Abstract
Three large consortia present comprehensive analyses that identify genetic factors influencing smoking initiation, intensity and cessation. The genetic architecture of these three phases of smoking behavior appears to be largely distinct.
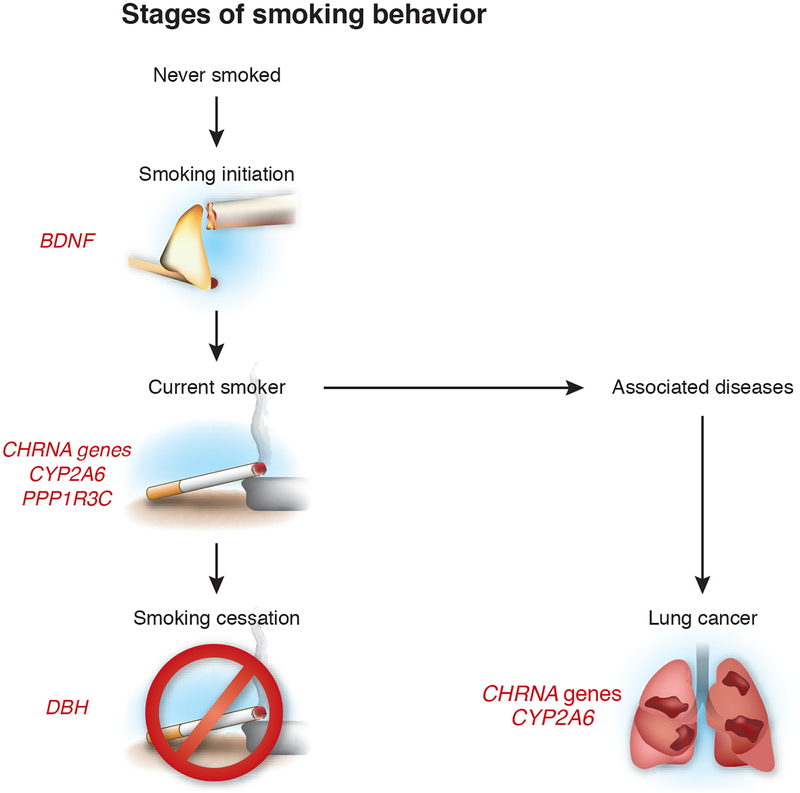
Nicotine dependence results from an interplay of neurobiological, environmental and genetic factors. Smoking stages are categorized into smoking initiation, current smoking and smoking cessation (Fig. 1). Genetic influences at each step in this process have been documented in numerous twin and family studies1. Patterns of smoking initiation reflect individual differences in sensitivity to nicotine, the availability of tobacco and social norms. For an individual who has become a habitual smoker, both genetic and psychosocial factors play a role in determining the intensity of smoking, known as smoking dependence, and the ability to quit (cease smoking). On pages 436, 441 and 448 of this issue, three collaborating groups2–4—the Oxford-GlaxoSmithKline (Ox-GSK)3, Tobacco and Genetics Consortium (TAG)4 and ENGAGE2 consortia—present results of combined analyses from over 140,000 individuals, bringing new insights into the genetic factors that influence smoking initiation, dependency and cessation. Although many different measures are available for assessing the degree to which smokers are dependent, the number of cigarettes smoked per day (CPD) is easily measured and features prominently in measures of dependency, and it therefore was used by all three consortia. The large sample sizes of the combined studies enabled the first identification in a genome-wide study of loci influencing smoking initiation and cessation and supported the discovery of new loci influencing smoking dependence.
Figure 1.
Smoking behavior as a multistep process with genetic influences. Nonsmoking individuals (those who have never smoked) may begin smoking (smoking initiation). At that point, they may become dependent upon cigarettes, and they are classified as having smoked (ever-smokers) once they have consumed 100 or more cigarettes. The TAG consortium4 report that variants at the BDNF locus influence smoking initiation. A region on chromosome 15 encompassing the nicotinic receptor subunit genes CHRNA5, CHRNA3 and CHRNB4 was associated with cigarettes consumed per day (CPD), a measure of smoking dependence2–4. The TAG and ENGAGE consortia also identified the CHRNA6–CHRNB3 cluster on chromosome 8p11, as well as loci found near CYP2A6 on 19q13 and in a region containing a noncoding RNA on chromosome 10q23, as associated with CPD2,4. For smoking cessation, the TAG consortium identified a region on chromosome 9 near the gene encoding dopamine β-hydroxylase (DBH)4. Further studies are needed to define the specific genes and causal variants in these regions.
