Abstract
Background:
Epigenetic variability is hypothesized as a regulatory pathway through which prenatal exposures may influence child development and health.
Objective:
We sought to examine the associations of residential proximity to roadways at birth and epigenome-wide DNA methylation. We also assessed associations of differential methylation with child cognitive outcomes.
Methods:
We estimated residential proximity to roadways at birth using a geographic information system (GIS) and cord blood methylation using Illumina’s HumanMethylation450-array in 482 mother–child pairs in Project Viva. We identified individual CpGs associated with residential-proximity-to-roadways at birth using robust linear regression []. We also estimated association between proximity-to-roadways at birth and methylation of the same sites in blood samples collected at age 7–11 y (). We ran the same analyses in the Generation R Study for replication (). In Project Viva, we investigated associations of differential methylation at birth with midchildhood cognition using linear regression.
Results:
Living closer to major roadways at birth was associated with higher cord blood (and—more weakly—midchildhood blood) methylation of four sites in LAMB2. For each halving of residential-proximity-to-major-roadways, we observed a 0.82% increase in DNA methylation at cg05654765 [95% confidence interval (CI): (0.54%, 1.10%)], 0.88% at cg14099457 [95% CI: (0.56%, 1.19%)], 0.19% at cg03732535 [95% CI: (0.11%, 0.28)], and 1.08% at cg02954987 [95% CI: (0.65%, 1.51%)]. Higher cord blood methylation of these sites was associated with lower midchildhood nonverbal cognitive scores. Our results did not replicate in the Generation R Study.
Conclusions:
Our discovery results must be interpreted with caution, given that they were not replicated in a separate cohort. However, living close to major roadways at birth was associated with cord blood methylation of sites in LAMB2—a gene known to be linked to axonal development—in our U.S. cohort. Higher methylation of these sites associated with lower nonverbal cognitive scores at age 7–11 y in the same children. https://doi.org/10.1289/EHP2034
Introduction
Residential proximity to major roadways in the United States is associated with traffic-related air and noise pollution, and with social-demographic disparities. The environmental justice literature describes such a phenomenon as “triple jeopardy,” whereby the combination of higher traffic exposures and adverse social-behavioral factors leads to disproportionate effects of traffic pollution on socially disadvantaged population subgroups (Boehmer et al. 2013; Gould and Sutton 2008). There is a growing public health concern that residential proximity to roadways during pregnancy may adversely affect offspring health. Although evidence remains mixed, several recent epidemiological studies have linked prenatal residential proximity to major roadways with a wide range of health problems, including reduced fetal growth (Fleisch et al. 2015), higher systolic blood pressure in newborns (van Rossem et al. 2015), as well as adverse respiratory health (Rice et al. 2014) and cognitive impairment during early and midchildhood (Harris et al. 2015; Yorifuji et al. 2016). However, the identification of intermediate biomarkers linking roadway proximity to health outcomes has been impeded due to their inability to capture and reflect fetal responses to environmental stressors at critical prenatal exposure windows.
Epigenetic marks, such as DNA methylation, undergo dynamic reprogramming during gametogenesis and early preimplantation of embryos (Feil and Fraga 2012; Reik 2007). Experimental evidence shows that the epigenome of mammalian embryonic cells is more susceptible to environmental stimuli than fully differentiated cells are (Dean et al. 1998; Doherty et al. 2000; Khosla et al. 2001; Young et al. 2001), possibly owing to the abundance of de novo DNA methyltransferases in these rapidly dividing pluripotent cells (Feil and Fraga 2012; Reik 2007). Epidemiological studies also revealed that an adverse intrauterine environment [such as changes in nutritional supply (Heijmans et al. 2008; Joubert et al. 2016b), smoking (Joubert et al. 2012; Joubert et al. 2016b; Lee et al. 2015; Markunas et al. 2014; Pilsner et al. 2009; Rzehak et al. 2016; Zhu et al. 2016), environmental chemical exposures (Broberg et al. 2014; Cardenas et al. 2015; Khosla et al. 2001; Kile et al. 2012; Kippler et al. 2013; Pilsner et al. 2009), ambient air pollution (Gruzieva et al. 2017; Herbstman et al. 2012; Kingsley et al. 2016; Tang et al. 2012), and stress (Cao-Lei et al. 2016; Liu et al. 2012; Vidal et al. 2014)] may result in epigenetic perturbation of the developing fetus, which in turn could be associated with increased risks of adverse health outcomes in later life. One classic example of such intergenerational epigenetic inheritance comes from the Hunger Winter Families Study (Lumey et al. 2007), in which Heijmans et al. (Heijmans et al. 2008) showed that individuals who were prenatally exposed to famine had persistent DNA hypomethylation of the imprinted insulin-like growth factor II (IGF2) gene in comparison with their unexposed, sex-matched siblings. These exposed individuals were also more likely to develop obesity and diabetes, perhaps secondary to epigenetic programming that resulted from a mismatch between the intrauterine and postnatal environments (Heijmans et al. 2008). Hence, DNA methylation may represent a biomarker of, or plausible mechanism by which, fetal programming of later-life diseases occurs.
Prenatal/birth cohort studies have examined factors such as maternal diet (Heijmans et al. 2008; Joubert et al. 2016a), smoking (Joubert et al. 2012, 2016b; Lee et al. 2015), and exposure to ambient air pollution (Gruzieva et al. 2017; Tang et al. 2012) during pregnancy as predictors of DNA methylation at birth. However, to our knowledge, associations between residential proximity to major roadways and fetal DNA methylation have not been evaluated. Furthermore, it is unknown if these epigenetic marks observed in cord blood persist into childhood or are predictive of developmental outcomes.
In this study, we sought to evaluate the association between residential proximity to roadways at birth and epigenome-wide DNA methylation in umbilical cord blood samples from newborns in Project Viva, a prenatal/birth cohort study in Massachusetts. We previously reported that residential proximity to major roadways was associated with midchildhood cognitive outcomes in this cohort (Harris et al. 2015). For the present study, we also examined whether epigenetic modifications associated with roadway proximity predicted cognitive function in midchildhood. We hypothesized that living closer to major roadways during pregnancy may be associated with epigenetic alterations in the fetus that would be manifested in cord blood at birth and that these alterations would influence later childhood cognitive outcomes.
Materials and Methods
Study Population
We performed this analysis in Project Viva, a prospective longitudinal birth cohort that enrolled pregnant women during 1999–2002 at their initial obstetric visit at Atrius Harvard Vanguard Medical Associates in eastern Massachusetts. We excluded participants with parity at the time of enrollment, with gestational age at enrollment, who were unable to answer questions in English, and who were intending to move away from the study area before delivery. A detailed description has been published previously (Oken et al. 2015). In brief, we collected information on social-demographic characteristics, lifestyle, medical history, and medications for mothers and children via in-person interviews or mailed questionnaires. We also collected blood at in-person research visits and assessed neurodevelopment of children at midchildhood (, , years). Of the 2,128 mother–child pairs enrolled in the cohort, 482 had complete information on residential proximity to major roadways at birth and cord blood DNA methylation measurements, and 415 participants had complete information on residential proximity to major roadways at birth and midchildhood peripheral white blood cell DNA methylation measurements. We obtained written informed consent from the mothers. All study protocols were reviewed and approved by the Institutional Review Boards of the participating institutions.
Residential Proximity to Major Roadway Measurements
We collected participants’ residential addresses at birth based on maternal self-reported questionnaires. Residential proximity to A1 (primary highway with limited access, i.e., interstate highways and some toll highways) and A2 (primary road without limited access, i.e., federal and state highways) roadways at birth was calculated using geocoded addresses of the participants and ArcGIS 10.1 Street Map™ North America (ESRI) (Fleisch et al. 2014; Harris et al. 2015). Specifically, we used ArcGIS geocoding to transform each residential address to a location on the Earth’s surface. We then used ArcGIS’s Spatial Join tools to calculate the straight-line distance from the geocoded address to the closest road type (A1, A2) for each participant in meters (the software assumed that the Earth is flat and calculates the Euclidean distance).
