Abstract
Two new species of Pleosporales, Anteaglonium rubescens (Anteagloniaceae) and Atrocalyx asturiensis (Lophiotremataceae), are described. Phylogenetic placement was determined by combined analyses of a DNA data matrix containing ITS, LSU, SSU, rpb2, and tef1. Anteaglonium rubescens is a stromatic fungus characterized by brown didymospores disarticulating within asci, and by the production of a red-orange to pink pigment produced in nature and in artificial culture. Atrocalyx asturiensis has massive ascomatal crests and brown phragmospores.
Keywords: Anteagloniaceae, Dothideomycetes, hysteriaceous fungi, Hysterodifractum, Lophiotremataceae, Ohleria, phylogenetic analysis, pyrenomycetes, Xenolophium
The genus Anteaglonium was erected by Mugambi & Huhndorf (2009a) for hysterothecial taxa with hyaline didymospores that do phylogenetically not belong to the Hysteriales, Mytilinidiales or Patellariales, but to the Anteagloniaceae, Pleosporales. In a comprehensive molecular study of fungi forming hysterothecia, Boehm et al. (2009) reached the same conclusions and included the four species A. abbreviatum, A. globosum, A. latirostrum and A. parvulum, in the genus Anteaglonium. According to Mugambi & Huhndorf (2009a) the genus can be distinguished from Glonium by its smaller ascomata that are either elongate or globose and its small ascospores that are less than 8 μm long except in A. latirostrum. However, in the latter species the ascomata are globose, often with raised laterally compressed apices, characters not present in Glonium. Anteaglonium latirostrum differs from the other species also in ascospores, which become 3–5-septate and pale brown at maturity. Another species, A. brasiliense was added by Carneiro de Almeida et al. (2014). They also described the new genus Hysterodifractum, which has ascospores that disarticulate in two-celled part-spores, similar to Ohleria (Jaklitsch & Voglmayr 2016). However, molecular phylogenetic analyses placed Hysterodifractum in Hysteriales (Carneiro de Almeida et al. 2014). The sixth species of Anteaglonium, A. thailandicum, was added by Jayasiri et al. (2016). Here we describe a new species of Anteaglonium, which morphologically differs from other species in several respects.
Phylogenetically close to the Anteagloniaceae are the Lophiotremataceae, which were differentiated from the Lophiostomataceae by Hirayama & Tanaka (2011) and only contained the single genus Lophiotrema until 2016 (Jaklitsch et al. 2016a). However, Hashimoto et al. (2017) described five new genera in the family including Atrocalyx and erected the three new related families Aquasubmersaceae, Cryptocoryneaceae, and Hermatomycetaceae. Here we describe a new species of Atrocalyx, which morphologically deviates considerably from other species of the genus, notably by brown phragmospores, which lack a gelatinous sheath.
Materials and methods
Isolates and specimens
All newly prepared isolates used in this study originated from ascospores of fresh specimens. Strain identifiers including NCBI GenBank accession numbers of gene sequences used to compute the phylogenetic trees are listed in Tab. 1. Strain acronyms other than those of official culture collections are used here primarily as strain identifiers throughout the work. Representative isolates have been deposited at the Westerdijk Fungal Biodiversity Centre, Utrecht, The Netherlands (CBS culture collection). Details of the specimens used for morphological investigations are listed in the Taxonomy section under the respective descriptions. Specimens have been deposited in the Herbarium of the Institute of Botany, University of Vienna (WU).
Tab. 1. Isolates and accession numbers used in the phylogenetic analyses. Isolates/sequences in bold were isolated/sequenced in the present study.
| Species | Family | Original no. | Specimen no.a | Strain no. | GenBank Accession no |
||||
|---|---|---|---|---|---|---|---|---|---|
| SSU | ITS | LSU | tef1 | rpb2 | |||||
| Amniculicola immersa | Amniculicolaceae | – | CBS H-20226H | CBS 123083 | GU456295 | – | FJ795498 | GU456273 | GU456358 |
| Amniculicola parva | Amniculicolaceae | – | CBS H-20227H | CBS 123092 | GU296134 | – | GU301797 | GU349065 | – |
| Anteaglonium abbreviatum | Anteagloniaceae | – | ANM 925.1 | – | – | – | GQ221877 | GQ221924 | – |
| Anteaglonium abbreviatum | Anteagloniaceae | – | GKM 1029 | – | – | – | GQ221878 | GQ221915 | – |
| Anteaglonium brasiliense | Anteagloniaceae | – | HUEFS 192250H | – | – | – | KF906410 | KF906410 | – |
| Anteaglonium globosum | Anteagloniaceae | – | ANM 925.2H | – | – | – | GQ221879 | GQ221925 | – |
| Anteaglonium globosum | Anteagloniaceae | – | SMH 5283 | – | – | – | GQ221911 | GQ221919 | – |
| Anteaglonium latirostrum | Anteagloniaceae | – | GKM 1119H | – | – | – | GQ221874 | GQ221937 | – |
| Anteaglonium latirostrum | Anteagloniaceae | – | GKM L100Nb | – | – | – | GQ221876 | GQ221938 | – |
| Anteaglonium parvulum | Anteagloniaceae | – | MFLU 16-0473 | MFLUCC 14-0815 | KU922912 | – | KU922911 | KU922919 | – |
| Anteaglonium parvulum | Anteagloniaceae | – | MFLU 16-0472 | MFLUCC 14-0817 | KU922914 | – | KU922913 | – | – |
| Anteaglonium parvulum | Anteagloniaceae | – | MFLU 16-0474 | MFLUCC 14-0821 | KU922916 | – | KU922915 | KU922921 | – |
| Anteaglonium parvulum | Anteagloniaceae | – | MFLU 16-0470 | MFLUCC 14-0823 | KU922918 | – | KU922917 | KU922922 | – |
