Abstract
Purpose
Radon is a risk factor for lung cancer and uranium miners are more exposed than the general population. A genome-wide interaction analysis was carried out to identify genomic loci, genes or gene sets that modify the susceptibility to lung cancer given occupational exposure to the radioactive gas radon.
Methods
Samples from 28 studies provided by the International Lung Cancer Consortium were pooled with samples of former uranium miners collected by the German Federal Office of Radiation Protection. In total 15,077 cases and 13,522 controls, all of European ancestries, comprising 463 uranium miners were compared. The DNA of all participants was genotyped with the OncoArray. We fitted single-marker and in multi-marker models and performed an exploratory gene-set analysis to detect cumulative enrichment of significance in sets of genes.
Results
We discovered a genome-wide significant interaction of the marker rs12440014 within the gene CHRNB4 (OR=0.26, 95%.CI: 0.11–0.60, p=0.0386 corrected for multiple testing). At least suggestive significant interaction of linkage disequilibrium blocks was observed at the chromosomal regions 18q21.23 (p=1.2×10−6), 5q23.2 (p=2.5×10−6), 1q21.3 (p=3.2×10−6), 10p13 (p=1.3×10−5) and 12p12.1 (p=7.1×10−5). Genes belonging to the Gene Ontology term „DNA dealkylation involved in DNA repair” (GO:0006307; p=0.0139) or the gene-family HGNC:476 „microRNAs” (p=0.0159) were enriched with LD-block-wise significance.
Conclusion
The well-established association of the genomic region 15q25 to lung cancer might be influenced by exposure to radon among uranium miners. Further, lung cancer susceptibility is related to the functional capability of DNA damage signalling via ubiquitination processes and repair of radiation-induced double-strand breaks by the single-strand annealing mechanism.
Keywords: GWAS, radon progeny, occupational exposure, gene-environment interaction, DNA repair
Introduction
You cannot see it; you cannot hear it and you cannot smell it; but be aware it is omnipresent in indoor and outdoor air and contaminates many underground mines (Sethi et al. 2012). Radon is a radioactive noble gas released by the uranium decay chain. An increased risk for lung cancer (LC), the main cause of cancer-related death worldwide (Jemal et al. 2011; Siegel et al. 2016; Torre et al. 2016), caused by inhalation of radon has been consistently demonstrated in several studies of indoor-exposure in dwellings as well as for uranium miners (Darby et al. 2005; Grosche et al. 2006; National Research Council 1999; Sethi et al. 2012). It was estimated, that ionising radiation due to residential radon causes 3% to 15% of LC cases in the general population (Sethi et al. 2012). That is why radon is the second strongest risk factor for LC and among the top 4 environmental risks to public health in the United States (McColl et al. 2015; Sethi et al. 2012).
Pooled analyses of genome-wide association studies (GWASs) within the International Lung Cancer Consortium (ILCCO) have revealed that genomic variations at e.g. 5p15.33, 6p21–22 and 15q25 and further 42 LC susceptibility loci influence LC risk in European populations (Bosse and Amos 2018). In total 92 genes are postulated to be suspected causal genes for LC. Although the strongest genetic association with an odds ratio (OR) of 7.2 was reported for 15q25 in a familial form of LC, for sporadic LC an OR of only ~1.3 was observed, albeit highly significant (p = 3.08 × 10−103). However, “cumulative effects of loci have shown promising results to improve the discriminatory performance of risk prediction models”(Bosse and Amos 2018) Nevertheless, genes can be associated to several traits and contribute to the functional efficacy of multiple interlocked biological processes. One may assume that for example nicotine dependency or DNA repair play a role in an individual’s susceptibility to developing LC (Brennan et al. 2011; Romero-Laorden and Castro 2017). For example, some genetic variants in CHRNA5 on chromosome 15q25.1 increase the risk for smoking-related disorders such as LC and chronic obstructive pulmonary disease (COPD) but are also associated with delayed smoking cessation (Amos et al. 2008; Chen et al. 2015b). Taken together, the harming mechanisms of smoking consist at least in parts of a complex interplay between tobacco exposure, previous diseases and genetics. However, smoking is the most important but an avoidable risk factor.
Exposure to radon is ubiquitous and not self-inflicted, but can be reduced in homes and buildings; the related biological defence mechanisms are complex (McColl et al. 2015). DNA damage, induced by radioactive alpha particles emitted by radon progenies, is considered as pivotal mechanism of carcinogenesis in the lung (Sethi et al. 2012). A heritable component in the capacity to repair DNA-damage was demonstrated (Rosenberger et al. 2012). Ionizing radiation induces oxidation of DNA bases and generates single-strand breaks (SSBs) and double-strand breaks (DSBs) (Rosenberger et al. 2012). An individual’s capacity to repair DSBs is recognised as a risk factor or an effect modifier in LC (Ishida et al. 2014; Ridge et al. 2013). DSBs capacity determining genes are widely investigated as susceptibility genes for lung cancer.(Chen et al. 2015a; Kazma et al. 2012) The interaction of radon with some genes belonging to biological mechanisms other than DNA damage response was also investigated with candidate gene approaches in either high dose exposed uranium miners (SIRT1; P53; CDKN2A and MGMT; IL6) or low dose exposed humans in dwellings (GSTM, GSTT and EPHX1; P53) (Leng et al. 2013; Leng et al. 2016; Ruano-Ravina et al. 2014; Vahakangas et al. 1992; Yngveson et al. 1999). Nevertheless, it is still unclear which genomic dispositions make one susceptible to radiation-induced LC.
The uranium miners of the former German Wismut mining company, with about 400 000 employees, form a large population with documented radiation exposure. In 2009 the German Federal Office of Radiation Protection (Bundesamt für Strahlenschutz, BfS) started to build up the German Uranium Miners Bio- and Databank (GUMB) with DNA from blood and/or tissue samples from LC cases and healthy controls of former uranium miners of this company. Exposure estimations and data are captured in the same way as for a large cohort study of the same population and includes an estimate of the cumulative occupational exposure to radon progeny (Kreuzer et al. 2010b; Walsh et al. 2010).
This work was conducted as collaboration between the Transdisciplinary Research of Cancer in Lung and the International Lung Cancer Consortium (TRICL/ILCCO), the German Federal Office of Radiation Protection (BfS) and the University Medical Centre Göttingen. We merged phenotypic and genotypic information from TRICL/ILCCO and from BfS. Genotypes were yielded by the OncoArray, to perform a genome-wide search for radon x gene interaction –without restricting the investigation to any presumed mode of action.
Materials and methods
The participating studies of TRICL/ILCCO are individually described in the supplement of McKay et al. (2017), Table 1 and Supplementary Table I (Online Resource 1). The LC cases of the BfS sample collection were recruited for a study investigating indoor-radon exposure between 1990 und 1997 (Brüske-Hohlfeld et al. 2006). The cancer-free BfS controls are former uranium miners recruited from 2009–2012, who continuously participated in health surveillance program of the German Social Accident Insurance and are long term survivors (Pesch et al. 2015). This control samples stored in German Uranium Miners Biobank (GUMB) of the BfS were drawn from these miners, which were either very high (>750 Working Level Months, WLM) or low (≤50 WLM) radiation exposed in a targeted and no-representative ratio of 2:1 (Pesch et al. 2015). The method how radon exposures was measured is given elsewhere (Kreuzer et al. 2010b) (see Scaling residential and occupational radon exposure, Online Resource 1).
Table 1.