DNA Methylation Measurements
Umbilical cord blood samples were stored immediately after delivery in a dedicated refrigerator (4°C) and shipped to the laboratory for sample processing within 24 h. Samples were processed on the same day. We collected white blood cell (WBC) pellets from whole blood samples using centrifugation. Umbilical vein cord blood DNA was extracted using the Qiagen Puregene® Kit (Qiagen, N.V.) and bisulfite converted using the EZ DNA Methylation-Gold™ Kit (Zymo Research). Samples were randomly allocated to chips and plates and analyzed using Infinium® HumanMethylation450 BeadChip (Illumina, Inc.) that interrogates CpG sites simultaneously at a single nucleotide resolution, covering 99% of the RefSeq genes. For quality control, we removed samples that were technical replicates, samples with low quality (i.e., if of the probe had a detection ), and samples with genotype mismatch (through comparing replicate samples from the same child) or sex mismatch (through comparing self-reported gender with genetic information from sex chromosomes). At the probe level, we removed low quality probes with nonsignificant detection p-values () (459), probes on sex chromosomes (11,648), single nucleotide polymorphisms (SNP)-associated probes (58,579), nonCpG probes (2,994), as well as nonspecific (3,399) and cross-reactive probes (28,196) (Chen et al. 2013). To further reduce the influence of nearby SNPs, we removed probes within of a known SNP with minor allele frequency (66,094) [Bioconductor Illumina 450K Probe Variants.db (Genome Build 37) (1000 Genomes Project Consortium et al. 2012)]. After exclusion, we had 314,208 probes that passed quality control in 482 cord blood samples. In the preprocessing step, we applied the normal-exponential out-of-band (“noob”) method for background correction and dye bias adjustment (Triche et al. 2013). We further normalized our sample using Beta Mixture Quantile Dilation (BMIQ) to adjust the distribution of type 2 design probes into a statistical distribution characteristic of type 1 design probes (Teschendorff et al. 2013). We used an empirical Bayes method (ComBat in the Bioconductor sva package; version 3.7) to adjust for batch effects resulting from technical variability (Johnson et al. 2007). Further, we plotted our exposure of interest—distance to A1/A2 road at birth by batch. We also performed a one-way analysis of variance (ANOVA) of distance to A1/A2 road at birth by batch, and observed marginal significance p-value (0.06). Because batch was marginally associated with our exposure and may be associated with methylation values (confounder), we adjusted for batch to avoid biases in our results. We used sample plate (96-well) and scanner as a batch covariate and adjusted for in ComBat. We applied the same quality control (at sample level and at probe level), normalization and batch effect adjustment protocols to process peripheral blood samples at the midchildhood in-person clinical visits (age 7–11). We retained 314,208 probes that passed quality control in 460 peripheral blood samples. We report the degree of methylation for individual CpG sites as beta values, which were expressed as the percentage of methylated cytosines over the sum of methylated and unmethylated cytosines at the 5C position.
In Project Viva, we have 42 replication samples (32 replicates for one child and 16 replicates for another child). For the first child, there are 30 replicates for cord blood DNA methylation, 1 for early childhood and 1 for midchildhood peripheral blood DNA methylation. Mean [interquartile range (IQR)] for the pairwise mean-centered correlation (Spearman) was 0.979 (0.009) for all 32 technical replicates. For the second child, we had 14 replicates for cord blood DNA methylation, 1 for early childhood and 1 for midchildhood peripheral blood DNA methylation. Mean (IQR) for the pairwise mean-centered correlation (Spearman) was 0.979 (0.010) for all 16 technical replicates.
Cellular heterogeneity is a significant source of DNA methylation variation. Thus, we adjusted for estimated cell proportions in cord blood and in peripheral blood to reduce confounding by shifts in cell populations. Distinct from adult peripheral blood, cord blood of newborns contains nRBCs in addition to various WBCs. We estimated cord blood cell proportions using a validated cord blood reference panel developed by Bakulski et al. (Bakulski et al. 2016) to capture the distinct nucleated cellular signature in the cord blood. Specifically, actual cord blood cell counts were measured using florescence activated cell sorting (FACS). Differences between DNA methylation patterns for each sorted cell type were assessed using principle component analysis (PCA) and F-test statistics. Pairwise t-tests were used to identify methylation sites that most distinguished a specific cell type (i.e., the top 100 probes with the greatest absolute values of magnitude of methylation difference for each cell type with a ). These cord blood cell estimates were then validated with two independent cohorts with cord blood samples (Bakulski et al. 2016).
We estimated peripheral blood cell proportion using the method of Houseman (Houseman et al. 2012). In brief, Houseman’s estimations used a method adapted from regression calibration to identify changes in the distribution of WBC subpopulations associated with case–control status using DNA methylation signatures (Houseman et al. 2012). This method has also been validated with external datasets (Houseman et al. 2012).
Cognitive Outcomes
During the midchildhood in person visits, trained personnel from Project Viva administered a battery of cognitive tests at in-person visits, including the Kaufman Brief Intelligence Test, second edition (KBIT-2) (Kaufman and Kaufman 2004), the visual memory index of the Wide Range Assessment of Memory and Learning, second edition (WRAML2) (Sheslow and Adams 2003) and the visual-motor subtest of the Wide Range Assessment of Visual Motor Abilities (WRAVMA) (Adams and Sheslow 1995). The KBIT-2 yields verbal and nonverbal subscores. The verbal test measures crystalized intelligence, which includes an individual’s vocabulary and depth and breadth of general knowledge; the nonverbal test measures fluid intelligence, which includes an individual’s ability to perceive relationships and complete visual analogies (Kaufman and Kaufman 2004). The visual memory index of WRAML2 measures picture memory and design memory (Sheslow and Adams 2003), whereas the visual-motor subtest of WRAVMA captures both fine motor skills and visuospatial perception (Adams and Sheslow 1995). In the present study, we analyzed the verbal (KBIT-2 verbal test) and nonverbal (KBIT-2 nonverbal test) components of KBIT-2, picture memory (WRAML2 picture memory standard score), design memory (WRAML2 design memory standard score), and the summary of the two (WRAML2 visual memory standard score). We also analyzed WRAVMA standard score.
Statistical Analysis
Prenatal residential proximity to major roadways and DNA methylation.
CpG-by-CpG analysis.
We evaluated the association of residential proximity to major roadways at birth with epigenome-wide DNA methylation at each individual CpG site using robust linear regression to down-weight the influence of extreme methylation values and reduce the problem associated with heteroscedasticity. We natural log-transformed residential proximity to major roadways, as the influence of distance to roadways is thought to increase exponentially when moving closer to the roadway. Model estimates are expressed as percentage of differential methylation level for a halving of distance to major roadways. In each model, we adjusted for the following covariates selected a priori: maternal characteristics [age at enrollment (continuous), prepregnancy body mass index (weight in in , continuous), race/ethnicity (white, black, or others (Asian/Hispanic/American Indian/more than one race or race that did not fall into any of these categories), smoking status (never, former, or during pregnancy), education level (college/graduate level or not college/graduate level)], neighborhood income [census tract median household income at enrollment (continuous)], child characteristics [gestational age at birth (continuous), sex (female or male), season of birth (spring, summer, fall, winter)], and estimated cord blood cell proportions [percentages of monocytes, CD8T cells, CD4T cells, NK cells, B cells, and nucleated red blood cells (nRCB)] (Bakulski et al. 2016; Cardenas et al. 2016). A CpG site was considered to be epigenome-wide significant if it reached the FDR-corrected p-value of based on the method of Benjamini and Hochberg (Benjamini and Hochberg 1995). We checked for technical artifacts by calculating the genomic inflation factor () and plotting the q-q plot. The genomic inflation factor was calculated based on the chi-square goodness of fit test and was defined as the median of the resulting chi-square test statistics (observed) divided by the expected median of the chi-square distribution (expected). Lambda equals to 1 indicates that there is no inflation or systematic bias. A value close to 1 indicates that there is little inflation.
DNA methylation regional analysis.
As a secondary analysis, we applied a regional-based approach to identify differentially methylated regions associated with prenatal residential proximity to major roadways. We used an existing package from Bioconductor titled DMRcate (Peters et al. 2015). In brief, DMRcate identifies and characterizes differentially methylated regions spatially across the genome using a tunable Gaussian kernel-smoothing method. DMRcate first fits individual linear regressions and calculates a t statistic for each CpG site using limma, and then uses unsigned weights (limma’s ) to pass the kernel estimator. We set the Gaussian kernel bandwidth for smoothed function estimation to be 1,000 nucleotides. We used Benjamini-Hochberg (BH) method for p-value adjustment for the significance test (Benjamini and Hochberg 1995). We set p-value cutoff to determine differential methylation region to be .
Sensitivity analyses.
In the sensitivity analyses, we repeated analyses of CpG sites associated with proximity with FDR-corrected () after stratifying by sex and white or nonwhite race/ethnicity (where nonwhite comprises black and “other” race/ethnicity).