| Anteaglonium rubescens | Anteagloniaceae | OR | WU 36962 | – | – | MG912909 | MG912909 | MG912913 | MG912917 |
| Anteaglonium rubescens | Anteagloniaceae | OR1 | WU 36960H | CBS 143911 | – | MG912910 | MG912910 | MG912914 | MG912918 |
| Anteaglonium rubescens | Anteagloniaceae | OR2 | WU 36963 | – | – | MG912911 | MG912911 | MG912915 | MG912919 |
| Anteaglonium thailandicum | Anteagloniaceae | – | MFLU 16-0471H | MFLUCC 14-0816 | KU922910 | – | KU922909 | KU922920 | – |
| Antealophiotrema brunneosporum | incertae sedis | – | CBS H-20222H | CBS 123095 | LC194298 | LC194474 | LC194340 | LC194382 | LC194419 |
| Aquasubmersa japonica | Aquasubmersaceae | KT 2813 | HHUF 30468p | MAFF 245218 | LC061581 | LC061591 | LC061586 | LC194383 | LC194420 |
| Aquasubmersa mircensis | Aquasubmersaceae | MFLU 111001H | MFLUCC 11-0401 = IFRDCC 2572 | JX276956 | JX276954 | JX276955 | – | – | |
| Atrocalyx acutisporus | Lophiotremataceae | KT 2436 | HHUF 30504H | MAFF 245613 = NBRC 112316 | LC194299 | LC194475 | LC194341 | LC194386 | LC194423 |
| Atrocalyx asturiensis | Lophiotremataceae | OF | WU 36964H | CBS 143912 | – | MG912912 | MG912912 | MG912916 | MG912920 |
| Atrocalyx bambusae | Lophiotremataceae | – | MFLU 11-0150H | MFLUCC 10-0558 | KX672159 | KX672154 | KX672162 | KX672161 | KX672149 |
| Atrocalyx lignicola | Lophiotremataceae | – | CBS H-20221H | CBS 122364 | LC194300 | LC194476 | LC194342 | LC194387 | LC194424 |
| Byssolophis sphaerioides | incertae sedis | – | – | IFRDCC 2053 | GU456296 | – | GU456318 | GU456263 | GU456348 |
| Crassimassarina macrospora | Lophiotremataceae | KT 1764 | HHUF 29084H | JCM 13096 = MAFF 239606 | LC194302 | LC194478 | LC194344 | LC194389 | LC194426 |
| Cryptoclypeus oxysporus | Lophiotremataceae | KT 2772 | HHUF 30507H | MAFF 245614 = NBRC 112317 | LC194303 | LC194479 | LC194345 | LC194390 | LC194427 |
| Cryptoclypeus ryukyuensis | Lophiotremataceae | KT 3534 | HHUF 30509H | MAFF 245615 = NBRC 112318 | LC194305 | LC194481 | LC194347 | LC194392 | LC194429 |
| Cryptocoryneum akitaense | Cryptocoryneaceae | KT 3019 | HHUF 30477H | MAFF 245365 = NBRC 111758 | LC194306 | LC096154 | LC194348 | LC096136 | LC194430 |
| Cryptocoryneum brevicondensatum | Cryptocoryneaceae | yone 152 | HHUF 30478H | MAFF 245366 = NBRC 111759 | LC194307 | LC096155 | LC194349 | LC096137 | LC194431 |
| Cryptocoryneum condensatum | Cryptocoryneaceae | KT 2892 | HHUF 30479H | MAFF 245367 = NBRC 111760 | LC194311 | LC096159 | LC194353 | LC096141 | LC194435 |
| Cryptocoryneum japonicum | Cryptocoryneaceae | KT 3300 | HHUF 30482H | MAFF 245370 = NBRC 111761 | LC194314 | LC096162 | LC194356 | LC096144 | LC194438 |
| Cryptocoryneum longicondensatum | Cryptocoryneaceae | KT 2913 | HHUF 30486p | MAFF 245374 = NBRC 111762 | LC194318 | LC096166 | LC194360 | LC096148 | LC194442 |
| Cryptocoryneum paracondensatum | Cryptocoryneaceae | KT 3241 | HHUF 30489H | MAFF 245377 = NBRC 111763 | LC194321 | LC096169 | LC194363 | LC096151 | LC194445 |
| Flammeascoma bambusae | Anteagloniaceae | − | MFLU 11–0143H | MFLUCC 10-0551 | KP753952 | KP744440 | KP744485 | − | − |
| Flammeascoma lignicola | Anteagloniaceae | − | MFLU 10-0061H | MFLUCC 10-0128 | KT324584 | KT324582 | KT324583 | KT324585 | KT324586 |
| Galeaticarpa aomori | Lophiotremataceae | KT 2563 | HHUF 30505H | MAFF 245618 = NBRC 112319 | LC194324 | LC194482 | LC194366 | LC194393 | LC194448 |
| Hermatomyces iriomotens | Hermatomycetaceae | KH 361 | HHUF 30518H | MAFF 245730 = NBRC 112471 | LC194325 | LC194483 | LC194367 | LC194394 | LC194449 |
| Hermatomyces tectonae | Hermatomycetaceae | − | MFLU 15-3437H | MFLUCC 14-1140 | KU712465 | KU144917 | KU764695 | KU872757 | KU712486 |
| Hermatomyces thailandica | Hermatomycetaceae | − | MFLU 15-3440H | MFLUCC 14-1143 | KU712468 | KU144920 | KU764692 | KU872754 | KU712488 |
| Lepidosphaeria nicotiae | Testudinaceae | − | − | CBS 101341 | − | − | DQ678067 | DQ677910 | DQ677963 |
| Lophiostoma arundinis | Lophiostomataceae | − | − | CBS 621.86 | DQ782383 | AJ496633 | DQ782384 | DQ782387 | DQ782386 |
| Lophiostoma crenatum | Lophiostomataceae | − | − | CBS 629.86 | DQ678017 | − | DQ678069 | DQ677912 | DQ677965 |
| 'Lophiotrema' boreale | incertae sedis | − | − | CBS 114422 = JCM 14136 | LC194333 | LC194491 | LC194375 | LC194402 | LC194457 |
| Lophiotrema eburnoides | Lophiotremataceae | KT 1424-1 | HHUF 30079H | JCM 17826 = MAFF 242970 | LC001706 | LC001709 | LC001707 | LC194403 | LC194458 |
| Lophiotrema fallopiae | Lophiotremataceae | KT 274 | HHUF 30506H | MAFF 245612 | LC149911 | LC149913 | LC149915 | LC194404 | LC194459 |
| Lophiotrema neoarundinaria | Lophiotremataceae | KT 856 | HHUF 27547 | MAFF 239461 | AB524455 | AB524786 | AB524596 | AB539109 | AB539096 |
| Lophiotrema neohysterioides | Lophiotremataceae | KH 17 | HHUF 30511 | MAFF 245619 | LC194334 | LC194493 | LC194376 | LC194406 | LC194461 |
| Lophiotrema nucula | Lophiotremataceae | − | − | CBS 627.86 = JCM 14132 | AB618703 | LC194497 | AB619021 | LC194410 | LC194465 |
| Lophiotrema vagabundum | Lophiotremataceae | − | F-634236 | CBS 113975 = JCM 14138 | AB618707 | LC194502 | AB619025 | LC194415 | LC194470 |
| Polyplosphaeria fusca | Tetraplosphaeriaceae | − | HHUF 29399H | JCM 13175 = MAFF 239685 | AB524463 | AB524789 | AB524604 | − | − |
| Pseudoastrosphaeriella bambusae | Pseudoastrosphaeriellaceae | − | MFLU 11-0155H | MFLUCC 11-0205 | KT955455 | − | KT955475 | KT955437 | KT955414 |