Source studies
| Acronym | Study name | Institution | PI (principal investigator) |
Country | Design | Participants in this analysis |
Time span of recruitment |
|---|---|---|---|---|---|---|---|
| CARET | The Carotene and Retinol Efficacy Trial | Fred Hutchinson Cancer Research Center (FHCRC) | G. Goodman, J. Doherty, C. Chen | USA | Cohort | 1065 | Recruitment 1985–1996 |
| BioVU | The Vanderbilt Lung Cancer Study | Vanderbilt University | M. Aldrich | USA | Hosp. CC | 1 160 | 2007-ongoing |
| HLCS | Harvard Lung Cancer Study | Harvard School of Public Health, Mass General Hospital | D. Christiani | USA | Hosp. CC | 1 605 | 1992–2004 |
| ATBC | The Alpha-Tocopherol, Beta-Carotene Cancer Prevention | National Cancer Institute (NCI) | D. Albanes | Finland | Cohort | 1 683 | 1985–1993 |
| PLCO | The Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial | National Cancer Institute (NCI) |
N. Caporaso | USA | Cohort | 2 231 | 1992–2001 |
| MSH-PMH | Princess Margaret Hospital Early Detection Study | Mount Sinai Hospital (MSH), Princess Margaret Hospital (PMH) | R.J. Hung, G. Liu | Canada | Hosp. CC | 2 295 | 2008–2012 |
| LCRI-DOD | Population based case-control study of lung cancer in Appalachian Kentucky | Markey Cancer Center | S. Arnold | USA | Pop. CC | 220 | 2012- ongoing |
| TAMPA | Tampa Lung Cancer Study | H. Lee Moffitt Cancer Center | P. Lazarus | USA | Hosp. CC | 242 | 1999–2003 |
| NELCS | New England Lung Cancer Study | Dartmouth College of Medicine | A. Andrew | USA | Pop. CC | 329 | 2005–2007 |
| TLC | Total Lung Cancer: Molecular Epidemiology of Lung Cancer Survival | Moffitt Cancer Center, Tampa | M.B. Schabath | USA | case only | 419 | 2012-- ongoing |
| MEC | Multi Ethnic Cohort Study | University of Hawaii (USC) | L. Le Marchand, Ch. Haiman | USA | Cohort | 430 | Recruitment 1993–1996 |
| Canada | Pan-Candadian screening study | University Health Network (UHN), British Columbia Cancer Agency (BCCA) | S. Lam, G. Liu | Canada | screening cohort | 656 | 2004–2011, 2008–2013 |
| EAGLE | Environment and Genetics in Lung Cancer Study Etiology | National Cancer Institute (NCI) | M.T. Landi | Italy | Pop.CC | 3 494 | 2002–2005 |
| Copenhagen | Copenhagen Lung Cancer Study | University of Copenhagen | S. E. Bojesen | Denmark | 1 823 | ||
| CAPUA | Cancer de Pulmon en Asturias | University of Oviedo | A. Tardon | Spain | Hosp.CC | 1 399 | 2002–2012 |
| GLC | German Lung Cancer Study | University of Göttingen, Deutsches Krebsforschungszentrum Heidelberg (DKFZ) DKFZ-part |
H. Bickeböller, A. Risch | Germany | Mixed CC | 1 014 | 1998–2013 |
| GLC-500K | German Lung Cancer Study | University of Göttingen, Helmholtz Zentrum München (HMGU), HMGU-part | H. Bickeböller, A. Risch, H.-E. Wichmann | Germany | Mixed CC | 949 | 1998–2013 |
| Nijmegen | The Nijmegen Lung Cancer Study | Radboud University Medical Centre | B. Kiemeney | The Netherlands | Pop.CC | 816 | 2002–2008 |
| ReSoLucent | Resource for the Study of Lung Cancer Epidemiology in North Trent | University of Sheffield, | M.D. Teare | UK | Mixed CC | 750 | 2005–2014 |
| Norway | Norway National Institute of Occupational Health Study | National Institute of Occupational Health (NIOH) | A. Haugen | Norway | Pop.CC | 725 | 1986–2005 |
|
LLP-2008,
LLP-2013 |
Roy Castle Lung Study (Liverpool Lung Cancer Project) |
University of Liverpool | J.K. Field | UK | Cohort | 200 675 |
1999–2007, 1999–2011 |
| NSHDC | Northern Sweden Health and Disease Cohort | Umeå University | M. Johansson | Sweden | Cohort | 473 | 1985-ongoing |
| MDCS | The Malmö Diet and Cancer Study | Lund University | H. Brunnsstöm | Sweden | Cohort | 325 | 1991–1996 |
| NICCC-LCA | Israel Lung Cancer Study | Carmel Medical Center & Technion | G. Rennert | Israel | Pop.CC | 1149 | 2008-ongoing |
| L2 | The IARC L2 Study | International Agency for Research on Cancer (IARC) | P. Brennan | Central Europe | Pop./Hosp. CC | 2009 | 2005–2013 |
| Wismut | Case-control study on lung cancer in former Wismut uranium miners (cases) | Helmholtz Zentrum München (HMGU) | H.-E. Wichmann, L. Kreienbrock, | Germany | case sample | 58 | 1990–1995 |
| GUMB | Biobank of healthy former Wismut uranium miners (controls) | Bundesamt für Strahlenschutz (BfS) | M. Gomolka | Germany | sample collection | 405 | 2009-ongoing |
Study population
The analysed sample consisted of 28 599 study participants with European ancestry and valid information on age at diagnosis/interview, sex and smoking status (15 077 cases : 13 522 controls); 463 thereof are former uranium miners of the Wismut mining company (61 cases : 402 controls), 949 are from the German Lung Cancer Study (471 cases : 478 controls), the remaining are from 25 studies of TRICL/ILCCO (14 545 cases : 12 642 controls) (see Table 1 and Supplementary Table I, Online Resource 1). 49 of 15 077 (0.3%) LC cases and 259 of 13 522 cancer-free controls (1.9%) had been occupationally exposed by a high cumulative dose exposure to radon and its progeny (WLM>50). It is unlikely that a cumulative lifetime exposure solely due to an exposure by indoor or other environmental radon sums up to more than 50 WLM. Thus, we categorised occupational radon exposure into ≤50 (“unexposed”) and >50 WLM (“exposed”), a threshold for significant elevated relative LC-risk (Kreuzer et al. 2010a). All TRICL/ILCCO participants were assigned to the exposure categories ≤50 WLM. Misclassification would be conservative. A detailed justification is given in the supplement (see Online Resource 1).
Genotyping and QC
The Infinium OncoArray-500K was used for high-throughput genotyping. Quality control (QC) was performed following the approach previously described for the OncoArray (Amos et al. 2017). To validate the European ancestry of the participants the probability of being Caucasian based on a set of 159 ancestry- and PCA-informative markers was estimated (Huckins et al. 2014; Kosoy et al. 2009; Setsirichok et al. 2012) applying the program ADMIXTURE (Alexander et al. 2009). 407 117 markers entered the analysis, after excluding markers of low quality genotyping or a minor allele frequencies (MAF) <1%. These remaining markers could be clustered into 103 983 blocks (67 161 LD blocks and 36 822 hot spots; for definition see Online Resource 1).
