Abstract
Worldwide Salmonella enterica infections result in substantial morbidity and mortality and are the major cause of infant bacteremia in Sub-Saharan Africa. Diseases caused by Salmonella are treatable with antibiotics, but successful antibiotic treatment has become difficult due to antimicrobial resistance and collateral effects on the microbiome. An effective vaccine together with public health efforts may be a better strategy to control these infections. Protective immunity against Salmonella depends primarily on CD4 T-cell-mediated immune responses; therefore, identifying relevant T-cell antigens is necessary for Salmonella vaccine development. We previously used a dendritic-cell-based immunoproteomics approach in our laboratory to identify T-cell antigens. The testing of these antigens as vaccine candidates against Chlamydia infection in mice yielded positive results. We applied this technology in the present study by infecting murine bone-marrow-derived dendritic cells from C57BL/6 mice with Salmonella enterica strain SL1344, followed by immunoaffinity isolation of MHC class I and II molecules and elution of bound peptides. The sequences of the peptides were identified using tandem mass spectrometry. We identified 87 MHC class-II- and 23 MHC class-I-binding Salmonella-derived peptides. Four of the 12 highest scoring class-II-binding Salmonella peptides stimulated IFN-γ production by CD4+ T cells from the spleens of mice with persistent Salmonella infection. We conclude that antigens identified by MHC immunoproteomics will be useful for Salmonella immunobiology studies and are potential Salmonella vaccine candidates. Data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD004451.
Keywords: Salmonella, immunoproteomics, MHC, T cell, epitope, peptide, antigen, vaccine
INTRODUCTION
Salmonella enterica infections are associated with a variety of diseases ranging from self-limiting diarrhea to life-threatening bacteremia. Nontyphoidal Salmonella is a common cause of gastroenteritis in healthy individuals and a leading cause of lethal bloodstream infections in elderly, immunocompromised patients and children, especially in the developing world.1,2 In Sub-Saharan Africa nontyphoidal Salmonella is the most common cause of invasive bacterial infection in infants less than 1 year of age with an associated case fatality of 20–25%.3 Diseases caused by Salmonella are treatable with antibiotics, but successful antibiotic treatment has become difficult due to widespread antimicrobial resistance and because of its effects on the host microbiome. An effective vaccine, especially when combined with public health efforts to improve food and water safety, appears to be the best strategy to reduce the incidence of these diseases.4 No vaccine is currently available against nontyphoidal Salmonella.
Protective immunity against Salmonella depends on a wide range of innate and adaptive immune mechanisms.5 As this facultative intracellular pathogen preferentially infects and resides within macrophages, cell-mediated immune (CMI) responses are essential in the host clearance of Salmonella infections. Antigen-presenting cells such as dendritic cells (DCs) are at the center of initiation of CMI responses. DCs capture antigens of microbes entering the periphery of the host and transport to regional lymph nodes, where they present processed peptides (epitopes) on MHC class I or class II molecules to naiv̈e CD8+ or CD4+ T cells, respectively.6 CD4+ T cells largely mediate protective T-cell immunity to Salmonella via secreted cytokines, and much of their function is secondary to T-helper 1 (Th1) cell phenotype-secreting IFN-γ and TNF-α.7 The role of CD8+ T cells in adaptive immunity is smaller but appreciable and also appears dependent on cytokine secretion.8 Activated T cells undergo proliferation and produce IFN-γ, which, in turn, activates macrophages to generate effector molecules such as nitric oxide that damage heme-containing proteins, providing a major contribution to the control and containment of Salmonella infections.9 B cells and antibodies, on the contrary, are not essential to the control of primary Salmonella infection in mice. However, B-cell-deficient mice exhibit increased susceptibility to Salmonella reinfection, and antibodies provide passive transfer of immunity in T-cell- primed mice.10
It is evident from past experience that the development of vaccines against pathogens that require protective CMI is more difficult than for pathogens that require protective antibodies.11 Intracellular pathogens are among some of the worst scourges affecting humans today, including malaria, tuberculosis, and HIV. To date, there are only a few vaccines available against intracellular pathogens, such as capsular polysaccharide vaccine (ViCPS) and live attenuated oral vaccine (Ty21a) against S. typhi and Bacille Calmette Guerin (BCG) vaccine against tuberculosis. These vaccines provide limited efficacy at best. The reason behind the difficulty in making effective vaccines against intracellular pathogens is partly due to the lack of knowledge about antigens that induce strong T-cell-mediated immune responses.
Because T-cell responses mainly recognize protein antigens, protective vaccine candidates ought to be found within the proteome of the organism. Thanks to genomics, the potential proteome for Salmonella has been fully inferred.12 Using the available genomic information, the T-cell immune proteome of any microbial pathogen can be surveyed by an approach called immunoproteomics in which peptides presented by MHC molecules are identified by tandem mass spectrometry (MS/ MS).13 Identifying peptides that bind MHC molecules allows identifications of microbial proteins capable of entering the presentation pathways, and it is such proteins that may be useful in vaccine development. While the concept of using MS/ MS to identify MHC-bound peptides is not new, in the past the sensitivity and accuracy of mass spectrometers severely limited their applicability to infectious diseases. Now, however, there are commercially available mass spectrometers with proven practical sensitivity limits near 1 fmol.14,15 The recent advancement of MS/MS detection technology to 1 fmol level now allows the identification of MHC-bound peptides of microbial origin that can be reasonably be purified.
The immunoproteomic approach has been used successfully in our laboratories to analyze the immunoproteome of Chlamydia.16–19 Further study in our laboratory demonstrated that 11 out of 13 chlamydial proteins identified via the immunoproteomic approach turned out to be protective in challenge studies.20–22 In this study, we used the immunopro- teomic approach to identify 87 MHC class II and 23 MHC class I Salmonella enterica-derived peptides. Several of these peptides stimulated IFN-γ production by T cells from mice persistently infected with Salmonella.