| Pseudoastrosphaeriella longicolla | Pseudoastrosphaeriellaceae | − | MFLU 11-0207H | MFLUCC 11-0171 | − | − | KT955476 | KT955438 | KT955420 |
| Pseudoastrosphaeriella thailandensis | Pseudoastrosphaeriellaceae | − | MFLU 11-0145H | MFLUCC 10-0553 | KT955456 | − | KT955477 | KT955439 | KT955411 |
| Pseudocryptoclypeus yakushimensis | Lophiotremataceae | KT 2186 | HHUF 30503H | MAFF 245622 = NBRC 112320 | LC194338 | LC194504 | LC194380 | LC194417 | LC194472 |
| Pseudolophiotrema elymicola | incertae sedis | KT 1450 | HHUF 28984H | JCM 13090 = MAFF 239600 | LC194339 | LC194505 | LC194381 | LC194418 | LC194473 |
| Pseudotetraploa curviappendiculata | Tetraplosphaeriaceae | − | HHUF 28582H | JCM 12852 = MAFF 239495 | AB524467 | AB524792 | AB524608 | − | − |
| Quadricrura septentrionalis | Tetraplosphaeriaceae | − | HHUF 28781P | CBS 125429 | AB524474 | AB524799 | AB524615 | − | − |
| Tetraploa sasicola | Tetraplosphaeriaceae | − | HHUF 27566H | JCM 13167 = MAFF 239677 | AB524490 | AB524807 | AB524631 | − | − |
| Triplosphaeria maxima | Tetraplosphaeriaceae | − | HHUF 29390H | JCM 13172 = MAFF 239682 | AB524496 | AB524812 | AB524637 | − | − |
| Ulospora bilgramii | Testudinaceae | − | − | CBS 101364 | DQ678025 | − | DQ678076 | DQ677921 | DQ677974 |
| Verruculina enalia | Testudinaceae | − | − | BCC 18402 | GU479771 | − | GU479803 | GU479864 | GU479836 |
H = holotype; P = paratype.
Culture preparation, growth rate determination and phenotype analysis
Cultures were prepared and maintained as described previously (Jaklitsch 2009) except that 2 % malt extract agar (MEA; 2 % w/v malt extract, 2 % w/v agar-agar; Merck, Darmstadt, Germany) was used as the isolation medium. Cultures used for the determination of growth rates and study of asexual morph micro-morphology were grown on CMD, 2 % MEA or potato dextrose agar (PDA, 39 g/l; Merck, Darmstadt, Germany) at 22–25 °C in darkness. Microscopic observations were made in tap water except where noted. Morphological analyses of microscopic characters were carried out as described earlier (Jaklitsch 2009). Data were gathered using a Nikon Coolpix 995 or Coolpix 4500 or a Nikon DS-U2 digital camera and measured with NIS-Elements D v. 3.0, or with a Zeiss Axiocam 506 colour digital camera and measured with Zeiss ZEN Blue Edition software. Methods of microscopy included stereomicroscopy using an Olympus SZ 60 or Nikon SMZ 1500 and Nomarski differential interference contrast (DIC) using the compound microscopes Nikon Eclipse E600 or Zeiss Axio Imager.A1. For certain images of ascomata the stacking software Zerene Stacker v. 1.04 (Zerene Systems LLC, Richland, WA, USA) was used. Measurements are reported as maximum and minimum in parentheses and the range representing the mean plus and minus the standard deviation of a number of measurements given in parentheses.
DNA extraction and sequencing methods
The extraction of genomic DNA was performed as reported previously (Voglmayr & Jaklitsch 2011, Jaklitsch et al. 2012) using the DNeasy Plant Mini Kit (QIAgen GmbH, Hilden, Germany). The following loci were amplified and sequenced: the complete internally transcribed spacer region (ITS1-5.8S-ITS2) and a ca. 900 bp fragment of the large subunit nuclear ribosomal DNA (nLSU rDNA), amplified and sequenced as a single fragment with primers V9G (de Hoog & Gerrits van den Ende 1998) and LR5 (Vilgalys & Hester 1990); a ca. 1.1 kb fragment of the RNA polymerase II subunit 2 (rpb2) with primers fRPB2-5f and fRPB2-7cr (Liu et al. 1999); and a ca. 1.3–1.5 kb fragment of the translation elongation factor 1-alpha (tef1) with primers EF1-728F (Carbone & Kohn 1999) and TEF1LLErev (Jaklitsch et al. 2005) or EF1-2218R (Rehner & Buckley 2005). PCR products were purified using an enzymatic PCR cleanup (Werle et al. 1994) as described in Voglmayr & Jaklitsch (2008). DNA was cycle-sequenced using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit v. 3.1 (Applied Biosystems, Warrington, UK) with the same primers as in PCR; in addition, primers ITS4 (White et al. 1990), LR2R-A (Voglmayr et al. 2012) and LR3 (Vilgalys & Hester 1990) were used for the ITS-LSU region. In some cases the tef1 was cycle-sequenced with internal primers TEF1_INTF (forward; Jaklitsch 2009) and TEF1_INT2 (reverse; Voglmayr & Jaklitsch 2017). Sequencing was performed on an automated DNA sequencer (3730xl Genetic Analyzer, Applied Biosystems).
Analysis of sequence data
For the phylogenetic analyses, a combined matrix of ITS-LSU, SSU, rpb2 and tef1 sequences was produced. For this, GenBank sequences of selected families of Pleosporales were selected from Hashimoto et al. (2017) and supplemented with GenBank nucleotide sequences of some additional taxa. All alignments were produced with the server version of MAFFT (www.ebi.ac.uk/Tools/mafft), checked and refined using BioEdit version 7.0.9.0 (Hall 1999). Due to the lack of tef1 intron sequence data for most species, only the exon was included for this marker. The combined matrix contained 5054 nucleotide characters, i.e. 2105 from the ITS-LSU, 1001 from the SSU, 1027 from rpb2, and 921 from tef1.
Maximum likelihood (ML) analyses were performed with RAxML (Stamatakis 2006) as implemented in raxmlGUI 1.3 (Silvestro & Michalak 2012), using the ML + rapid bootstrap setting and the GTRGAMMAI substitution model with 1000 bootstrap replicates. The matrix was partitioned for the individual gene regions, and substitution model parameters were calculated separately for them.