Merging samples
The crude odds ratio (OR) for the occupational radon exposure within participants of the BfS sample collection was OR=2.25. Because naïvely adding the TRICL/ILCCO participants would biased this association to OR=0.17, we down-weighted the cases of TRICL/ILCCO by the factor 1 : 13.6. In this way we avoided this unjustified inversion of the crude association, and still use all available information for analysis. However, we have fixed the marginal risk of a radon exposure at the point estimate from the BfS sample collection (for a more detailed explanation see Online Resource 1).
Statistical analysis
We fitted two models to individual data and also carried out a gene-set analysis (GSA) to search for accumulated significance in pre-defined groups of genes for pathways and gene families of interest. All calculations, data handling and image acquire were performed using PLINK 1.9 (Purcell et al. 2007) and SAS 9.4 of the SAS Institute Inc., Cary. NC. USA.
Single-marker interaction analysis
We first performed single-marker interaction analysis fitting the log-additive model:
| [1] |
where D is the disease status (D=1: LC patient; D=0: control); G is minor allele count at marker m; E is the exposure category (0: ≤50 WLM, 1: WLM>50); PS is a propensity score comprising the probability being a case explained by age, sex and smoking. To adjust for genomic population stratification we calculated the principal components (PC) of genotypes. Only the first four PCs were included in the statistical modelling, because the fifth PC was significantly correlated with the disease status. The remaining inflation factor (median of the χ²-distribution for unadjusted association) was λ~1.1, which is acceptable close to 1.0 (Yang et al. 2011).
The data at hand are not a representative data set of a well-defined source population. Thus the effect estimate of interaction, expressed as odds ratio, is potentially proportionally biased. Therefore, the tilde is added to indicate that a weighted sample was used for estimation (see Merging samples). However, estimating OR is not our main interest, rather than testing the null hypothesis, which is still valid (Mukherjee et al. 2008; Stenzel et al. 2015).
With as global level of significance, we use as Bonferroni-corrected, genome-wide level of significance. A suggestive level of significance was set to. Significance was determined according the Hybrid 2-step (H2) method of Murcray et al. (2011). All markers were first grouped into four classes: (a) disease-gene (DxG) effect only, (b) environmental-gene (ExG) effect only, (c) both or (d) none. Correction for multiple testing was performed within these groups, however under accounting for a tuning parameter that can take values between 0.5 to 1–10−20 (for a more detailed explanation see Online Resource 1).
Multi-marker interaction analysis
We also searched for the best fitting model of each LD block, allowing all markers of a block to enter the model (denoted as complete model). We then applied a backward selection with the best model chosen according to Akaike’s information criteria (AIC), requiring at least one interaction with a marker (denoted as AIC-best model) (for a more detailed explanation see Online Resource 1).
Gene-set analysis
We applied a Gene-Set Enrichment Analysis (GSEA), based on the p-values obtained from the multi-marker interaction analysis (Subramanian et al. 2005). For GSA we assigned markers to genes according to ENSEMBL (Cunningham et al. 2015), and genes to gene sets according to Gene Ontology (GO) and the Human Genome Nomenclature Committee (HGNC) (Ashburner et al. 2000; Gray et al. 2015). In addition, the gene set of homeobox (HOX) genes in regulatory networks with respect to LC was defined based on literature (Bhatlekar et al. 2014). In total 119 gene sets were considered for analysis. Due to the subjective and in parts data driven selection of gene sets, the GSA was performed as explorative data analysis. The global level of significance of α=0.05 was used. For a list of all investigated gene sets, along with literature references and further detailed explanations see Online Resource 1.
Results
Of 20 study participants each, 9 are from North America (43%), 9 from Europe (46%) and 2 from Israel or Russia (11%). 63% of the total sample were man, 37% were women. The median age was 63 years. 20% of the participants never smoked during their lifetime; 33% were former smokers and 42% were current smokers at the time the entered the particular study (see Supplementary Table I, Online Resource 1). The proportion of never smokers as well as of current smokers where higher in uranium miners (unexposed: 36%, respectively 51%; exposed: 26%, respectively 55%). However, investigating all available former Wismut employees Kreuzer et al. (2010a) concluded, that “... there was [only] a low correlation between smoking and cumulative radon exposure. Thus, it is unlikely that smoking is a major confounder [for the estimation of radon-related risk of lung cancer]”. Radon exposure among the 308 exposed spread from 51 to 1479 WLM (mean 966 WLM). The second smallest observed value within exposed LC cases was 335 WLM, corresponding to about 2850 Bq/m3, which is a very unlikely level of elevated indoor radon exposure.
Single-marker interaction analysis
For three markers we achieved suggestive significant gene-radon (GxE; gene-environment) interaction when applying a Bonferroni correction for multiple testing. Two of them, rs6891344 and rs11747272, are near each other at chromosome 5q23.2 but belong to different LD blocks. We estimated an interaction effect of (95% CI: 2.2–7.0) and (95% CI: 2.0–5.7). Both can be assigned to the gene CSNK1G3 (casein kinase 1 gamma 3), which encodes a member of a family of serine/threonine protein kinases that phosphorylate caseins and other acidic proteins. The third marker, rs10911725, is located in an inter-genetic region of chromosome 1q25.3 (see Table 2 and Supplementary Figure 1, Online Resource 1)
Table 2.
Markers with genome-wide significance or suggestive significance for gene-radon (GxE) interaction
| Marker | Chr. | Position | Block No. |
(95%-CI)* | p-value1 |
pmt-value² |
||
|---|---|---|---|---|---|---|---|---|
| G | E | GxE | GxE | |||||
| rs10911725 | 1 | 185395182 | 5078 | 1.02 (0.89–1.16) |
6.70 (4.30–10.4) |
0.21 (0.11–0.42) |
5.3×10−6 | 0.5515 |
| rs6891344 | 5 | 123136656 | 33135 | 0.96 (0.84–1.10) |
1.57 (0.96–2.55) |
3.91 (2.18–6.99) |
2.7×10−6 | 0.2832 |
| rs11747272 | 5 | 123179990 | 33137 | 0.97 (0.86–1.10) |
1.23 (0.69–2.19) |
3.35 (1.98–5.68) |
4.3 ×10−6 | 0.4504 |
| rs6495309 | 15 | 78915245 | 82002 | 0.99 (0.87–1.13) |
4.05 (2.75–5.98) |
0.35 (0.16–0.76) |
0.0072 | 0.2387 |
| rs28534575 | 15 | 78923845 | 82002 | 1.00 (0.89–1.12) |
4.15 (2.80–6.14) |
0.36 (0.17–0.75) |
0.0060 | 0.1964 |
| rs12440014 | 15 | 78926726 | 82003 |
0.99 (0.88–1.12) |
4.43 (3.00–6.55) |
0.26 (0.11–0.60) |
0.0012 | 0.0386 |
| rs1316971 | 15 | 78930510 | 82005 | 0.97 (0.84–1.13) |
4.09 (2.78–6.02) |
0.32 (0.14–0.72) |
0.0052 | 0.1722 |
| rs17487514 | 15 | 78953785 | 82008 | 1.02 (0.89–1.17) |
1.81 (1.06–3.07) |
2.01 (1.19–3.39) |
0.0071 | 0.2325 |
| rs6495314 | 15 | 78960529 | 82008 | 1.02 (0.91–1.13) |
1.62 (0.86–3.05) |
1.87 (1.12–3.12) |
0.0145 | 0.4779 |
uncorrected p-value (genome-wide significant if < 0.5×10−7, suggestive significant if < 1×10−5);
p-value corrected for multiple testing (genome-wide significant if < 0.05, suggestive significant if < 1); using the Hybrid 2-step (H2) method of Murcray et al. (2011) with 1-ρ=1×10−16; chr: chromosome; position: position on the chromosome [bp]; G: genotypic, log-additive main effect; E: main effect of radon exposure; GxE: gene-radon interaction; OR: This is not an unbiased estimate owing to sampling and merging of samples, hence useful only to compare the strength of effects
Applying the Hybrid 2-step (H2) method and choosing the parameter ρ=1–1×10−16 (the screening weight is almost completely set to the genetic-disease (GxD) marginal effects), we could detect a genome-wide significant interaction for marker rs12440014. The H2-corrected p-value of pmt=0.03856 would correspond to a fictive uncorrected p-value of p* = 0.03856 / 103 983 LD block = 3.7 × 10−7with an estimated odds ratio (95% CI: 0.11–0.60). This marker, and five closely related markers with suggestive significance (rs6495309, rs28534575, rs1316971, rs17487514 and rs6495314), are located on chromosome 15q25.1, nearby or within the gene CHRNB4 encoding the cholinergic receptor nicotinic beta 4 subunit. This is a well-known LC region; however the strongest association was observed 69 kb upstream nearby the gene CHRNA5 (ORG=1.29; p=3.6×10−101) (McKay et al. 2017). The marker rs12440014 was found associated with LC by McKay, et al. (p=1.6×10−51; OR=0.81), but no genetic (G) main effect was seen in our analysis (=0.99, 95% CI: 0.88–1.12). Changing the tuning parameter ρ diminishes the significance of all these markers (see Supplementary Figure 2, Online Resource 1) (McKay et al. 2017).