EXPERIMENTAL PROCEDURES
Salmonella
Salmonella enterica serovar typhimurium Strain SL1344 was grown overnight in Luria−Bertani (LB) medium and diluted in phosphate-buffered saline, and bacterial concentration was determined by plating on LB agar plates. The whole genome of this strain is already sequenced, and the transcriptional landscape is available in public domains.12
Mice
Female C57BL/6 (H2-Kb, Db, and I-Ab) mice (6 to 8 weeks old) were purchased from Charles River Canada (Saint Constant, Canada). The mice were maintained and used in strict accordance with University of British Columbia guidelines for animal care. Female 129X1/SvJ (H2-Kb, Db, and I-Ab) mice (6 to 8 weeks old) were purchased from the Jackson Laboratory (Bar Harbor, Maine) and maintained under the supervision of research animal resource division at the University of Minnesota School of Medicine.
Generation of BMDCs
Bone-marrow-derived dendritic cells (BMDCs) were generated as previously described.23 In brief, bone marrow cells flushed from the femurs of female C57BL/6 mice were cultured in Falcon Petri dishes at 4 × 107 cells in 50 mL of DC medium. DC medium was IMDM supplemented with 10% FCS, 0.5 mM 2-ME, 4 mM L-glutamine, 50 μg/mL gentamicin, and 5% of culture supernatant of murine GM-CSF-transfected plasmacy- toma X63-Ag8 and 5% of culture supernatant of murine IL-4 transfected plasmacytoma X63-Ag8, which contained 10 ng/mL GM-CSF and 10 ng/mL IL-4, respectively. On day 3, half of culture supernatants were removed and fresh DC medium was added. On day 5, nonadherent cells (purity of >50% CD11c+) were harvested and cultured in fresh DC medium for Salmonella infection.
Optimization of Salmonella Infection of BMDCs
BMDCs were infected with Salmonella at multiplicities of infection (MOI) of 0,1.25, 2.5, 5, and 10. After incubation for 0, 0.5, 1, or 2 h, which allow the cells to phagocytize the bacteria, gentamicin (final concentration 20 μg/mL) was added to the culture to stop extracellular replication of Salmonella in the culture media. Then BMDC/Salmonella cultures were incubated for an additional 14 h. Cell viability (%) and expression of CD11c, MHC I, and MHC II in the BMDC cultures were measured by flow cytometry using fixable viability stain 510 (BD Biosciences), PE-anti mouse CD11c (clone HL3, BD Biosciences), FITC-anti mouse H-2Kb (clone AF6–88.5, BD Biosciences), and V450-anti mouse I-A/I-E (clone M5/ 114.15.2, eBioscience).
Purification of MHC-Bound Peptides
BMDCs were infected with Salmonella for 0.5 h at 2.5 MOI and were incubated for an additional 14 h. Six billion infected BMDCs were washed with PBS twice, harvested in batches, and stored at −80 °C. Different batches of BMDCs infected with Salmonella were pooled and solubilized in lysis buffer (1% 3- [(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate, 150 mM NaCl, 20 mM Tris-HCl, pH 8, 0.04% sodium azide, protease inhibitors). Lysates were centrifuged and supernatant containing MHC molecules bound to peptides was transferred to tubes containing mAb-bound rPAS (mAb AF6–88.5.3 or HB158 specific to H2-Kb class I allele, Y-3P or HB183 specific to I-A class II allele) and rotated at 4 °C for 2 to 3 h. Beads were spun down and transferred to a tube, washed twice with lysis buffer, and transferred to a Bio Rad Poly prep chromatography column with 20 mM Tris-HCl, pH 8.0, 150 mM NaCl. Beads were washed with the following buffers: 1 time 20 mM Tris-HCl, pH 8.0, 150 mM NaCl; 2 times 20 mM Tris-HCl, pH 8.0, 1 M NaCl; 3 times 20 mM Tris-HCl, pH 8.0. Peptide-bound MHC molecules were eluted from rPAS by adding 4 bed volumes of 0.2 N acetic acid, and 1/10th volume of 1.6 N acetic acid was then added to the elute to further separate the peptides from MHC molecules. Elute containing MHC class I/class II molecules and the peptides was transferred to a filter unit with 10,000 Da cutoff and centrifuged. The flow through containing the peptides eluted from class I and class II molecules was processed for mass spectrometry sequencing. The MHC class-I- and class-II-bound peptides were further purified, concentrated, filtered, and desalted using STop And Go Extraction tips.24 Peptides were then analyzed by LC−MS/ MS using an LTQ-Orbitrap Velos (Thermo Electron) online coupled to Agilent 1200 Series nanoflow HPLCs using nanospray ionization sources (Proxeon Biosystems, Odense, Denmark). Analytical columns were packed into 15 cm long, 50 μm inner diameter fused silica emitters using 3 μm diameter ReproSil Pur C18 AQ beads (Dr. Maisch, www.Dr-Maisch.com), joint with 2 cm long, 100 μm inner diameter fused silica trap column packed with 5 μm diameter Aqua C-18 beads (Phenomenex, www.phenomenex.com), and a 20 μm inner diameter fused silica gold-coated spray tip with 6 μm diameter opening. LC buffer A consisted of 0.5% acetic acid and buffer B consisted of 0.5% acetic acid and 80% acetonitrile in water. Gradients were run from 10% B to 32% B over 51 min, then from 32% B to 40% B in the next 5 min, then increased to 100% B over 2 min period, held at 100% B for 2.5 min, and then dropped to 0% B for another 20 min to recondition the column. The Velos was set to acquire a full range scan at 60 000 resolution in the Orbitrap, from which the 10 most intense multiply charged ions per cycle were isolated for fragmentation in the LTQ. Centroided fragment peak lists were processed with Proteome Discoverer v. 1.2 (ThermoFisher Scientific). The search was performed with Mascot algorithm v. 2.4 against a database composed of the protein sequences from the mouse and Salmonella proteome. The databases were downloaded from Uniprot on November 21, 2014. The estimated false discovery rate (FDR) here was below 2%. Even though 1% FDR rate is typically applied for proteins, empirically, setting a 1% FDR protein cutoff was too stringent for our approach because some peptides eliminated at a 1% FDR but included at a 2% FDR scored positive in the T-cell assay. For a peptide to be assigned to a particular protein, the peptide must have an exact match to the protein sequence.