In addition, we used multivariate robust linear regression to evaluate whether differential methylation of specific CpG sites in association with residential proximity to roadways at birth () was still evident in peripheral blood samples collected from a subset of the children in midchildhood (7–11 years of age, ). In the regression model, we additionally adjusted for children’s age at blood draw and blood cell proportions estimated from an adult reference panel (Houseman et al. 2012). We examined the correlations between residential proximity to major roadways and estimated cell proportions to assess whether our observed associations might be due to changes in actual DNA methylation levels or changes in cell proportions at birth and in midchildhood.
Replication
We performed an epigenome-wide replication in The Generation R study—a prospective birth cohort based in Rotterdam, Netherlands. All study protocols were reviewed and approved by the Institutional Review Boards of the participating institutions. Cord blood DNA methylation was measured using Illumina’s Infinium® HumanMethylation450 BeadChip (Illumina Inc.). We removed eight low quality samples (i.e., sample call rate or poor bisulfite conversion), two sex mismatch samples, and one sample that retracted informed consent. We performed background adjustment, between-array normalization, and dye bias correction using DASES, which is a modified normalization pipeline based on DASEN (Pidsley et al. 2013). We report the degree of methylation for individual CpG sites as beta values, which were expressed as the percentage of methylated cytosines over the sum of methylated and unmethylated cytosines at the 5C position. A subset of 641 mother–child pairs had complete information on residential proximity to major roadways and cord blood DNA methylation measurements. We natural log-transformed residential proximity to major roadways; model estimates are expressed as percentage of differential methylation level per halving of distance to major roadways. In each model, we controlled for the following set of covariates: maternal age at enrollment (continuous), maternal prepregnancy body mass index [, continuous], maternal smoking status (never, ever during pregnancy), maternal education level (college/graduate level or not college/graduate level), gestational age (continuous), child’s sex (female or male), season of birth (spring, summer, fall, winter), estimated cell proportions for cord blood using the Houseman’s method and batch effects (dummy variables for plate).
DNA methylation and cognition.
Because of LAMB2’s well-defined role in axonal development and neurotransmission, and because we previously found associations of residential proximity to roadways and cognitive outcomes, we fit multivariate linear regression models to examine the association between LAMB2 related CpG sites () in the cord blood with each cognitive test measured at midchildhood, which includes KBIT-2 verbal test (), KBIT-2 nonverbal test (), visual-motor subtest of WRAVMA (), design memory (), and picture memory () of WRAML2 and WRAML visual standard score (). Model estimates are expressed as the difference in cognitive test scores with a 10% increase in DNA methylation values. We adjusted for maternal characteristics [age at enrollment (continuous), race/ethnicity (white, black, or others), smoking status (never, former, or during pregnancy), education level (college/graduate level or not college/graduate level)] and child characteristics [age at cognitive test (continuous), sex (female or male)].
Blood–Brain DNA Methylation Comparison
Given the potential function in neuronal development of the gene identified in our main analysis, we leveraged a publicly available database in the Gene Expression Omnibus (GEO) repository and evaluated the interindividual methylation variation across blood and brain for the top CpG sites we identified (http://epigenetics.essex.ac.uk/bloodbrain/). A detailed protocol has been published elsewhere (Hannon et al. 2015). In brief, DNA methylation was analyzed using Infinium® HumanMethylation450 BeadChip (Illumina, Inc.) in matched DNA samples from both premortem whole blood and postmortem brain tissues from the same 122 adult participants. The four brain regions assessed include prefrontal cortex, entorhinal cortex, superior temporal gyrus, and cerebellum. Pearson correlation was calculated for each blood–brain CpG pair to reflect the covariations of DNA methylation across blood and brain.
All analyses were performed in SAS® version 9.4 (SAS Institute, Inc.) and R version 3.2.4 (R Core Team).
Results
Study Population
A total of 482 mother–child pairs in Project Viva had complete information on residential proximity to major roadways at birth and cord blood DNA methylation measurements (Table 1). Among the 482 pairs, 11% of mothers smoked during pregnancy, and 21% were former smokers. Sixty-six percent of the mothers completed college or graduate school education. Median neighborhood household income was , based on census-tract information. Roughly half the offspring were female (48%), and mean gestational age was 39.7 wk at birth. The study population was predominantly white (71%).
Table 1.
Characteristics of study population in Project Viva and the Generation R study.
| Variablea | Project viva | The Generation R study | p value |
|---|---|---|---|
| () | () | ||
| Maternal characteristics | |||
| Age at enrollment (years), mean (SD) | 32.1 (5.4) | 31.6 (4.1) | 0.078 |
| Pre-pregnancy BMI (), mean (SD) | 24.7 (5.3) | 24.3 (4.2) | 0.159 |
| Maternal race, N (%) | |||
| White | 342 (71%) | 641 (100%) | |
| Black | 57 (12%) | — | |
| Otherb | 83 (17%) | — | |
| Maternal smoking status, N (%) | |||
| Smoking during pregnancy | 54 (11%) | 160 (25%) | |
| Former | 102 (21%) | 481 (75%)c | |
| Never | 326 (68%) | ||
| Maternal education, N (%) | 0.849 | ||
| College or graduate level | 319 (66%) | 420 (65.5%) | |
| Not college or graduate level | 163 (34%) | 221 (34.5%) | |
| Median neighborhood household income (), mean (SD) | 57.0 (20.9) | — | — |
| Distance to A1&2 roadways at birth (meters), mean (SD) | 1881.2 (1911.4) | 351 (201) | |
| Child characteristics | |||
| Child’s sex, N (%) | 1.000 | ||
| Female | 230 (48%) | 307 (48%) | |
| Male | 252 (52%) | 334 (52%) | |
| Gestational age (weeks), mean (SD) | 39.7 (1.6)d | 40.2 (1.5) | |
| Date of birth by season, N (%) | 0.244 | ||
| Spring | 123 (26%) | 158 (25%) | |
| Summer | 119 (25%) | 173 (27%) | |
| Fall | 111 (23%) | 168 (26%) | |
| Winter | 129 (27%) | 142 (22%) |
Note: We included maternal alcohol consumption for the first trimester during pregnancy in the analysis of cord blood DNA methylation and cognitive test scores (Table 4 Model 2), is glass per week ().
Data were complete for all variables unless otherwise indicated.
Other races included Hispanics, Asian, children with more than one race and children whose race was not specified in those categories.
In the Generation R Study, women who never smoked and women who did not smoke during pregnancy were in the same category.
For Project Viva, 455 out of 482 children (94%) were born after 37 weeks of gestation.
Prenatal Residential Proximity to Major Roadways and DNA Methylation
CpG-by-CpG analysis.
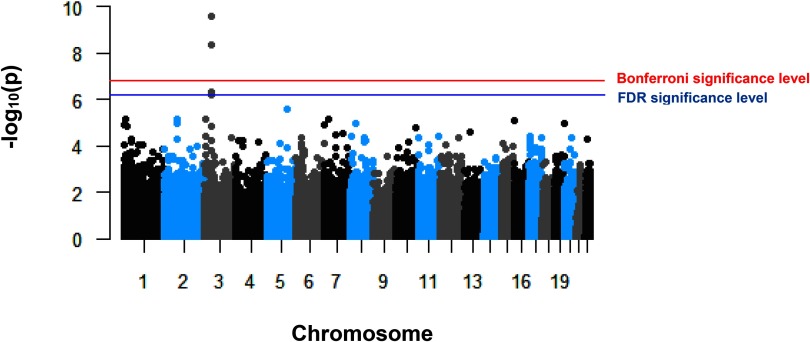
In the primary analysis, we performed a CpG-by-CpG analysis and examined the association of residential proximity to major roadways at birth with epigenome-wide DNA methylation in cord blood (Figure 1) (see Excel Table S1 for complete numeric data). Genomic inflation factor was 0.97, and there was no evidence of early deviation from the q-q plot (Figure S1). Four CpG sites within the same gene, LAMB2, were differentially methylated in association with living closer to major roadways at birth () (Table 2) (see Table S1 for descriptive data on methylation levels and Figure S2 for scatter plots of methylation and roadway proximity at the four sites). For each halving of residential proximity to major roadways, we estimated a 0.82% increase in DNA methylation at cg05654765 [95% confidence interval (CI): (0.54%, 1.10%), ], 0.88% at cg14099457 [95% CI: (0.56%, 1.19%), ], 0.19% at cg03732535 [95% CI: (0.11%, 0.28%), ], and 1.08% at cg02954987 [95% CI: (0.65%, 1.51%), ]. For example, in comparison with a mother who lived away from a major roadway, a mother who lived away had 0.82% higher methylation value at cg05654765 loci. Interestingly, all those CpG sites were located within of the transcription start site (TSS) of LAMB2 and spanned a distance.
