain-specific. Likewise, two Lactobacillus reuteri strains derived either from rat or human have been shown to increase the thickness of the inner mucus layer in a mouse model of chemically induced dextran sodium sulfate (DSS) colitis—an effect that was paralleled by reduced expression of inflammatory markers and increased expression of tight-junction proteins [94].
A similar mucus-thickness-promoting effect has recently been observed in rats after application of the Bifidobacterium pseudolongum strain Patronus, which has been isolated from the stool of antibiotic-treated rats [95]. Microbiota analyses in these rats revealed a relative decrease of A. muciniphila, which is, despite its specific mucus-degrading capacity [39, 96], considered to be a beneficial intestinal inhabitant [43, 97]. Thus, it is unclear whether the improved mucus thickness is due to increased abundance of Bifidobacterium or due to reduced mucus degradation by A. muciniphila. Of note, and in contrast to expectations, administration of A. muciniphila to mice fed a high-fat diet led to increased colonic mucus thickness [43]. Since such an outcome was not observed in mice fed a control diet, the effect of A. muciniphila on mucus degradation, and consequently mucus thickness, may be context- and especially diet-dependent. In addition, and in favor of a direct mucus-modulating capacity of distinct Bifidobacterium strains, we have recently shown that application of a probiotic human-derived B. longum strain could prevent the mucus growth defect in mice fed a WSD [83]. Moreover, enriching low-fiber WSD with the dietary fiber inulin, which is known to promote growth of Bifidobacterium [98, 99], prevented mucus penetrability in our ex vivo mucus analysis. Thus, although the molecular mechanism is so far unclear, it is possible that distinct metabolic or structural components of specific Bifidobacterium and/or Lactobacillus strains prevent intestinal disease by modulating the function of the mucus layer.
Gut microbiota not only affect the physiology of the intestine, but also signal to distant organs, thereby affecting whole-body metabolism [29, 83]. As such, gut bacteria have been identified as a contributing factor to metabolic diseases, and a defective intestinal mucosal barrier may be the crucial interface between host and microbes [100–104]. Indeed, translocation of the bacterial endotoxin lipopolysaccharide (LPS) from the gut induces adipose tissue inflammation and obese humans and mice have increased levels of LPS in their plasma (Figure 2) [102, 104, 105].
Without doubt, consumption of a WSD with its high content of sugar and fat contributes to metabolic diseases but, at the same time, impairment of intestinal mucus function has been observed in several studies, as described above. In addition, common dietary emulsifiers such as carboxymethylcellulose and polysorbate-80 have been shown to induce low-grade inflammation, metabolic syndrome and colitis in a susceptible mouse model via modulation of the gut microbiota [106]. Even in humans, a recent study observed an increased number of unidentified gut bacteria in the mucus of patients with impaired glucose metabolism when compared with a control group [5]. While this association suggests a critical role of an intact mucus barrier for glucose metabolism, further studies are required to demonstrate that a dysfunctional mucus layer does indeed contribute to metabolic alterations in humans.
Conclusion and perspective
The intestinal mucus layer separates the gut microbiota from the host and accumulating evidence suggests that a breakdown of this anti-infective barrier contributes to diseases such as colitis and metabolic diseases. While alterations in the gut-microbiota composition have been associated with these diseases, the molecular details are yet to be identified.
In recent years, several studies have discovered that the gut microbiota affects physiology and function of the mucus layer. After previous, mostly associative, studies observed correlations between the mucus structure and specific gut microbial communities, current analyses investigate the contribution of isolated bacteria on mucus function. These studies revealed that microbial modulation of mucus function is not defined on a species level, but that microbial characterization requires at least strain-level resolution. As the gut microbiota is a complex community that is influenced by environmental factors, these external contributors also need to be considered. Here, the composition of the diet, and specifically the abundance of dietary fiber, is a critical factor that affects how individual gut microbiota interacts with the mucus layer.
Targeting the gut microbiota to improve mucosal barrier function is a major aim that drives research in this area. The intestinal mucus layer as the crucial interface between host and microbes is therefore a promising target that has only recently come into focus. Accordingly, identification of the molecular mechanism—why individual intestinal bacteria elicit beneficial, barrier-strengthening effects while other, closely related strains do not—could be exploited for the development of future next-generation probiotics. Moreover, enriching beneficial microbiota by targeted dietary interventions may provide an additional approach to treat or even prevent gastrointestinal diseases in the future.
