Abstract
Background:
Cardiovascular diseases (CVDs) are leading cause of death and primary cause of morbidity and mortality in diabetic population. Epicardial adipose tissue (EAT) covers the heart’s surface and is a source of biomolecules regulating heart and blood vessel physiology. The protective activation of the unfolded protein response (UPR) and autophagy allows the cardiomyocyte reticular network to restore energy and/or nutrient homeostasis and to avoid cell death. However, an excessive or prolonged UPR activation can trigger cell death. UPR activation is an early event of diabetic cardiomyopathies and deregulated autophagy is associated with CVDs.
Results:
An upregulation of UPR markers (glucose-regulated protein 78KDa, glucose-regulated protein 94KDa, inositol-requiring enzyme 1α, protein kinase RNA-like ER kinase and CCAAT/-enhancer-binding protein homologous protein (CHOP) gene) in EAT compared to subcutaneous adipose tissue (SAT), was observed as well as the UPR-related apoptosis marker caspase-4/procaspase-4 ratio but not in CHOP protein levels. Additionally, levels of ubiquitin and ubiquitinated proteins were decreased in EAT. Moreover, upregulation of autophagy markers (5' adenosine monophosphate-activated protein kinase, mechanistic target of rapamycin, Beclin 1, microtubule-associated protein light chain 3-II, lysosome-associated membrane protein 2, and PTEN-induced putative kinase 1) was observed, as well as an increase in the apoptotic Bim but not the ratio between Bim and the anti-apoptotic Bcl-2 in EAT. Diabetic patients show alterations in UPR activation markers but not in autophagy or apoptosis markers.
Conclusion:
UPR and autophagy are increased in EAT compared to SAT, opening doors to the identification of early biomarkers for cardiomyopathies and novel therapeutic targets.
Keywords: Epicardial adipose tissue, endoplasmic reticulum stress, autophagy, apoptosis, heart failure
1. Introduction
The global epidemic of cardiovascular disease (CVD) remains the leading cause of death across the world [1]. In Western countries, the most common aetiological factors of heart failure (HF) are Coronary Artery Disease and hypertension, but can also be cardiomyopathy, valvular heart disease, pericardial disease as well as other factors [2, 3]. CVD is the primary cause of morbidity and mortality among the diabetic population [4]. Both experimental and clinical evidence suggest that diabetic subjects are predisposed to a distinct cardiomyopathy, named diabetic cardiomyopathy, independent of concomitant macro- and microvascular disorders [4]. Diabetic cardiomyopathy is a heart muscle-specific disease that increases the risk of HF and mortality in diabetic patients independent of vascular pathology [5]. Overweight and obesity are associated with a cluster of cardiometabolic risk factors, such as high blood pressure, hyperlipidemia and insulin resistance, all of which can significantly increase the risk of both type 2 diabetes and CVD [6].
Epicardial adipose tissue (EAT) is an adipose tissue storage that covers up to 80% of the heart’s surface, and represents about 20% of the heart’s total weight [7]. This fat depot is a major source of bioactive molecules, including compartmentalized production of cytokines and hormones, acting therefore as a localized active metabolic organ [8]. Moreover, it regulates heart and blood vessel physiology, via paracrine and vasocrine mechanisms [9]. The content of adipokines, pro- and anti-inflammatory cytokines are different in epicardial and subcutaneous adipocytes. For instance, epicardial adipocytes of coronary heart disease patients have more leptin, TNF-α, and IL-1 and less adiponectin, IL-10, and FGF-β than subcutaneous adipocytes [10]. EAT can also act as an important energy reservoir for cardiomyocytes, which greatly depend on fatty acid oxidation as their primary energy source [11]. Therefore, EAT is considered to be a real visceral adipose tissue depot [9]. Although this cardiac fat depot is needed for cardiomyocyte function, it has been suggested that its increased thickness greatly enhances the risk for CVD and the metabolic syndrome (MS) [12], and therefore it has been recognized as a new pharmacological target for primary and secondary prevention strategies.
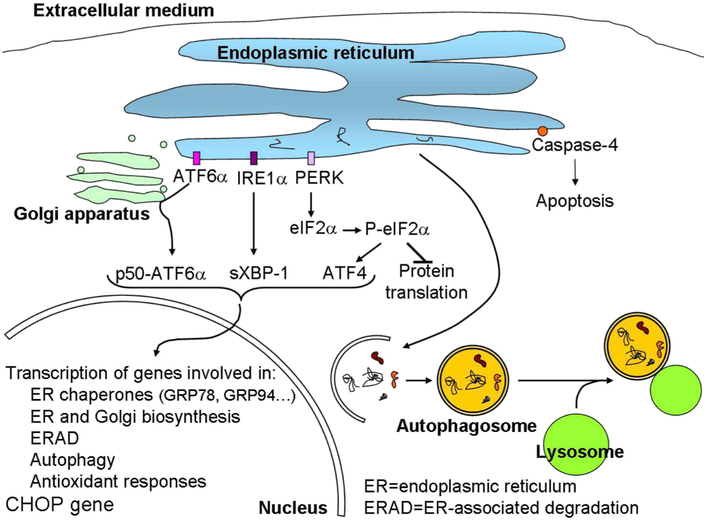
Adequate protein turnover is essential for cardiac homeostasis [13]. Different protein quality control mechanisms are involved in the maintenance of protein homeostasis, including molecular chaperones and the autophagy pathway [14]. In addition, endoplasmic reticulum (ER) stress has been linked to both physiological and pathological states in the cardiovascular system; such as myocardial infarction, oxygen starvation (hypoxia), fuel starvation, ischemia, pressure overload, dilated cardiomyopathy, hypertrophy, and HF [15, 16]. The ER stress coping response (e.g., the Unfolded Protein Response, UPR) is composed of discrete pathways that are controlled by a collection of common regulatory components that may function as a single entity involved in reacting to ER stress [15] (Figure 1). These corrective strategies allow the cardiomyocyte reticular network to restore energy and/or nutrient homeostasis and to avoid cell death [15]. Flowever, it can eventually trigger cell death if ER stress is excessive or prolonged [17]. Previous studies have demonstrated the important role of ER stress in diabetes-induced cardiac cell death [17]. In fact, a unique role has been speculated for ER stress in the pathogenesis of diabetes mellitus (DM) and its complications [18]. Recent studies have shown that ER stress is an early event associated with diabetic cardiomyopathies, and may be triggered by hyperglycemia, free fatty acids (FFAs) and inflammation [18].
Figure 1.
Proteostasis regulation. Accumulation of unfolded proteins in the ER lumen activates ER membrane-associated ATF6α, IRE1α, and PERK. Consequently, transcription factors p55 (cleaved ATF6α), sXBP-1, and ATF4 translocate to the nucleus and induce the transcription of several genes involved in: i) organelle biosynthesis; ii) processing of unfolded proteins, such as ER chaperones; iii) degradation of unfolded proteins through proteasome and autophagy; iv) antioxidant response. The phosphorylation of eIF2α by PERK also inhibits general protein translation to decrease the amount of proteins that follow the secretory pathway. This process is known as the Unfolded Protein Response (UPR). An excessive UPR can induce apoptosis through activation of caspase-4 and increased expression of CHOP protein. Moreover, proteins that are not repaired in the ER or degraded by the proteasome or dysfunctional organelles can be degraded by autophagy. In this process a double membrane is formed around proteins that need to be degraded forming an autophagosome that fuses with lysosomes to degrade its content.
Furthermore, autophagy plays a vital role in cardiac physiology and homeostasis [19, 20] (figure 1). However, dysregulated autophagy is associated with a variety of CVD [21], many of which culminate in HF [19]. Increases in autophagy have been observed in various CVD, such as atherosclerosis, cardiac ischemia/reperfusion, cardiomyopathy, HF and hypertension [22], but it is still unclear whether enhanced autophagy is an adaptive or maladaptive response to stress [20, 23]. Studies indicate that autophagy contributes to cardiac ischemia, hypertension and diabetes by interaction with ER and mitochondria [22]. Moreover, it has been suggested that ER stress may induce autophagy in the diabetic heart [24-27].
ER stress has been observed in human adipose tissue (AT) in numerous studies, indicating that ER stress may play a crucial role in AT disorders, including diabetes and obesity [28-32], inflammation and apoptosis [33-35]. Similarly, expression of autophagy genes is upregulated, and autophagy is likely activated, in AT of obese patients, which is linked with adipose tissue dysfunction [36]. Moreover, previous studies have reported a reciprocal functional interaction among autophagy and ER stress in adipose tissue and adipocytes, supporting an adaptive role between autophagic dysfunction and ER stress in human adipose tissue-related diseases [37]. However, these cellular quality control mechanisms have never been studied in EAT from HF patients, with and without diagnosed DM.
Considering that EAT is a potential cardiovascular risk factor, drastically influencing the constitution of the muscular wall of the heart and its vessels, we sought to evaluate ER stress and autophagy in EAT. We hypothesize that the quality control mechanisms, such as ER stress and autophagy are upregulated in EAT compared to SAT since EAT is a highly bioactive tissue in continuous metabolic and direct cross talk with the cardiomyocyte. By unravelling these cellular and molecular mechanisms, we expect to further contribute to the characterization of this cardiac fat depot, thus moving the field forward.
2. Experimental procedures
2.1. Adipose tissue donors
The study included 95 subjects with HF; 56 were nondiabetic (NDM; 44 male patients), and 39 had DM (28 male patients). Demographic, clinical and biochemical characteristics are shown in Table 1. HOMA-IR was calculated as fasting insulin (mU/L) × fasting glucose (mM)/22.5; HOMA-β was calculated as (20 × fasting insulin (mU/L))/(fasting glucose (mM) - 3.5); and QUICKI was calculated as 1/Log(fasting insulin (mU/L) + Log(fasting glucose (mg/dL)). Paired subcutaneous adipose tissue (SAT) and EAT biopsies were obtained from the same patients during elective coronary artery bypass grafting [coronary artery disease (CAD)], valvular replacement, or valvuloplasty [non-coronary artery disease (NCAD)] surgery. EAT was extracted from the heart's proximal right artery and SAT from the sternum region [38]. Written consent was obtained from patients before enrolling in the study. The study was approved by the Ethics Committee of the Coimbra University Hospital Centre (CHUC). Studies were carried out in accordance with the Declaration of Helsinki.
Table 1.