Figure 1.
Manhattan plot of the association between distance to roadway at birth and epigenome wide DNA methylation in cord blood in Project Viva ().
Table 2.
Associations between a 50% reduction in residence proximity to major roadways at birth and methylation of CpG sites in LAMB2 with significant differential methylation in cord blood DNA () and corresponding associations with methylation of the same sites in peripheral blood samples at age 7–11 ().
| CpG | Gene | Cord blooda | Peripheral blood at midchildhoodb | ||||
|---|---|---|---|---|---|---|---|
| Estimate (95% CI) | P-value | FDRc | Bonferroni | Estimate (95% CI) | P-value | ||
| cg05654765 | LAMB2 | 0.82% (0.54%, 1.10%) | 0.37% (0.01%, 0.76%) | 0.0542 | |||
| cg14099457 | LAMB2 | 0.88% (0.56%, 1.19%) | 0.53% (0.07%, 1.00%) | 0.0244 | |||
| cg03732535 | LAMB2 | 0.19% (0.11%, 0.28%) | 0.22% (0.01%, 0.45%) | 0.0578 | |||
| cg02954987 | LAMB2 | 1.08% (0.65%, 1.51%) | 0.56% (0.02%, 1.11%) | 0.0412 | |||
Estimates of differential methylation in cord blood are from linear regression models adjusted for maternal [age at enrollment (continuous), prepregnancy body mass index (BMI) (continuous), race (white/black/others), smoking status (smoking during pregnancy/former/never), education level (college or graduate/not college or graduate)], neighborhood [median household income (continuous)], children [child’s sex (female/male), gestational age (continuous), season (DOB)], and estimated cell proportions (percentage of monocyte, percentage of CD8T cell, percentage of CD4T cell, percentage of NK cell, percentage of B cell, percentage of nucleated red blood cells).
Models of differential methylation in peripheral blood (age 7–10 y) are adjusted for maternal age at enrollment, pre-pregnancy BMI, race/ethnicity, smoking during pregnancy, education, median neighborhood household income, child’s sex, gestational age at birth, season of birth (spring, summer, fall, winter), child’s age at blood donation, and proportions of blood cells estimated using the Houseman method (Houseman et al. 2012) (monocytes, CD8 cells, CD4 cells, NK cells, B cells).
FDR correspond to Benjamini-Hochberg (BH) adjusted p-value ().
DNA methylation regional (DMR) analysis.
As a secondary analysis, we conducted a regional analysis to identify differentially methylated regions in cord blood in relation to prenatal residential proximity to major roadways at birth (Table 3). We identified three genes with differentially methylated regions, located in or near genes LAMB2 (chr3:49,170,496–49,171,343; TSS1500–first exon), C2orf65 (chr2:74,875,227–74,875,387; TSS200–TSS1500), and PPP1R2P1 (chr6: 32,847,112–32,847,845; gene body). The four FDR-significant methylation sites that we identified from the CpG-by-CpG analysis were also included in the LAMB2 region. Table 3 shows the CpGs included in each of the DMRs.
Table 3.
Associations between a 50% reduction in residence proximity to major roadways at birth and methylation of CpG sites (cord blood) in gene regions with Gaussian kernel bandwidth of 1,000 nucleotide and a p-value cutoff of ().
| CHR | Gene | CpG | Gene group | CHR location | % change |
|---|---|---|---|---|---|
| 3 | LAMB2 | cg01919208 | 1st exon | 49170496 | 0.47% |
| 3 | LAMB2 | cg02954987 | TSS200 | 49170599 | 1.04% |
| 3 | LAMB2 | cg05654765 | TSS200 | 49170727 | 0.81% |
| 3 | LAMB2 | cg14099457 | TSS200 | 49170794 | 0.90% |
| 3 | LAMB2 | cg03732535 | TSS1500 | 49171051 | 0.20% |
| 3 | LAMB2 | cg01638175 | TSS1500 | 49171179 | |
| 3 | LAMB2 | cg05323436 | TSS1500 | 49171185 | |
| 3 | LAMB2 | cg02497019 | TSS1500 | 49171343 | |
| 2 | C2orf65 | cg20945085 | TSS200 | 74875227 | 0.21% |
| 2 | C2orf65 | cg02109484 | TSS200 | 74875253 | 0.36% |
| 2 | C2orf65 | cg17019053 | TSS200 | 74875263 | 0.36% |
| 2 | C2orf65 | cg00273449 | TSS200 | 74875268 | 0.20% |
| 2 | C2orf65 | cg17105755 | TSS200 | 74875317 | 0.44% |
| 2 | C2orf65 | cg02611848 | TSS1500 | 74875387 | 0.35% |
| 6 | PPP1R2P1 | cg02269978 | Body | 32847585 | 0.49% |
| 6 | PPP1R2P1 | cg24639679 | Body | 32847590 | 0.65% |
| 6 | PPP1R2P1 | cg19628603 | Body | 32847596 | 0.53% |
| 6 | PPP1R2P1 | cg00797104 | Body | 32847605 | 0.42% |
| 6 | PPP1R2P1 | cg17352045 | Body | 32847702 | 0.55% |
| 6 | PPP1R2P1 | cg00892368 | Body | 32847704 | 0.58% |
Note: Model adjusted for maternal [age at enrollment (continuous), pre-pregnancy BMI (continuous), race (white/black/others), smoking status (smoking during pregnancy/former/never), education level (college or graduate/not college or graduate)], neighborhood [median household income (continuous)], children [child’s sex (female/male), gestational age (continuous), season (DOB)], and estimated cell proportions (percentage of monocyte, percentage of CD8T cell, percentage of CD4T cell, percentage of NK cell, percentage of B cell, percentage of nucleated red blood cell). .
Sensitivity analyses.
We repeated analyses of the FDR-significant CpG sites near LAMB2 after stratifying by sex and by race/ethnicity (white or nonwhite) (Table S2). Methylation at each CpG site was positively associated with roadway proximity in males and females, and in white and nonwhite children. Although estimates were imprecise and differences between strata were not significant (interaction ), the percentage increase in methylation associated with a 50% reduction in roadway proximity was larger in nonwhite vs. white children, and in female vs. males, at all four sites. Effect estimates were in the same direction for the four FDR-significant CpG sites near LAMB2 in each of the restricted analyses in comparison with the full subcohort (the associations attenuated in the white and male strata).
To evaluate whether the association of those differentially methylated CpG sites at LAMB2 persisted in midchildhood, we evaluated the association between residential proximity at birth and peripheral blood DNA methylation in blood samples collected when participants were 7–11 y old. Associations between roadway proximity at birth and methylation in midchildhood samples differed from corresponding associations with methylation at birth but remained positive for each CpG sites, with each halving of prenatal residential proximity to major roadways associated with increased DNA methylation: 0.37% (95% CI: 0.01, 0.76%) at cg05654765 (vs. 0.82% in cord blood); 0.53% (95% CI: 0.07, 1.00%) at cg14099457 (vs. 0.88% in cord blood); 0.22% (95% CI: 0.01, 0.45%) at cg03732535 (vs. 0.19% in cord blood); and 0.56% (95% CI: 0.02, 1.10%) at cg02954987 (vs. 1.08% in cord blood) (Table 2). Cord blood and midchildhood methylation levels were positively correlated for each of the FDR-significant CpG sites, with Spearman correlation coefficients ranging from 0.53 (for cg03732535) to 0.83 (for cg05654765) (Figure S3). Residential proximity to major roadways at birth was not correlated with proportions of individual blood cell subtypes in cord blood or midchildhood peripheral blood samples (Spearman correlations ranging from to 0.04 and from to 0.03, respectively) (Figure S4).
Replication
The Generation R Study population used for the replication analysis () included white women only (in comparison with 71% of the Project Viva population), and participants lived closer to major roadways (mean distance vs. for Project Viva), and were more likely to have smoked during pregnancy (25% vs. 11%) (Table 1). None of the LAMB2 CpG sites that were significantly associated with major roadway proximity at birth in Project Viva were associated with major roadway proximity in the Generation R Study (Table 4). Estimated differences in methylation with a 50% reduction in distance to major roadways ranges from a decrease of [95% CI: (, 0.43%), ] for cg02954987 to an increase of 0.06% [95%, CI: (, 0.24%), ] for cg05654765.
Table 4.