Acknowledgements
B.O.S. thanks Rosie Perkins for valuable editorial feedback on the manuscript and Anna Hallén for excellent assistance with the graphical illustrations.
Funding
B.O.S. is supported by a Long-Term Fellowship from the Human Frontier Science Program (LT000109/2014).
Conflict of interest statement: none declared.
References
- 1. Schroeder BO, Bäckhed F.. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med 2016;22:1079–89. [DOI] [PubMed] [Google Scholar]
- 2. Rodríguez-Piñeiro AM, Bergström JH, Ermund A. et al. Studies of mucus in mouse stomach, small intestine, and colon. II. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am J Physiol Gastrointest Liver Physiol 2013;305:G348–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Swidsinski A, Loening-Baucke V, Theissig F. et al. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut 2007;56:343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johansson MEV, Gustafsson JK, Holmén-Larsson J. et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 2014;63:281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chassaing B, Raja SM, Lewis JD. et al. Colonic microbiota encroachment correlates with dysglycemia in humans. Cell Mol Gastroenterol Hepatol 2017;4:205–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Birchenough GMH, Nyström EEL, Johansson MEV. et al. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 2016;352:1535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Allen A, Hutton DA, Pearson JP.. The MUC2 gene product: a human intestinal mucin. Int J Biochem Cell Biol 1998;30:797–801. [DOI] [PubMed] [Google Scholar]
- 8. Axelsson MAB, Asker N, Hansson GCO.. Glycosylated MUC2 monomer and dimer from LS 174T cells are water-soluble, whereas larger MUC2 species formed early during biosynthesis are insoluble and contain nonreducible intermolecular bonds. J Biol Chem 1998;273:18864–70. [DOI] [PubMed] [Google Scholar]
- 9. Asker N, Axelsson MAB, Olofsson S-O. et al. Dimerization of the human MUC2 mucin in the endoplasmic reticulum is followed by a N-glycosylation-dependent transfer of the mono- and dimers to the Golgi apparatus. J Biol Chem 1998;273:18857–63. [DOI] [PubMed] [Google Scholar]
- 10. Godl K, Johansson MEV, Lidell ME. et al. The N terminus of the MUC2 mucin forms trimers that are held together within a trypsin-resistant core fragment. J Biol Chem 2002;277:47248–56. [DOI] [PubMed] [Google Scholar]
- 11. Ambort D, Johansson MEV, Gustafsson JK. et al. Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proc Natl Acad Sci USA 2012;109:5645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johansson MEV, Hansson GC.. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol 2016;16:639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McGuckin MA, Hasnain SZ.. Goblet cells as mucosal sentinels for immunity. Mucosal Immunol 2017;10:1118–21. [DOI] [PubMed] [Google Scholar]
- 14. Meldrum OW, Yakubov GE, Bonilla MR. et al. Mucin gel assembly is controlled by a collective action of non-mucin proteins, disulfide bridges, Ca2+-mediated links, and hydrogen bonding. Sci Rep 2018;8:5802.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ermund A, Schütte A, Johansson MEV. et al. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastrointest Liver Physiol 2013;305:G341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schütte A, Ermund A, Becker-Pauly C. et al. Microbial-induced meprin β cleavage in MUC2 mucin and a functional CFTR channel are required to release anchored small intestinal mucus. Proc Natl Acad Sci USA 2014;111:12396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meyer-Hoffert U, Hornef MW, Henriques-Normark B. et al. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut 2008;57:764–71. [DOI] [PubMed] [Google Scholar]
- 18. Bevins CL, Salzman NH.. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 2011;9:356–68. [DOI] [PubMed] [Google Scholar]
- 19. Vaishnava S, Yamamoto M, Severson KM. et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 2011;334:255–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Loonen LM, Stolte EH, Jaklofsky MT. et al. REG3γ-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol 2014;7:939–47. [DOI] [PubMed] [Google Scholar]
- 21. Moor K, Diard M, Sellin ME. et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 2017;544:498–502. [DOI] [PubMed] [Google Scholar]
- 22. Chu H, Pazgier M, Jung G. et al. Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 2012;337:477–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol 2003;3:331–41. [DOI] [PubMed] [Google Scholar]