Demographic, clinical and biochemical characteristics of the study population
| Non-diabetic patients |
Diabetic patients |
p value | |
|---|---|---|---|
| N | 56 (58.95%) | 39 (41.05%) | |
| Male, % | 78.6 | 71.8 | 0.4500 |
| Age, y (mean ± SEM) | 66.20 ± 1.52 | 68.03 ± 1.31 | 0.3920 |
| BMI, Kg/m2 (mean ± SEM) | 25.95 ± 0.42 | 27.41 ± 0.37 | 0.032* |
| Risk factors, % | |||
| Hypertension | 69.6 | 84.6 | 0.0940 |
| Dyslipidemia | 67.9 | 74.4 | 0.4940 |
| Blood pressure, mm/Hg (mean ± SEM) | |||
| Systolic | 130.5 ± 2.56 | 132.6 ± 3.36 | 0.4480 |
| Diastolic | 71.63 ± 1.80 | 72.33 ± 2.16 | 0.9320 |
| Medications, % | |||
| Antiplatelet | 75.00 | 76.92 | 0.8296 |
| Antiarrhythmic | 16.07 | 15.38 | 0.9280 |
| Anticoagulant | 19.64 | 20.51 | 0.9169 |
| Antidiabetic | |||
| Insulin | ----- | 20.51 | ----- |
| Metformin | ----- | 20.51 | ---- |
| Alpha-glucosidase inhibitor | ----- | 5.13 | ----- |
| DPP4 inhibitor | ----- | 0.00 | ----- |
| DPP4 inhibitor + Biguanide | ----- | 33.33 | ----- |
| Sulfonylurea | ----- | 7.69 | ----- |
| Sulfonylurea + Biguanide | ----- | 33.33 | ----- |
| Antihypertension/anti-angina pectoris | |||
| ACEI | 58.93 | 58.97 | 0.9964 |
| Angiotensin receptor blocker | 5.36 | 12.82 | 0.1900 |
| β blocker | 57.14 | 71.79 | 0.1453 |
| Calcium channel blocker | 26.79 | 15.38 | 0.1877 |
| Organic nitrate | 21.43 | 23.08 | 0.8489 |
| Others | 8.93 | 2.56 | 0.2097 |
| Diuretic | 67.86 | 76.92 | 0.3352 |
| Electrolyte – KCl | 32.14 | 35.90 | 0.7033 |
| Statins | 60.71 | 74.36 | 0.1800 |
| Non-CAD Patients | |||
| N | 22 (39.29%) | 13 (33.33%) | |
| Male, % | 68.2 | 53.8 | 0.3964 |
| Age, y (mean ± SEM) | 67.82 ± 2.21 | 71.08 ± 1.72 | 0.3120 |
| BMI, Kg/m2 (mean ± SEM) | 25.55 ± 0.82 | 28.00 ± 0.55 | 0.045* |
| NHYA Functional Classification of Heart Failure, % | |||
| I; I-II | 14.29; 9.52 | 7.69; 0 | 0.5933; 0.26 |
| II; II-III | 38.10; 19.05 | 61.54; 7.69 | 0.14; 0.39 |
| III; III-IV | 4.76; 14.29 | 23.08; 0 | 0.095; 0.1638 |
| IV | 0 | 0 | N/A |
| CCS Functional Classification of Angina, % | |||
| I; I-II | 19.05; 14.29 | 23.08; 15.38 | 0.72; 0.59 |
| II; II-III | 42.86; 4.76 | 53.85; 0 | 0.33; 0.43 |
| III; III-IV | 9.52; 9.52 | 7.69; 0 | 0.89; 0.26 |
| IV | 0 | 0 | N/A |
| Operation type, % | |||
| Valvular replacement | 72.73 | 84.62 | 0.4200 |
| Valvuloplasty | 18.18 | 23.08 | 0.7200 |
| LVEF, % (≥ 50%) | 86.36 | 69.23 | 0.2200 |
| LVSF, % (≥ 27%) | 90.91 | 84.62 | 0.5700 |
| Heart rate, bpm (mean ± SEM) | 84.10 ± 4.41 | 76.62 ± 4.85 | 0.3440 |
| CAD Patients | |||
| N | 34 (60.71%) | 26 (66.66%) | |
| Male, % | 85.3 | 80.8 | 0.6000 |
| Age, y (mean ± SEM) | 65.15 ± 2.06 | 66.50 ± 1.70 | 0.6300 |
| BMI, Kg/m2 (mean ± SEM) | 26.21 ± 0.45 | 27.12 ± 0.47 | 0.2550 |
| NHYA Functional Classification of Heart Failure, % | |||
| I; I-II | 30.30; 12.12 | 15.38; 7.69 | 0.20; 0.6 |
| II; II-III | 33.33; 18.18 | 50; 7.69 | 0.16; 0.26 |
| III; III-IV | 6.06; 0 | 19.23; 0 | 0.22; N/A |
| IV | 0 | 0 | N/A |
| CCS Functional Classification of Angina, % | |||
| I; I-II | 36.36; 6.06 | 7.69; 15.38 | 0.01*; 0.22 |
| II; II-III | 45.45; 3.03 | 53.85; 7.69 | 0.46; 0.40 |
| III; III-IV | 9.09; 0 | 15.38; 0 | 0.43; N/A |
| IV | 0 | 0 | N/A |
| Operation type, % | |||
| 1 vessel disease | 17.65 | 23.08 | 0.6000 |
| 2 vessels disease | 23.53 | 15.38 | 0.4300 |
| 3 vessels disease | 47.06 | 50.00 | 0.8200 |
| 4 vessels disease | 0 | 0 | N/A |
| Aneurysm | 11.76 | 11.54 | 0.9700 |
| LVEF, % (≥ 50%) | 73.53 | 61.54 | 0.3225 |
| LVSF, % (≥ 27%) | 73.53 | 61.54 | 0.3225 |
| Heart rate, bpm (mean ± SEM) | 79.33 ± 4.53 | 79.62 ± 4.75 | 0.9390 |
| Metabolic biochemical parameters | |||
| Fasting glucose (mg/dL) | 99.68 ± 2.13 | 131.82 ± 6.90 | 0,000*** |
| Fasting insulin (mU/L) | 4.65 ± 0.42 | 9.96 ± 2.95 | 0.758 |
| Fasting c-peptide (ng/mL) | 1.44 ± 0.10 | 1.57 ± 0.15 | 0.787 |
| HOMA-IR, units | 1.19 ± 0.12 | 3.25 ±1.03 | 0.068 |
| HOMA-β, units | 50.07 ± 4.54 | 63.61 ± 19.95 |
0.017* |
| QUICKI, units | 0.39 ± 0.01 | 0.37 ± 0.01 | 0.068 |
| Kidney biochemical parameters, mg/dL | |||
| Urea | 20.87 ± 0.95 | 23.51 ± 1.81 | 0.485 |
| Creatinine | 0.93 ± 0.02 | 0.97 ± 0.05 | 0.749 |
| Liver biochemical parameters, U/L | |||
| GOT | 24.87 ± 1.29 | 23.31 ± 1.14 | 0.461 |
| GPT | 24.70 ± 2.43 | 24.82 ± 1.76 | 0.244 |
| GGT | 32.52 ± 3.66 | 49.82 ± 11.15 | 0.575 |
| ALP | 70.48 ± 2.75 | 73.28 ± 4.04 | 0.637 |
| LDH | 196.40 ± 6.49 | 185.00 ± 6.00 | 0.133 |
| Non-CAD Patients | |||
| INR, units | 1.31 ± 0.08 | 1.27 ± 0.09 | 0.727 |
| CK, U/L | 91.95 ± 11.23 | 130.20 ± 27.35 | 0.272 |
| CAD Patients | |||
| INR, units | 1.12 ± 0.02 | 1.18 ± 0.04 | 0.313 |
| CK, U/L | 98.13 ± 17.18 | 82.68 ± 10.07 | 0.515 |
Quantitative measurements are presented as mean ± SEM, and qualitative parameters are presented as percentage. For normally distributed data, a parametric t-test was performed, whereas for non-normally distributed data the Mann-Whitney nonparametric test was used. For categorical variables the X2 test was used. A p value inferior to 0.05 was considered statistically different.
P<0.05 and
P≤0.001.
ACEI, Angiotensin-converting-enzyme inhibitor; ALP, alkaline phosphatase; BMI, body mass index; CAD, coronary artery disease; CCS, Canadian Cardiovascular Society; CK, creatine kinase; GGT, γ-glutamyl transferase; GOT, glutamic oxaloacetic transaminase; GPT, glutamic pyruvic transaminase; HOMA-β, HOMA-β-cell function; HOMA-IR, homeostatic model assessment-insulin resistance index;INR, international normalized ratio; LDH, lactate dehydrogenase; LVEF, left ventricle ejection fraction; LVSF, left ventricle shortening fraction; N/A, not assigned; NCAD, non-coronary artery disease; NHYA, New York Heart Association; QUICKI, quantitative insulin sensitivity check index.
2.2. Blood tests
Fasting blood glucose levels were measured before surgery began (post-anaesthesia) with an Accu-Chek Performa glucometer (Roche Diagnostics, Indianapolis, IN). Fasting whole blood, plasma and serum were collected before surgery began (post-anaesthesia) and stored at −80°C for metabolic assays. C-peptide and ultrasensitive insulin ELISA kits were obtained from Mercodia (Uppsala, Sweden).
2.3. Chemicals
RNeasy MiniKits were from Qiagen Sciences (Germantown, MD). High Capacity cDNA Reverse Transcriptase kits were from Applied Biosystems (Forest City, CA). PCR primers (Table 2) were designed using the Beacon Designer software and synthesized by IDT-Integrated DNA Technologies (BVBA, Leuven, Belgium). SYBR Green Supermix was from Quanta Biosciences (Gaithersburg, MD). The prestained Precision Plus Protein All Blue Standard was purchased from Bio-Rad (Hercules, CA, USA). All other reagents were from Sigma (St. Louis, MO).
Table 2.
Primers used in qRT-PCR
| Protein name | Primer name | Sequence | |
|---|---|---|---|
| 5' adenosine monophosphate-activated protein kinase (AMPK) | h-AMPK | forward reverse |
TCTGTAAGAATGGAAGGCTGGA GGACCACCATATGCCTGTGA |
| Beclin (BECN1) | h-Beclin | forward reverse |
TGGCACAATCAATAACTTC CAAGCAGCATTAATCTCAT |
| B-cell lymphoma 2 (Bcl-2) | h-Bcl2 | forward reverse |
ATTGGTGAGTCGGATCGCAG TCCACAAAAGTATCCCAGCCG |
| Bcl-2-like protein 11 (BCL2L11) or Bim | h-Bim | forward reverse |
GCTACCAGATCCCCGCTTTT CCTGCCTCATGGAAGCCATT |
| Glucose-regulated protein, 78kDa (GRP78) | h-GRP78 | forward reverse |
GAACGTCTGATTGGCGATGC TCAACCACCTTGAACGGCAA |
| Glucose-regulated protein, 94kDa (GRP94) | h-GRP94 | forward reverse |
TGACCCAAGAGGAAACAC AGGTAATCAGATGCTTCTTCT |
| Growth arrest and DNA-damage-inducible gene 153 (GADD153) | h-GADD153 | forward reverse |
CTGAGCGTATCATGTTAA TACACTTCCTTCTTGAAC |
| Inositol-requiring enzyme 1α (IRE1α) | h-IRE1α | forward reverse |
CAACTACTTGAGGAATTACTG AATCAGCAGGAATCACAT |
| Lysosome-associated membrane protein 2 (Lamp2) | h-Lamp2 | forward reverse |
CTACTACAACACCTACTC TAATAACTGAAGCAACCT |
| Mammalian target of rapamycin (mTOR) | h-mTOR | forward reverse |
CGATGGCCAGGGATCTCTTC GGTCTGTGTGACTTCAGCGA |
| PTEN-induced putative kinase 1 (PINK1) | h-PINK1 | forward reverse |
CAGCAAGAGACCATCTGCCC AGCCAGCCAACCATCTTGTC |
| Protein kinase RNA-like endoplasmic reticulum kinase (PERK) | h-PERK | forward reverse |
TCACTTCTTGAATCTTCTTA TTCCATTTCGTCACTATC |
| Sequestosome 1 (SQSTM1) or ubiquitin-binding protein p62 | h-SQSTM1_1 h-SQSTM1_2 |
forward reverse forward reverse |
GGCTCGTGCGAGTCGG CCCCGTCCTCATCCTTTCTC CATTGCGGAGCCTCATCTCC TCCCCGTCCTCATCCTTTCT |
2.4. Adipose tissue gene expression
SAT and EAT gene expression was analyzed as previously described [38]. Briefly, RNA from SAT and EAT was isolated using the RNeasy MiniKit, and the concentration was determined by OD260 measurement using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). cDNA was synthesized using the Applied Biosystems High Capacity cDNA Reverse Transcriptase kit. Reverse transcription-polymerase chain reaction was then performed in a Bio-Rad iCycler iQ5 (Hercules, CA, USA) using β-actin as the housekeeping gene for normalization since as in previous studies [38]. Subsequently, the expression of each gene was normalized to control (SAT NDM) and calculated as a relative fold change (2−ΔΔCt method). The presence of specific gene products was confirmed with the melting curve analysis and electrophoresis to confirm product size.
2.5. Adipose tissue protein expression
SAT and EAT biopsies were homogenized in lysing buffer for total protein extraction, as previously described [39]. Total protein extracts were then subjected to SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes (Millipore Chemicon, MA, USA) and immunoblotted with primary antibodies: anti-Glucose-Regulated Protein (GRP)78/HSPA5 and anti-lysosome-associated membrane protein 2 (Lamp2) were from Thermo Fisher Scientific (Waltham, MA, USA), anti-gp96/HSP90B1/GRP94, anti-Beclin1, anti-microtubule-associated protein light chain 3 (LC3), anti-p62 and anti-PTEN-induced putative kinase 1 (PINK1) were from Novus Biologicals (Littleton, CO, USA), anti-Actin (I-19), anti-5' adenosine monophosphate-activated protein kinase (AMPK)α1 and P-AMPKα (Thr 172) were from Santa Cruz Biotecnology (Dallas, TX, USA), anti-caspase-4 and anti-ubiquitin were from Cell Signaling Technology, Inc. (Danvers, MA, USA) and anti-mechanistic target of rapamycin (mTOR) was form Millipore Chemicon. Secondary antibodies (Santa Cruz Biotecnology) for GRP78, GRP94, Beclin 1, LC3, Lamp2 and p62 were linked to alkaline phosphatase and complexes were detected using Enhanced chemiFluorescence (ECF) reagent (Amersham Pharmacia Biotech, Buckinghamshire, UK). Secondary antibodies (Thermo Fisher Scientific) for caspase-4, ubiquitin, AMPα1, P-AMPKα (Thr 172), mTOR and PINK1 were linked to horseradish peroxidase (HRP) and complexes were detected using Enhanced chemiLuminescence (ECL, Thermo Fisher Scientific). Relative protein expression was normalized to β-actin expression using β-actin antibody (Sigma), that was selected based on our previous results demonstrating that it does not change under these conditions Subsequently, the values were also normalized to control (SAT NDM).