Maximum parsimony (MP) bootstrap analysis of the combined matrix was performed with PAUP v. 4.0a159 (Swofford 2002), implementing 1000 bootstrap replicates with 5 rounds of replicates of heuristic search with random addition of sequences and subsequent TBR branch swapping (MULTREES option in effect, steepest descent option not in effect) during each bootstrap replicate. In all MP analyses molecular characters were unordered and given equal weight; analyses were performed with gaps treated as missing data; the COLLAPSE command was set to minbrlen.
Results
Molecular phylogeny
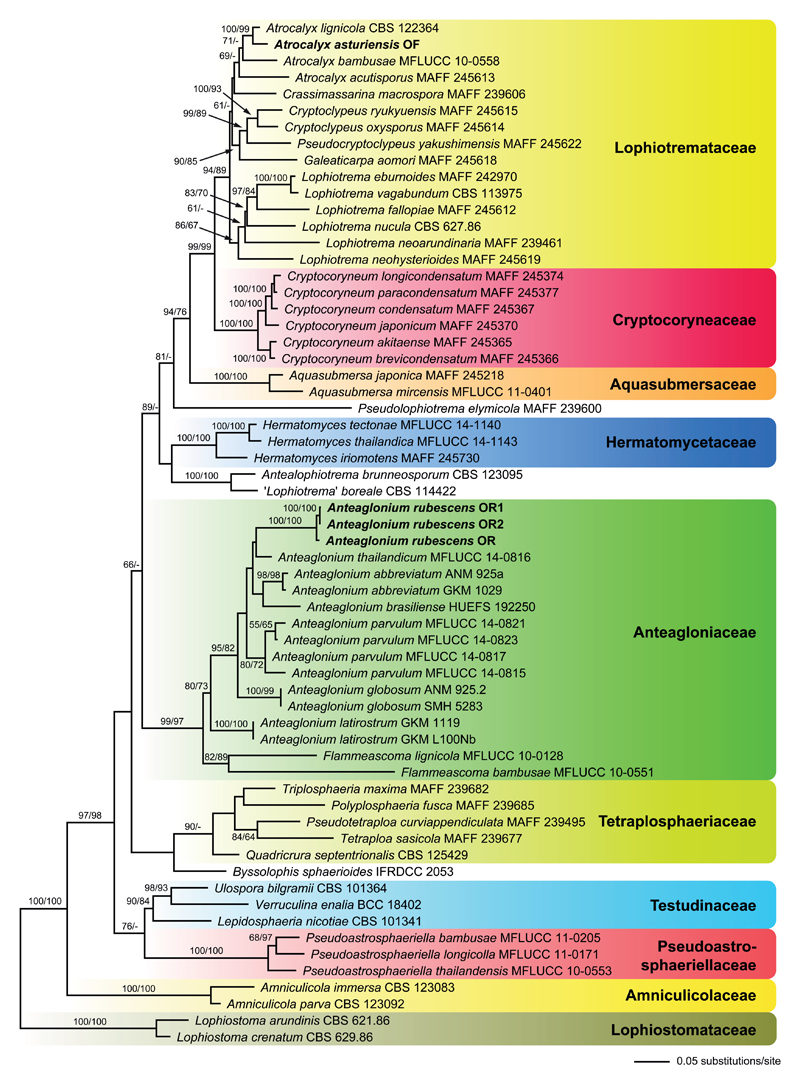
Of the 5054 nucleotide characters of the combined matrix, 1122 are parsimony informative (367 of ITS-LSU, 46 of SSU, 443 of rpb2, and 266 of tef1). The best tree revealed by ML analyses (-lnL = 34887.9896) is shown as phylogram in Fig. 1. Tree topologies are similar to those obtained by Hashimoto et al. (2017) and are therefore not described in detail here, except for the results relevant for the two new species described herein. The Anteagloniaceae receive high support (99 % ML and 97 % MP), containing the two monophyletic genera Anteaglonium and Flammeascoma. Anteaglonium rubescens is placed within the moderately supported Anteaglonium clade, but its closest relatives remain unresolved due to lack of significant bootstrap support of most nodes within Anteaglonium.
Fig. 1.
Phylogram of the best ML tree (lnL = −34887.9896) revealed by RAxML from an analysis of the combined ITS-LSU-SSU-rpb2-tef1 matrix of selected Pleosporales, showing the phylogenetic position of Anteaglonium rubescens and Atrocalyx asturiensis (in bold). ML and MP bootstrap support above 50% are given at the first and second positions, respectively, above or below the branches. Familial classification is according to Hashimoto et al. (2017).
The Lophiotremataceae are highly supported (94 % ML, 89 % MP), but the Atrocalyx clade receives only weak (69 %; ML) or no (MP) support. Atrocalyx asturiensis is revealed as sister species to A. lignicola with maximum (ML) or high (99 %, MP) support.
Taxonomy
Anteagloniaceae
Anteaglonium rubescens Jaklitsch & Voglmayr, sp. nov. – Figs. 2, 3.
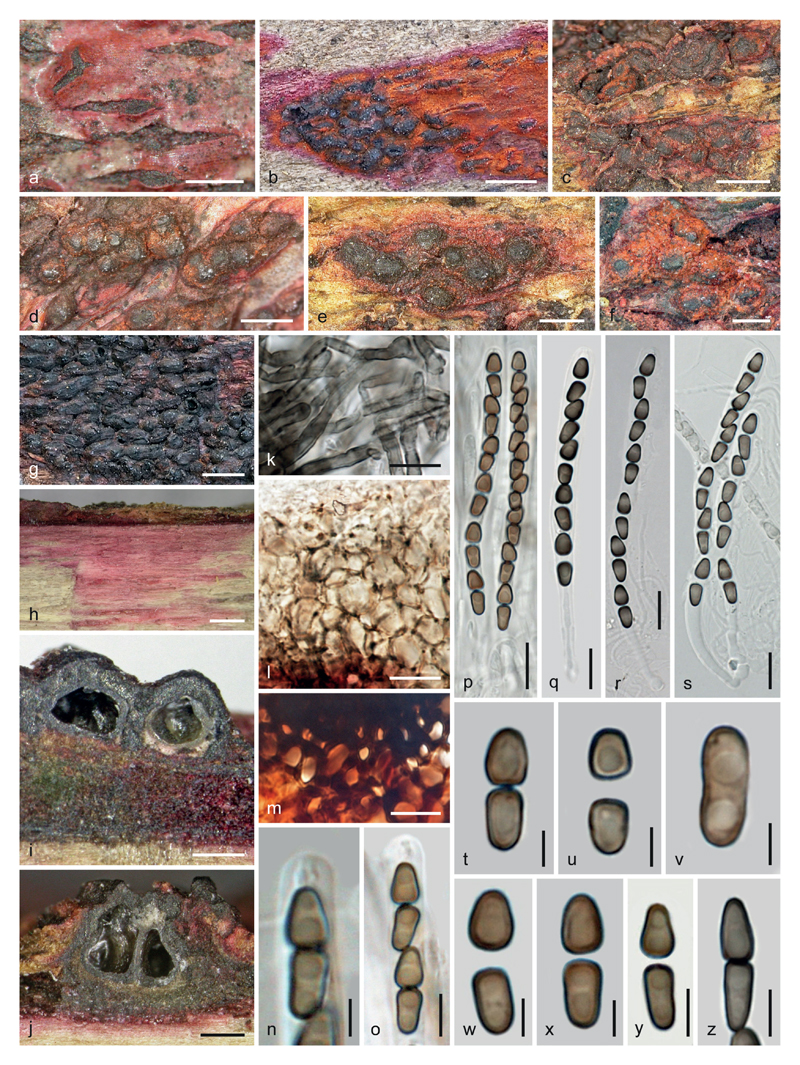
Fig. 2.