Synthetic Peptides
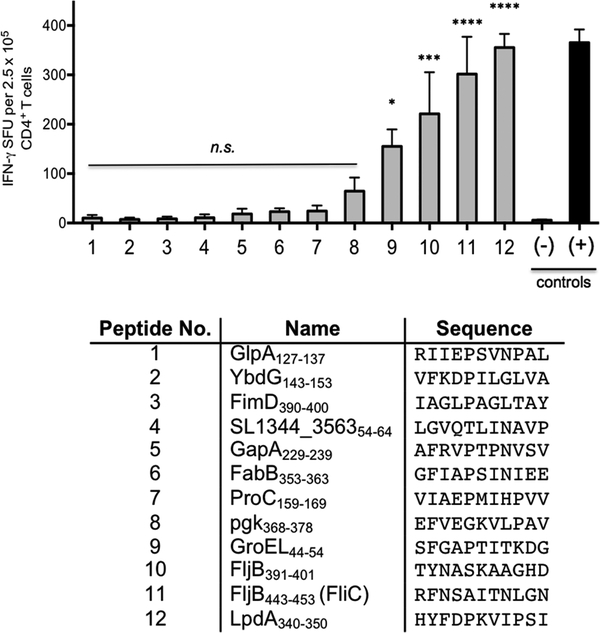
We selected 12 of the highest-scoring MHC class-II-binding peptides (GlpA127 –137, YbdG143 –153, FimD390–400, SL1344_356354–64, GapA229–239, FabB353–363, ProC159–169, pgk368 –378, GroEL44 –54, FljB391–401, FljB443–453, and LpdA340–350) for immunological analysis, and the core for each peptide was predicted using a previously published I-Ab binding matrix.25 These 11-mer peptides were synthesized for ex vivo restimulation of CD4+ T cells.
ELISPOT Assay
129X1/SvJ mice were infected with 108 colony forming units of Salmonella enterica serovar Typhimurium strain SL1344 by oral gavage to establish a chronic persistent infection. After 60 days of infection, CD4+ T cells from spleens and mesenteric lymph nodes were isolated by positive selection using anti-CD4+ magnetic beads (Miltenyi biotech), and infection status was confirmed by plating a portion of the flow through onto MacConkey agar with 100 μg/mL streptomycin. Purified CD4+ T cells were cultured overnight with irradiated splenocytes from naiv̈e syngeneic mice and candidate peptides at a concentration of 20 μM and analyzed for the presence of IFN-γ-producing cells by ELISPOT. The ELISPOT was adapted from a published protocol.26 In brief, 96-well MultiscreenHTS filter plates (EMD Millipore) were coated with unlabeled antimouse IFN-γ (clone AN-18; eBioscience). After washing, the cells and peptides were added to each well in triplicate and incubated overnight at 37 °C. The following day, plate-bound cytokines were detected with biotinylated anti- IFN-γ (Clone R4–6A2; eBioscience), followed by streptavidin- alkaline phosphatase (Life Technologies) and developed with BCIP/NBT substrate (Life Technologies). Spots were counted using an ImmunoSpot S6Micro Analyzer and ImmunoSpot software (Cellular Technology Limited)
Statistical Analysis
ELISPOT data was analyzed using one-way ANOVA with Tukey’s post hoc test.
RESULTS
Optimization of Salmonella Infection of Murine BMDCs
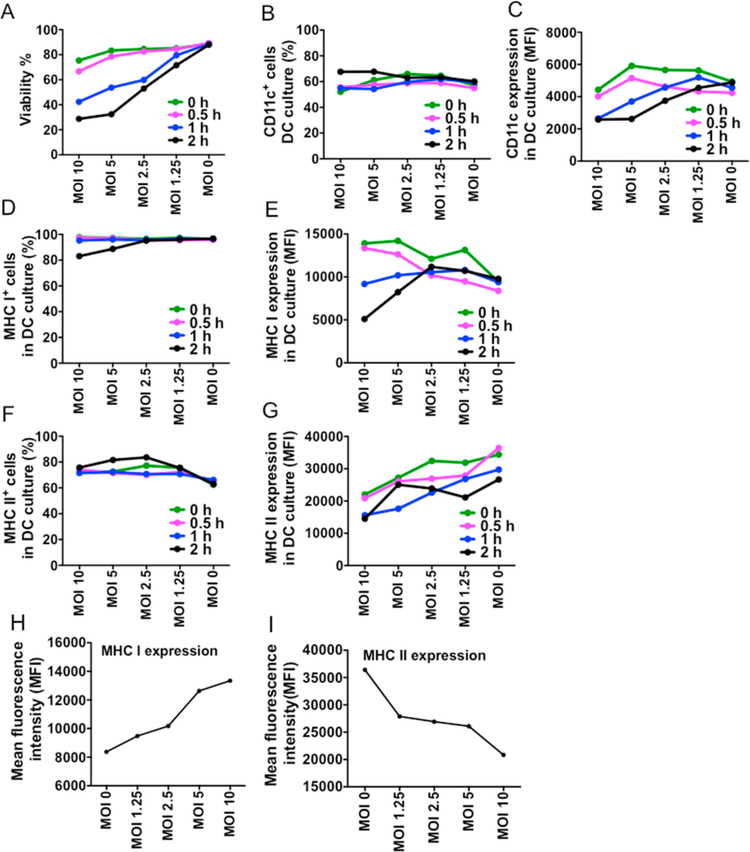
To determine the optimal conditions for antigen presentation, BMDCs were infected with Salmonella at different multiplicities of infection (MOI) and incubated for different time periods to allow the cells to phagocytize the bacteria. Cell viability and expression of CD11c, MHC I, and MHC II in the BMDC cultures were measured (Figure 1). On the basis of the results, the optimal MOI of Salmonella for BMDC infection was determined as 2.5, and the incubation time before the addition of gentamicin was determined as 0.5 h.
Figure 1.
Optimization of conditions for Salmonella peptide presentation. (A) Cell viability (%) in the BMDC culture infected with different multiplicities of infection (MOIs) of Salmonella at different incubation times. (B,C) CD11c expression of BMDCs infected with different MOIs of Salmonella at different incubation times. (D,E) MHC I expression of BMDCs infected with different MOIs of Salmonella at different incubation times. (F,G) MHC II expression of BMDCs infected with different MOIs of Salmonella at different incubation times. (H,I) MHC class I and class II expression of BMDCs infected with different MOIs of Salmonella when gentamicin is added at 0.5 h after Salmonella is added to BMDCs.