Differentially methylation site in cord blood in relation to per halving of distance to major roadways in the Generation R study ().
| CHR | CpG | Gene | Gene group | Estimate (95% CI) | P-value |
|---|---|---|---|---|---|
| 3 | cg05654765 | LAMB2 | TSS200 | (, 0.24%) | 0.68 |
| 3 | cg14099457 | LAMB2 | TSS200 | (, 0.28%) | 0.82 |
| 3 | cg03732535 | LAMB2 | TSS1500 | 0.08% (, 0.22%) | 0.25 |
| 3 | cg02954987 | LAMB2 | TSS200 | 0.09% (, 0.43%) | 0.61 |
Note: Model adjusted for maternal [age at enrollment (continuous), pre-pregnancy BMI (continuous), race (white/black/others), smoking status (smoking during pregnancy/former/never), education level (college or graduate/not college or graduate)], neighborhood [median household income (continuous)], children [child’s sex (female/male), gestational age (continuous), season (DOB)], and estimated cell proportions (percentage of monocyte, percentage of CD8T cell, percentage of CD4T cell, percentage of NK cell, percentage of B cell, percentage of nucleated red blood cells). .
Cord Blood DNA Methylation and Cognitive Test Scores
We estimated associations between cognitive test scores during midchildhood and a 10% increase in methylation at the four LAMB2 CpG sites that were significantly associated with roadway proximity at birth (Table 5). KBIT-2 nonverbal cognitive test scores () were inversely associated with methylation at all four of the sites evaluated, with an average reduction of points [95% CI (, ), ] for cg05654765, points [95% CI (, 0.00), ] for cg14099457, points [95% CI (; ), ] for cg03732535, and points [95% CI (, 0.11), ] for cg02954987 (). In general, associations with other cognitive test scores were close to the null.
Table 5.
Estimated differences in cognitive test scores at age7–11 y in association with a 10% increase in methylation of LAMB2 CpG sites in cord blood.
| CpGd | Model 1a | Model 2b,c | ||||
|---|---|---|---|---|---|---|
| N | Estimate (95% CI) | P-value | N | Estimate (95% CI) | P-value | |
| KBIT-2 verbal test | ||||||
| cg05654765 | 415 | 0.70 (, 2.78) | 0.51 | 382 | 0.49 (, 2.67) | 0.66 |
| cg14099457 | 415 | 0.70 (, 2.58) | 0.46 | 382 | 0.67 (, 2.62) | 0.50 |
| cg03732535 | 415 | 0.88 (, 6.62) | 0.76 | 382 | 1.04 (, 7.10) | 0.74 |
| cg02954987 | 415 | 0.54 (, 1.81) | 0.41 | 382 | 0.41 (, 1.71) | 0.54 |
| KBIT-2 nonverbal test | ||||||
| cg05654765 | 418 | (, ) | 0.03 | 385 | (, ) | 0.04 |
| cg14099457 | 418 | (, 0.00) | 0.05 | 385 | (,) | 0.05 |
| cg03732535 | 418 | (,) | 0.002 | 385 | (, ) | 0.007 |
| cg02954987 | 418 | (, 0.11) | 0.07 | 385 | (, 0.07) | 0.06 |
| WRAML2 picture memory standard score | ||||||
| cg05654765 | 417 | 0.02 (, 0.50) | 0.93 | 384 | (, 0.44) | 0.79 |
| cg14099457 | 417 | 0.06 (, 0.49) | 0.80 | 384 | (, 0.45) | 0.98 |
| cg03732535 | 417 | (, 1.25) | 0.90 | 384 | (, 1.13) | 0.69 |
| cg02954987 | 417 | (, 0.26) | 0.80 | 384 | (, 0.26) | 0.79 |
| WRAML2 design memory standard score | ||||||
| cg05654765 | 416 | 0.13 (, 0.56) | 0.57 | 383 | 0.06 (, 0.51) | 0.80 |
| cg14099457 | 416 | 0.16 (, 0.55) | 0.42 | 383 | 0.13 (, 0.54) | 0.54 |
| cg03732535 | 416 | 0.95 (, 2.14) | 0.12 | 383 | 0.76 (, 2.02) | 0.24 |
| cg02954987 | 416 | 0.20 (, 0.46) | 0.14 | 383 | 0.15 (, 0.43) | 0.27 |
| WRAML2 visual memory standard score | ||||||
| cg05654765 | 415 | 0.15 (, 0.83) | 0.65 | 382 | 0.00 (, 0.71) | 0.99 |
| cg14099457 | 415 | 0.22 (, 0.83) | 0.48 | 382 | 0.13 (, 0.76) | 0.70 |
| cg03732535 | 415 | 0.88 (, 2.74) | 0.35 | 382 | 0.48 (, 2.47) | 0.63 |
| cg02954987 | 415 | 0.16 (, 0.57) | 0.44 | 382 | 0.11 (, 0.54) | 0.61 |
| WRAVMA standard score | ||||||
| cg05654765 | 414 | 0.90 (, 3.52) | 0.50 | 381 | 0.94 (, 3.69) | 0.50 |
| cg14099457 | 414 | 0.29 (, 2.65) | 0.81 | 381 | 0.18 (, 2.65) | 0.89 |
| cg03732535 | 414 | 3.33 (, 10.55) | 0.37 | 381 | 3.86 (, 11.54) | 0.32 |
| cg02954987 | 414 | 0.61 (, 2.21) | 0.45 | 381 | 0.43 (, 2.08) | 0.60 |
Model 1: Adjusted for maternal age at enrollment, race/ethnicity (white, black, other), smoking during pregnancy (any, former, never), education (any college, no college), child’s age at testing (continuous, in years), and child’s sex.
Model 2: Additionally adjusted for maternal alcohol consumption during the first trimester of pregnancy (servings per day, continuous) and median neighborhood household income.
Model 2 had 33 more missing values than model 1 due to the additional adjustment of alcohol consumption and median household income.
The CpG sites evaluated had significantly higher methylation in cord blood () in association with a 50% reduction in residential proximity to major roadways at birth.
Blood–Brain DNA Methylation Comparison
We examined the range and covariation of the four FDR-significant CpG sites near/within LAMB2 in matched pre-mortem whole blood and postmortem brain tissues from the same adult individuals () (Hannon et al. 2015). Methylation levels were higher and more variable in DNA extracted from blood than in brain tissue samples (Figure S5). Methylation levels at cg05654765, cg14099457, and cg02954987 were moderately correlated between DNA extracted from blood and brain tissue samples for the prefrontal cortex, entorhinal cortex, and superior temporal gyrus (Pearson correlations 0.59–0.73), whereas correlations between blood and brain tissue methylation at the same sites were weaker for cg03732535 (Pearson correlations 0.16–0.25). DNA methylation at all four of the CpG sites was only weakly correlated between blood and brain tissue samples from the cerebellum (Pearson correlations –0.17) (Figure S5).
Discussion
Residential proximity to major roadways at birth was positively associated (epigenome-wide FDR significant) with cord blood DNA methylation at four CpG sites near/within LAMB2 among participants in Project Viva (Boston, Massachusetts, USA). These associations were generally consistent between males and females and between children of white and nonwhite mothers. Regional analyses also indicated differentially methylated regions associated with prenatal residential proximity in the LAMB2 region, and in two additional regions near C3orf65 and PPP1R2P1 genes. In addition, proximity to major roadways during pregnancy also was positively associated with methylation of the same sites in DNA extracted from peripheral blood samples collected from a subset of the same children at 7–11 years of age. However, cord blood methylation at the LAMB2 CpG sites was not associated with proximity to major roadways during pregnancy among participants in the Generation R Study population (Rotterdam, Netherlands). Methylation of the four LAMB2 CpG sites that were differentially methylated in association with roadway proximity at birth was also associated with lower KBIT-2 nonverbal cognitive test scores in a subset of Project Viva children at age 7–11 y. This finding is partly consistent with a previous analysis of associations between roadway proximity at birth and cognitive test scores in a larger group of Project Viva participants () (Harris et al. 2015), including inverse associations between proximity and KBIT-2 nonverbal scores and null findings for WRAML2 design memory and picture memory scores. However, in contrast with findings from roadway proximity from the previous study, methylation at the LAMB2 CpG sites was not associated with lower KBIT-2 verbal scores or lower WRAVMA visual motor subscale scores.