2.6. Histochemical analysis
The immunohistochemical study was performed on EAT and SAT samples. Immunohistochemical staining was performed with Bond Polymer Refine Detection™ (DS9800; Leica Biosystems, Newcastle Ltd, United Kingdom) according to manufacturer’s instructions on BOND-MAX™ (Leica Biosystems, Newcastle Ltd, United Kingdom). Primary antibodies against Beclin 1 and LC3 at a dilution of 1:400 for 60 minutes were applied to the sections and incubated at room temperature. In parallel, known positive and negative controls were used. The intensity of the staining was graded semi-quantitatively on a four-point scale (0; 1+, 2+, 3+). The percentage of immunostained cells was also registered. A final score was obtained multiplying the intensity by the percentage of cells with immunohistochemical expression and the cut off considered was 10% positive cells. Protein immunostaining was observed on a microscope Nikon H600L with Digital Camera DXM 1200F (Nikon, Germany). Analysis of stained adipose tissue sections was performed by an experienced pathologist.
2.7. Statistical analyses
Statistical tests were performed using the SPSS software (version 24), with a significance level of 0.05. Outliers, shown as black circles in box-and-whisker plots in all of the figures, lie more than one and a half times the interquartile range (IQR), that is, below Q1 − 1.5 * IQR or above Q3 + 1.5 * IQR.
To check whether the data followed a normal distribution the Kolmogorov-Smirnov test was used. When the data followed a normal distribution, we used the Levene's test to check if the variance was homogeneous. For table 1, when the data followed a normal distribution the unpaired t-test was used. When the data did not follow normal distribution, the Mann-Whitney non-parametric test was used. To assess the role of tissue type or disease status on quantitative variables, non-parametric multivariate ANOVA tests were performed. The results are reported using the X2 test, with the correspondent degrees of freedom.
3. Results
3.1. Characterization of AT donors
From the 95 patients enrolled in this study, 56 were non-diabetic (NDM) and 39 were diabetic (DM) patients. 22 of the NDM had NCAD, and most presented level II in the New York Heart Association (NYHA) functional classification of HF, level II in the Canadian Cardiovascular Society (CCS) functional classification of angina, left ventricle ejection fraction (LVEF) ≥ 50%, and left ventricle shortening fraction (LVSF) ≥ 27%, as observed in Table 1. The remaining 34 NDM subjects had CAD, and most had level I and II in the NYHA functional classification, level II in the CCS functional classification of angina, LVEF ≥ 50%, and LVSF ≥ 27%. Among the NDM CAD patients, most of them presented three-vessel CAD, as indicated in Table 1. Of the 39 DM patients, 13 had NCAD, and presented mostly level II in the NYHA functional classification, level II in the CCS functional classification of angina, LVEF ≥ 50%, and LVSF ≥ 27%. The remaining 26 NDM patients had CAD and presented mostly level II in the NYHA functional classification, level II in the CCS functional classification of angina, LVEF ≥ 50%, and LVSF ≥ 27%, as described in Table 1. Among the DM CAD patients, most of them presented three-vessel CAD. Moreover, DM subjects exhibited a significantly higher body mass index (BMI) and fasting blood glucose (Table 1) compared with NDM subjects. In addition, Table 1 also describes the various medications taken by the study cohort.
The fasting c-peptide, insulin and glucose levels were measured in the blood of anesthetised patients. The anaesthesia used was propofol, thiopental, pancuronium bromide, remifentanil, fentanil, midazolam, sevoflurane and morphine. Since some of these drugs may alter insulin secretion and insulin sensitivity [40-48], the HOMA-IR, HOMA-β and QUICKI ratios in this condition do not represent the normal ratios of these patients. While, it is possible to observe a non-significant increase in HOMA-IR, a significant increase in the HOMA-β index was observed in diabetic patients compared to non-diabetic patients (Table 1).
3.2. Gene and protein expression of ER stress-related genes in SAT and EAT from HF patients
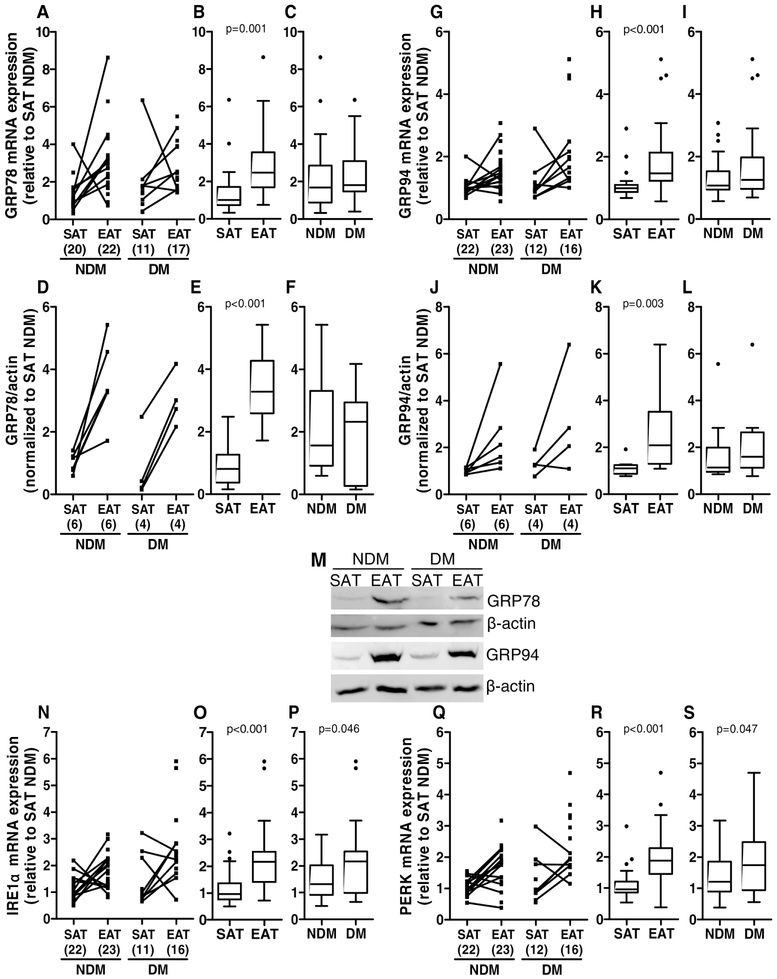
Relative gene and protein expression levels were determined for GRP78 and GRP94 in SAT and EAT from diabetic and non-diabetic HF patients. GRP78 is a major ER protein chaperone critical for protein quality control in the ER [49]. GRP94 is a key downstream chaperone that mediates the ER unfolded protein response (UPR), crucial for maintaining protein homeostasis [50-52]. Both GRP78 and GRP94 gene (p=0.001 and p<0.001, respectively) and protein levels (p<0.001 and p<0.010, respectively) were significantly increased in EAT compared to SAT (medians: EAT GRP78 mRNA=2.468 versus SAT GRP78 mRNA=1.005, EAT GRP78 protein=3.284 versus SAT GRP78 protein=0.805, EAT GRP94 mRNA=1.473 versus SAT GRP94 mRNA=0.992, EAT GRP94 protein=2.088 versus SAT GRP94 protein=1.101), but were not significantly different between diabetics and non-diabetic subjects (medians: DM GRP78 mRNA=1.802 versus NDM GRP78 mRNA=1.685, DM GRP78 protein=2.323 versus NDM GRP78 protein=1.561, DM GRP94 mRNA=1.256 versus NDM GRP94 mRNA=1.073, DM GRP94 protein=1.594 versus NDM GRP94 protein=1.135, Figure 2A-M). Relative gene expression levels of inositol-requiring enzyme 1α (IRE1α) and protein kinase RNA-like endoplasmic reticulum kinase (PERK) were also determined in SAT and EAT form diabetic and non-diabetic HF patients. IRE1α and PERK are ER membrane proteins that are activated during ER stress [53]. IRE1α and PERK gene levels were significantly increased (p<0.001) in EAT compared to SAT (mRNA medians: EAT IRE1α=2.165 versus SAT IRE1α=0.960 and EAT PERK=1.883 versus SAT PERK=0.958) and in diabetics compared to non-diabetics (mRNA medians: DM IRE1α=2.165 versus NDM IRE1α=1.318, DM PERK=1.745 versus NDM PERK=1.206, Figure 2N-S).
Figure 2.
ER stress-related gene and protein expression in SAT and EAT from HF patients with or without DM. GRP78 gene (A-C) and protein (D-F and M) expression, GRP94 gene (G-I) and protein (J-L and M) expression, IRE1α gene expression (N-P), and PERK gene expression (Q-S) were analysed by qRT-PCR and Western blot, respectively. The number of patients is indicated in parentheses in the x axis (A, D, G, J, N and Q). Gene and protein expression was calculated relative to SAT NDM after normalization to the reference gene or protein β-actin. To assess the role of tissue type or disease status, nonparametric multivariate ANOVA tests were performed. A p value inferior to 0.05 was considered statistically different.
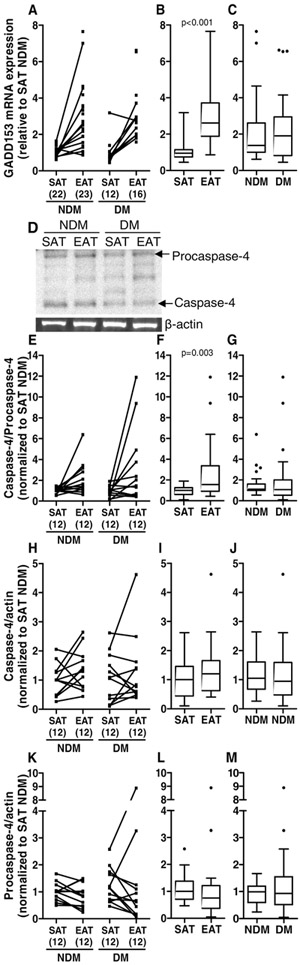
Furthermore, an excessive or prolonged ER stress induces the transcription of the apoptotic CCAAT/-enhancer-binding protein homologous protein (CHOP, the same as growth arrest and DNA damage 153, GADD153) gene and the activation of the ER membrane caspase-4 [53, 54]. CHOP mRNA expression levels were significantly increased (p<0.001) in EAT (median=2.618) compared to SAT (median=0.951) but not in diabetic (median=1.911) compared to non-diabetic (medians=1.375) patients (Figure 3A-C). The protein levels of procaspase-4 and caspase-4 were not significantly altered between tissues (medians: EAT procaspase-4=0.749 versus SAT procaspase-4=1.000, EAT caspase-4=1.201 versus SAT caspase-4=1.000) or diabetics versus non-diabetic subjects (medians: DM procaspase-4=0.930 versus NDM procaspase-4=0.992, DM caspase-4=0.950 versus NDM caspase-4=1.052), nor the ratio between caspase-4 and procaspase-4 in DM (median=1.072) compared to NDM (median=1.113) patients. Although, the ratio caspase-4/procaspase-4 was significantly increased (p<0.010) in EAT (median=1.560) compared to SAT (median=1.000) from the same subjects (Figure 3D-M).
Figure 3.
ER stress-related apoptotic gene and protein expression in SAT and EAT from HF patients with or without DM. GADD153 (the same as CHOP) gene expression (A-C) and caspase-4 (D and H-J) and procaspase-4 (D and K-M) protein expression were analysed by qRT-PCR and Western blot, respectively. The ratio between caspase-4 and procaspase-4 protein levels was also calculated (E-G). The number of patients is indicated in parentheses in the x axis (A, E, H and K). Gene and protein expression was calculated relative to SAT NDM after normalization to the reference gene or protein β-actin. To assess the role of tissue type or disease status, nonparametric multivariate ANOVA tests were performed. A p value inferior to 0.05 was considered statistically different.
When, analysing the group of genes, GRP78, GRP94, IRE1α, PERK and CHOP, there were significant differences when evaluating the disease status (X2=32.227 and p=0.040), as well as between the tissue types compared (X2=32.227 and p<0.001). Regarding the protein group evaluated as a whole, GRP78 and GRP94 did not show significant differences in the disease status (X2=2.565 and p=0.277). However, there were significant differences between the tissue types compared (X2=4.801 and p<0.001). The group caspase-4/procaspase-4, caspase-4 and procaspase-4 showed no significant differences in both the disease status (X2=7.439 and p=0.059) and the tissue types (X2=3.87 and p=0.276).