Identification of Salmonella-Derived MHC-Bound Peptides
To identify Salmonella-derived MHC-bound peptides, BMDCs were infected with Salmonella under the optimal conditions as described above. Infected BMDCs were lysed, and the MHC-bound peptides were isolated using MHC allele-specific monoclonal antibody affinity columns. MHC-bound peptides were eluted using low pH, and the eluted peptides were then separated from high-molecular-weight material using 10 kDa molecular weight cutoff filters. The identity of the purified MHC-bound peptides was determined using nanoflow liquid chromatography/tandem mass spectrometry and a hybrid linear trapping quadrupole/Fourier transform-ion cyclotron resonance mass spectrometer. Bioinformatic analysis was used to assign peptides to mouse or Salmonella origin. Among the 2508 MHC-bound peptides identified in this study, 1891 were MHC class-II (I-Ab)-binding and 617 were MHC class-I (H2−Kb or Db)-binding peptides (Tables 1–3 and Supplemental Table S1). Around 4% of the 2508 total peptides were derived from Salmonella proteins. Among the 110 Salmonella-derived peptides, 87 MHC class-II-binding peptides mapped to 54 distinct epitopes were derived from 53 unique source proteins, and the remaining 23 MHC class-I-binding peptides were derived from 23 unique source proteins of Salmonella.
Table 1.
Summary of Salmonella and Murine-Derived MHC Class-I- and -II-Bound Peptides, Epitopes, And Source Proteins Identified in This Study
| peptides | epitopes | proteins | ||
|---|---|---|---|---|
| mouse-derived | class I | 594 | ||
| class II | 1804 | |||
| Salmonella-derived | class I | 23 | 23 | 23 |
| class II | 87 | 54 | 53 |
Table 3.
MHC Class-I-Bound Salmonella Peptides Identified Using Immunoproteomics
| protein | name | peptide | start residue | end residue |
|---|---|---|---|---|
| Peptide Score >40 | ||||
| E1WBH0 | hypothetical regulatory protein | AADILFPAI | 347 | 355 |
| E1WBJ8 | hypothetical membrane protein | AYSVFSTV | 109 | 116 |
| Peptide Score 30–40 | ||||
| E1WDT4 | probable PTS system permease | GLIGGMLASLCGA | 199 | 211 |
| H8WUJ6 | conjugative transfer, oriT nicking-unwinding protein | FESAYVAL | 1425 | 1432 |
| E1WEA3 | HTH-type transcriptional activator RhaS | AFQFLAGLDQL | 91 | 101 |
| E1WA96 | formate hydrogenlyase subunit 3 | SIAMLVGLMAMAALPPLNGF | 369 | 388 |
| Peptide Score 20–30 | ||||
| E1WCY7 | glycerol-3-phosphate transport system permease protein | ALVDYVVR | 95 | 102 |
| E1WA24 | hypothetical glycosyltransferase | GPPLYGLL | 7 | 14 |
| E1WAT3 | D-serine/D-alanine/glycine transporter | IMFYVFALIVIMSVTPWSSVVP | 257 | 278 |
| E1WEW0 | LysR family regulatory protein | LGIPPMVGM | 95 | 103 |
| H8WV41 | conjugal transfer protein | MTTTEHPV | 1 | 8 |
| E1WIY0 | exopolyphosphatase | AMERGLSCL | 61 | 69 |
| E1WFF7 | glyceraldehyde-3-phosphate dehydrogenase | FRVPTPNVSVVD | 231 | 242 |
| E1WEV7 | hypothetical glutathione S transferase | IGRFKPAD | 79 | 86 |
| H8WUT4 | thiol:disulfide interchange protein | SIKSPAVNDMVALQERLF | 149 | 166 |
| E1WH34 | colanic acid biosynthesis protein | IDIRHLLN | 372 | 379 |
| E1WCR6 | phosphoenolpyruvate-protein phosphotransferase | TSIMARSLELPAIVGTGSV | 190 | 208 |
| E1WBK7 | glutaminase | SLVQLEMEQGIPRNPFI | 100 | 116 |
| E1WIY9 | host colonisation factor | ETGAIFTLNGDLINMGTMTSGSSSSTPGNT | 1537 | 1566 |
| E1WHJ3 | glycine dehydrogenase (decarboxylating) | IPASAHGTNPA | 600 | 610 |
| E1WH53 | hypothetical outer membrane assembly protein | EIGTILKAFNYPISLTGKMSLVGD | 410 | 433 |
| E1W8W8 | hypothetical lyase | WGALQLAAR | 291 | 299 |
| E1WCM6 | UPF0115 protein YfcN | ALFRQLMVGTRKIKQDTI | 13 | 30 |
Properties of MHC Class-I- and Class-II-Bound Peptides
As reported by others, class-I-bound peptides varied between 8 and 18 amino acids, with most being 8 or 9 amino acids in length. Because of varying degrees of proteolytic processing, the length of MHC class II peptides was much more heterogeneous and ranged between 11 and 26 amino acids (Tables 2 and 3). Many of these epitopes were represented by different sequence length variants. Four Salmonella proteins (GapA, GroEL, LpdA, MoeB) generated multiple overlapping peptides due to differential proteolytic processing at the open ends of the MHC II binding cleft. Strikingly GapA generated 31 overlapping peptides (Figure 2).
Table 2.