Multi marker interaction analysis
The “inflation factor” of the χ²-test statistics for unadjusted association was λ~1.0 for the complete as well as the AIC-best models, indicating no distracting influence of residual population stratification or model selection.
For one block (no. 91734) on chromosome 18q21.32 we observed a suggestive significant gene-radon (GxE) interaction (p=2.6×10−6), when all five markers of the block (rs1346830, rs11659206, rs7237496, rs9946324) were included in the model. However, fitting the model results in a strong increase in the estimated association strength of the radon (E) main effect (=197 instead of ~2.25). At the same time the GxE interaction of the marker rs1346830 was estimated with =0.09 (95% CI: 0.03–0.22; p=2.0×10−7). Potentially, strong collinearity between the marker and the exposure results in such extreme point estimates. Hence, the estimated ORs are untrustworthy and no marker can be highlighted. This block is also merely surrounded by two uncharacterized gens (LOC107985187, LOC105372156) and two pseudogenes (CTBP2P3 / ENSG00000267153, RP11–325K19.2 / ENSG00000267382).
Allowing for marker selection (AIC-best model) revealed a suggestive gene-radon (GxE) interaction within the block no. 33137 on chromosome 5q23.2. This interaction is related to the single marker rs11747272 discussed above (see Table 3).
Table 3.
Regions with genome-wide or suggestive significant gene-radon (GxE) interaction
| LD-block | Chr. | p-value1 | Hybrid 2-step (H2) method | ||||
|---|---|---|---|---|---|---|---|
| Gene | GxE | Min. pmt-value2 |
ρ of min. pmt-value |
Modified min. p-value |
Range of ρ with pmt<13 |
||
| 2271 | 1p21.3 | UBE2U | 3.2×10−6 | 0.0563 | 0.9999 | 5.4 ×10−7 | 0.5 to 1–10−17 |
| 33135 | 5q23.2 | CSNK1G3 | 2.5×10−6 | 0.2585 | 0.5 | 2.5 ×10−6 | 0.5 to 1–10−17 |
| 58899 | 10p13 | CUBN | 1.3 ×10−5 | 0.1878 | 0.5 | 1.8 ×10−6 | 0.5 to 0.6 |
| 69267 | 12p12.1 | SOX5 | 7.1 ×10−5 | 0.9875 | 0.5 | 9.5 ×10−6 | 0.5 |
| 91734 | 18q21.32 | -- | 1.2×10−6 | 0.0214 | 0.9999 | 2.1 ×10−7 | 0.5 to 1–10−17 |
uncorrected p-value for gene-radon (GxE) interaction of the AIC-best model (genome-wide significant if < 0.5×10−7, suggestive significant if < 1×10−5);
p-value corrected for multiple testing (genome-wide significant if < 0.05, suggestive significant if < 1) with tuning parameter ρ; Chr: chromosome; GxE: gene-radon interaction;
corresponding to suggestive significance
When the Hybrid 2-step (H2)-method was applied, genome-wide significance for the block no. 91734 on chromosome 18q21 was achieved, consistent across a wide range of the tuning parameter ρ. Additional, we observed suggestive gene-radon (GxE) interaction of the blocks no. 2271 on chromosome 1p21.3 and the blocks no. 33135 and no. 33137 on chromosome 5q23.2 (see Figure 1 and Supplementary Figure 3, Online Resource 1).
Figure 1: Manhattan plot of p-values of AIC-best models corrected with the Hybrid 2-step (H2) method with ρ=0.5.
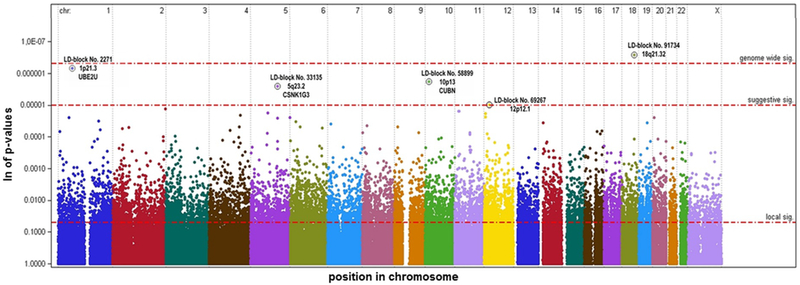
Each point represents the significance of a gene-radon (GxE) interaction within a LD block., p-value are modified according to Hybrid 2-step (H2) method of Murcray et al. (2011)
The block no. 2271 on chromosome 1p21.3 contains in total 10 markers, 7 of these remained in the AIC-best model; three of these with a local significant GxE interaction, while no marker carried a genetic (G) main effect. The strongest LC risk increasing effect was observed for marker rs2029868, with an estimated OR=22.6 for each minor allele (95% CI: 1.7–109; p=0.0001). The block covers the gene UBE2U (ubiquitin conjugating enzyme E2 U), a member of the gene family UBE2.
Setting ρ=0.5 of the Hybrid 2-step (H2)-method, the block no. 58899 on chromosome 10p13 (pmt=0.1878) and the block no. 69267 on chromosome 12p12.1 (pmt=0.9875) advanced to suggestive significance (see Figure 1; more details are given in the Online Resource 1).
Gene-set analysis
In total 148 sets of genes were considered for the analysis; 29 too small or duplicate sets were excluded (see Supplementary Table IV, Online Resource 1); hence 119 gene sets entered the GSA. Among them are 95 sets build according to GO terms, 23 HGNC gene families and one set was built on basis of the literature. These sets contained 6 to 3946 genes (median: 46) and cover 5 to 7237 LD blocks (median: 67).
For two gene sets we observed local significance (see Table 4), further two were borderline significant.
Table 4.