Anteaglonium rubescens, sexual morph. a–g. Stromata in face view. h. Wood with purple stain. i, j. Ascomata and stromata in vertical section. k. Subiculum. l. Peridial cells in vertical section. m. Stroma cells in vertical section near subiculum. n, o. Ascus apices with ascospores. p–s. Asci. t–z. Ascospores (t. before disarticulation; v. aberrant, unicellular, non-disarticulating). m, q–s, z. in 3 % KOH. a. WU 36962. b, g, k–m, q, t–v, z. WU 36963. c–f, h–j, n–p, w–y. WU 36960. r, s. WU 36961. b, g. photographs by J. Martin. Scale bars: a, e–g 0.5 mm; b–d, h 1 mm; i, j 200 μm; k–m, p–s 10 μm; n, t–x 3 μm; o, y, z 5 μm.
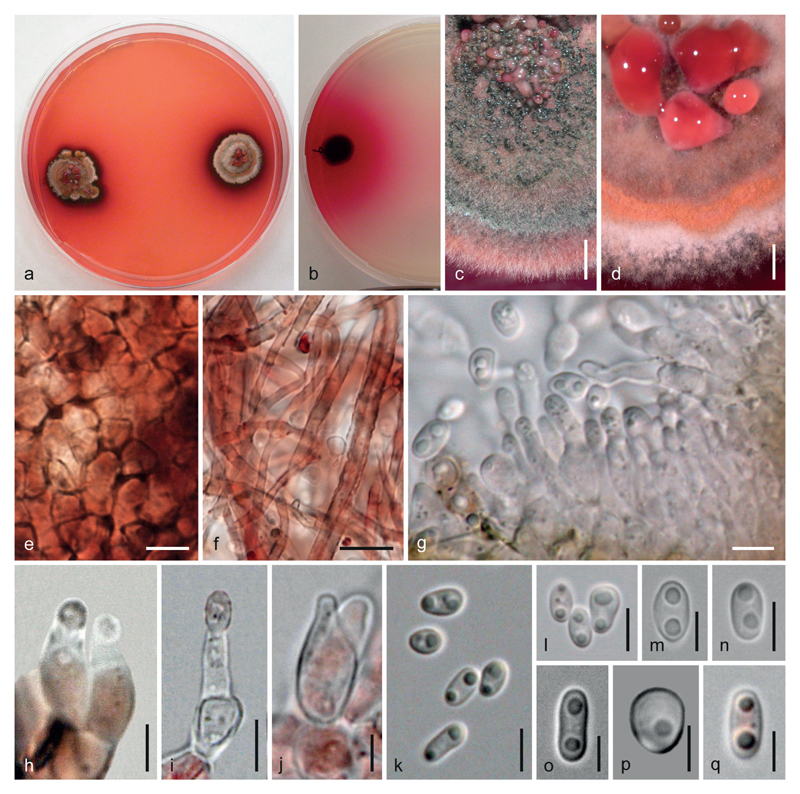
Fig. 3.
Anteaglonium rubescens, asexual morph in culture at 22–25 °C. a, b. Cultures. c, d. Colonies (sections) with pycnidia and conidial drops. e. Pycnidial wall. f. Pycnidial hyphae. g. Short conidiophores and phialides. h, i. Phialides (note thickened collarette in the left phialide in h). j. Prophialide. k–q. Conidia. e, f, h. in 3 % KOH. a, d–f, h, p. CBS 143911. b, c, g, i–o, q. OR. a, b, d–f, h–j, m–p. On/from MEA. c, g, k, l, q. On/from PDA. a, d. After 60 d; b, i, j, m–o. 37 d; c, g, k, l, q. 29 d; e, f, h, p. 22 d. Scale bars: c, d 1.5 mm; e, g, i, k, l, n 5 μm; f 10 μm; h, j, m, o–q 3 μm.
MycoBank no.: MB 824180
Etymology. – rubescens means reddening, due to the reddening of natural and artificial substrates.
Holotype. – GREECE. Corfu, Kanakades, on twigs of Pistacia lentiscus, soc. Xylaria xylarioides and a coelomycete, 20 April 2012, leg. W. Jaklitsch & H. Voglmayr (WU 36960; culture CBS 143911 = OR1).