To our knowledge, this is the first study to report evidence of epigenetic modifications in association with residential proximity to major roadways at birth. Specifically, our epigenome-wide analysis indicated that living closer to major roadways was associated with significantly higher cord blood DNA methylation in four CpG sites located near the transcription start site of LAMB2. In addition, methylation of the same CpGs in cord blood DNA was associated with lower nonverbal intelligence scores at 7–11 years of age. LAMB2, located on chromosome 3p21, encodes for a subtype of extracellular matrix glycoprotein and is involved in multiple biological processes, including cell adhesion, differentiation, signaling, and neurite outgrowth. The role of the LAMB2 gene product at the synapse has been extensively studied since the early 1990s (Hall 1995; Hunter et al. 1989; Martin et al. 1995; Noakes et al. 1995). LAMB2 gene product, a now-well-recognized synaptic component of the basal lamina, is often enriched at the neuromuscular junction of the motor nerve terminals. Experimental evidence shows that the LAMB2 gene product helps to recruit presynaptic constituents and facilitates the release of neurotransmitters at the synaptic cleft (Hunter et al. 1989). In LAMB2 knock-out mice, Sanes and et al. observed a lack of presynaptic specialization, meaning that instead of being clustered near the neuromuscular junction, the synaptic vesicles are distributed evenly in the presynaptic nerve terminal (Porter et al. 1995). This lack of presynaptic specialization distribution subsequently led to reduced neurotransmitter release by exocytosis and reduced the firing of action potentials (Porter et al. 1995). Moreover, the LAMB2 gene product also helps to confine growing nerve terminals to the appropriate location (Hall 1995). Motor neurons cultured on LAMB2 stripes showed inhibition of neurite extension, supporting a role of LAMB2 in facilitating the attachment of presynaptic nerve terminals to the postsynaptic nerve terminals during synaptogenesis (Lieth et al. 1992).
Given its essential role in assuring the precise joining of presynaptic and postsynaptic nerve structures, we further evaluated the association between LAMB2 DNA methylation and cognitive function at midchildhood. All four LAMB2 FDR-significant CpG sites were associated with KBIT-2 nonverbal cognitive test scores (i.e., higher DNA methylation associated with lower cognitive function). The nonverbal section of the KBIT test measures the ability to perceive relationships and complete visual analogies and assesses multiple domains, including visual-spatial ability and fluid reasoning (Kaufman and Kaufman 2004). We did not observe associations of LAMB2 DNA methylation with other neurocognitive measurements in this study (which includes the KBIT-2 verbal test, the visual-motor subtest of WRAVMA, and the visual memory index of WRAML2).
In adult human samples, DNA methylation often exhibits tissue specificity. Thus, a key question arises as to whether DNA methylation in cord blood reflects neurological development of the fetus. One plausible explanation would be that during the early stage of embryogenesis, there is a rapid erasure and reestablishment of DNA methylation pattern in undifferentiated stem cells, and exposure to environmental insults at this early stage of embryogenesis may result in stable epigenetic modifications that could propagate across different cell lineages (such as hematopoietic and neural stem cells) (Feng et al. 2010; Smith et al. 2012). Such a pattern may be maintained through de novo DNA methyltransferases during subsequent successive somatic cell divisions (Petronis 2010). However, additional experiments would be required to further elucidate such a hypothesis.
Although maternal residential proximity to major roadways has not been studied with respect to fetal DNA methylation, several epidemiology studies have reported associations between prenatal ambient air pollution and DNA methylation in children. For example, a recently published epigenome-wide meta-analysis showed that prenatal exposure to the gaseous pollutant nitric dioxide () was associated with differential offspring DNA methylation in three CpG sites at mitochondrial related genes (Gruzieva et al. 2017). Maternal exposure to benzo[a]pyrene (BaP)—a representative of the ubiquitous urban air pollutant polycyclic aromatic hydrocarbons—has also been associated with hypermethylation of interferon gamma () (Tang et al. 2012), and lower global DNA methylation in cord blood (Herbstman et al. 2012). Further, a number of studies reported associations between traffic pollution and placental DNA methylation (Janssen et al. 2013, 2015; Kingsley et al. 2016). For example, Janssen et al. showed decreased placental LINE-1 DNA methylation associated with living closer to major roadways (Janssen et al. 2013). In a separate study, the same group demonstrated increased placental mitochondrial DNA methylation associated with higher particulate matter () concentrations (Janssen et al. 2015). In the Rhode Island Child Health Study (RICHS), Kingsley et al. reported lower placental LINE-1 DNA methylation and seven epigenome-wide significant CpG sites associated with living closer to major roadways (Kingsley et al. 2016). Unfortunately, the associations we observed between distance to major roadways and cord blood DNA methylation did not appear to be the top hits for the other air pollution studies. Part of the reason maybe that distance to major roadways not only captures traffic-related air pollution, but also reflects noise and socioeconomic status (SES). Further, our results may be less generalizable to other parts of the world, where residential proximity to major roadways captures a different set of risk factors, or may present an exposure matrix on different scales. Future research is warranted to examine distance to major roadway and cord blood DNA methylation in a similar exposure setting. Alternatively, there is also the possibility that our findings were due to chance or systematic error.
In vertebrates, CpGs are not evenly distributed across the genome. In fact, functional CpGs are often clustered into regions identified as CpG islands or CpG shores near or within promoter regions of a gene (Jones 2012). A group of closely related methylation sites often function in concert to alter gene expression (i.e., each individual CpG site may not achieve the epigenome-wide FDR significance, but there may be an additive effect in which each CpG site adds an increment, and together they exhibit an effect). However, such associations may not always be captured in the CpG-by-CpG analysis. Therefore, as a secondary analysis, we conducted a regional analysis and identified two additional differentially methylated regions associated with residential proximity to major roadways, which include Chromosome 2 open reading frame 65 (C2orf65) and Protein Phosphatase 1 Regulatory Inhibitor Subunit 2 Pseudogene (PPP1R2P1) (regions identified were within CpG islands for both genes). PPP1R2P1 is a pseudo-gene located on chromosome 6p21, and it is related to Protein Phosphatase-1 (PP1)—one of the main serine/threonine phosphatases in human. C2orf65 gene is located on chromosome 2p13. The function of C2orf65 gene remains unclear.
For the present study, we measured DNA methylation in umbilical cord blood and peripheral blood samples composed of complex cell mixtures. Therefore, methylation may vary among participants due to differences in cell counts or proportions. To address this limitation, in the statistical analysis, we adjusted for peripheral WBC proportions based on the method developed by Houseman (Houseman et al. 2012), and cord blood cell proportions based on the method developed from the MeDALL study (Bakulski et al. 2016) to reduce confounding by cell type fractions. Residential proximity to major roadways at birth was not correlated with proportions of blood cell subtypes, which suggests that confounding due to an association between proximity and cell subtype distributions was unlikely. However, measuring DNA methylation in samples of individual cell types would be preferred to avoid this potential limitation.
Our discovery results need to be interpreted with caution, in that none of the CpG sites near LAMB2 replicated at in the Generation R Study—an independent birth cohort based in Rotterdam, Netherlands. Other than the possibility that our findings are due to chance, there may be several reasons for the lack of replication that are related to potential noncomparability of exposures: a) More so in the northeast United States than in Netherlands, residential proximity to major roadways may reflect socioeconomic disparities in adverse neighborhood-level exposures. Although we adjusted for maternal education and neighborhood median household income, we cannot rule out the possibility of residual confounding by unmeasured socioeconomic factors; b) In the northeastern United States, there is a wider exposure range (i.e., residential proximity to major roadways in our study population ranged from ) than in Rotterdam, where people tend to live clustered within the city. For example, most of the study participants in The Generation R Study live within of a major road; c) Also, automobile exhaust in the United States mainly comes from gasoline combustions, whereas diesel exhaust contributes to a significant part of traffic pollution in Netherlands. The differing fuel sources may give rise to different emission profiles. Any of these differences between the two study locations, alone or in combination, may have been responsible for the discrepancies in the results. In conclusion, our inability to replicate findings from Project Viva in the Generation R Study may reflect differences in exposures resulting from residential proximity, or from other differences between the two populations, though the possibility that our findings were due to chance must also be acknowledged.