- 24. Johansson MEV, Phillipson M, Petersson J. et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA 2008;105:15064–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gustafsson JK, Ermund A, Johansson MEV. et al. An ex vivo method for studying mucus formation, properties, and thickness in human colonic biopsies and mouse small and large intestinal explants. Am J Physiol-Gastrointest Liver Physiol 2012;302:G430–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nyström EEL, Birchenough GMH, van der Post S. et al. Calcium-activated chloride channel regulator 1 (CLCA1) controls mucus expansion in colon by proteolytic activity. EBioMedicine 2018;33:134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lamas B, Richard ML, Leducq V. et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016;22:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zelante T, Iannitti RG, Cunha C. et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013;39:372–85. [DOI] [PubMed] [Google Scholar]
- 29. Koh A, De Vadder F, Kovatcheva-Datchary P. et al. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 2016;165:1332–45. [DOI] [PubMed] [Google Scholar]
- 30. Donia MS, Fischbach MA.. Small molecules from the human microbiota. Science 2015;349:1254766.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dodd D, Spitzer MH, Treuren WV. et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017;551:648–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okumura R, Kurakawa T, Nakano T. et al. Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature 2016;532:117–21. [DOI] [PubMed] [Google Scholar]
- 33. Bergström JH, Birchenough GMH, Katona G. et al. Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proc Natl Acad Sci USA 2016;113:13833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Propheter DC, Chara AL, Harris TA. et al. Resistin-like molecule β is a bactericidal protein that promotes spatial segregation of the microbiota and the colonic epithelium. Proc Natl Acad Sci USA 2017;114:11027–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Szentkuti L, Riedesel H, Enss M-L. et al. Pre-epithelial mucus layer in the colon of conventional and germ-free rats. Histochem J 1990;22:491–7. [DOI] [PubMed] [Google Scholar]
- 36. Kandori H, Hirayama K, Takeda M. et al. Histochemical, lectin-histochemical and morphometrical characteristics of intestinal goblet cells of germfree and conventional mice. Exp Anim 1996;45:155–60. [DOI] [PubMed] [Google Scholar]
- 37. Bry L, Falk PG, Midtvedt T. et al. A model of host-microbial interactions in an open mammalian ecosystem. Science 1996;273:1380–3. [DOI] [PubMed] [Google Scholar]
- 38. Earle KA, Billings G, Sigal M. et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 2015;18:478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Desai MS, Seekatz AM, Koropatkin NM. et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 2016;167:1339–53.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bergström A, Kristensen MB, Bahl MI. et al. Nature of bacterial colonization influences transcription of mucin genes in mice during the first week of life. BMC Res Notes 2012;5:402.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johansson MEV, Jakobsson HE, Holmén-Larsson J. et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe 2015;18:582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Johansson MEV, GC Hansson. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH In: McGuckin MA, Thornton DJ (eds). Mucins: Methods and Protocols. Totowa, NJ: Humana Press, 2012, 229–35. [DOI] [PubMed] [Google Scholar]
- 43. Everard A, Belzer C, Geurts L. et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA 2013;110:9066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zoetendal EG, Akkermans AD, De Vos WM.. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol 1998;64:3854–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qin J, Li R, Raes J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eckburg PB, Bik EM, Bernstein CN. et al. Diversity of the human intestinal microbial flora. Science 2005;308:1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hold GL, Pryde SE, Russell VJ. et al. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol Ecol 2002;39:33–9. [DOI] [PubMed] [Google Scholar]
- 48.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jakobsson HE, Rodríguez-Piñeiro AM, Schütte A. et al. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep 2015;16:164–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Breitbart M, Hewson I, Felts B. et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 2003;185:6220–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barr JJ, Auro R, Furlan M. et al. Bacteriophage adhering to mucus provide a non–host-derived immunity. Proc Natl Acad Sci USA 2013;110:10771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Norman JM, Handley SA, Baldridge MT. et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015;160:447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang W, Jovel J, Halloran B. et al. Metagenomic analysis of microbiome in colon tissue from subjects with inflammatory bowel diseases reveals interplay of viruses and bacteria. Inflamm Bowel Dis 2015;21:1419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Marcobal A, Southwick AM, Earle KA. et al. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology 2013;23:1038–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martens EC, Chiang HC, Gordon JI.. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 2008;4:447–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martens EC, Neumann M, Desai MS.. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat Rev Microbiol 2018;16:457–70. [DOI] [PubMed] [Google Scholar]