Significant results of the gene-set enrichment analysis
| Gene set ID |
Description | Number of genes |
Number of markers |
Number of „driving”- genes |
Number of „driving”- LD blocks |
pGS-value |
|---|---|---|---|---|---|---|
| GO:0006307 | DNA dealkylation involved in DNA repair | 10 | 90 | 7 | 21 | 0.0139 |
| HGNC:476 | microRNAs | 1,776 | 147 | 38 | 44 | 0.0159 |
| GO:0006637 | acyl-CoA metabolic process | 23 | 36 | 11 | 20 | 0.0538 |
| GO:0016020 | membrane (cellular-component) | 1,896 | 5,903 | 90 | 178 | 0.0558 |
The most significant gene set was „DNA dealkylation involved in DNA repair” (GO:0006307;pGS = 0.0139). It consists of 10 genes and comprises genotyped markers in 90 LD blocks. For 15 of these 90 LD blocks (16%) at least local significant interactions were observed in the multi-marker analysis, in contrast to 6404 out of all remaining 90 768 LD-blocks (7%). This set hosts 7 „driving”-genes assigned to 21 „driving”-LD blocks. The most significant LD-block (no. 84619, p=0.0005 for gene-radon interaction) is located within the gene FTO (fat mass and obesity-associated protein) on chromosome 16q12.2, also known as ALKBH9 (alpha-ketoglutarate dependent dioxygenase).
Within the GO hierarchy of terms, GO:0006307 is a direct subtopic of DNA repair (GO:0006281) which yielded a pGS=1.0, as well as of DNA dealkylation (GO:0035510), which was not tested. The second best subtopic of DNA repair (GO:0006281) was the double-strand break repair via single-strand annealing (GO:0045002) with a pGS =0.1574, which was rank 8 within all tested gene sets. For comparison, e.g. double-strand break repair (GO:0006302) attained rank 75 with pGS =0.834.
The other significant set was the gene family HGNC:476 „microRNAs” (pGS=0.0159), which consists by definition of 1776 very short, non-coding genes, but markers were genotyped for only 147 of these genes. This set hosts in total 38 „driving” genes assigned to 44 „driving” LD-blocks, spread over all chromosomes.
The gene sets „acyl-CoA metabolic process” (GO:0006637, pGS=0.0538) and “Membrane” (GO:0016020, pGS=0.0558) were borderline significant.
Discussion
Lung cancer has a complex disease mechanism, in particular with respect to the interaction of environmental and genetic factors. Environmental exposure to the radioactive noble gas radon is considered as the second strongest risk factor for LC in the general population; but the occupational exposure of former uranium miners, e.g. of the Wismut mining company, can be tens of times higher. We conducted a genome-wide gene-radon interaction analysis on LC using data of 28599 samples from 27 studies in men and women of European descent. Although heterogeneity in genetic susceptibility across histological subtypes of LC was demonstrated, the informative sample (n=463 miners, comprising 49 exposed LC cases) is too small to stratify the analysis by histological subtypes or by smoking behaviour (McKay et al. 2017). We performed three types of analyses: single-marker and multi-marker interaction analyses and gene-set enrichment analysis on top of the latter. We determined significance according to the Hybrid 2-step (H2) method of Murcray et al. (2011). In brief, markers or regions with marginal effect in a disease-gene (DxG) or an environmental-gene (ExG) model are given a higher a-priori weight for the final test on gene-environmental (GxE) interaction on then disease (D).
We detected a genome-wide significant gene-radon interaction for marker rs12440014 (pmt=0.03856) located within the gene CHRNB4 on chromosome 15q25.1, a well-known LC susceptibility region (Bosse and Amos 2018; Sakoda et al. 2011). Previously this intronic marker was described as associated with LC (p=2.8×10−52; OR=0.80; 95% CI: 0.78–0.83) in Caucasians (McKay et al. 2017). In our analysis we observed no significant genetic main effect (=0.99, 95% CI: 0.88–1.12), but a lower LC risk for carriers of the minor allele among the occupationally radon exposed miners (≥50 WLM), compared to non-carriers or not occupationally exposed individuals, respectively (=0.26 per minor allele, 95% CI: 0.11–0.60). The region 15q25.1 hosts three genes (CHRNA5, CHRNA3 and CHRNB4) that encode nicotinic acetylcholine receptor (nAChR) subunits. Due to strong linkage disequilibrium in this region, the observed interaction may possibly only mark interactions of functional variants in neighbouring genes. It is also believed that the association of this region with lung cancer cannot be reduced to a single variant, but is modified by age and smoking (Sakoda et al. 2011). In vitro studies examining the functional role of the genes at 15q24–25.1 in human lung tissue notified an involvement of CHRNA3 and CHRNA5 in lung carcinogenesis. An up-regulation of CHRNA5 and a down-regulation of CHRNA3 in lung adenocarcinoma as compared with the normal lung was observed (Falvella et al. 2009). However CHRNA3 is not required to maintain cancer cell proliferation (Liu et al. 2009).
The marker rs16969968 assigned to CHRNA3 was most detailed discussed as associated with smoking quantity and nicotine dependence, suggesting that this variant confers risk of LC through its effect on tobacco addiction. Interestingly, no modification of risk was found across smoking categories or histological subtypes of LC (Bosse and Amos 2018; Sakoda et al. 2011). Also, evidence exists that nAChRs can be directly associated with lung carcinogenesis owing to the complexity of nAChR function in the brain (Papke 2014). This is of interest, since a sub-multiplicative interaction between radon and smoking in causing LC was speculated independently in several uranium miner cohorts and case-control studies (Leuraud et al. 2011; National Research Council 1999; Schubauer-Berigan et al. 2009). The estimated excess relative risk (ERR) per WLM was higher for never than for current smokers (e.g. ERR/WLM=0.012 for never and long-term ex-smokers vs. ERR/WLM=0.007 for short-term ex- and current smokers (Leuraud et al. 2011)). This resulted in a small decrease of the point estimate of the relative risk for current smokers compared to never smokers from RR=6.70 (unadjusted on radon exposure) to RR=6.41 (adjusted on radon exposure). However, the difference was statistically not significant. Accordingly, a protective effect of smoking against radon induced LC was hypothesized and justified by thicker mucus layer and increased mucus velocities. Contrary, Baias et al. (2010) calculated the local radiation dose due to inhaled radon progeny in bronchial target cells to be twice as high in heavy smokers compares to never smokers. However, the apparent “LC protection by smoking” perhaps results from interaction in opposite direction of genes at chromosome 15q25.1 with smoking- respectively radon-induced LC.
Furthermore, the risk of LC for homozygous carriers of the minor allele of two markers within 15q25.1 (rs8034191, rs1051730) was estimated as at least five-fold higher in subjects who had a familial history of LC (Liu et al. 2008). We have discovered LC-risk stratification within this genomic region with respect to radon. Thus the observed familial risk of the region 15q25.1 may in part be caused by a common environmental radon exposure, albeit at a lower level than the occupational exposure of former uranium miners.