MHC Class-II-Bound Salmonella Peptides Identified Using Immunoproteomics
| protein | name | peptide | start residue | end residue |
|---|---|---|---|---|
| Peptide Score >40 | ||||
| E1WFF7 | glyceraldehyde-3-phosphate dehydrogenase | TGMAFRVPTPNVSVVDLT (31 peptides) | 227 | 244 |
| E1WCL8 | 3-oxoacyl-[acyl-carrier-protein] synthase I | LEHGFIAPSINIEELD | 350 | 365 |
| E1WA22 | flagellin | IDGKTYNASKAAGHDF | 387 | 402 |
| AVQNRFNSAITNLGNTVN | 439 | 456 | ||
| Peptide Score 30–40 | ||||
| E1W826 | dihydrolipoyl dehydrogenase | AGKKHYFDPKVIPSIAY (2 peptides) | 336 | 352 |
| E1WHK9 | phosphoglycerate kinase | LEFVEGKVLPAVA | 367 | 379 |
| E1WCC4 | glycerol-3-phosphate dehydrogenase | DPQQARIIEPSVNPALIG | 122 | 139 |
| E1WD28 | L-asparginase | GELGVQTLINAVPE | 52 | 65 |
| E1W8P7 | pyrroline-5-carboxylate reductase | GGAEVIAEPMIHPVVG | 155 | 170 |
| E1WF20 | 60 kDa chaperonin (GroEL protein) | GPKGRNVVLDKSFGAPTITKDGV (2 peptides) | 32 | 55 |
| Peptide Score 20–30 | ||||
| E1WFF1 | uncharacterized protein | KEPENSSIYSKMRVYDG | 324 | 340 |
| E1WE27 | 5-methyltetrahydro-pteroyltriglutamate--homocysteine methyltransferase | LLEEVDEALALG | 143 | 154 |
| E1W970 | hypothetical membrane protein | FKDPILGLVAGIQLSANDML | 144 | 163 |
| E1WH74 | fructose-bisphosphate aldolase class I | DLGDAVRTAVINKRAGGMGL | 298 | 317 |
| E1WFF7 | glyceraldehyde-3-phosphate dehydrogenase | KLTGMAFRVPTPNVSVVD | 225 | 242 |
| GMAFRVPTPNVS | 228 | 239 | ||
| E1WHM6 | biosynthetic arginine decarboxylase | ELMAVLAHAGMTRSVI | 133 | 148 |
| E1WC68 | TonB protein | PVVEPEPEPEPEPIPEPPK | 67 | 85 |
| E1W8I9 | hypothetical permease/MSF transporter | ALSMLIICMMVGAFLTLI | 302 | 319 |
| E1WIA3 | RNA polymerase sigma-54 factor | LDTADALEQKEMPEELPL | 67 | 84 |
| E1WCH9 | hypothetical membrane protein | AIAAWFMLLF | 398 | 407 |
| E1W9R3 | galactose-1-phosphate uridylyltransferase | VGYEMLAETQRDLTAEQAAERL | 314 | 335 |
| E1WGX3 | glycerol dehydratase large subunit | FIARYGINLNRAEEVMAM | 65 | 82 |
| E1W953 | outer membrane usher protein FimD | GTVIAGLPAGLTAYGGTQLA | 387 | 406 |
| E1WD27 | glutathione reductase | VPKKVMWHAAQIREAIH | 48 | 64 |
| E1W7R3 | Na(+)/H(+) antiporter NhaA | GVSLQGVTIDGLTSML | 273 | 288 |
| E1W9Y1 | molybdopterin biosynthesis MoeB protein | INPHITITPVNARLDDDAM (2 peptides) | 97 | 115 |
| E1WEE9 | transcriptional repressor | PTAVFCHSDVMAL | 246 | 258 |
| E1WBX7 | hypothetical exported protein | RMPQHDPALLRVQNISSSEL | 706 | 725 |
| E1WHT5 | hydrogenase-2 large subunit | DDVGPYEQSLVGTPIADPAK | 513 | 532 |
| E1WE91 | hypothetical 1 formate dehydrogenase-O, major subunit | NIRTMAMIQLLLGNMGMAGGGVNALR | 420 | 445 |
| E1WDV3 | aspartate--ammonia ligase | RGEMPQTIGGGIGQSRL | 284 | 300 |
| E1W898 | UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase) | CMIGGASVINGHMEI | 265 | 279 |
| E1WDR4 | chromosomal replication initiator protein DnaA | KADENDIRLPG | 308 | 318 |
| E1WBS8 | gamma-aminobutyraldehyde dehydrogenase | PALAAGNCVVIKPSEIT | 168 | 184 |
| E1WHM1 | hypothetical oxidoreductase | LTGVLMGANFSNHIVRM | 238 | 254 |
| E1W9N8 | TolQ protein | AFIALGAVKQATLQMVAPGIAEAL | 152 | 175 |
| E1W8V3 | cytochrome o ubiquinol oxidase subunit II | VAKAKQSPNTMNDMAAFEKVAMPSEYN | 235 | 261 |
| E1W9H9 | Pts system, N-acetylglucosamine-specific IIABC component | KAMVTINPEINMGVLAGIITG | 85 | 105 |
| E1WID9 | possible membrane transport protein | LMPLVFAAALGGNLSLIGA | 135 | 153 |
| E1WER9 | maltose transport inner membrane protein | LQLKETDALPGGERANLRII | 157 | 176 |
| E1WDG8 | DNA ligase B | KYPPVAQVAQVSAIQFSVG | 305 | 323 |
| E1W9T0 | uncharacterized protein | GSGLVAVP | 81 | 88 |
| E1W7W4 | 4-hydroxythreonine-4-phosphate dehydrogenase | LVVQLAQRAWPIELVVCADGA | 21 | 41 |
| E1W7F5 | sodium/proline symporter | KQYGWLDLYEIIPGFI | 445 | 460 |
| E1WEV1 | large repetitive protein | SAWTDGDYTLTVTVKDEAGN | 1988 | 2007 |
| E1W9E9 | leucine--tRNA ligase | YPSGRLHMGHVRNYT | 43 | 57 |
| H8WV08 | conjugal transfer protein | MLHDSINKLGEAIKKDEE | 121 | 138 |
| E1WJB0 | exodeoxyribonuclease VIII | LRPAMDFAKRIIAEDRED | 315 | 332 |
| E1WB21 | superfamily I DNA helicase | RKDLKTAETKIYTLHGV | 1003 | 1019 |
| E1WEI8 | pantothenate kinase | MHRLVKFVSDLKSGVP | 153 | 168 |
| E1WGH6 | tyrosine-specific transport protein | FSVTLGLLIGLWALMCYTA | 33 | 51 |
| E1WG19 | hypothetical exported protein | RKNASLFGNVLMGLGLVVMVVGVGYS | 3 | 28 |
| E1WHQ4 | hypothetical membrane protein | GINGAVAKIQQL | 78 | 89 |
Figure 2.