3.3. Ubiquitination in SAT and EAT from HF patients
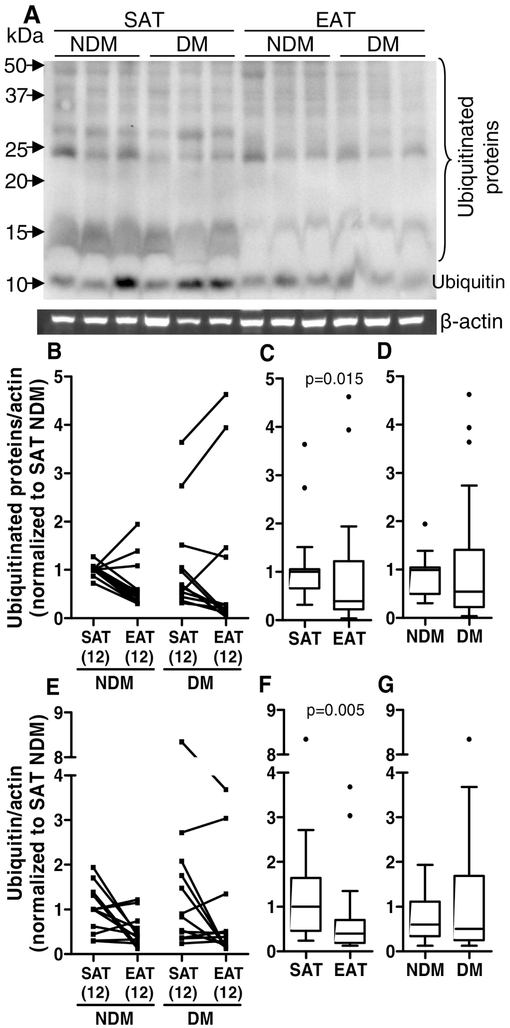
The unfolded or misfolded proteins that are not corrected in the ER can be translocated to the cytosol and ubiquitinated for subsequent degradation in the proteasome or by autophagy [55]. The levels of ubiquitin and ubiquitinated proteins in SAT and EAT were measured. The levels of ubiquitin and ubiquitinated proteins were significantly decreased (p<0.010 and p<0.05, respectively) in EAT compared to SAT (medians: EAT ubiquitin=0.395 versus SAT ubiquitin=1.000; EAT ubiquitinated proteins=0.392 versus SAT ubiquitinated proteins=1.000), but were not altered in diabetic compared to non-diabetic patients (medians: DM ubiquitin=0.502 versus NDM ubiquitin=0.598; DM ubiquitinated proteins=0.546 versus NDM ubiquitinated proteins=0.994, Figure 4).
Figure 4.
Protein ubiquitination in SAT and EAT from HF patients with or without DM. Ubiquitinated (A-D) and ubiquitin (D and E-G) protein levels were quantified by Western blot. The SAT and EAT samples in the represented blot (A) are from the same subjects in the same order. The number of patients is indicated in parentheses in the x axis (B and E). Protein expression was calculated relative to SAT NDM after normalization to the reference protein β-actin. To assess the role of tissue type or disease status, nonparametric multivariate ANOVA tests were performed. A p value inferior to 0.05 was considered statistically different.
In the group analysis of ubiquitin and ubiquitinated proteins, there are no significant differences in both the disease status (X2=3.483 and p=0.175) and the tissue types compared (X2=1.677 and p=0.432).
3.4. Gene and protein expression of autophagy-related genes in SAT and EAT from HF patients
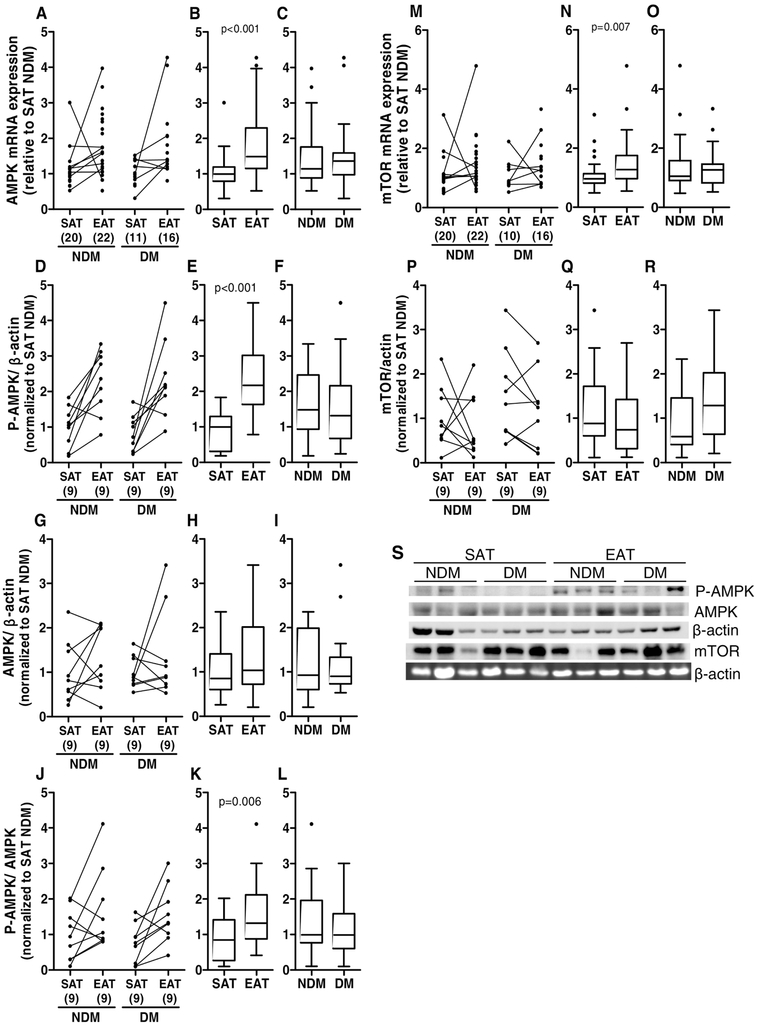
AMPK is an energy sensor and is involved in many pathways in order to regulate cellular metabolism to maintain energy homeostasis. mTOR, in the mTOR complex 1 (mTORC1), inhibits autophagy and AMPK induces autophagy through the inactivation of mTORC1 and direct activation of uncoordinated family member-51-like kinase 1 (ULK1) [56, 57]. Relative gene and protein expression levels for AMPKα and mTOR were analysed. Moreover, the activated form of AMPKα, P-AMPKα (Thr 172), was also analysed and the ratio between P-AMPK and AMPK was calculated. AMPKα and mTOR mRNA levels were significantly increased (p<0.001 and p<0.010, respectively) in EAT compared to SAT (medians: EAT AMPK mRNA=1.491 versus SAT AMPK mRNA=0.997 and EAT mTOR mRNA=1.273 versus SAT mTOR mRNA=0.956), while there were no significant differences between non-diabetic and diabetic subjects (medians: DM AMPK mRNA=1.362 versus NDM AMPK mRNA=1.143, DM mTOR mRNA=1.266 versus NDM mTOR mRNA=1.053, Figure 5A-C and M-O). AMPKα and mTOR protein levels were not significantly altered between tissue type (medians: EAT AMPK protein=0.937 versus SAT AMPK protein=0.886, EAT mTOR protein =0.737 versus SAT mTOR protein =0.882) or between diabetic and non-diabetic subjects (medians: DM AMPK protein=0.901 versus NDM AMPK protein=0.927, DM mTOR protein=1.284 versus NDM mTOR protein=0.585, Figure 5G-I and P-S). However, the P-AMPKα protein levels was significantly increased (p<0.001) in EAT compared to SAT but not in diabetic compared to non-diabetic patients (medians: EAT P-AMPK =2.150 versus SAT P-AMPK= 0.986; DM P-AMPK =1.313 versus NDM P-AMPK=1.480). Moreover, the ratio P-AMPK/AMPK that shows the levels of phosphorylation of the existing total AMPK was also increased (p=0.006) in EAT compared to SAT and not in diabetic compared to non-diabetic patients (medians: EAT P-AMPK/AMPK =1.332 versus SAT P-AMPK/AMPK =0.765; DM P-AMPK/AMPK =0.984 versus NDM P-AMPK/AMPK =0.996 Figure 5D-F, J-L and S). The group analysis of AMPK/actin, P-AMPK/actin and P-AMPK/AMPK was significantly altered in tissue types compared (X2=19.04 p<0.001) but not in the disease status (X2=0.77 p=0.857).
Figure 5.
AMPK and mTOR gene and protein expression and P-AMPK levels in SAT and EAT from HF patients with or without DM. AMPK gene (A-C) and protein (G-I and S) expression, P-AMPK (Thr 172) levels (D-F and S) and mTOR gene (M-O) and protein (P-S) expression were analysed by qRT-PCR and Western blot, respectively. Moreover, the ratio P-AMPK/AMPK was also calculated (J-L). The number of patients is indicated in parentheses in the x axis (A, D, G, J, M and P). Gene and protein expression was calculated relative to SAT NDM after normalization to the reference gene or protein β-actin. To assess the role of tissue type or disease status, nonparametric multivariate ANOVA tests were performed. A p value inferior to 0.05 was considered statistically different.
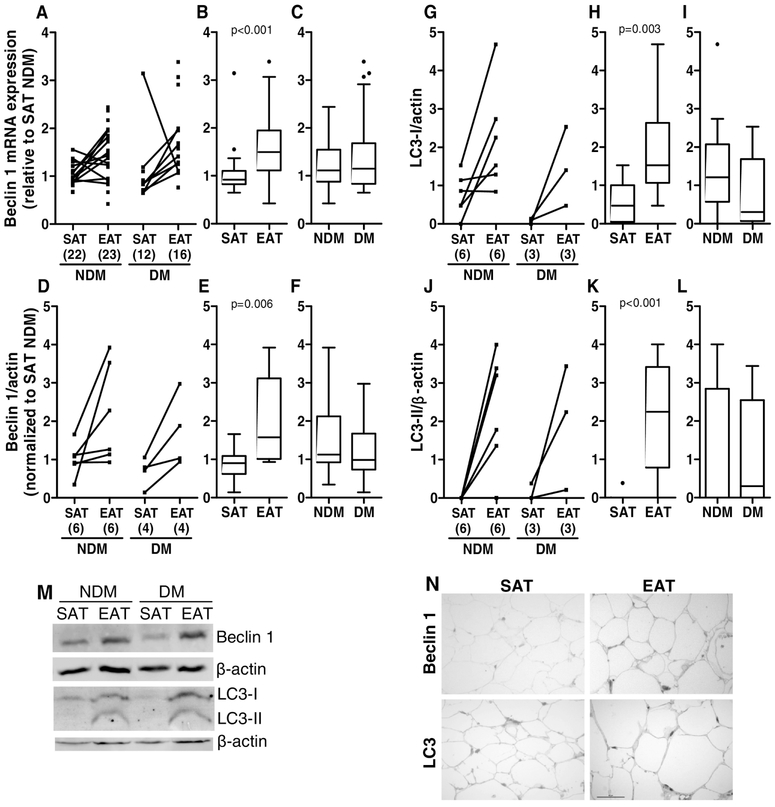
Moreover, relative gene expression levels for Beclin 1 (BECN1) and relative protein expression levels for Beclin1 and microtubule associated protein 1 light chain 3 (LC3) were determined in SAT and EAT. Beclin1 plays a critical role in the regulation of both autophagy and cell death [58], whereas LC3 protein conversion (LC3-I to LC3-II) has been used to monitor autophagy since the amount of LC3-II clearly correlates with the number of autophagosomes [59]. BECN1 mRNA and protein levels were significantly increased (p<0.001 and p<0.010, respectively) in EAT compared to SAT from the same subjects (medians: EAT BECN1 mRNA=1.495 versus SAT BECN1 mRNA=0.921 and EAT BECN1 protein=1.573 versus SAT BECN1 protein=0.902), while there were no significant differences between diabetic and non-diabetic subjects (medians: DM BECN1 mRNA=1.149 versus NDM BECN1 mRNA=1.113 and DM BECN1 protein=0.986 versus NDM BECN1 protein=1.126, Figure 6A-F and M). Since the active form of the LC3 protein is the lipidated form (LC3-II), we only evaluated the protein expression of this protein. LC3-I and LC3-II protein levels were significantly increased (p<0.010 and p<0.001, respectively) in EAT compared to SAT (medians: EAT LC3-I=1.526 versus SAT LC3-I=0.474 and EAT LC3-II=2.243 versus SAT LC3-II=0.000), while there were no significant differences between diabetic and non-diabetic subjects (medians: DM LC3-I=0.304 versus NDM LC3-I=1.215 and DM LC3-II=0.296 versus NDM LC3-II=0.000, Figure 6G-M). The group analysis of LC3-I and LC3-II confirmed that there were no significant differences in the disease status (X2=4.012 and p=0.135), there were however, significant differences between the tissue types compared (X2=12.087 and p=0.002). In order to further validate the data obtained by Western blotting, we evaluated Beclin1 and LC3 proteins by immunostaining in SAT and EAT histological sections from HF patients. We detect only positive staining for both proteins in EAT, but not in SAT (Figure 6N).
Figure 6.