The most significant gene-radon interaction outside suspected LC susceptibility regions (Bosse and Amos 2018) was observed for UBE2U (1p21.3), a gene of the family of ubiquitin-conjugating enzymes UBE2, also known as E2 enzymes. The coded enzyme performs a central step in the ubiquitination reaction that targets a protein for degradation, a major factor for life and death of proteins (van Wijk and Timmers 2010). Protein ubiquitination is a pivotal regulatory reaction promoting the cellular responses to DNA damage (Guo et al. 2017; Kazma et al. 2012). UBE2U was recently identified as a positive regulator of TP53BP1, which promotes the formation of ionizing radiation-induced foci and thereby chromatin responses at DSBs in human cell lines (Guo et al. 2017). E2 ligases are in general involved in multiple biological processes, for example UBE2T (1q32.1) promotes efficient DNA repair; UBE2B (5q31.1) is involved in UV mutagenesis, and UBE2N (12q22) is implicated in post-replication DNA repair following UV and ionizing radiations. UBE2N was associated with LC by a candidate gene approach (p=7×10−6) (Kazma et al. 2012). The strong involvement of the human E2 ubiquitin- and ubiquitin-like conjugating enzymes in DNA damage signalling and DNA-repair processes confirms mechanistically the plausibility for the observed gene-radon interaction of UBE2U resulting in an increased radiations sensitivity for individuals bearing this genetic make-up.
In a review of DNA repair and cancer risk Romero-Laorden and Castro (2017) recently stated that defects in DNA repair genes are the genetic events most commonly involved in hereditary cancers. Once the DNA is damaged 16 or more repair mechanisms can be engaged, and a substantial cross-talk between these pathways exist (Ciccia and Elledge 2010). Exposure of a cell to a dose of 1 Gy of X-rays can cause more than 1000 base lesions, about 1000 single-strand breaks (SSBs) and 30–40 double-strand DNA breaks (DSBs) (Ward 1988). DSBs, the most harmful lesions, are repaired by an intricate network of multiple DNA repair pathways; inter alia single-strand annealing (SSA), non-homologous end-joining (NHEJ) or homologous recombination (HR) (Ciccia and Elledge 2010). Seven of the 93 genes suspected to affect susceptibility to LC are DNA repair genes: BRCA2, CHEK2, GTF2H4, MSH5, PMS1, RAD52, XRCC4. Only the first (HR and SSA), the second last (HR and SSA) and the last (NHEJ) belong to DSB repair pathways.
Because gene-radon interaction with a long-term occupational exposure to radon was investigated, we expected findings to be related to DNA repair, in particular DSB (Robertson et al. 2013). To our surprise we did not achieve cumulative significance overall on DNA-repair genes (GO:0006281, p=1.0), nor for DSB repair (GO:0006302, p=0.8340) or SSB repair (GO:0000012, p=0.9204). This missing significance may be attributed to the nature of the applied test in the GSA. The power for broadly defined gene sets of interest is low, because these contain too many not associated genes. For the more specifically defined pathway SSA of DSB repair we achieved a stronger, albeit not significant signal for association (GO:0045002, p=0.1574). Local significance was achieved for genes involved in DNA dealkylation involved in DNA repair (G0:0006307, p=0.0139), a reaction to DNA damage caused by free radicals and other reactive species generated by metabolism which results in alkylated bases. Ionizing radiation induces this type of DNA damage by indirect radiation reactions through the induction of ROS. Bases can become oxidized, alkylated, or hydrolysed through interactions with these agents (Dexheimer 2013). These lesions are repaired through base excision repair.
To our knowledge, this is the first genome-wide investigation for radon exposure x gene interaction with respect to LC. We have combined samples of disparate sizes from several sources, resulting in an extreme relation of 1 exposed to about 90 unexposed individuals. The most informative subsample consists of only 463 former uranium miners but with carefully determined occupational exposure to radon. To have enough power for the genome-wide analysis, we had to include such a large amount of controls. This should be seen as a necessity rather than a disadvantage, given the small available sample of occupational radon-exposed lung cancer cases. We were further forced to make some assumptions, e.g. no participant of a TRICL/ILCCO study was substantial long-term exposed to radon (WLM < 50) However, even long-term but low-dose exposure to radon, occupational (Kreuzer et al. 2015) as well as residential (Darby et al. 2005), was previously associated with a small increase in lung cancer risk. Thus, the small risk of misclassifying few of the many participants of a TRICL/ILCCO study is more likely for cases than for controls. Hence the allocation made is conservative in terms of statistical testing.
We also needed to fix the marginal odds ratio for radon exposure to the value observed within the miners. Subgroup analysis by histological cancer type could not be performed owing to the small number of exposed cases, in particular those with reliable records.
The risk of confounding effect due to smoking on radon-associated risk for lung cancer was previously investigated in a case-control study nested in the cohort of German uranium miners. The estimation of radon-related lung cancer risks was robust against model fitting with and without smoking. Consequently, smoking does not act as a major confounder (Schnelzer et al. 2010). Potential confounding due to other mining related exposures was also examined within the German uranium miners cohort (Kreuzer et al. 2010a; Preston et al. 2003). The correlation between measured radon exposure with external gamma radiation, long-lived radionuclides or arsenic was low; the correlation with fine dust or silica dust was moderate. The influence of adjustment for these potential confounders on the exposure–response relationship was only modest. Hence major confounding by these other occupational risk factors can be excluded (Walsh et al. 2010).
The reported study was restricted to Caucasian populations to minimize population stratification. Although the miners came from a small area in the middle of Germany, no differing genetic background compared to the TRICL/ILCCO samples from Russia to Hawaii was found. The results may not be generalized to other ethnicities because of different genetic background. It should also be noted that within the small sample of miners, controls are long-term survivors with a disproportionally high sampling of high radon exposed subjects. To discover further susceptibility genes for radon-related lung cancer or to assess the usefulness of determining the susceptibility of a subject, e.g. genetic testing requires further study.
Conclusion
We could demonstrate that the well-established association of the genomic region 15q25 might be influenced in parts by exposure to radon among uranium miners. Further, the susceptibility to lung cancer is related to the functional capability of DNA damage signalling via ubiquitination processes and repair of radiation-induced double strand breaks by the single-strand annealing mechanism.
Supplementary Material
Acknowledgments
Funding: This investigation, respectively the data sources used, were funded by following bodies:
Federal Ministry for the Environment, Nature Conservation and Nuclear Safety (Bundesministerium für Umwelt, Naturschutz und Reaktorsicherheit, BMU) (3608S04532 to G. Johnen and B. Pesch for sample collection of the German Uranium Miners Bio- and Databank (GUMB), 3614S10013, 3614S10014, 3615S32253 to H. Bickeböller for planning, genotyping, quality control and analysis of this project)
National Institutes of Health (U19-CA148127, CA148127S1, 1U19CA148127–02 to Transdisciplinary Research for Cancer in Lung)
Cancer Care Ontario Research (to R.J. Hung for ILCCO data harmonization)
FIS-FEDER/Spain (FIS-01/310, FIS-PI03– 0365, FIS-07-BI060604 for the CAPUA study)
FICYT/Asturias (FICYT PB02–67 and FICYT IB09–133 for the CAPUA study)
National Institute of Health / National Cancer Institute (UM1 CA167462 to G.E. Goodman for the CARET study)
National Institute of Health R01 (CA111703 to C. Chen, 5R01 CA151989–01A1 to J.A. Doherty, both for the CARET study)
Roy Castle Lung Cancer Foundation (for the Liverpool Lung project)
National Cancer Institute (CA092824, CA090578, CA074386; for the Harvard Lung Cancer Study)
Canadian Cancer Society Research Institute (020214 for the MSH-PMH study)
Ontario Institute of Cancer and Cancer Care Ontario Chair Award (to R.J. Hung and G.Liu for the MSH-PMH study)
Alan Brown Chair and Lusi Wong Programs at the Princess Margaret Hospital Foundation (for the MSH-PMH study)
Norwegian Cancer Society/Norwegian Research Council (for the Norway study)
James & Esther King Biomedical Research Program (09KN-15, for the TLC study)
National Institutes of Health Specialized Programs of Research Excellence (SPORE) Grant (P50 CA119997, for the TLC study)
Cancer Center Support Grant (CCSG) at the H. Lee Moffitt Cancer Center and Research Institute (P30-CA76292, for the TLC study)
Vanderbilt University Medical Center’s BioVU (1S10RR025141–01, for the Vanderbilt Lung Cancer Study)
National Center for Advancing Translational Sciences (UL1TR000445 for the Vanderbilt Lung Cancer Study)
National Cancer Institute (K07CA172294 to Aldrich for the Vanderbilt Lung Cancer Study)
National Genome Research Institute (U01HG004798, to Crawford for the Vanderbilt Lung Cancer Study)
Chief Physician Johan Boserup and Lise Boserup Fund / Danish Medical Research Council and Herlev Hospital ( for the Copenhagen General Population Study)
National Center for Research Resources (P20RR018787 for the NELCS study)
Department of Defense / Congressionally Directed Medical Research Program, US Army Medical Research and Materiel Command Program (10153006 (W81XWH-11–1-0781 for NELCS study within the Kentucky Lung Cancer Research Initiative)
International Agency for Research on Cancer (for coordination the IARC L2 study)
National Institutes of Health (R01-DE12206, P01-CA68384, R01-DE13158 for the Tampa Lung Cancer Study)
Terry Fox Research Institute and the Canadian Partnership Against Cancer (for the Pan-Canadian Early Detection of Lung Cancer Study)
Footnotes
Compliance with ethical standards
Supplementary materials and methods, supplementary figures I to III, supplementary tables I to IV and further discussion of the gene-radon interaction at 10p13 and 12p12.1can be found at Online Resource 1.