GapA overlapping peptides as an example of differential proteolytic cleavage on a class II molecule.
We identified two epitopes derived from Salmonella flagellin: IDGKTYNASKAAGHDF (derived from FljB, 387–402) and AVQNRFNSAITNLGNTVN (derived from FliC, 428–442 or FljB, 439–456). The epitope AVQNRFNSAITNLGNTVN is identical to an I-Ab epitope previously identified by our laboratory via epitope mapping27 (Figure 3).
Figure 3.
Position of flagellin MHC class-II-bound epitopes within the full-length sequence of FljB protein. The two epitopes identified in this study are highlighted, and the I-Ab epitope previously identified by McSorley et al.27 is underlined.
Using an I-Ab consensus sequence we previously deduced for Chlamydia class-II-binding peptides (Figure 4), we were able to fit in the anchor residues of pockets 1, 4, and 6 of the Salmonella Gap protein-derived peptides (Figure 2) to the same amino acids F, P, and P, respectively.
Figure 4.
Logo plot I-Ab class II core peptides based on estimated alignment, created with the WebLogo program.31 Aromatic amino acids are in green, acidic in red, basic in blue, polar in orange, and hydrophobic in pink.
The cellular locations of the 53 Salmonella proteins that gave rise to the 87 MHC class II peptides identified in this study were analyzed, and the results suggested that they originated from both cytosol and membrane compartments.
In Vitro Recognition of Salmonella MHC Class-II-Binding Peptides by Immune T Cells
To validate the immunological relevance of the identified MHC class-II-binding Salmonella peptides, we evaluated whether MHC class II Salmonella peptides are recognized by CD4+ T cells from persistently infected mice. For these experiments we selected the 12 highest-scoring MHC class-II-binding peptides identified from infected DC in vitro (Figure 5). It has been shown that the vast majority of peptide:MHC class-II-specific CD4+ T cells differentiate into IFN-γ producing Th1 cells during Salmonella infection.28 Therefore, we used an IFN-γ ELISPOT to evaluate the antigen-specific CD4+ T cell response to validate Salmonella peptides in persistently infected mice. Purified CD4+ T cells from mice with persistent Salmonella infection were stimulated in vitro overnight with Salmonella peptides, and the frequency of IFN-γ-secreting Th1 cells was determined (Figure 5). Our results showed that 4 out of the 12 peptides (Figure 5; peptides 9–12) elicited IFN-γ production in >0.06% of total CD4+ T cells and stimulated significantly stronger responses when compared with negative controls. However, the remaining eight peptides eluted from SL1344- infected DCs (Figure 5; peptides 1–8) did not stimulate significant IFN-γ secretion from memory Th1 cells. On the basis of the 12 selected peptides tested, these results show that 30% of the MHC class-II-bound Salmonella peptides were able to prime and maintain corresponding Salmonella peptide:MHC class-II-specific CD4+ T cells during persistent infection, as measured by antigen-specific IFN-γ production.
Figure 5.
Recognition of Salmonella enterica serovar Typhimurium MHC class-II-bound peptides eluted from I-Ab molecules in immune 129 × 1/sv/J mice, as identified by IFN-γ ELISPOT assay. 11-mer peptides were selected from among the highest-scoring peptides eluted from Salmonella-infected BMDCs in vitro. Heat-killed Salmonella (+) and no peptide (−) were used as controls. ELISPOT results were analyzed using one-way ANOVA, and p values were generated by applying Tukey’s posthoc test to comparisons of all experimental groups (* = p < 0.05, *** = p < 0.001, and **** = p < 0.0001). This Figure is representative of three different experiments.
DISCUSSION
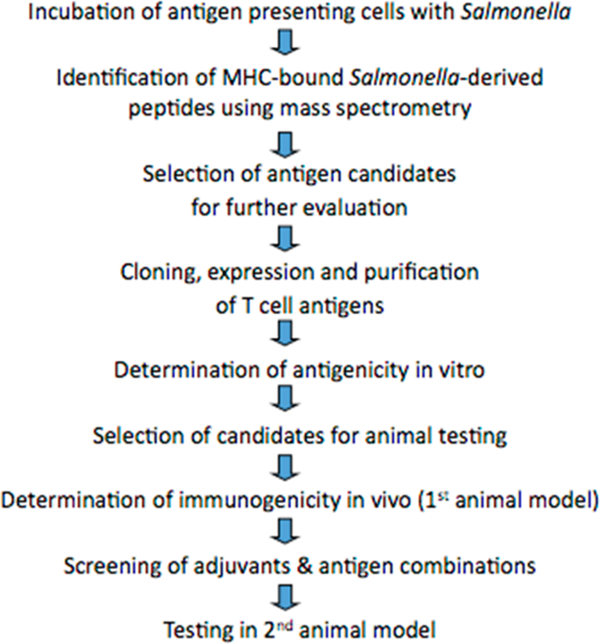
The immunoproteomic approach has been used successfully in our laboratory over the last 8 years to analyze the immunoproteome of Chlamydia.16–19 In vivo vaccine challenge studies performed in mice using the identified T-cell antigens proved that Chlamydia T-cell antigens identified by immuno- proteomics can be successfully used as T-cell protein-based subunit vaccine against Chlamydia infection.20–22 Our Chlamydia study provides direct support for the discovery of T-cell-based subunit molecular vaccines against other intra- cellular pathogens, including Salmonella (schematically de- scribed in Figure 6).
Figure 6.
Schematic depiction of the sequence of steps involved in the immunoproteomic approach for T-cell vaccine development.
This study identified 110 Salmonella-derived MHC-bound peptides, 87 presented by MHC class II molecules and 23 presented by MHC class I molecules on DCs infected with Salmonella. The experiment shows for the first time that Salmonella peptides can be successfully isolated from MHC molecules of professional antigen presenting cells and analyzed by MS/MS. The identified peptides were immunologically relevant because Salmonella-specific CD4+ T cells from mice persistently infected with Salmonella recognized a subset of them. These results demonstrate the utility of immunoproteo- mic platform in reliably identifying peptides presented to CD4+ T cells from a persistent phagosomal pathogen.