Beclin 1 and LC3 gene and protein expression in SAT and EAT from HF patients with or without DM. Beclin 1 gene (A-C) and protein (D-F and M) expression and LC3-I (G-I and M) and LC3-II (J-L and M) protein levels were analysed by qRT-PCR and Western blot, respectively. The number of patients is indicated in parentheses in the x axis (A, D, G and J). Gene and protein expression was calculated relative to SAT NDM after normalization to the reference gene or protein β-actin. To assess the role of tissue type or disease status, nonparametric multivariate ANOVA tests were performed. A p value inferior to 0.05 was considered statistically different. Representative histochemical images of Beclin1 and LC3 (N) antibodies staining in SAT and EAT. Magnification, x400. Scale bar, 50 μm.
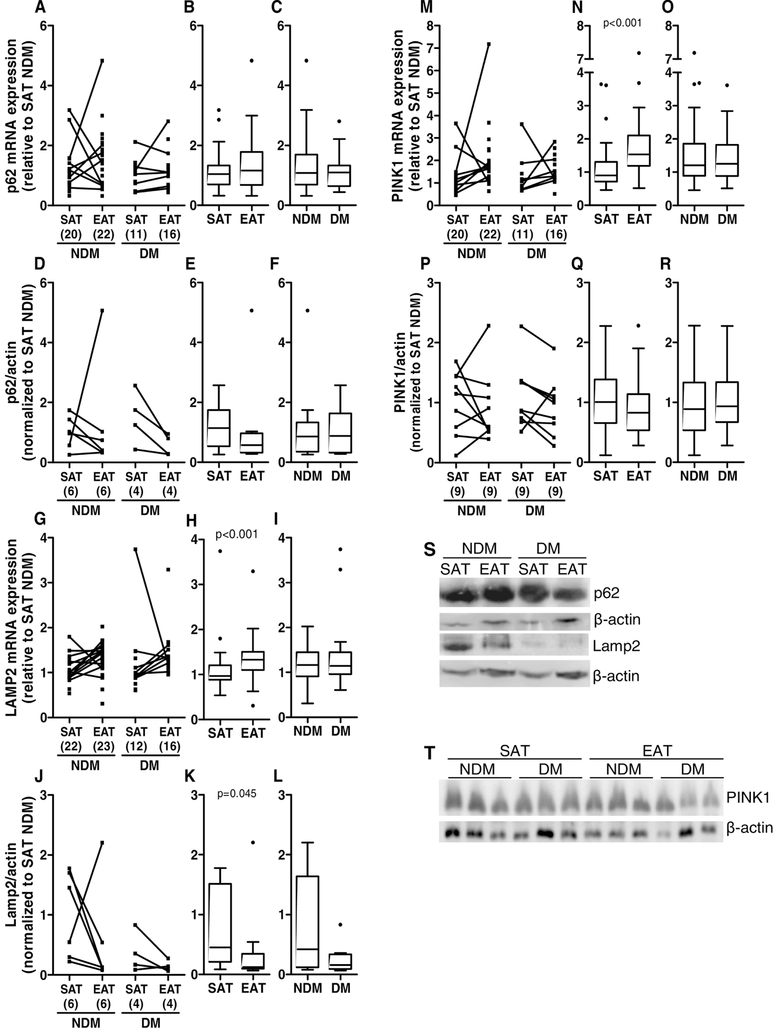
Moreover, p62 binds ubiquitinated proteins to the autophagosomes in formation for further degradation [55]. Lamp2 is a lysosome membrane protein involved in chaperone-mediated autophagy [60]. PINK1 accumulates in damaged mitochondria and is used as a mitophagy marker [55]. The mRNA and protein levels of p62 were not significantly altered in EAT or SAT (medians: EAT p62 mRNA=1.157 versus SAT p62 mRNA=1.045; EAT p62 protein=0.473 versus SAT p62 protein=1.142), or in diabetic compared to non-diabetic patients (medians: DM p62 mRNA=1.095 versus NDM p62 mRNA=1.080; DM p62 protein=0.876 versus NDM p62 protein=0.856; Figure 7A-F and S). In addition, although Lamp2 mRNA significantly increased (p<0.001) in EAT compared to SAT (medians: EAT Lamp2 mRNA=1.327 versus SAT Lamp2 mRNA=0.970), Lamp2 protein levels decreased (p<0.05, medians: EAT Lamp2 protein=0.123 versus SAT Lamp2 protein=0.452). In diabetic patients compared to non-diabetic patients Lamp2 mRNA and protein levels were not significantly altered (medians: DM Lamp2 mRNA=1.147 versus NDM Lamp2 mRNA=1.169; DM Lamp2 protein=0.157 versus NDM Lamp2 protein=0.420; Figure 7G-L and S). Furthermore, mRNA levels of PINK1 were significantly increased (p<0.001) in EAT compared to SAT (medians: EAT PINK1 mRNA=1.534 versus SAT PINK1 mRNA=0.900), but not PINK1 protein levels between tissues (medians: EAT PINK1 protein=0.827 versus SAT PINK1 protein=1.007) or mRNA and protein levels in diabetic compared to non-diabetic patients (DM PINK1 mRNA=1.248 versus NDM PINK1 mRNA=1.203; DM PINK1 protein=0.934 versus NDM PINK1 protein=0.887, Figure 7M-R and T).
Figure 7.
p62, Lamp2 and PINK1 gene and protein expression in SAT and EAT from HF patients with or without DM. p62 gene (A-C) and protein (D-F and S) expression, Lamp2 gene (G-I) and protein (J-L and S) expression and PINK1 gene (M-O) and protein (P-R and T) expression and were analysed by qRT-PCR and Western blot, respectively. The number of patients is indicated in parentheses in the x axis (A, D, G, J, M and P). Gene and protein expression was calculated relative to SAT NDM after normalization to the reference gene or protein β-actin. To assess the role of tissue type or disease status, nonparametric multivariate ANOVA tests were performed. A p value inferior to 0.05 was considered statistically different.
3.5. Gene expression of apoptosis-related genes in SAT and EAT from HF patients
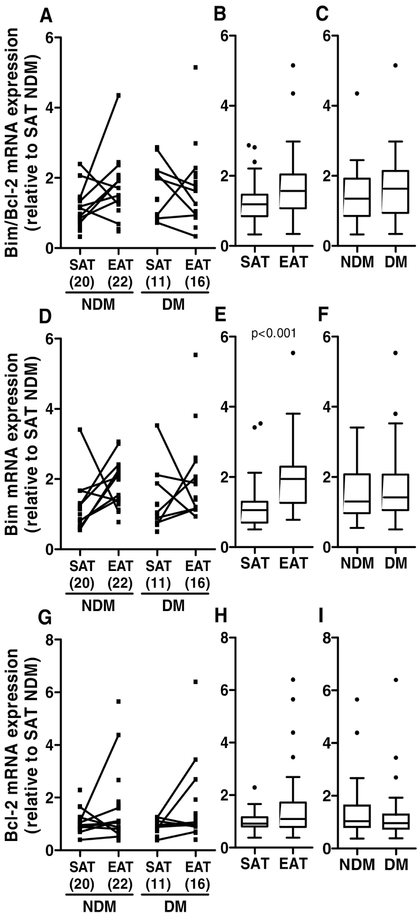
Since the deregulation of proteostasis can lead to an increase of apoptosis, the mRNA levels of the anti-apoptotic B-cell lymphoma 2 (Bcl-2) and of the apoptotic B-cell lymphoma 2-like protein 11 (Bim) were evaluated. Bim mRNA expression was significantly increased (p<0.001) in EAT compared to SAT (medians: EAT Bim=1.942 versus SAT Bim=1.052), but not in diabetic compared to non-diabetic patients (medians: DM Bim=1.418 versus NDM Bim=1.298, Figure 8D-F). On the other hand, Bcl-2 mRNA expression was not significantly altered between tissue type or disease status (medians: EAT Bcl-2=1.088 versus SAT Bcl-2=0.919; DM Bcl-2=0.959 versus NDM Bcl-2=1.04; Figure 8G-I). Although there are many factors that induce or prevent apoptosis, the ratio between the pro-apoptotic Bim, and anti-apoptotic Bcl-2 levels can be related with the degree of apoptosis in a specific tissue. The ratio Bim/Bcl-2 was not significantly altered in EAT compared to SAT (medians: EAT Bim/Bcl-2=1.570 versus SAT Bim/Bcl-2=1.187), or in diabetic compared to non-diabetic patients (medians: DM Bim/Bcl-2=1.633 versus NDM Bim/Bcl-2=1.349, Figure 8A-C). In regard to the grouped analysis of Bim/Bcl-2, Bim and Bcl-2 there are no statistical differences in the disease status (X2=2.73 and p=0.435). However, when analysing the different tissue types, the statistical tests revealed significant differences between EAT and SAT (X2=11.635 and p=0.009).
Figure 8.
Apoptosis related gene expression in SAT and EAT from HF patients with or without DM. Bim (D-F) and Bcl-2 (G-I) gene expression was measured by qRT-PCR. The ratio between Bim and Bcl-2 gene expression was also calculated (A-C). The number of patients is indicated in parentheses in the x axis (A, D and G). Gene expression was calculated relative to SAT NDM after normalization to the reference gene β-actin. To assess the role of tissue type or disease status, nonparametric multivariate ANOVA tests were performed. A p value inferior to 0.05 was considered statistically different.
4. Discussion
This study demonstrates for the first time that both the UPR and the autophagy pathways are significantly increased in EAT compared to SAT.
The ER is an organelle involved in protein folding, calcium homeostasis, and lipid biosynthesis [61]. Various factors that interfere with ER function lead to accumulation of unfolded proteins, including oxidative stress, ischemia, disturbance of calcium homeostasis, and overexpression of normal and/or incorrectly folded proteins. Under physiological conditions, the N-termini of PERK, IRE1α and ATF6 transmembrane ER proteins are held by the ER chaperone GRP78 [62]. When misfolded proteins accumulate, GRP78 releases these transmembrane signalling proteins, allowing PERK and IRE1α oligomerization and ATF6 translocation to the Golgi and consequently launching the UPR [62, 63]. The UPR is the induction of signal transduction events to reduce the accumulation of unfolded proteins by increasing ER resident chaperones, including GRP78 and GRP94 that promote correct protein folding, inhibiting protein translation, and accelerating the degradation of unfolded/misfolded proteins via a ubiquitin-proteasome system termed ER-associated degradation (ERAD) [61, 64-66] However, when the ER stress is excessively prolonged it can lead to apoptosis, for instance, increasing CHOP expression or activating the human ER membrane-resident caspase-4 [53]. (Figure 1). Recently, UPR and/or ER-initiated apoptosis (increased expression of GRP78, GRP94, CHOP, caspase-4, etc) have been implicated in the pathophysiology of various human diseases, including CVD, such as cardiac hypertrophy, HF, atherosclerosis, ischemic heart disease, dilated cardiomyopathy, ischemic cardiomyopathy, left ventricular hypertrophy and diabetic heart disease [17, 61, 67-77]. Accordingly, CHOP-dependent cell death pathway may be involved in the transition from cardiac hypertrophy to HF [17]. Therefore, therapeutic interventions that target molecules of the UPR component and reduce ER stress will be considered promising strategies to treat CVD [78].
In agreement with the literature, we observed a significant increase in UPR markers in EAT, including the GRP78 and GRP94 gene and proteins, indicating significant molecular alterations in this pathway in EAT. Moreover, we observed a significant increase in IRE1α and PERK gene levels in EAT and in diabetic patients. Obesity can trigger ER stress and induce UPR activation [29]. In this study, although the average of diabetic patients was not obese, their BMI was significantly increased compared to non-diabetic patients. So, the significantly increase of IRE1α and PERK mRNA in diabetic patients with a p value near to 0.05 (0.046 and 0.047 respectively) and without alterations in other UPR activation markers could in part be due to the increased BMI in this patients.
Furthermore, CHOP upregulation is a common point of convergence for all 3 arms of the UPR and its gene is transcript when the ER stress is excessive or prolonged, clearly illustrating the importance of this transcription factor [79], therefore CHOP gene expression levels were evaluated. CHOP mRNA levels were more elevated in EAT than in SAT, however no significant differences were observed in its correspondent pro-apoptotic protein levels (data not shown). Additionally, the pro-apoptotic Bim and the anti-apoptotic Bcl-2 are some of the downstream signalling pathways of CHOP leading to apoptosis. Bim gene expression was significantly increased in EAT. However, the ratio Bim/Bcl-2 gene expression was not significantly altered. On the other hand, IRE1α can stimulate apoptosis effectors caspase-4 which promotes ER-mediated apoptosis upon an excessive or prolonged ER stress [62, 80]. The ratio between caspase-4 and procaspase-4 was significantly increased in EAT. Nevertheless, the group analysis of caspase-4, procaspase-4 and caspase-4/procaspase-4 ratio showed no significantly alterations. So, we cannot say that ER stress-mediated cell death is being activated in this tissue but we can conclude that EAT has an increased tendency for apoptosis. Therefore, through an increase of several ER stress markers, we have confirmed that the UPR was activated in EAT compared to SAT.