Ethical approval: The sampling of blood from the Wismut miners was approved by the Bavarian Medical Association (Bayerische Landesärztekammer) #08082 and the German Federal Commissioner for data protection and freedom of information. This research was approval from the Dartmouth Committee for Protection of Human Subjects on 7/30/2014 with id STUDY00023602.
Informed consent: Informed consent was obtained from all individual participants included in the study.”
Conflict of interest: The authors declare that they have no conflict of interest.
References
- Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome research 19(9):1655–64 doi: 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, et al. (2017) The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 26(1):126–135 doi: 10.1158/1055-9965.EPI-16-0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, et al. (2008) Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 40(5):616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25(1):25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baias PF, Hofmann W, Winkler-Heil R, Cosma C, Duliu OG (2010) Lung dosimetry for inhaled radon progeny in smokers. Radiation protection dosimetry 138(2):111–8 doi: 10.1093/rpd/ncp183 [DOI] [PubMed] [Google Scholar]
- Bhatlekar S, Fields JZ, Boman BM (2014) HOX genes and their role in the development of human cancers. Journal of Molecular Medicine 92(8):811–823 doi: 10.1007/s00109-014-1181-y [DOI] [PubMed] [Google Scholar]
- Bosse Y, Amos CI (2018) A Decade of GWAS Results in Lung Cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 27(4):363–379 doi: 10.1158/1055-9965.EPI-16-0794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan P, Hainaut P, Boffetta P (2011) Genetics of lung-cancer susceptibility. The lancet oncology 12(4):399–408 doi: 10.1016/s1470-2045(10)70126-1 [DOI] [PubMed] [Google Scholar]
- Brüske-Hohlfeld I, et al. (2006) Lung cancer risk among former uranium miners of the WISMUT Company in Germany. Health physics 90(3):208–16 doi: 10.1097/01.HP.0000175443.08832.84 [DOI] [PubMed] [Google Scholar]
- Chen HJ, et al. (2015a) Contribution of Genotype of DNA Double-strand Break Repair Gene XRCC3, Gender, and Smoking Behavior to Lung Cancer Risk in Taiwan. Anticancer research 35(7):3893–9 [PubMed] [Google Scholar]
- Chen LS, et al. (2015b) CHRNA5 Risk Variant Predicts Delayed Smoking Cessation and Earlier Lung Cancer Diagnosis-A Meta-Analysis. J Natl Cancer Inst 107(5) doi: 10.1093/jnci/djv100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Molecular cell 40(2):179–204 doi: 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, et al. (2015) Ensembl 2015. Nucleic Acids Res 43(Database issue):D662–9 doi: 10.1093/nar/gku1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby S, et al. (2005) Radon in homes and risk of lung cancer: collaborative analysis of individual data from 13 European case-control studies. BMJ 330(7485):223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexheimer TS (2013) DNA Repair Pathways and Mechanisms In: Mathews LA, Cabarcas SM, Hurt EM (eds) DNA Repair of Cancer Stem Cells. Springer; Netherlands, Dordrecht, p 19–32 [Google Scholar]
- Falvella FS, et al. (2009) Transcription deregulation at the 15q25 locus in association with lung adenocarcinoma risk. Clinical cancer research : an official journal of the American Association for Cancer Research 15(5):1837–42 doi: 10.1158/1078-0432.CCR-08-2107 [DOI] [PubMed] [Google Scholar]
- Gray KA, Yates B, Seal RL, Wright MW, Bruford EA (2015) Genenames.org: the HGNC resources in 2015. Nucleic Acids Res 43(Database issue):D1079–85 doi: 10.1093/nar/gku1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosche B, Kreuzer M, Kreisheimer M, Schnelzer M, Tschense A (2006) Lung cancer risk among German male uranium miners: a cohort study, 1946–1998. British journal of cancer 95(9):1280–7 doi: 10.1038/sj.bjc.6603403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, An L, Ng HM, Sy SM, Huen MS (2017) An E2-guided E3 Screen Identifies the RNF17-UBE2U Pair as Regulator of the Radiosensitivity, Immunodeficiency, Dysmorphic Features, and Learning Difficulties (RIDDLE) Syndrome Protein RNF168. The Journal of biological chemistry 292(3):967–978 doi: 10.1074/jbc.M116.758854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckins LM, et al. (2014) Using ancestry-informative markers to identify fine structure across 15 populations of European origin. European journal of human genetics : EJHG 22(10):1190–200 doi: 10.1038/ejhg.2014.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida M, et al. (2014) Smoking cessation reverses DNA double-strand breaks in human mononuclear cells. PLoS One 9(8):e103993 doi: 10.1371/journal.pone.0103993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2):69–90 [DOI] [PubMed] [Google Scholar]
- Kazma R, et al. (2012) Lung cancer and DNA repair genes: multilevel association analysis from the International Lung Cancer Consortium. Carcinogenesis 33(5):1059–64 doi: 10.1093/carcin/bgs116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy R, et al. (2009) Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Human mutation 30(1):69–78 doi: 10.1002/humu.20822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer M, Fenske N, Schnelzer M, Walsh L (2015) Lung cancer risk at low radon exposure rates in German uranium miners. British journal of cancer 113(9):1367–9 doi: 10.1038/bjc.2015.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer M, Grosche B, Schnelzer M, Tschense A, Dufey F, Walsh L (2010a) Radon and risk of death from cancer and cardiovascular diseases in the German uranium miners cohort study: follow-up 1946–2003. Radiation and environmental biophysics 49(2):177–85 doi: 10.1007/s00411-009-0249-5 [DOI] [PubMed] [Google Scholar]
- Kreuzer M, Schnelzer M, Tschense A, Walsh L, Grosche B (2010b) Cohort profile: the German uranium miners cohort study (WISMUT cohort), 1946–2003. Int J Epidemiol 39(4):980–7 doi: 10.1093/ije/dyp216 [DOI] [PubMed] [Google Scholar]
- Leng S, et al. (2013) Genetic variation in SIRT1 affects susceptibility of lung squamous cell carcinomas in former uranium miners from the Colorado plateau. Carcinogenesis 34(5):1044–50 doi: 10.1093/carcin/bgt024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng S, et al. (2016) Radon Exposure, IL-6 Promoter Variants, and Lung Squamous Cell Carcinoma in Former Uranium Miners. Environmental health perspectives 124(4):445–51 doi: 10.1289/ehp.1409437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuraud K, et al. (2011) Radon, smoking and lung cancer risk: results of a joint analysis of three European case-control studies among uranium miners. Radiat Res 176(3):375–87 [DOI] [PubMed] [Google Scholar]