All previous studies that search for T-cell antigens have been based on the consensus T-cell epitope prediction approach where potential epitopes were first identified using bioinformatic methods, followed by testing of each antigen for a positive immune response with the pathogen of interest. There have been studies previously undertaken to identify Salmonella MHC class II epitopes using this type of bioinformative approach.29,30 Somewhat surprisingly, several of these previously predicted epitopes were not identified in our study. It is important to note, however, that our immunoproteomic approach identifies peptides that result from in vivo physiological processing and presentation in comparison with most of the previous epitope discovery methods that have used bioinformatical approach. In fact, Lee et al. reported that none of the epitopes predicted by bioinformatics approach in their study were naturally presented.29 Even though the bioinformatic approach allows systematic testing of antigens for their ability to provide protection, the number of bioinformatically identified peptides usually exceeds the ability to test, and only a minority are experimentally confirmed to be T-cell antigens.32 The advantage of using the immunoproteomics over the bioinformatics approach is that the positive validation rate is higher for the immunoproteomic approach. This was evident in our Chlamydia studies, where 11 out of 13 T-cell antigens tested turned out to be protective in mouse model.21 This higher positive validation rate of the peptides identified via the immunoproteomic approach could be due to the fact that peptides are directly identified as they are presented by APCs under natural physiological conditions.
Even though each microbial protein can be potentially presented to the immune system, only a limited number of proteins are typically presented to the immune system via MHC molecules. In this study only 76 (1.6%) of 4743 Salmonella proteins generated MHC-binding peptides. Immu- nodominance is a complex phenomenon that can be influenced by both external and internal factors.33 For uncertain reasons, immunodominance is much more characteristic of T-cell than B-cell responses. There may be selection mechanisms that limit immune responses to a few select antigens. One possibility is that some proteins may be better processed by the complex antigen processing and presentation pathways.34 Selection mechanisms at the microbial protein level might include variables such as membrane localization, sequence, structure, and protease cleavage sites, among others. The possibility also exists, though, that our mass spectrometry survey of class II peptides is nonsaturating. Thus immunodominant epitope identification may vary from experiment to experiment.
The major challenge in performing immunoproteomics is the cost and the amount of work involved in each experiment. To determine the number of BMDCs used in our experiments, we assumed that each antigen-presenting cell presents as few as one molecule of a particular antigen, yet in practice the current sensitivity of mass spectrometers has only been in the range of ∼6 billion molecules or ∼10 fmol. To collect five to ten billion antigen-presenting DCs requires approximately 50 to 100 mice to be sacrificed, which is already at the limits of this approach, so even assuming minimal losses in each purification step, this would still only yield microbial peptides at or below the sensitivity limit. An additional complication is that the terminal residues of MHC-presented peptides are not as predictable as the cut sites generated by proteases such as trypsin, which are commonly used in the identification of peptides by MS/MS. Because of this unpredictability, all 20 amino acids must be considered as potential terminal residues, greatly increasing the possibility of misidentifying the peptide.35 This increased rate of false-positive identifications can be partially compensated by increasing the accuracy of mass measurements. Nonetheless, sensitivity and accuracy are often traded off in mass spectrometers. Now, however, there are commercially available mass spectrometers (e.g., LTQ-FT and LTQ-Orbitrap from ThermoScientific) with proven (i.e., when analyzing complex samples) sensitivity limits near 1 fmol and that are able to measure peptide masses to within one part per million.14,15,36
This recent advancement of increased sensitivity in mass spectrometry overlaps with the amount of pathogen-derived MHC-bound peptides that we can purify from APCs infected with pathogens.
Success in developing T-cell vaccines based on immunopro- teomics will have significant implications for other intracellular pathogens. In principle, the discovery methods used here can be applied to many of the most important uncontrolled infectious diseases such as Mycobacterium tuberculosis with potentially major public health impact.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Grants R01AI103760 and P01AI035296 (M.K.J.).
Footnotes
The authors declare no competing financial interest.
Data have been deposited to the ProteomeXchange Con- sortium via the PRIDE partner repository with the data set identifier PXD004451.
REFERENCES
- (1).Crump JA; Sjolund-Karlsson M; Gordon MA; Parry CM Epidemiology, Clinical Presentation, Laboratory Diagnosis, Antimicro- bial Resistance, and Antimicrobial Management of Invasive Salmonella Infections. Clin. Microbiol. Rev 2015, 28, 901–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Gordon MA Salmonella infections in immunocompromised adults. J. Infect 2008, 56, 413–22. [DOI] [PubMed] [Google Scholar]
- (3).Ao TT; Feasey NA; Gordon MA; Keddy KH; Angulo FJ; Crump JA Global burden of invasive nontyphoidal Salmonella disease, 2010(1). Emerging Infect. Dis 2015, 21, 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mastroeni P; Rossi O Immunology, epidemiology and mathematical modelling towards a better understanding of invasive non-typhoidal Salmonella disease and rational vaccination approaches. Expert Rev. Vaccines 2016, 1–11. [DOI] [PubMed] [Google Scholar]
- (5).McSorley SJ Immunity to intestinal pathogens: lessons learned from Salmonella. Immunol Rev. 2014, 260, 168–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Sundquist M; Rydstrom A; Wick MJ Immunity to Salmonella from a dendritic point of view. Cell. Microbiol 2004, 6, 1–11. [DOI] [PubMed] [Google Scholar]