Furthermore, another key component of the ER stress response pathway is ERAD [64, 65, 81, 82]. Previous studies reported the involvement of both processes in the development of ischemic heart diseases [83]. Furthermore, hyperubiquitination of proteins have been previously observed in human dilated cardiomyopathy [84]. Besides the UPR, the ERAD is induced in order to activate degradation of unfolded or misfolded proteins via the ubiquitin-proteasome system [85, 86]. The accumulation of unfolded proteins induced by ER dysfunction can surpass the capacity of the proteasome to degrade them, as previously observed in a mouse model of ischemic heart disease [83]. To test this, we examined the accumulation of ubiquitinated proteins in the EAT compared to SAT, from diabetic and non-diabetic patients. However, the immunoreactivity for ubiquitin and ubiquitinated proteins was significantly decreased in EAT and was not altered in diabetic patients. The group analyses of ubiquitin and ubiquitinated proteins revealed no significant alterations in tissue type or disease status. This could be due to the preserved function of the proteasome or to an increase of autophagy activation, since ubiquitinated proteins can also be degraded by autophagy [55]. However, we observed an increase in some of the UPR activation markers in diabetic subjects, but we did not observe an increase in autophagy markers in diabetic compared to non-diabetic patients. Thus, it seems that in diabetic patients, the ERAD is sufficient to degrade the unfolded/misfolded proteins originated from the slight increase in ER stress.
A plethora of stress conditions, including oxidative stress, metabolic dysfunction and ER stress, are observed to activate autophagy as a pro-survival pathway [87]. In addition, Raptor, a regulatory-associated protein of mTOR, as well as other proteins form the complex mTORC1, that inhibit autophagy, are hyper-activated under diabetes conditions [88]. mTOR also plays a major role in promoting adipocyte formation and lipid synthesis in response to feeding and insulin [88]. Moreover, AMPK is a well-known kinase that induces autophagy through the inhibition of mTOR [89]. The activity of AMPK is decreased in adipose tissue from obese and insulin resistance subjects [90]. Since AMPK regulates several important metabolic pathways, some authors suggest that AMPK could be an important target to treat obesity, insulin resistance, type 2 diabetes and CVD [56, 91]. Since mTOR protein expression was not significantly altered, but the active form of AMPK (P-AMPK Thr 172) was increased in EAT compared to SAT it may be possible that autophagy is activated in EAT. To further investigate this question, other autophagy markers were analysed. Beclin 1 induces the formation of the autophagosome leading to the induction of macroautophagy [92, 93]. However, Beclin 1 forms a complex with different types of proteins that can activate or inactivate Beclin 1. Bcl-2, for instance, inactivates Beclin 1 and Bim activates it [93, 94]. The IRE1 arm of UPR, increased in EAT compared to SAT, can lead to JNK activation and increase phosphorylation of Bcl-2, which promotes its dissociation from Beclin 1 [95, 96], resulting in autophagy induction [97, 98]. The PERK arm of UPR, also increased in EAT compared to SAT, can also lead to the activation of autophagy [93]. We observed a significant rise in Beclin 1 gene and protein levels in EAT compared to SAT. Since the gene expression of Bim was increased in EAT, this could activate even more Beclin 1. Moreover, during macroautophagy, the lipidation of LC3-I to form LC3-II is essential for the formation of autophagosomes [99]. The increase in LC3-II reflects either increased autophagosome formation due to increases in autophagic activity, or to reduced turnover of autophagosomes [100]. We observed a significant increase of LC3-I in EAT which may indicate that EAT has more capacity for macroautophagy than SAT. Moreover, the levels of LC3-II in EAT were also significantly increased, and together with the increase of Beclin 1 and P-AMPK, these results are suggestive of an increased autophagy in EAT compared to SAT, in agreement with previous studies [101-103]. As mentioned before, EAT exhibits increased ER stress and, as such, increase in misfolded or dysfunctional proteins. However, we did not find an increase in immunoreactivity for ubiquitin in EAT but rather a decrease. In addition, an increase in autophagy induction in this tissue was observed, which leads us to believe that this increase in the autophagic process in EAT might have a protective/survival role, since it promotes the degradation of dysfunctional proteins. However, we do not know if this is the case in normal healthy EAT since we used EAT from HF patients. Moreover, p62 binds ubiquitin and LC3 and it is involved in the recruitment of ubiquitinated proteins during the formation of the autophagosome [99]. Since p62 is degraded during macroautophagy, its levels may decrease when macroautophagy increases [99]. Although there was a tendency of p62 protein to decrease in EAT, we observed no significant changes in p62 gene or protein levels in agreement with previous studies that have also reported that p62 levels do not always correlate with macroautophagy activation [104]. Furthermore, Lamp2 is an important lysosome membrane protein involved in chaperone-mediated autophagy [99]. While the Lamp2 gene expression was significantly increased in EAT, its protein levels were significanty decreased in EAT compared to SAT. This could indicate an increased degradation of Lamp2, leading to a decreased chaperone-mediated autophagy, or decreased lysosome levels that could lead to autophagosome accumulation and the consequent LC3-II increase that we have observed. However, the amount of ubiquitinated proteins did not increased in EAT, on the contrary it decreased, even in the presence of strong ER stress. This leads us to believe that macroautophagy is not dysfunctional and the increase of LC3-II protein levels in EAT is really due to an increase in macroautophagy activity. In addition, PINK1 is involved in mitophagy [99] and its gene expression increased in EAT showing that the degradation of mitochondria by autophagy could be increased in EAT compared to SAT. However, the PINK1 protein levels were not significantly altered.
Autophagy is a normal and a common process observed in the heart and it is induced in response to stress, such as ischemia and during HF [92]. However, accumulation of protein aggregates and dysfunctional organelles is commonly observed in a variety of heart diseases, including hemodynamic stress-induced cardiac hypertrophy, chronic myocardial infarction, and dilated cardiomyopathy [105, 106], suggesting that activation of autophagy may not be sufficient in these conditions to maintain cardiomyocytes alive. A recent report revealed that enhancing autophagy ameliorates desmin-related and inherited cardiomyopathies that result in severe HF due to protein aggregation, as well as myofibrillar disarray [107]. Thus, autophagy plays an important role in protecting normal cardiac function by maintaining optimal protein quality control, which that might be what is happening at the level of the EAT depot under these conditions. Although activation of autophagy is generally protective in the heart in many conditions, it can also induce cell death and cardiac dysfunction in a specific context-dependent manner. For example, ischemia/reperfusion [108] and pressure overload [109-113] upregulate autophagy and these could cause detrimental effects in the heart.
Furthermore, the diabetic state enhances both ER stress and autophagy in cardiovascular complications [114, 115]. However, we only observed diabetes-mediated increase in IRE1α and PERK gene expression. On the other hand, same of the medications taken by patients could decrease possible metabolic differences between diabetic and non-diabetic patients. For instance, metformin and resveratrol are described to have anti-ER stress and anti-apoptotic effects in adipose tissue or adipocytes exposed to high glucose [116].
As a whole, our results indicate that patients, with and without diagnosed diabetes, have increased ER stress in EAT compared to SAT. Therefore, this increase in ER stress may lead to an increase in autophagy which, in this specific case, may have a protective/survival function, since we did not observe a significant increase in ubiquitinated proteins. Thus, there is a balance between mechanisms that induce the death of epicardial adipocytes through alterations in ER and processes that aim to repair the damage caused by the dysfunction in this organelle. Thus, EAT is sensitive to cardiac pathology and develops cellular signalling in response to the functional and/or structural changes in the heart. However, the balance between pro-apoptotic and survival mechanisms might be established in this fat depot due to its micro environment in order to not aggravate cardiac pathology.
One of the limitations of this study, similarly to our previous report [38], is the lack of healthy control subjects matched for age, sex, and BMI, which as prevented us from comparing the results with the subjects studied. Although diabetic patients have a more severe heart disease than non-diabetic patients, the presence of DM just slightly increased ER stress, probably because diabetic patients take drugs to control DM, and that the possible effects mediated by the presence of DM are, somewhat, overshadowed by their antidiabetic medication. Thus, to evaluate the effects of DM per se, studies should be undertaken in a population of diabetic subjects not taking antidiabetic drugs. Moreover, the amount of tissue biopsy that can be collected is very small. Therefore, it is not possible to perform all the experiments in the same tissue, and instead, we used tissues form different patients in different experiments, but always used paired EAT and SAT biopsies from the same subject in each experiment [38].
5. Conclusion
Although more studies are required to clarify the underlying molecular mechanisms regulating epicardial adipose tissue ER stress and autophagy in HF patients and eventually in healthy EAT, our results identify important new markers of the UPR and autophagy pathways that are activated in EAT compared to SAT. Since we do not have EAT from healthy subjects to compare these results with, our study may indicate that activation of the ER stress and autophagy pathways in EAT might be associated with heart diseases, as it is the case in this population, but it could also be that this is a specific metabolic phenotype of this particular fat depot, which in turn may be of importance in identifying early biomarkers for cardiomyopathies and possible therapeutic targets.
Highlights.
Epicardial adipose tissue has increased endoplasmic reticulum stress compared to subcutaneous adipose tissue from the same patients
Diabetic patients have slightly increase of endoplasmic reticulum stress in adipose tissue compared to non-diabetic patients
Epicardial adipose tissue has increased autophagy activation compared to subcutaneous adipose tissue from the same patients
6. Acknowledgements
This work was supported by the Portuguese Society of Diabetology/GIFT, the European Foundation for the Study of Diabetes, the Portuguese Foundation for Science and Technology (FCT fellowships: SFRH/BPD/26837/2006,SFRH/PBD/101030/2014, PTDC/SAU-OSM/ 104124/2008, EXCL/DTP-PIC/0069/2012), Strategic Project POCI-01-0145-FEDER-007440 funded by FEDER through Operational Programme Competitiveness Factors-COMPETE. HealthyAging2020 CENTRO-01-0145-FEDER-000012-N2323. EC was partially supported by P30-AG-028718, RO1-AG-033761 and P20GM109096.
Abbreviations
- AMPK
5' adenosine monophosphate-activated protein kinase
- AT
adipose tissue
- BECN1
beclin 1
- Bcl-2
B-cell lymphoma 2
- Bim
Bcl-2-like protein 11 (BCL2L11)
- BMI
body mass index
- CAD
coronary artery disease
- CCS
Canadian Cardiovascular Society
- CHOP
CCAAT-enhancer-binding protein homologous protein
- CVD
cardiovascular diseases
- DM
diabetes mellitus
- ECF
enhanced chemiFluorescence
- ECL
Enhanced chemiLuminescence
- EAT
epicardial adipose tissue
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- FFA
free fatty acid
- GADD153
growth arrest and DNA damage 153
- GRP
glucose-regulated protein
- HF
heart failure
- IRE1α
inositol-requiring enzyme 1α
- Lamp2
lysosome-associated membrane protein 2
- LC3
microtubule-associated protein light chain 3
- LVEF
left ventricle ejection fraction
- LVSF
left ventricle shortening fraction
- MS
metabolic syndrome
- mTOR
mechanistic target of rapamycin
- mTORC1
mTOR complex 1
- NCAD
non-coronary artery disease
- NDM
non-diabetic
- NYHA
New York Heart Association
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- PINK1
PTEN-induced putative kinase 1
- PVDF
polyvinylidene difluoride
- p62
ubiquitin-binding protein p62
- SAT
subcutaneous adipose tissue
- ULK1
uncoordinated family member-51-like kinase 1
- UPR
unfolded protein response
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement
The authors declare that this work was conducted without financial or commercial relationships that could be considering as a potential conflict of interest.