Our study has a number of strengths. For example, the longitudinal nature of this cohort allows us to examine prospectively the association between residential proximity to roadways at birth and DNA methylation or cognitive test scores measured at midchildhood. In addition, we performed not only CpG-by-CpG analysis, but also explored regional analysis when we studied the association between residential proximity to roadways at birth and cord blood DNA methylation. The consistency of the findings between CpG-by-CpG analysis using robust linear regression and in regional analysis using DMRcate indicates the signals we identified near/within the LAMB2 gene were robust to the methods of analyses. Our study also has several limitations. As already noted, we were not able to replicate our findings in the Generation R Study population. Second, we do not have genetic information for our study participants. Therefore, it is difficult to rule out the possibility that the observed associations were primarily driven by SNP or genetic background. Nevertheless, in the data preprocessing steps, we removed SNP probes and probes within of a known SNP. We also did not identify SNPs close to our FDR-significant CpG sites in the UCSC Genome Browser [Human Feb. 2009 (GRCh37/hg19) Assembly]. However, it remains plausible that a SNP at a distance away from the methylation site could have an influence. Third, we measured DNA methylation levels in blood, and we do not have information on other biologically relevant tissues (such as the brain). Therefore, further experimental evidence is warranted to verify the biological significance of LAMB2 DNA methylation in blood samples. Further, blood DNA methylation levels (of the same loci) often vary with age and time (Bjornsson et al. 2008). Hence, we need to interpret with caution when we compare methylation levels from our birth cohort (cord blood/childhood peripheral blood) with values from the publically available dataset, in which their blood DNA methylation levels were assessed before death (age range 64–99) (Hannon et al. 2015). Note also that no race/ethnicity information was reported in the publicly available dataset. DNA methylation may be influenced by near (cis)/or distant (trans) SNPs, and we could not rule out the possibility that the difference in race/ethnicity compositions may introduce unwanted biological variabilities that may hinder a direct comparison. In addition, potential misclassification of exposure may arise due to use of the maternal address at birth as a proxy for exposure to major roadways throughout pregnancy. Our results also may not be generalizable to other areas of the world, where residential proximity to major roadways may capture a different set of risk factors. Finally, both the verbal and the nonverbal subset of the KBIT-2 test serve as global measures of cognitive functioning and development and do not assess specific functional domains in the brain, which means that two children with the same KBIT-2 score could have different patterns of performance.
Conclusion
Among U.S. children in Project Viva, living closer to major roadways at birth was associated with higher methylation of CpG sites in the LAMB2 gene in cord blood samples at birth, and in peripheral blood samples collected from the same children at 7–11 years of age. In addition, methylation of the same CpG sites in cord blood was associated with lower mean scores on a test of nonverbal cognitive function in midchildhood, consistent with effects on the LAMB2 gene, which is involved in neurological function and development. However, we were unable to replicate associations between residential proximity at birth and methylation of the LAMB2 CpG sites in cord blood in the Generation R Study, conducted in Netherlands, and although this inability to replicate associations might reflect differences in exposures or susceptibility factors, additional research is needed to determine whether our findings can be confirmed in other study populations.
Supplementary Material
Acknowledgments
The authors would like to thank Project Viva staff and participants and Dr. M. Mazumdar. The Generation R Study is conducted by the Erasmus Medical Center (MC) in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University Rotterdam, the Municipal Health Service Rotterdam area, Rotterdam, the Rotterdam Homecare Foundation, Rotterdam and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond (STAR-MDC), Rotterdam, Netherlands. We gratefully acknowledge the contribution of children and parents, general practitioners, hospitals, midwives and pharmacies in Rotterdam. The study protocol was approved by the Medical Ethical Committee of the Erasmus MC, University Medical Centre, Rotterdam, Netherlands. The generation and management of the Illumina 450K methylation array data (EWAS data) for the Generation R Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, Erasmus University Medical Center, Rotterdam, Netherlands. We thank M. Verbiest, M. Jhamai, S. Higgins, M. Verkerk, and Dr. L. Stolk for their help in creating the EWAS database.
References
- Adams W, Sheslow D. 1995. Wide range assessment of visual motor abilities (WRAVMA). Lutz, FL:Psychological Assessment Resources, Inc. [Google Scholar]
- Bakulski KM, Feinberg JI, Andrews SV, Yang J, Brown S, L McKenney S, et al. 2016. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics 11(5):354–362, PMID: 27019159, 10.1080/15592294.2016.1161875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. 2008. Intra-individual change over time in DNA methylation with familial clustering. JAMA 299(24):2877–2883, PMID: 18577732, 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer TK, Foster SL, Henry JR, Woghiren-Akinnifesi EL, Yip FY, Centers for Disease Control. 2013. Residential proximity to major highways – United States, 2010. MMWR Suppl 62(3):46–50, PMID: 24264489. [PubMed] [Google Scholar]
- Broberg K, Ahmed S, Engström K, Hossain MB, Jurkovic Mlakar S, Bottai M, et al. 2014. Arsenic exposure in early pregnancy alters genome-wide DNA methylation in cord blood, particularly in boys. J Dev Orig Health Dis 5(4):288–298, PMID: 24965135, 10.1017/S2040174414000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao-Lei L, Veru F, Elgbeili G, Szyf M, Laplante DP, King S. 2016. DNA methylation mediates the effect of exposure to prenatal maternal stress on cytokine production in children at age 13 1/2 years: Project Ice Storm. Clin Epigenetics 8:54, PMID: 27182285, 10.1186/s13148-016-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A, Allard C, Doyon M, Houseman EA, Bakulski KM, Perron P, et al. 2016. Validation of a DNA methylation reference panel for the estimation of nucleated cells types in cord blood. Epigenetics 11(11):773–779, PMID: 27668573, 10.1080/15592294.2016.1233091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A, Koestler DC, Houseman EA, Jackson BP, Kile ML, Karagas MR, et al. 2015. Differential DNA methylation in umbilical cord blood of infants exposed to mercury and arsenic in utero. Epigenetics 10(6):508–515, PMID: 25923418, 10.1080/15592294.2015.1046026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. 2013. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium Humanmethylation450 microarray. Epigenetics 8(2):203–209, PMID: 23314698, 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean W, Bowden L, Aitchison A, Klose J, Moore T, Meneses JJ, et al. 1998. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development 125(12):2273–2282, PMID: 9584126. [DOI] [PubMed] [Google Scholar]
- Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. 2000. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod 62(6):1526–1535, PMID: 10819752, 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- Feil R, Fraga MF. 2012. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 13(2):97–109, PMID: 22215131, 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. 2010. Epigenetic reprogramming in plant and animal development. Science 330(6004):622–627, PMID: 21030646, 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleisch AF, Rifas-Shiman SL, Koutrakis P, Schwartz JD, Kloog I, Melly S, et al. 2015. Prenatal exposure to traffic pollution: associations with reduced fetal growth and rapid infant weight gain. Epidemiology 26(1):43–50, PMID: 25437317, 10.1097/EDE.0000000000000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleisch AF, Gold DR, Rifas-Shiman SL, Koutrakis P, Schwartz JD, Kloog I, et al. 2014. Air pollution exposure and abnormal glucose tolerance during pregnancy: The Project Viva Cohort. Environ Health Perspect 122 (4):378–383, PMID: 24508979, 10.1289/ehp.1307065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491(7422):56–65, PMID: 23128226, 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould HJ, Sutton BJ. 2008. IgE in allergy and asthma today. Nat Rev Immunol 8(3):205–217, PMID: 18301424, 10.1038/nri2273. [DOI] [PubMed] [Google Scholar]
- Gruzieva O, Xu CJ, Breton CV, Annesi-Maesano I, Antó JM, Auffray C, et al. 2017. Epigenome-wide meta-analysis of methylation in children related to prenatal NO2 air pollution exposure. Environ Health Perspect 125(1):104–110, PMID: 27448387, 10.1289/EHP36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall ZW. 1995. Laminin beta 2 (S-laminin): a new player at the synapse. Science 269(5222):362–363, PMID: 7618101, 10.1126/science.7618101. [DOI] [PubMed] [Google Scholar]