Description. – Stromata erumpent from wood or immersed in or erumpent from bark as effused dark brown to black zones or crusts, running parallel to the wood surface for several cm or sometimes erumpent as stripes up to 1.8 mm long; in places encasing one to several ascomata, often considerably thickened at the sides and particularly at the apices of the latter, sometimes absent at their bases; parts containing ascomata forming erumpent to superficial roundish pustules with convex or flat, rounded, angular or slightly elongate laterally compressed apices, less commonly with variable black papillae of 25–195 μm diam; in places erumpent as hysterothecioid domes ca. 200–900 μm long and up to ca. 150 μm thick; pseudoparenchymatous, consisting of dark brown, thick-walled cells (2.8)3.2–6(10) μm (n = 40) diam. Hysterothecioid domes dark brown to black, smooth and with a sunken longitudinal slit containing several inconspicuous ostiolar apices; when fresh sometimes with longitudinal crests or striae. Stromata often covered or underlain by brown or brightly orange or red to pink or purple subiculum of septate, thick-walled, smooth to warted, 3–6 μm wide hyphae encrusted with amorphous pigment remaining unchanged in 3 % KOH. Subiculum merging into dark reddish brown pseudoparenchyma of thick-walled cells becoming gradually smaller toward stroma tissue; at the stroma surface sometimes becoming decomposed into amorphous particles; subiculum often also present between wood and bark as compressed tissue. Pigment diffusing and staining wood and bark in patches with pink to purple or violaceous tones. – Ascomata (256)265–367(425) μm diam, (230)285–393(460) μm high (n = 13), globose to subglobose or subconical, individually or collectively encased by stromatic tissue. – Ostioles minute, rounded in section, openings scarcely visible. – Peridium 10–55 μm thick, laterally sometimes up to 90 μm, often appearing thicker due to coalescence with stromatic tissue, pseudoparenchymatous, consisting of rather thin-walled cells (3)4.5–10.5(16) μm (n = 53) diam., red when young, turning pale to dark brown, often surrounded by a whitish to brown, brittle or hard cellular tissue followed by dark stromatic tissue. – Hamathecium trabeculate, consisting of numerous richly branched, 1–2.5(3) μm wide pseudoparaphyses embedded in a gel matrix. – Asci (65)72–87(95) × (4.3)4.7–5.7(6.2) μm (n = 32), cylindrical, bitunicate, fissitunicate, with a refractive ocular chamber, a stipe up to ca. 30 μm long, a simple or knob-like base, containing 4–8 ascospores in uniseriate, sometimes partly biseriate arrangement. – Ascospores 2-celled, dark brown, darkening in 3% KOH, thick-walled, smooth, biconical or dimorphic, cells disarticulating, distal cells broader, subglobose to wedge-shaped, (4.0)4.8–6.0(7.0) × (3.2)3.5–4.0(4.5) μm, l/w (1.2)1.3–1.6(1.9) (n = 75), proximal cell wedge-shaped or oblong, (4.4)5.2–6.7(7.6) × (2.7)3.0–3.6(4.0) μm, l/w (1.3)1.5–2.0(2.6) (n = 75), each containing 1–2 guttules; calculated entire ascospore length (8.5)10–12.5(14.5) μm, l/w (2.4)2.7–3.4(4.0) μm (n = 75).
Cultures. – Growth slow and variable. On MEA colony radius 11–13 mm after 2 months at 22–25 °C; colony thick, surface first whitish, turning greyish, brown to black, often becoming zonate with white margin, orange zones and centre covered with rosy or reddish conidial drops, reverse black; odour indistinct to unpleasant; after a few days pigment diffusing into agar and discolouring surroundings of the colony and later the whole agar intensely pink or red to red-orange. On CMD colony radius up to 5–6 mm after 1 month; colony thick, red, dark brown to black, zonate, with hyaline to reddish conidial drops in the centre; agar turning pink to red. On PDA colony radius ca. 10 mm after 1 month; colony surface dark brown to black, velvety, reverse black; very intense pigment diffusing into agar.
Asexual morph. – Pycnidia formed on CMD, MEA and PDA, scattered to mostly tightly aggregated and confluent in the colony centre, more or less globose with slightly papillate central ostiole, 130–350 μm diam., sometimes with broadened base and often collapsing apically, dark olive-brown to black, often covered with a mat of white or reddish aerial hyphae or by conidial drops. – Pycnidial wall pseudoparenchymatous, consisting of (3)4–7(9) μm (n = 30) wide, thin-walled, inhomogenously pigmented, dark red or dark brown cells incrusted with red pigment, surrounded by red to brown hyphae. Interior lined with a palisade of hyaline to pinkish, lageniform to ampulliform or subglobose, less commonly cylindrical phialides (3.7)5.0–7.0(8.3) × (2.0)2.4–3.7(5.0) μm (n = 37), usually with broad, often thickened collarette; formed on roundish cells and short simple cylindrical conidiophores with roundish base cells. – Conidia very variable, subglobose to globose or ellipsoid to oblong, (2.7)3.5–5.5(7.0) × (2.0)2.3–3.5(4.5) μm, l/w (1.0)1.2–2.0(2.8) (n = 86), 1-celled, with broadly rounded ends, hyaline to pink, with 1–2(3) guttules, smooth, sometimes with 1–2 lateral protuberances.
Habitat. – On wood and bark of various trees and shrubs.
Distribution. – Southern Europe (Greece, Spain), possibly also southern USA (Florida).
Other material examined. – SPAIN. Canary Islands, Tenerife, Anaga mountains, Chinobre, pista cortada, on Laurus novocanariensis, 26 June 2002, leg. W. Jaklitsch W.J. 1908 (WU 36961); Chinobre, La Ensillada, on Laurus novocanariensis, soc. cf. Dothidotthia sp., Xylaria xylarioides, 16 December 2010, leg. H. Voglmayr & W. Jaklitsch (WU 36962; culture OR); Basque Country, Gipuzkoa, Tolosa, in a garden, 30TWN7577, on wood of Rhamnus cathartica, on/soc. black stromatic pyrenomycete, the latter also colonized by Tubeufia cerea and Bisporella sulfurina, 22 June 2016, leg. P.M. Pasaban, comm. J. Martin (WU 36963; culture OR2).
Notes. – Ascomata may sometimes appear larger, particularly wider, due to difficult optical differentiation of stromatic tissue and peridium; only the true peridium was included in measurements. Red or purple colours in specimens are fading with time, particularly in bark: in the specimen WU 36962 the bark was intensely pink coloured one month after collection, six years later the pink colour was gone and had partly turned brown. A possible occurrence of A. rubescens in the USA was deduced from personal communication of W.J. with Margaret Barr in 2002, who reported a specimen collected in Coconut Grove, Miami, Florida (Thaxter 5687, FH; not examined) with virtually identical morphology and tentatively named it Ohleria rubescens.
Lophiotremataceae
Atrocalyx asturiensis Jaklitsch, J. Fourn. & Voglmayr, sp. nov. – Fig. 4.