- 57. Slomiany BL, Murty VL, Slomiany A.. Isolation and characterization of oligosaccharides from rat colonic mucus glycoprotein. J Biol Chem 1980;255:9719–23. [PubMed] [Google Scholar]
- 58. Thomsson KA, Holmén-Larsson JM, Ångström J. et al. Detailed O-glycomics of the Muc2 mucin from colon of wild-type, core 1- and core 3-transferase-deficient mice highlights differences compared with human MUC2. Glycobiology 2012;22:1128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Karlsson NG, Herrmann A, Karlsson H. et al. The glycosylation of rat intestinal Muc2 mucin varies between rat strains and the small and large intestine. J Biol Chem 1997;272:27025–34. [DOI] [PubMed] [Google Scholar]
- 60. Robbe C, Capon C, Coddeville B. et al. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J 2004;384:307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sommer F, Adam N, Johansson MEV. et al. Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS One 2014;9:e85254.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rausch P, Rehman A, Künzel S. et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci USA 2011;108:19030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Watkins WM, Biochemistry and genetics of the ABO, Lewis, and P blood group systems In: Harris H, Hirschhorn K (eds). Advances in Human Genetics. Boston, MA: Springer US, 1980, 1–136. [DOI] [PubMed] [Google Scholar]
- 64. Kelly RJ, Rouquier S, Giorgi D. et al. Sequence and expression of a candidate for the human secretor blood group α(1, 2)fucosyltransferase gene (FUT2). J Biol Chem 1995;270:4640.. [DOI] [PubMed] [Google Scholar]
- 65. McGovern DPB, Jones MR, Taylor KD. et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum Mol Genet 2010;19:3468–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wacklin P, Mäkivuokko H, Alakulppi N. et al. Secretor genotype (FUT2 gene) is strongly associated with the composition of bifidobacteria in the human intestine. PLoS One 2011;6:e20113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Turpin W, Bedrani L, Espin-Garcia O. et al. FUT2 genotype and secretory status are not associated with fecal microbial composition and inferred function in healthy subjects. Gut Microbes 2018;9:357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Davenport ER, Goodrich JK, Bell JT. et al. ABO antigen and secretor statuses are not associated with gut microbiota composition in 1, 500 twins. BMC Genomics 2016;17:941.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hooper LV, Xu J, Falk PG. et al. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci USA 1999;96:9833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Meng D, Newburg DS, Young C. et al. Bacterial symbionts induce a FUT2-dependent fucosylated niche on colonic epithelium via ERK and JNK signaling. Am J Physiol Gastrointest Liver Physiol 2007;293:G780–7. [DOI] [PubMed] [Google Scholar]
- 71. Wrzosek L, Miquel S, Noordine M-L. et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitziiinfluence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol 2013;11:61.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bhattacharya T, Ghosh TS, Mande SS.. Global profiling of carbohydrate active enzymes in human gut microbiome. PLoS One 2015;10:e0142038.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lombard V, Golaconda Ramulu H, Drula E. et al. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 2014;42:D490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xu J, Bjursell MK, Himrod J. et al. A genomic view of the human-bacteroides thetaiotaomicron symbiosis. Science 2003;299:2074–6. [DOI] [PubMed] [Google Scholar]
- 75. Salyers AA, West SE, Vercellotti JR. et al. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl Environ Microbiol 1977;34:529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Salyers AA, Vercellotti JR, West SE. et al. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol 1977;33:319–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sonnenburg ED, Sonnenburg JL.. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab 2014;20:779–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bjursell MK, Martens EC, Gordon JI.. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, bacteroides thetaiotaomicron, to the suckling period. J Biol Chem 2006;281:36269–79. [DOI] [PubMed] [Google Scholar]