8. References
- [1].Dominic EA, Ramezani A, Anker SD, Verma M, Mehta N, Rao M, Mitochondrial cytopathies and cardiovascular disease, Heart (British Cardiac Society), 100 (2014) 611–618. [DOI] [PubMed] [Google Scholar]
- [2].Lip GY, Gibbs CR, Beevers DG, ABC of heart failure: aetiology, BMJ (Clinical research ed, 320 (2000) 104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].McMurray JJ, Stewart S, Epidemiology, aetiology, and prognosis of heart failure, Heart (British Cardiac Society), 83 (2000) 596–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Huynh K, Bernardo BC, McMullen JR, Ritchie RH, Diabetic cardiomyopathy: mechanisms and new treatment strategies targeting antioxidant signaling pathways, Pharmacology & therapeutics, 142 (2014) 375–415. [DOI] [PubMed] [Google Scholar]
- [5].Kobayashi S, Liang Q, Autophagy and mitophagy in diabetic cardiomyopathy, Biochimica et biophysica acta, 1852 (2015) 252–261. [DOI] [PubMed] [Google Scholar]
- [6].Pourshahidi LK, Wallace JM, Mulhern MS, Horigan G, Strain JJ, McSorley EM, Magee PJ, Bonham MP, Livingstone MB, Indices of adiposity as predictors of cardiometabolic risk and inflammation in young adults, J Hum Nutr Diet, 29 (2016) 26–37. [DOI] [PubMed] [Google Scholar]
- [7].Rabkin SW, Epicardial fat: properties, function and relationship to obesity, Obes Rev, 8 (2007) 253–261. [DOI] [PubMed] [Google Scholar]
- [8].Iacobellis G, Malavazos AE, Corsi MM, Epicardial fat: from the biomolecular aspects to the clinical practice, The international journal of biochemistry & cell biology, 43 (2011) 1651–1654. [DOI] [PubMed] [Google Scholar]
- [9].Salazar J, Luzardo E, Mejias JC, Rojas J, Ferreira A, Rivas-Rios JR, Bermudez V, Epicardial Fat: Physiological, Pathological, and Therapeutic Implications, Cardiology research and practice, 2016 (2016)1291537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gruzdeva OV, Akbasheva OE, Dyleva YA, Antonova LV, Matveeva VG, Uchasova EG, Fanaskova EV, Karetnikova VN, Ivanov SV, Barbarash OL, Adipokine and Cytokine Profiles of Epicardial and Subcutaneous Adipose Tissue in Patients with Coronary Heart Disease, Bulletin of experimental biology and medicine, 163 (2017) 608–611. [DOI] [PubMed] [Google Scholar]
- [11].Iacobellis G, Barbaro G, The double role of epicardial adipose tissue as pro- and anti-inflammatory organ, Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme, 40 (2008) 442–445. [DOI] [PubMed] [Google Scholar]
- [12].Cikim AS, Topal E, Harputluoglu M, Keskin L, Zengin Z, Cikim K, Ozdemir R, Aladag M, Yologlu S, Epicardial adipose tissue, hepatic steatosis and obesity, Journal of endocrinological investigation, 30 (2007) 459–464. [DOI] [PubMed] [Google Scholar]
- [13].Henning RH, Brundel B, Proteostasis in cardiac health and disease, Nat Rev Cardiol, 14 (2017) 637–653. [DOI] [PubMed] [Google Scholar]
- [14].Schlossarek S, Frey N, Carrier L, Ubiquitin-proteasome system and hereditary cardiomyopathies, Journal of molecular and cellular cardiology, 71 (2014) 25–31. [DOI] [PubMed] [Google Scholar]
- [15].Groenendyk J, Agellon LB, Michalak M, Coping with endoplasmic reticulum stress in the cardiovascular system, Annual review of physiology, 75 (2013) 49–67. [DOI] [PubMed] [Google Scholar]
- [16].Teng X, Qi YF, Tang CS, [Endoplasmic reticulum stress and heart diseases], Sheng li ke xue jin zhan [Progress in physiology], 40 (2009) 106–110. [PubMed] [Google Scholar]
- [17].Xu J, Zhou Q, Xu W, Cai L, Endoplasmic reticulum stress and diabetic cardiomyopathy, Experimental diabetes research, 2012 (2012) 827971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang L, Zhao D, Ren J, Yang J, Endoplasmic reticulum stress and protein quality control in diabetic cardiomyopathy, Biochimica et biophysica acta, 1852 (2015) 209–218. [DOI] [PubMed] [Google Scholar]
- [19].Li DL, Hill JA, Cardiomyocyte autophagy and cancer chemotherapy, Journal of molecular and cellular cardiology, 71 (2014) 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jimenez RE, Kubli DA, Gustafsson AB, Autophagy and mitophagy in the myocardium: therapeutic potential and concerns, British journal of pharmacology, 171 (2014) 1907–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ma S, Wang Y, Chen Y, Cao F, The role of the autophagy in myocardial ischemia/reperfusion injury, Biochimica et biophysica acta, 1852 (2015) 271–276. [DOI] [PubMed] [Google Scholar]
- [22].Mei Y, Thompson MD, Cohen RA, Tong X, Autophagy and oxidative stress in cardiovascular diseases, Biochimica et biophysica acta, 1852 (2015) 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ren J, Taegtmeyer H, Too much or not enough of a good thing--The Janus faces of autophagy in cardiac fuel and protein homeostasis, Journal of molecular and cellular cardiology, 84 (2015) 223–226. [DOI] [PubMed] [Google Scholar]
- [24].Younce CW, Wang K, Kolattukudy PE, Hyperglycaemia-induced cardiomyocyte death is mediated via MCP-1 production and induction of a novel zinc-finger protein MCPIP, Cardiovascular research, 87 (2010) 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang B, Zhang Y, La Cour KH, Richmond KL, Wang XM, Ren J, Mitochondrial aldehyde dehydrogenase obliterates endoplasmic reticulum stress-induced cardiac contractile dysfunction via correction of autophagy, Biochimica et biophysica acta, 1832 (2013) 574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang L, Hu N, Jiang S, Zou Y, Yang J, Xiong L, Ren J, Heavy metal scavenger metallothionein attenuates ER stress-induced myocardial contractile anomalies: role of autophagy, Toxicology letters, 225 (2014) 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Appenzeller-Herzog C, Hall MN, Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling, Trends in cell biology, 22 (2012) 274–282. [DOI] [PubMed] [Google Scholar]
- [28].Wu J, Kaufman RJ, From acute ER stress to physiological roles of the Unfolded Protein Response, Cell Death Differ, 13 (2006) 374–384. [DOI] [PubMed] [Google Scholar]
- [29].Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS, Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes, Science, 306 (2004) 457–461. [DOI] [PubMed] [Google Scholar]
- [30].Furukawa S, Fujita T, Shimabukuro M, Iwaki M, Yamada Y, Nakajima Y, Nakayama O, Makishima M, Matsuda M, Shimomura I, Increased oxidative stress in obesity and its impact on metabolic syndrome, The Journal of clinical investigation, 114 (2004) 1752–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hosogai N, Fukuhara A, Oshima K, Miyata Y, Tanaka S, Segawa K, Furukawa S, Tochino Y, Komuro R, Matsuda M, Shimomura I, Adipose tissue hypoxia in obesity and its impact on adipocytokine dysregulation, Diabetes, 56 (2007) 901–911. [DOI] [PubMed] [Google Scholar]
- [32].Lazar MA, How obesity causes diabetes: not a tall tale, Science, 307 (2005) 373–375. [DOI] [PubMed] [Google Scholar]
- [33].Boden G, Duan X, Homko C, Molina EJ, Song W, Perez O, Cheung P, Merali S, Increase in endoplasmic reticulum stress-related proteins and genes in adipose tissue of obese, insulin-resistant individuals, Diabetes, 57 (2008) 2438–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sharma NK, Das SK, Mondal AK, Hackney OG, Chu WS, Kern PA, Rasouli N, Spencer HJ, Yao-Borengasser A, Elbein SC, Endoplasmic reticulum stress markers are associated with obesity in nondiabetic subjects, The Journal of clinical endocrinology and metabolism, 93 (2008) 4532–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gregor MF, Yang L, Fabbrini E, Mohammed BS, Eagon JC, Hotamisligil GS, Klein S, Endoplasmic reticulum stress is reduced in tissues of obese subjects after weight loss, Diabetes, 58 (2009) 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Maixner N, Bechor S, Vershinin Z, Pecht T, Goldstein N, Haim Y, Rudich A, Transcriptional Dysregulation of Adipose Tissue Autophagy in Obesity, Physiology (Bethesda, Md, 31 (2016) 270–282. [DOI] [PubMed] [Google Scholar]
- [37].Li H, Zhou B, Xu L, Liu J, Zang W, Wu S, Sun H, The reciprocal interaction between autophagic dysfunction and ER stress in adipose insulin resistance, Cell cycle (Georgetown, Tex, 13 (2014) 565–579. [DOI] [PubMed] [Google Scholar]
- [38].Burgeiro A, Fuhrmann A, Cherian S, Espinoza D, Jarak I, Carvalho RA, Loureiro M, Patricio M, Antunes M, Carvalho E, Glucose uptake and lipid metabolism are impaired in epicardial adipose tissue from heart failure patients with or without diabetes, Am J Physiol Endocrinol Metab, 310(2016)E550–E564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pereira MJ, Palming J, Rizell M, Aureliano M, Carvalho E, Svensson MK, Eriksson JW, The immunosuppressive agents rapamycin, cyclosporin A and tacrolimus increase lipolysis, inhibit lipid storage and alter expression of genes involved in lipid metabolism in human adipose tissue, Molecular and cellular endocrinology, 365 (2013) 260–269. [DOI] [PubMed] [Google Scholar]
- [40].Sato K, Kitamura T, Kawamura G, Mori Y, Sato R, Araki Y, Yamada Y, Glucose use in fasted rats under sevoflurane anesthesia and propofol anesthesia, Anesthesia and analgesia, 117 (2013) 627–633. [DOI] [PubMed] [Google Scholar]
- [41].De Oliveira JC, Ludemann Camargo R, Barella LF, Chaves Souto Branco R, Gravena C, Grassiolli S, Torrezan R, Cezar P De Freitas Mathias, Anesthetic-induced transient hyperglycemia and insulin resistance do not depend on the sympathoadrenal axis, Minerva endocrinologica, 38 (2013) 379–388. [PubMed] [Google Scholar]
- [42].Magnusson J, Nybell-Lindahl G, Tranberg KG, Clearance and action of insulin during general or epidural anaesthesia, Clinical nutrition (Edinburgh, Scotland: ), 5 (1986) 159–165. [DOI] [PubMed] [Google Scholar]
- [43].Cok OY, Ozkose Z, Pasaoglu H, Yardim S, Glucose response during craniotomy: propofol-remifentanil versus isoflurane-remifentanil, Minerva anestesiologica, 77 (2011) 1141–1148. [PubMed] [Google Scholar]
- [44].Schricker T, Galeone M, Wykes L, Carli F, Effect of desflurane/remifentanil anaesthesia on glucose metabolism during surgery: a comparison with desflurane/epidural anaesthesia, Acta anaesthesiologica Scandinavica, 48 (2004) 169–173. [DOI] [PubMed] [Google Scholar]
- [45].Acar D, Erkilic EK, Gumus T, Sahin D, Dincel AS, Kanbak O, The Effects of Different Anaesthetic Techniques on Surgical Stress Response During Inguinal Hernia Operations, Turkish journal of anaesthesiology and reanimation, 43 (2015) 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Windelov JA, Pedersen J, Holst JJ, Use of anesthesia dramatically alters the oral glucose tolerance and insulin secretion in C57Bl/6 mice, Physiological reports, 4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ueoka N, Hikasa Y, Effects in cats of atipamezole, flumazenil and 4-aminopyridine on stress-related neurohormonal and metabolic responses induced by medetomidine, midazolam and ketamine, Journal of feline medicine and surgery, 17 (2015) 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Salarinasab S, Nourazarian A, Nikanfar M, Abdyazdani N, Kazemi M, Feizy N, Rahbarghazi R, Impact of morphine on the expression of insulin receptor and protein levels of insulin/IGFs in rat neural stem cells, Neuroscience letters, 660 (2017) 147–154. [DOI] [PubMed] [Google Scholar]
- [49].Wang M, Wey S, Zhang Y, Ye R, Lee AS, Role of the unfolded protein response regulator GRP78/BiP in development, cancer, and neurological disorders, Antioxid Redox Signal, 11 (2009) 2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hong F, Mohammad Rachidi S, Lundgren D, Han D, Huang X, Zhao H, Kimura Y, Hirano H, Ohara O, Udono H, Meng S, Liu B, Li Z, Mapping the Interactome of a Major Mammalian Endoplasmic Reticulum Heat Shock Protein 90, PLoS One, 12 (2017) e0169260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yang Y, Li Z, Roles of heat shock protein gp96 in the ER quality control: redundant or unique function?, Molecules and cells, 20 (2005) 173–182. [PubMed] [Google Scholar]
- [52].Li X, Zhang K, Li Z, Unfolded protein response in cancer: the physician's perspective, Journal of hematology & oncology, 4 (2011) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hiramatsu N, Chiang WC, Kurt TD, Sigurdson CJ, Lin JH, Multiple Mechanisms of Unfolded Protein Response-Induced Cell Death, Am J Pathol, 185 (2015) 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Di Fazio P, Ocker M, Montalbano R, New drugs, old fashioned ways: ER stress induced cell death, Current pharmaceutical biotechnology, 13 (2012) 2228–2234. [DOI] [PubMed] [Google Scholar]
- [55].Wang X, Terpstra EJ, Ubiquitin receptors and protein quality control, Journal of molecular and cellular cardiology, 55 (2013) 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Day EA, Ford RJ, Steinberg GR, AMPK as a Therapeutic Target for Treating Metabolic Diseases, Trends in endocrinology and metabolism: TEM, 28 (2017) 545–560. [DOI] [PubMed] [Google Scholar]