- Hannon E, Lunnon K, Schalkwyk L, Mill J. 2015. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics 10(11):1024–1032, PMID: 26457534, 10.1080/15592294.2015.1100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MH, Gold DR, Rifas-Shiman SL, Melly SJ, Zanobetti A, Coull BA, et al. 2015. Prenatal and childhood traffic-related pollution exposure and childhood cognition in the Project Viva Cohort (Massachusetts, USA). Environ Health Perspect 123(10):1072–1078, PMID: 25839914, 10.1289/ehp.1408803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. 2008. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A 105(44):17046–17049, PMID: 18955703, 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbstman JB, Tang D, Zhu D, Qu L, Sjödin A, Li Z, et al. 2012. Prenatal exposure to polycyclic aromatic hydrocarbons, benzo[a]pyrene-DNA adducts, and genomic DNA methylation in cord blood. Environ Health Perspect 120(5):733–738, PMID: 22256332, 10.1289/ehp.1104056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86, PMID: 22568884, 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DD, Shah V, Merlie JP, Sanes JR. 1989. A laminin-like adhesive protein concentrated in the synaptic cleft of the neuromuscular junction. Nature 338(6212):229–234, PMID: 2922051, 10.1038/338229a0. [DOI] [PubMed] [Google Scholar]
- Janssen BG, Byun HM, Gyselaers W, Lefebvre W, Baccarelli AA, Nawrot TS. 2015. Placental mitochondrial methylation and exposure to airborne particulate matter in the early life environment: an ENVIRONAGE birth cohort study. Epigenetics 10(6):536–544, PMID: 25996590, 10.1080/15592294.2015.1048412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BG, Godderis L, Pieters N, Poels K, Kiciński M, Cuypers A, et al. 2013. Placental DNA hypomethylation in association with particulate air pollution in early life. Part Fibre Toxicol 10:22, PMID: 23742113, 10.1186/1743-8977-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1):118–127, PMID: 16632515, 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(7):484–492, PMID: 22641018, 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Joubert BR, den Dekker HT, Felix JF, Bohlin J, Ligthart S, Beckett E, et al. 2016a. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun 7:10577, PMID: 26861414, 10.1038/ncomms10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. 2016b. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet 98(4):680–696, PMID: 27040690, 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert BR, Håberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, et al. 2012. 450k epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect 120(10):1425–1431, PMID: 22851337, 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman AS, Kaufman NL. 2004. Kaufman Brief Intelligence Test (KBIT-2), 2nd Edition Bloomington, MN:Pearson, Inc. [Google Scholar]
- Khosla S, Dean W, Brown D, Reik W, Feil R. 2001. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod 64(3):918–926, PMID: 11207209, 10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- Kile ML, Baccarelli A, Hoffman E, Tarantini L, Quamruzzaman Q, Rahman M, et al. 2012. Prenatal arsenic exposure and DNA methylation in maternal and umbilical cord blood leukocytes. Environ Health Perspect 120(7):1061–1066, PMID: 22466225, 10.1289/ehp.1104173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley SL, Eliot MN, Whitsel EA, Huang YT, Kelsey KT, Marsit CJ, et al. 2016. Maternal residential proximity to major roadways, birth weight, and placental DNA methylation. Environ Int 92–93:43–49, PMID: 27058926, 10.1016/j.envint.2016.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippler M, Engström K, Mlakar SJ, Bottai M, Ahmed S, Hossain MB, et al. 2013. Sex-specific effects of early life cadmium exposure on DNA methylation and implications for birth weight. Epigenetics 8(5):494–503, PMID: 23644563, 10.4161/epi.24401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Richmond R, Hu P, French L, Shin J, Bourdon C, et al. 2015. Prenatal exposure to maternal cigarette smoking and DNA methylation: epigenome-wide association in a discovery sample of adolescents and replication in an independent cohort at birth through 17 years of age. Environ Health Perspect 123(2):193–199, PMID: 25325234, 10.1289/ehp.1408614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieth E, Cardasis CA, Fallon JR. 1992. Muscle-derived agrin in cultured myotubes: expression in the basal lamina and at induced acetylcholine receptor clusters. Dev Biol 149(1):41–54, PMID: 1309458, 10.1016/0012-1606(92)90262-F. [DOI] [PubMed] [Google Scholar]
- Liu Y, Murphy SK, Murtha AP, Fuemmeler BF, Schildkraut J, Huang Z, et al. 2012. Depression in pregnancy, infant birth weight and DNA methylation of imprint regulatory elements. Epigenetics 7(7):735–746, PMID: 22677950, 10.4161/epi.20734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumey LH, Stein AD, Kahn HS, van der Pal-de Bruin KM, Blauw GJ, Zybert PA, et al. 2007. Cohort profile: the Dutch Hunger Winter Families Study. Int J Epidemiol 36(6):1196–1204, PMID: 17591638, 10.1093/ije/dym126. [DOI] [PubMed] [Google Scholar]
- Markunas CA, Xu Z, Harlid S, Wade PA, Lie RT, Taylor JA, et al. 2014. Identification of DNA methylation changes in newborns related to maternal smoking during pregnancy. Environ Health Perspect 122(10):1147–1153, PMID: 24906187, 10.1289/ehp.1307892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PT, Ettinger AJ, Sanes JR. 1995. A synaptic localization domain in the synaptic cleft protein laminin beta 2 (s-laminin). Science 269(5222):413–416, PMID: 7618109, 10.1126/science.7618109. [DOI] [PubMed] [Google Scholar]
- Noakes PG, Gautam M, Mudd J, Sanes JR, Merlie JP. 1995. Aberrant differentiation of neuromuscular junctions in mice lacking s-laminin/laminin beta 2. Nature 374(6519):258–262, PMID: 7885444, 10.1038/374258a0. [DOI] [PubMed] [Google Scholar]
- Oken E, Baccarelli AA, Gold DR, Kleinman KP, Litonjua AA, De Meo D, et al. 2015. Cohort profile: Project Viva. Int J Epidemiol 44(1):37–48, PMID: 24639442, 10.1093/ije/dyu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, V Lord R, et al. 2015. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 8:6, PMID: 25972926, 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A. 2010. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature 465(7299):721–727, PMID: 20535201, 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. 2013. A data-driven approach to preprocessing Illumina 450k methylation array data. BMC Genomics 14:293, PMID: 23631413, 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Hu H, Ettinger A, Sánchez BN, Wright RO, Cantonwine D, et al. 2009. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect 117(9):1466–1471, PMID: 19750115, 10.1289/ehp.0800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter BE, Weis J, Sanes JR. 1995. A motoneuron-selective stop signal in the synaptic protein s-laminin. Neuron 14(3):549–559, PMID: 7695901, 10.1016/0896-6273(95)90311-9. [DOI] [PubMed] [Google Scholar]
- Reik W. 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447(7143):425–432, PMID: 17522676, 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Rice MB, Rifas-Shiman SL, Oken E, Gillman MW, Ljungman PL, Litonjua AA, et al. 2014. Exposure to traffic and early life respiratory infection: a cohort study. Pediatr Pulmonol 50(3):252–259, PMID: 24678045, 10.1002/ppul.23029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzehak P, Saffery R, Reischl E, Covic M, Wahl S, Grote V, et al. 2016. Maternal smoking during pregnancy and DNA-methylation in children at age 5.5 years: epigenome-wide-analysis in the European Childhood Obesity Project (CHOP)-Study. PLoS One 11(5):e0155554, PMID: 27171005, 10.1371/journal.pone.0155554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheslow DA, Adams W. 2003. Wide Range Assessment of Memory and Learning (WRAML2):Administration and Technical Manual. 2nd Edition Lutz, FL: Psychological Assessment Resources, Inc. [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, et al. 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484(7394):339–344, PMID: 22456710, 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WY, Levin L, Talaska G, Cheung YY, Herbstman J, Tang D, et al. 2012. Maternal exposure to polycyclic aromatic hydrocarbons and 5'-CpG methylation of interferon-γ in cord white blood cells. Environ Health Perspect 120(8):1195–1200, PMID: 22562770, 10.1289/ehp.1103744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. 2013. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29(2):189–196, PMID: 23175756, 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triche TJ Jr., Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. 2013. Low-level processing of Illumina Infinium DNA methylation bead arrays. Nucleic Acids Res 41(7):e90, PMID: 23476028, 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossem L, Rifas-Shiman SL, Melly SJ, Kloog I, Luttmann-Gibson H, Zanobetti A, et al. 2015. Prenatal air pollution exposure and newborn blood pressure. Environ Health Perspect 123(4):353–359, PMID: 25625652, 10.1289/ehp.1307419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal AC, Benjamin Neelon SE, Liu Y, Tuli AM, Fuemmeler BF, Hoyo C, et al. 2014. Maternal stress, preterm birth, and DNA methylation at imprint regulatory sequences in humans. Genet Epigenet 6:37–44, PMID: 25512713, 10.4137/GEG.S18067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Soc Series B (Methodological) 57(1):289–300. [Google Scholar]
- Yorifuji T, Kashima S, Higa Diez M, Kado Y, Sanada S, Doi H. 2016. Prenatal exposure to traffic-related air pollution and child behavioral development milestone delays in Japan. Epidemiology 27(1):57–65, PMID: 26247490, 10.1097/EDE.0000000000000361. [DOI] [PubMed] [Google Scholar]
- Young LE, Fernandes K, McEvoy TG, Butterwith SC, Gutierrez CG, Carolan C, et al. 2001. Epigenetic change in IgF2r is associated with fetal overgrowth after sheep embryo culture. Nat Genet 27(2):153–154, PMID: 11175780, 10.1038/84769. [DOI] [PubMed] [Google Scholar]
- Zhu X, Li J, Deng S, Yu K, Liu X, Deng Q, et al. 2016. Genome-wide analysis of DNA methylation and cigarette smoking in a Chinese population. Environ Health Perspect 124(7):966–973, PMID: 26756918, 10.1289/ehp.1509834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.