- 79. Sonnenburg JL, Xu J, Leip DD. et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 2005;307:1955–9. [DOI] [PubMed] [Google Scholar]
- 80. Bergstrom KSB, Kissoon-Singh V, Gibson DL. et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog 2010;6:e1000902.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Collins JW, Keeney KM, Crepin VF. et al. Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol 2014;12:612–23. [DOI] [PubMed] [Google Scholar]
- 82. Paassen NB, Loonen LMP, Witte-Bouma J. et al. Mucin Muc2 deficiency and weaning influences the expression of the innate defense genes Reg3β, Reg3γ and Angiogenin-4. PLoS One 2012;7:e38798.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Schroeder BO, Birchenough GMH, Ståhlman M. et al. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe 2018;23:27–40.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sluis MV, der Koning BAED, Bruijn ACJMD. et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 2006;131:117–29. [DOI] [PubMed] [Google Scholar]
- 85. Pullan RD, Thomas GA, Rhodes M. et al. Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut 1994;35:353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Swidsinski A, Weber J, Loening-Baucke V. et al. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol 2005;43:3380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gevers D, Kugathasan S, Denson LA. et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014;15:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tamboli CP, Neut C, Desreumaux P. et al. Dysbiosis in inflammatory bowel disease. Gut 2004;53:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zuo T, Kamm MA, Colombel J-F. et al. Urbanization and the gut microbiota in health and inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2018;15:440–52. [DOI] [PubMed] [Google Scholar]
- 90. Halfvarson J, Brislawn CJ, Lamendella R. et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2017;2:17004.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hall AB, Yassour M, Sauk J. et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 2017;9:103.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Crost EH, Tailford LE, Gall GL. et al. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One 2013;8:e76341.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Morampudi V, Dalwadi U, Bhinder G. et al. The goblet cell-derived mediator RELM-β drives spontaneous colitis in Muc2-deficient mice by promoting commensal microbial dysbiosis. Mucosal Immunol 2016;9:1218–33. [DOI] [PubMed] [Google Scholar]
- 94. Ahl D, Liu H, Schreiber O. et al. Lactobacillus reuteri increases mucus thickness and ameliorates dextran sulphate sodium-induced colitis in mice. Acta Physiol (Oxf) 2016;217:300–10. [DOI] [PubMed] [Google Scholar]
- 95. Mangin I, Dossou-Yovo F, Lévêque C. et al. Oral administration of viable Bifidobacterium pseudolongum strain Patronus modified colonic microbiota and increased mucus layer thickness in rat. FEMS Microbiol Ecol 2018;94: doi: 10.1093/femsec/fiy177. [DOI] [PubMed] [Google Scholar]
- 96. Derrien M, Vaughan EE, Plugge CM. et al. Akkermansia muciniphila gen. Nov., sp. Nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 2004;54:1469–76. [DOI] [PubMed] [Google Scholar]
- 97. Plovier H, Everard A, Druart C. et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 2017;23:107–13. [DOI] [PubMed] [Google Scholar]
- 98. Gibson GR, Beatty ER, Wang X. et al. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 1995;108:975–82. [DOI] [PubMed] [Google Scholar]
- 99. Vandeputte D, Falony G, Vieira-Silva S. et al. Prebiotic inulin-type fructans induce specific changes in the human gut microbiota. Gut 2017;66:1968–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bäckhed F, Ding H, Wang T. et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 2004;101:15718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bäckhed F, Manchester JK, Semenkovich CF. et al. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA 2007;104:979–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Caesar R, Reigstad CS, Bäckhed HK. et al. Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut 2012;61:1701–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Amar J, Chabo C, Waget A. et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med 2011;3:559–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Cani PD, Amar J, Iglesias MA. et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007;56:1761–72. [DOI] [PubMed] [Google Scholar]
- 105. Creely SJ, McTernan PG, Kusminski CM. et al. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am J Physiol-Endocrinol Metab 2007;292:E740–7. [DOI] [PubMed] [Google Scholar]
- 106. Chassaing B, Koren O, Goodrich JK. et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015;519:92–6. [DOI] [PMC free article] [PubMed] [Google Scholar]