- [57].Kim J, Kundu M, Viollet B, Guan KL, AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1, Nat Cell Biol, 13 (2011) 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Valente G, Morani F, Nicotra G, Fusco N, Peracchio C, Titone R, Alabiso O, Arisio R, Katsaros D, Benedetto C, Isidoro C, Expression and clinical significance of the autophagy proteins BECLIN 1 and LC3 in ovarian cancer, BioMed research international, 2014 (2014) 462658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Mizushima N, Yoshimori T, How to interpret LC3 immunoblotting, Autophagy, 3 (2007) 542–545. [DOI] [PubMed] [Google Scholar]
- [60].Cuervo AM, Wong E, Chaperone-mediated autophagy: roles in disease and aging, Cell research, 24 (2014) 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Minamino T, Kitakaze M, ER stress in cardiovascular disease, Journal of molecular and cellular cardiology, 48 (2010) 1105–1110. [DOI] [PubMed] [Google Scholar]
- [62].Xu C, Bailly-Maitre B, Reed JC, Endoplasmic reticulum stress: cell life and death decisions, The Journal of clinical investigation, 115 (2005) 2656–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL, ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs, Mol Cell, 6 (2000) 1355–1364. [DOI] [PubMed] [Google Scholar]
- [64].Brodsky JL, McCracken AA, ER protein quality control and proteasome-mediated protein degradation, Seminars in cell & developmental biology, 10 (1999) 507–513. [DOI] [PubMed] [Google Scholar]
- [65].Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL, Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction, Nature, 384 (1996) 432–438. [DOI] [PubMed] [Google Scholar]
- [66].Harding HP, Zhang Y, Ron D, Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase, Nature, 397 (1999) 271–274. [DOI] [PubMed] [Google Scholar]
- [67].Gao G, Xie A, Zhang J, Herman AM, Jeong EM, Gu L, Liu M, Yang KC, Kamp TJ, Dudley SC, Unfolded protein response regulates cardiac sodium current in systolic human heart failure, Circ Arrhythm Electrophysiol, 6 (2013) 1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Doroudgar S, Thuerauf DJ, Marcinko MC, Belmont PJ, Glembotski CC, Ischemia activates the ATF6 branch of the endoplasmic reticulum stress response, J Biol Chem, 284 (2009) 29735–29745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Thuerauf DJ, Marcinko M, Gude N, Rubio M, Sussman MA, Glembotski CC, Activation of the unfolded protein response in infarcted mouse heart and hypoxic cultured cardiac myocytes, Circ Res, 99 (2006) 275–282. [DOI] [PubMed] [Google Scholar]
- [70].Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, Robbins J, Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis, Proceedings of the National Academy of Sciences of the United States of America, 101 (2004) 10132–10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Guan HS, Shangguan HJ, Shang Z, Yang L, Meng XM, Qiao SB, Endoplasmic reticulum stress caused by left ventricular hypertrophy in rats: effects of telmisartan, The American journal of the medical sciences, 342 (2011) 318–323. [DOI] [PubMed] [Google Scholar]
- [72].Okada K, Minamino T, Tsukamoto Y, Liao Y, Tsukamoto O, Takashima S, Hirata A, Fujita M, Nagamachi Y, Nakatani T, Yutani C, Ozawa K, Ogawa S, Tomoike H, Hori M, Kitakaze M, Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis, Circulation, 110 (2004)705–712. [DOI] [PubMed] [Google Scholar]
- [73].Dally S, Monceau V, Corvazier E, Bredoux R, Raies A, Bobe R, del Monte F, Enouf J, Compartmentalized expression of three novel sarco/endoplasmic reticulum Ca2+ATPase 3 isoforms including the switch to ER stress, SERCA3f, in non-failing and failing human heart, Cell Calcium, 45 (2009) 144–154. [DOI] [PubMed] [Google Scholar]
- [74].Li Y, Guo Y, Tang J, Jiang J, Chen Z, New insights into the roles of CHOP-induced apoptosis in ER stress, Acta biochimica et biophysica Sinica, 46 (2014) 629–640. [DOI] [PubMed] [Google Scholar]
- [75].Terai K, Hiramoto Y, Masaki M, Sugiyama S, Kuroda T, Hori M, Kawase I, Hirota H, AMP-activated protein kinase protects cardiomyocytes against hypoxic injury through attenuation of endoplasmic reticulum stress, Mol Cell Biol, 25 (2005) 9554–9575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Myoishi M, Hao H, Minamino T, Watanabe K, Nishihira K, Hatakeyama K, Asada Y, Okada K, Ishibashi-Ueda H, Gabbiani G, Bochaton-Piallat ML, Mochizuki N, Kitakaze M, Increased endoplasmic reticulum stress in atherosclerotic plaques associated with acute coronary syndrome, Circulation, 116 (2007) 1226–1233. [DOI] [PubMed] [Google Scholar]
- [77].Thorp E, Li G, Seimon TA, Kuriakose G, Ron D, Tabas I, Reduced apoptosis and plaque necrosis in advanced atherosclerotic lesions of Apoe−/− and Ldlr−/− mice lacking CHOP, Cell metabolism, 9 (2009) 474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Minamino T, Komuro I, Kitakaze M, Endoplasmic reticulum stress as a therapeutic target in cardiovascular disease, Circ Res, 107 (2010) 1071–1082. [DOI] [PubMed] [Google Scholar]
- [79].Logue SE, Cleary P, Saveljeva S, Samali A, New directions in ER stress-induced cell death, Apoptosis, 18 (2013) 537–546. [DOI] [PubMed] [Google Scholar]
- [80].Hitomi J, Katayama T, Eguchi Y, Kudo T, Taniguchi M, Koyama Y, Manabe T, Yamagishi S, Bando Y, Imaizumi K, Tsujimoto Y, Tohyama M, Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death, J Cell Biol, 165 (2004) 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Kaufman RJ, Orchestrating the unfolded protein response in health and disease, The Journal of clinical investigation, 110 (2002) 1389–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ron D, Walter P, Signal integration in the endoplasmic reticulum unfolded protein response, Nat Rev Mol Cell Biol, 8 (2007) 519–529. [DOI] [PubMed] [Google Scholar]
- [83].Azfer A, Niu J, Rogers LM, Adamski FM, Kolattukudy PE, Activation of endoplasmic reticulum stress response during the development of ischemic heart disease, Am J Physiol Heart Circ Physiol, 291 (2006) H1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Chung KK, Dawson VL, Dawson TM, The role of the ubiquitin-proteasomal pathway in Parkinson's disease and other neurodegenerative disorders, Trends Neurosci, 24 (2001) S7–S14. [DOI] [PubMed] [Google Scholar]
- [85].Kaufman RJ, Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls, Genes & development, 13 (1999) 1211–1233. [DOI] [PubMed] [Google Scholar]
- [86].Mori K, Tripartite management of unfolded proteins in the endoplasmic reticulum, Cell, 101 (2000) 451–454. [DOI] [PubMed] [Google Scholar]
- [87].Takagi H, Matsui Y, Sadoshima J, The role of autophagy in mediating cell survival and death during ischemia and reperfusion in the heart, Antioxid Redox Signal, 9 (2007) 1373–1381. [DOI] [PubMed] [Google Scholar]
- [88].Saxton RA, Sabatini DM, mTOR Signaling in Growth, Metabolism, and Disease, Cell, 168 (2017) 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ha J, Kim J, Novel pharmacological modulators of autophagy: an updated patent review (2012-2015), Expert opinion on therapeutic patents, 26 (2016) 1273–1289. [DOI] [PubMed] [Google Scholar]
- [90].Ruderman NB, Carling D, Prentki M, Cacicedo JM, AMPK, insulin resistance, and the metabolic syndrome, The Journal of clinical investigation, 123 (2013) 2764–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Bullon P, Marin-Aguilar F, Roman-Malo L, AMPK/Mitochondria in Metabolic Diseases, Exs, 107 (2016) 129–152. [DOI] [PubMed] [Google Scholar]
- [92].Maejima Y, Isobe M, Sadoshima J, Regulation of autophagy by Beclin 1 in the heart, Journal of molecular and cellular cardiology, 95 (2016) 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Song S, Tan J, Miao Y, Li M, Zhang Q, Crosstalk of autophagy and apoptosis: Involvement of the dual role of autophagy under ER stress, Journal of cellular physiology, (2017). [DOI] [PubMed] [Google Scholar]
- [94].Levine B, Sinha S, Kroemer G, Bcl-2 family members: dual regulators of apoptosis and autophagy, Autophagy, 4 (2008) 600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Wei Y, Sinha S, Levine B, Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation, Autophagy, 4 (2008) 949–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P, Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy, J Biol Chem, 284 (2009) 2719–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, Korsmeyer S, Packer M, May HI, Hill JA, Virgin HW, Gilpin C, Xiao G, Bassel-Duby R, Scherer PE, Levine B, Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis, Nature, 481 (2012) 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Wei Y, Pattingre S, Sinha S, Bassik M, Levine B, JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy, Mol Cell, 30 (2008) 678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Tanida I, Autophagosome formation and molecular mechanism of autophagy, Antioxid Redox Signal, 14 (2011) 2201–2214. [DOI] [PubMed] [Google Scholar]
- [100].Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA Jr., Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL, Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes, Autophagy, 4 (2008) 151–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Lai L, Chen J, Wang N, Zhu G, Duan X, Ling F, MiRNA-30e mediated cardioprotection of ACE2 in rats with Doxorubicin-induced heart failure through inhibiting cardiomyocytes autophagy, Life sciences, 169 (2017) 69–75. [DOI] [PubMed] [Google Scholar]
- [102].Li L, Tian J, Long MK, Chen Y, Lu J, Zhou C, Wang T, Protection against Experimental Stroke by Ganglioside GM1 Is Associated with the Inhibition of Autophagy, PLoS One, 11 (2016) e0144219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Gao H, Yang Q, Dong R, Hou F, Wu Y, Sequential changes in autophagy in diabetic cardiac fibrosis, Molecular medicine reports, 13 (2016) 327–332. [DOI] [PubMed] [Google Scholar]
- [104].Fonseca ACRG, Oliveira CR, Pereira CF, Cardoso SM, Loss of proteostasis induced by amyloid beta peptide in brain endothelial cells, BBA - Molecular Cell Research, 1843 (2014) 1150–1161. [DOI] [PubMed] [Google Scholar]
- [105].Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, Miller FJ Jr., Rothermel BA, Hill JA, Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy, Circulation, 117 (2008) 3070–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A, Sciarretta S, Del Re DP, Zablocki DK, Hsu CP, Lim DS, Isobe M, Sadoshima J, Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2, Nature medicine, 19 (2013) 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Bhuiyan MS, Pattison JS, Osinska H, James J, Gulick J, McLendon PM, Hill JA, Sadoshima J, Robbins J, Enhanced autophagy ameliorates cardiac proteinopathy, The Journal of clinical investigation, 123 (2013) 5284–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J, Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy, Circ Res, 100 (2007) 914–922. [DOI] [PubMed] [Google Scholar]
- [109].Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA, Cardiac autophagy is a maladaptive response to hemodynamic stress, The Journal of clinical investigation, 117 (2007) 1782–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Zhu H, He L, Beclin 1 biology and its role in heart disease, Current cardiology reviews, 11 (2015) 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Yin X, Peng C, Ning W, Li C, Ren Z, Zhang J, Gao H, Zhao K, miR-30a downregulation aggravates pressure overload-induced cardiomyocyte hypertrophy, Molecular and cellular biochemistry, 379 (2013) 1–6. [DOI] [PubMed] [Google Scholar]
- [112].Pan W, Zhong Y, Cheng C, Liu B, Wang L, Li A, Xiong L, Liu S, MiR-30-regulated autophagy mediates angiotensin II-induced myocardial hypertrophy, PLoS One, 8 (2013) e53950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Lin H, Li HF, Chen HH, Lai PF, Juan SH, Chen JJ, Cheng CF, Activating transcription factor 3 protects against pressure-overload heart failure via the autophagy molecule Beclin-1 pathway, Molecular pharmacology, 85 (2014) 682–691. [DOI] [PubMed] [Google Scholar]
- [114].Galán M, Kassan M, Choi SK, Partyka M, Trebak M, Henrion D, Matrougui K, A novel role for epidermal growth factor receptor tyrosine kinase and its downstream endoplasmic reticulum stress in cardiac damage and microvascular dysfunction in type 1 diabetes mellitus, Hypertension, 60 (2012) 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kubli DA, Gustafsson AB, Unbreak my heart: targeting mitochondrial autophagy in diabetic cardiomyopathy, Antioxid Redox Signal, 22 (2015) 1527–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Li A, Zhang S, Li J, Liu K, Huang F, Liu B, Metformin and resveratrol inhibit Drp1-mediated mitochondrial fission and prevent ER stress-associated NLRP3 inflammasome activation in the adipose tissue of diabetic mice, Molecular and cellular endocrinology, 434 (2016) 36–47. [DOI] [PubMed] [Google Scholar]