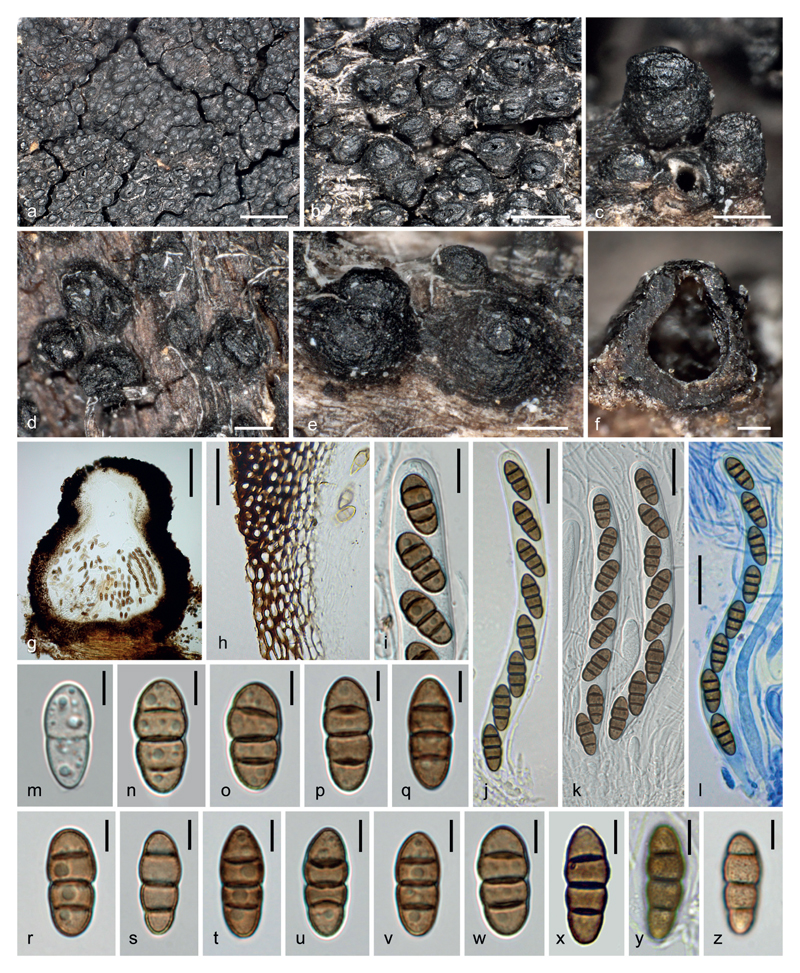
Fig. 4.
Atrocalyx asturiensis (WU 36964). a–e. Ascomata in face and side views. f, g. Ascomata in vertical section. h. Peridium. i. Ascus apex with ascospores. j–l. Asci (l. In Pelikan blue ink diluted in 1 % SDS). m–z. Ascospores (m. immature; y, z. with verruculose ornament). Scale bars: a 1 mm; b 500 μm; c–e 200 μm; f, g 100 μm; h, j–l 20 μm; i 10 μm; m–z 5 μm.
MycoBank no.: MB 824181
Etymology. – asturiensis due to its occurrence in Asturias, Spain.
Holotype. – SPAIN, Asturias, Lago del Valle, elev. ca. 1550 m, on decorticated stump of Cytisus sp., soc. Coniochaeta sp., 6 June 2017, leg. J. Fournier J.F. 17052 (WU 36964; culture CBS 143912 = OF).
Description. – Ascomata erumpent from bleached wood with their bases remaining more or less immersed, loosely to most often densely aggregated and in contact, rarely fusing by 2–3, then forming a black continuous crust to 15 mm in greatest dimension; subglobose to obpyriform, the base rounded to flattened, 320–380(430) μm high, (250)290–380(420) μm diam (n = 30); surface black, somewhat shiny, roughened by shallow wrinkles and low warts, sometimes encrusted with adherent wood fibres. Apex prominent, sharply delimited from the venter or integrated, variable in shape, obtusely rounded to crest-like, typically laterally flattened, ellipsoid in top view, with a longitudinal slit lined by two thick lips, rarely sub-circular in top view with a central rounded pore or Y-shaped, (90)120–170(210) μm high, 170–250(290) μm long, 125–210(240) μm wide (n = 30), less roughened than the venter to almost smooth, eventually turning shiny black. – Peridium dark brown, leathery, (22)38–45(55) μm thick at sides and base, pseudoparenchymatous, of two intergrading regions: outer region textura angularis of small, thick-walled isodiametric to slightly elongated cells 3–9 × 3–4.5 μm, wall dark brown, 1–1.8 μm thick; inner layer textura prismatica of less pigmented cells 8–11 × 2.5–5 μm, wall 0.5–0.8 μm thick; apical wall 70–80 μm thick, blackish brown, comprised of small cells similar to those of the outer layer but thicker-walled to occluded, with 2.5–3.5 μm thick cell walls, internally lined by subhyaline, thin- to thick-walled angular cells merging with the periphyses. – Hamathecium of narrow cellular pseudoparaphyses, basally 1.5–2 μm wide, tapering upwards to 1 μm, ramified and anastomosed above asci, with free ends, embedded in a gelatinous matrix. Asci (117)122–128(132) × 10–12(13.5) μm (n = 12), cylindrical, bitunicate, fissitunicate, with short stipe and furcate base, apically rounded with an ocular chamber, containing 6–8 obliquely uniseriate overlapping ascospores. – Ascospores (15.2)15.8–18.3(19.6) × (5.8)6.2–6.9(7.1) μm, l/w = (2.2)2.4–2.8(3.1) (n = 60), ellipsoid-fusiform with broadly rounded ends, straight, equally 3-euseptate, slightly constricted at the median septum, not to barely constricted at other septa, mid cells slightly swollen, not disarticulating, olivaceous brown to tobacco brown, wall verruculose, without mucilaginous sheath or appendages.
Asexual morph. – Not detected in cultures.
Habitat. – On wood and bark of Cytisus sp.
Distribution. – Southern Europe (Spain), only known from the type location.
Notes. – Atrocalyx asturiensis differs from other species of the genus by brown ascospores, which lack a mucilaginous sheath.
Discussion
Both species described here are difficult to place morphologically. Their phylogenetic positions are rather unexpected. Anteaglonium rubescens differs from all other species in the genus by brown didymospores, which disarticulate within asci, and by the production of a red-orange to pink pigment formed both in nature and in artificial culture. This pigment, which does not change its appearance in 3 % KOH, adds to other interesting substances such as anteagloniolides, which were extracted from cultures of a moss endophyte identified as Anteaglonium sp. and characterized by Xu et al. (2015).
Anteaglonium rubescens is clearly a stromatic fungus. As we did not study other species of the genus, this may be a special feature of this fungus or it was not interpreted or addressed correctly with other species earlier. In none of the earlier publications on the genus Anteaglonium the peridium was studied in detail, but elongate ascomata were interpreted as true hysterothecia. In A. rubescens the apical hysterothecioid dome often remains even when ascomata are decomposed or eaten by insects. A typical dome may contain 3 ascomata, which are separated by very delicate walls. In all specimens of A. rubescens black lines are present in the wood caused by xylariaceous fungi. In material from Tenerife the xylariaceous fungus is Xylaria xylarioides; in WU 36963 the Anteaglonium grows in part directly on stromata of an effused stromatic fungus. These tight associations suggest that A. rubescens may be fungicolous.
Jayasiri et al. (2016) described pycnidial asexual morphs of two Anteaglonium spp. However, the definition of the conidiogenous cells remained unclear, as they give a size of 3–5 × 2–3 μm for both conidiogenous cells and conidia of both species, whereas their illustrations rather indicate lageniform to ampulliform conidiogenous cells similar to those observed by us.