- Liu P, et al. (2008) Familial aggregation of common sequence variants on 15q24–25.1 in lung cancer. J Natl Cancer Inst 100(18):1326–30 doi: 10.1093/jnci/djn268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. (2009) Haplotype and cell proliferation analyses of candidate lung cancer susceptibility genes on chromosome 15q24–25.1. Cancer research 69(19):7844–50 doi: 10.1158/0008-5472.CAN-09-1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl N, et al. (2015) European Code against Cancer 4th Edition: Ionising and non-ionising radiation and cancer. Cancer epidemiology 39 Suppl 1:S93–100 doi: 10.1016/j.canep.2015.03.016 [DOI] [PubMed] [Google Scholar]
- McKay JD, et al. (2017) Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat Genet 49(7):1126–1132 doi: 10.1038/ng.3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee B, Ahn J, Gruber SB, Rennert G, Moreno V, Chatterjee N (2008) Tests for gene-environment interaction from case-control data: a novel study of type I error, power and designs. Genetic epidemiology 32(7):615–26 doi: 10.1002/gepi.20337 [DOI] [PubMed] [Google Scholar]
- Murcray CE, Lewinger JP, Conti DV, Thomas DC, Gauderman WJ (2011) Sample size requirements to detect gene-environment interactions in genome-wide association studies. Genetic epidemiology 35(3):201–10 doi: 10.1002/gepi.20569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (1999) Health Effects of Exposure to Radon: BEIR VI, Washington (DC) [PubMed]
- Papke RL (2014) Merging old and new perspectives on nicotinic acetylcholine receptors. Biochemical pharmacology 89(1):1–11 doi: 10.1016/j.bcp.2014.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch B, Johnen G, Lehnert M (2015) Aufbau einer Bioproben-Bank von ehemaligen Beschäftigten der SAG / SDAG Wismut – Pilotstudie. Ressortforschungsberichte zur kerntechnischen Sicherheit und zum Strahlenschutz. BfS - Bundesamt für Strahlenschutz. https://doris.bfs.de/jspui/handle/urn:nbn:de:0221-2015102213745. Accessed 29 June 2017
- Preston DL, Shimizu Y, Pierce DA, Suyama A, Mabuchi K (2003) Studies of mortality of atomic bomb survivors. Report 13: Solid cancer and noncancer disease mortality: 1950–1997. Radiat Res 160(4):381–407 [DOI] [PubMed] [Google Scholar]
- Purcell S, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–75 doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge CA, McErlean AM, Ginsberg MS (2013) Epidemiology of lung cancer. Seminars in interventional radiology 30(2):93–8 doi: 10.1055/s-0033-1342949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A, Allen J, Laney R, Curnow A (2013) The cellular and molecular carcinogenic effects of radon exposure: a review. International journal of molecular sciences 14(7):14024–63 doi: 10.3390/ijms140714024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Laorden N, Castro E (2017) Inherited mutations in DNA repair genes and cancer risk. Curr Probl Cancer 41(4):251–264 doi: 10.1016/j.currproblcancer.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Rosenberger A, et al. (2012) Heritability of radiation response in lung cancer families. Genes 3(2):248–60 doi: 10.3390/genes3020248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano-Ravina A, Pereyra MF, Castro MT, Perez-Rios M, Abal-Arca J, Barros-Dios JM (2014) Genetic susceptibility, residential radon, and lung cancer in a radon prone area. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 9(8):1073–80 doi: 10.1097/JTO.0000000000000205 [DOI] [PubMed] [Google Scholar]
- Sakoda LC, et al. (2011) Chromosome 15q24–25.1 variants, diet, and lung cancer susceptibility in cigarette smokers. Cancer Causes Control 22(3):449–61 doi: 10.1007/s10552-010-9716-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnelzer M, Hammer GP, Kreuzer M, Tschense A, Grosche B (2010) Accounting for smoking in the radon-related lung cancer risk among German uranium miners: results of a nested case-control study. Health physics 98(1):20–8 doi: 10.1097/HP.0b013e3181b8ce81 [DOI] [PubMed] [Google Scholar]
- Schubauer-Berigan MK, Daniels RD, Pinkerton LE (2009) Radon exposure and mortality among white and American Indian uranium miners: an update of the Colorado Plateau cohort. Am J Epidemiol 169(6):718–30 doi: 10.1093/aje/kwn406 [DOI] [PubMed] [Google Scholar]
- Sethi TK, El-Ghamry MN, Kloecker GH (2012) Radon and lung cancer. Clin Adv Hematol Oncol 10(3):157–64 [PubMed] [Google Scholar]
- Setsirichok D, et al. (2012) Small Ancestry Informative Marker panels for complete classification between the original four HapMap populations. Int J Data Min Bioinform 6(6):651–74 [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66(1):7–30 doi: 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- Stenzel SL, Ahn J, Boonstra PS, Gruber SB, Mukherjee B (2015) The impact of exposure-biased sampling designs on detection of gene-environment interactions in case-control studies with potential exposure misclassification. European journal of epidemiology 30(5):413–23 doi: 10.1007/s10654-014-9908-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. ProcNatlAcadSciUSA 102(43):15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre LA, Siegel RL, Jemal A (2016) Lung Cancer Statistics. Advances in experimental medicine and biology 893:1–19 doi: 10.1007/978-3-319-24223-1_1 [DOI] [PubMed] [Google Scholar]
- Vahakangas KH, et al. (1992) Mutations of p53 and ras genes in radon-associated lung cancer from uranium miners. Lancet 339(8793):576–80 [DOI] [PubMed] [Google Scholar]
- van Wijk SJ, Timmers HT (2010) The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 24(4):981–93 doi: 10.1096/fj.09-136259 [DOI] [PubMed] [Google Scholar]
- Walsh L, Tschense A, Schnelzer M, Dufey F, Grosche B, Kreuzer M (2010) The influence of radon exposures on lung cancer mortality in German uranium miners, 1946–2003. Radiat Res 173(1):79–90 doi: 10.1667/RR1803.1 [DOI] [PubMed] [Google Scholar]
- Ward JF (1988) DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Progress in nucleic acid research and molecular biology 35:95–125 [DOI] [PubMed] [Google Scholar]
- Yang J, et al. (2011) Genomic inflation factors under polygenic inheritance. European journal of human genetics : EJHG 19(7):807–12 doi: 10.1038/ejhg.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yngveson A, Williams C, Hjerpe A, Lundeberg J, Soderkvist P, Pershagen G (1999) p53 Mutations in lung cancer associated with residential radon exposure. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 8(5):433–8 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