- (7).Hess J; Ladel C; Miko D; Kaufmann SH Salmonella typhimurium aroA- infection in gene-targeted immunodeficient mice: major role of CD4+ TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J. Immunol 1996, 156, 3321–6. [PubMed] [Google Scholar]
- (8).Lee SJ; Dunmire S; McSorley SJ MHC class-I-restricted CD8 T cells play a protective role during primary Salmonella infection. Immunol. Lett 2012, 148, 138–43. DOI: 10.1021/acs.jproteome.6b00926 J. Proteome Res. 2017, 16, 298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mastroeni P; Villarreal-Ramos B; Hormaeche CE Role of T cells, TNF alpha and IFN gamma in recall of immunity to oral challenge with virulent salmonellae in mice vaccinated with live attenuated aro- Salmonella vaccines. Microb. Pathog 1992, 13, 477–91. [DOI] [PubMed] [Google Scholar]
- (10).Mastroeni P; Simmons C; Fowler R; Hormaeche CE; Dougan G Igh-6(−/−) (B-cell-deficient) mice fail to mount solid acquired resistance to oral challenge with virulent Salmonella enterica serovar typhimurium and show impaired Th1 T-cell responses to Salmonella antigens. Infect. Immun 2000, 68, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Rappuoli R; Aderem AA 2020 vision for vaccines against HIV, tuberculosis and malaria. Nature 2011, 473, 463–9. [DOI] [PubMed] [Google Scholar]
- (12).Kroger C; Dillon SC; Cameron AD; et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. U. S. A 2012, 109, E1277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).de Jong A Contribution of mass spectrometry to contemporary immunology. Mass Spectrom. Rev 1998, 17, 311–35. [DOI] [PubMed] [Google Scholar]
- (14).Beck S; Michalski A; Raether O; et al. The Impact II, a Very High-Resolution Quadrupole Time-of-Flight Instrument (QTOF) for Deep Shotgun Proteomics. Mol. Cell. Proteomics 2015, 14, 2014–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Scheltema RA; Hauschild JP; Lange O; et al. The Q Exactive HF, a Benchtop mass spectrometer with a pre-filter, high- performance quadrupole and an ultra-high-field Orbitrap analyzer. Mol. Cell. Proteomics 2014, 13, 3698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Karunakaran KP; Rey-Ladino J; Stoynov N; et al. Immunoproteomic discovery of novel T cell antigens from the obligate intracellular pathogen Chlamydia. J. Immunol. 2008, 180, 2459–65. [DOI] [PubMed] [Google Scholar]
- (17).Yu H; Karunakaran KP; Kelly I; Shen C; Jiang X; Foster LJ; Brunham RC Immunization with live and dead Chlamydia muridarum induces different levels of protective immunity in a murine genital tract model: correlation with MHC class II peptide presentation and multifunctional Th1 cells. J. Immunol 2011, 186, 3615–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).() Karunakaran KP; Yu H; Jiang X; et al. Outer membrane proteins preferentially load MHC class II peptides: Implications for a Chlamydia trachomatis T cell vaccine. Vaccine 2015, 33, 2159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Karunakaran KP; Yu H; Foster LJ; Brunham RC Development of a Chlamydia trachomatis T cell Vaccine. Hum. Vaccines 2010, 6, 676–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yu H; Jiang X; Shen C; Karunakaran KP; Brunham RC Novel Chlamydia muridarum T cell antigens induce protective immunity against lung and genital tract infection in murine models. J. Immunol 2009, 182, 1602–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Yu H; Karunakaran KP; Jiang X; Shen C; Andersen P; Brunham RC Chlamydia muridarum T Cell Antigens and Adjuvants That Induce Protective Immunity in Mice. Infect. Immun 2012, 80, 1510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yu H; Karunakaran KP; Jiang X; Brunham RC Evaluation of a multisubunit recombinant polymorphic membrane protein and major outer membrane protein T cell vaccine against Chlamydia muridarum genital infection in three strains of mice. Vaccine 2014, 32, 4672–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Su H; Messer R; Whitmire W; Fischer E; Portis JC; Caldwell HD Vaccination against chlamydial genital tract infection after immunization with dendritic cells pulsed ex vivo with nonviable Chlamydiae. J. Exp. Med 1998, 188, 809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ishihama Y; Rappsilber J; Mann M Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J. Proteome Res 2006, 5, 988–94. [DOI] [PubMed] [Google Scholar]
- (25).Zhu Y; Rudensky AY; Corper AL; Teyton L; Wilson IA Crystal structure of MHC class II I-Ab in complex with a human CLIP peptide: prediction of an I-Ab peptide-binding motif. J. Mol. Biol 2003, 326, 1157–74. [DOI] [PubMed] [Google Scholar]
- (26).Streeck H; Frahm N; Walker BD The role of IFN-gamma Elispot assay in HIV vaccine research. Nat. Protoc 2009, 4, 461–9. [DOI] [PubMed] [Google Scholar]
- (27).McSorley SJ; Cookson BT; Jenkins MK Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J. Immunol 2000, 164, 986–93. [DOI] [PubMed] [Google Scholar]
- (28).Nelson RW; McLachlan JB; Kurtz JR; Jenkins MK CD4+ T cell persistence and function after infection are maintained by low-level peptide:MHC class II presentation. J. Immunol 2013, 190, 2828–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lee SJ; McLachlan JB; Kurtz JR; et al. Temporal expression of bacterial proteins instructs host CD4 T cell expansion and Th17 development. PLoS Pathog. 2012, 8, e1002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Maybeno M; Redeker A; Welten SP; et al. Polyfunctional CD4+ T cell responses to immunodominant epitopes correlate with disease activity of virulent Salmonella. PLoS One 2012, 7, e43481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Schneider TD; Stephens RM Sequence Logos: A New Way to Display Consensus Sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Moutaftsi M; Peters B; Pasquetto V; et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol 2006, 24, 817–9. [DOI] [PubMed] [Google Scholar]
- (33).Sette A; Peters B Immune epitope mapping in the post- genomic era: lessons for vaccine development. Curr. Opin. Immunol. 2007, 19 (1), 106–110. [DOI] [PubMed] [Google Scholar]
- (34).Unanue ER; Turk V; Neefjes J Variations in MHC Class II Antigen Processing and Presentation in Health and Disease. Annu. Rev. Immunol 2016, 34, 265–97. [DOI] [PubMed] [Google Scholar]
- (35).Olsen JV; Ong SE; Mann M Trypsin cleaves exclusively C- terminal to arginine and lysine residues. Mol. Cell. Proteomics 2004, 3, 608–14. [DOI] [PubMed] [Google Scholar]
- (36).Olsen JV; de Godoy LM; Li G; et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 2005, 4, 2010–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.