The morphology of Atrocalyx asturiensis, particularly the massive ascomatal apices and brown ascospores, may, e.g., suggest Lophiostoma or Xenolophium (Hirayama & Tanaka 2011, Huhndorf 1993), which differ from Atrocalyx by long-stipitate clavate asci, or Ohleria (Jaklitsch & Voglmayr 2016), a member of Melanommataceae (Jaklitsch & Voglmayr 2017, Mugambi & Huhndorf 2009b) or Teichosporaceae (Jaklitsch et al. 2016b), but molecular analyses place the fungus in Lophiotremataceae. Hashimoto et al. (2017) described five new genera in the family Lophiotremataceae and erected three new related families. Judging from poor phylogenetic support of several clades within Lophiotremataceae, this splitting may seem excessive, on the other hand morphology and particularly asexual morphs were used as arguments for taxonomic conclusions. Here we recognize a new species of their genus Atrocalyx, which morphologically deviates by brown phragmospores, which are not surrounded by a gelatinous sheath. On the other hand, the well-developed crest and particularly the peridium of A. asturiensis as used for differentiation from Lophiotrema by Hashimoto et al. (2017) but also ascus morphology fit well with the generic concept.
Acknowledgements
We are indebted to Pedro Pasaban and Joaquin Martin for fresh material and images and Walter Till and Irmgard Greilhuber for managing collections at WU. The financial support by the Austrian Science Fund (FWF; project P25870-B16) is gratefully acknowledged.
References
- Boehm EWA, Mugambi GK, Miller AN, et al. A molecular phylogenetic reappraisal of the Hysteriaceae, Mytilinidiaceae and Gloniaceae (Pleosporomycetidae, Dothideomycetes) with keys to world species. Studies in Mycology. 2009;64:49–83. doi: 10.3114/sim.2009.64.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. [Google Scholar]
- Carneiro de Almeida DA, Pascholati Gusmão LF, Miller AN. A new genus and three new species of hysteriaceous ascomycetes from the semiarid region of Brazil. Phytotaxa. 2014;176:298–308. [Google Scholar]
- de Hoog GS, Gerrits van den Ende AHG. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hashimoto A, Matsumura M, Hirayama K, Tanaka K. Revision of Lophiotremataceae (Pleosporales, Dothideomycetes): Aquasubmersaceae, Cryptocoryneaceae, and Hermatomycetaceae fam. nov. Persoonia. 2017;39:51–73. doi: 10.3767/persoonia.2017.39.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama K, Tanaka K. Taxonomic revision of Lophiostoma and Lophiotrema based on reevaluation of morphological characters and molecular analyses. Mycoscience. 2011;52:401–412. [Google Scholar]
- Huhndorf SM. Neotropical ascomycetes 3. Reinstatement of the genus Xenolophium and two new species from French Guiana. Mycologia. 1993;85:490–502. [Google Scholar]
- Jaklitsch WM. European species of Hypocrea – Part I. Studies in Mycology. 2009;63:1–91. doi: 10.3114/sim.2009.63.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Komon M, Kubicek CP, et al. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia. 2005;97:1365–1378. doi: 10.3852/mycologia.97.6.1365. [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM, Stadler M, Voglmayr H. Blue pigment in Hypocrea caerulescens sp. nov. and two additional new species in sect. Trichoderma. Mycologia. 2012;104:925–941. doi: 10.3852/11-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Voglmayr H. Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Studies in Mycology. 2016;85:35–64. doi: 10.1016/j.simyco.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch W, Baral H-O, Lücking R, et al. Syllabus of Plant Families - A. Engler's Syllabus der Pflanzenfamilien Part 1/2: Ascomycota. 13th edn. Borntraeger; Berlin: 2016a. [Google Scholar]
- Jaklitsch WM, Olariaga I, Voglmayr H. Teichospora and the Teichosporaceae. Mycological Progress. 2016b;15:31. doi: 10.1007/s11557-016-1171-2. (1–20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Voglmayr H. Three former taxa of Cucurbitaria and considerations on Petrakia in the Melanommataceae. Sydowia. 2017;69:81–95. doi: 10.12905/0380.sydowia69-2017-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasiri SC, Jones EBG, Kang J-C, et al. A new species of genus Anteaglonium (Anteagloniaceae, Pleosporales) with its asexual morph. Phytotaxa. 2016;263:233–244. [Google Scholar]
- Liu YL, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Mugambi GK, Huhndorf SM. Parallel evolution of hysterothecial ascomata in ascolocularous fungi (Ascomycota, Fungi) Systematics and Biodiversity. 2009a;7:453–464. [Google Scholar]
- Mugambi GK, Huhndorf SM. Molecular phylogenetics of Pleosporales: Melanommataceae and Lophiostomataceae recircumscribed (Pleosporomycetidae, Dothideomycetes, Ascomycota) Studies in Mycology. 2009b;64:103–121. doi: 10.3114/sim.2009.64.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- Silvestro D, Michalak I. raxmlGUI: a graphical frontend for RAxML. Organisms Diversity & Evolution. 2012;12:335–337. [Google Scholar]
- Stamatakis E. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* 4.0b10: phylogenetic analysis using parsimony (*and other methods) Sinauer Associates; Sunderland Massachusetts: 2002. [Google Scholar]
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Jaklitsch WM. Prosthecium species with Stegonsporium anamorphs on Acer. Mycological Research. 2008;112:885–905. doi: 10.1016/j.mycres.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Voglmayr H, Jaklitsch WM. Molecular data reveal high host specificity in the phylogenetically isolated genus Massaria (Ascomycota, Massariaceae) Fungal Diversity. 2011;46:133–170. [Google Scholar]
- Voglmayr H, Jaklitsch WM. Corynespora, Exosporium and Helminthosporium revisited – new species and generic reclassification. Studies in Mycology. 2017;87:43–76. doi: 10.1016/j.simyco.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Rossman AY, Castlebury LA, Jaklitsch WM. Multigene phylogeny and taxonomy of the genus Melanconiella (Diaporthales) Fungal Diversity. 2012;57:1–44. [Google Scholar]
- Werle E, Schneider C, Renner M, et al. Convenient single-step one tube purification of PCR products for direct sequencing. Nucleic Acids Research. 1994;22:4354–4355. doi: 10.1093/nar/22.20.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenies. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. Academic Press; San Diego: 1990. pp. 315–322. [Google Scholar]
- Xu Y-M, Mafezoli J, Oliveira MCF, et al. Anteaglonialides A–F and palmarumycins CE1–CE3 from Anteaglonium sp. FL0768, a fungal endophyte of the spikemoss Selaginella arenicola. Journal of Natural Products. 2015;78:2738–2747. doi: 10.1021/acs.jnatprod.5b00717. [DOI] [PubMed] [Google Scholar]