Genetics by smoking stages
The TAG consortium4 performed data harmonization of smoking phenotypes from across 17 participating studies and subsequently used a meta-analysis to integrate the results with those of the studies from the other two consortia. The authors present evidence that the region containing the gene encoding brain-derived neurotrophic factor (BDNF) is associated with smoking initiation4. Identification of variants near BDNF as contributing to smoking initiation coincides with the increasing recognition of the protein as important both in neurobiological processes such as response to social stress5 and in moderating anxiety6. BDNF binds to NTRK2, variants of which were previously associated with smoking initiation and dependence7.
High-affinity binding of nicotine to nicotinic acetylcholine receptors (NAChR) results in increases in the levels of several neurotransmitters, including dopamine, in the reward circuits of the brain. NAChRs have an important role in modulating nicotine dependence and smoking behavior. The strongest associations with CPD have been reported for the 15q25 cluster that encompasses the CHRNA3, CHRNA5 and CHRNB4 genes, which encode NAChR receptors. The new studies2–4 further replicated this association and better defined the association from among these receptors at this locus. Previous efforts at refining association at the CHRNA3–CHRNA5–CHRNA4 locus have been complicated by strong association, or linkage disequilibrium, among the markers in this region in European-descended populations. Liu et al.3, representing the Ox-GSK study, report a genome-wide meta-analysis on smoking quantity, defined by categorizing CPD, from over 41,000 individuals, including 18,591 ‘ever-smokers’ (smokers and former smokers). They used imputation based on data from the 1000 Genomes project to refine the association signal at the 15q25 locus associated to CPD. Liu et al.3 demonstrated the increased resolution made possible by amassing data from a large number of participants and imputing from the 1000 Genomes Project. They resolved association at a previously unreported SNP (rs55853698) affecting mRNA transcription of CHRNA5. By conditioning on this SNP and analyzing additional SNPs in the region, they also identified a second significant SNP in CHRNA3, rs6495308.
Aside from the NAChR cluster on chromo-some 15q26, the ENGAGE2 and TAG4 consortia identified CPD associations with additional NAChR receptor subunit genes, CHRNB3 and CHRNA6. The Ox-GSK study also found suggestive evidence that CHRNA2 associates with CPD3. The nicotinic acetylcholine receptor is formed from different combinations of five subunits. The association of CHRNA6 with smoking behavior was reported previously in a candidate gene study8 and is consistent with the gene’s high expression in dopamine-releasing neurons9,10. Elucidating the complex interactions among the nicotinic receptor subunits and their effects in modulating smoking behaviors presents a ripe area for further research into the neurobiology of smoking dependence. In the TAG consortium data, an additional locus at 10q25 in a region of noncoding RNA of unknown function associated with CPD4.
An intriguing association reported from the TAG consortium4 was between current versus former smoking and variation in the region of DBH, encoding dopamine β-hydroxylase, which catalyzes the conversion of dopamine to norepinephrine. Although DBH is an excellent candidate locus to influence the ability of individuals to quit smoking, further analysis will be required to replicate the association in this region and to identify the causal gene and functional variants influencing smoking cessation. DBH represents a particularly interesting candidate gene because pharmacologic agents that modulate nicotine receptors in the dopaminergic pathway, such as varenicline, can be highly effective in assisting individuals to achieve smoking cessation. Further pharmacogenetic studies to evaluate the effectiveness of smoking cessation treatments, as well as nicotine withdrawal, in relation to SNP variations may help in the tailoring of cessation treatments to individual needs.
Genetics of nicotine metabolism
Smoking provides a highly efficient mechanism for rapidly delivering nicotine to the brain via the lungs with extraordinary speed (within 10 seconds), and the dose can be individually titrated by the smoker. However, the effects of nicotine are strongly influenced by its complex metabolism. Polymorphisms of cytochrome p450 2A6 (encoded by CYP2A6) strongly influence the catabolism of nicotine into inactive metabolites11, and individuals with rapid metabolism require higher levels of smoking to maintain the same nicotine level than do individuals whose genotypes confer slower metabolism. The TAG4 and ENGAGE2 studies both identified variation in the CYP2A6 region associating with CPD. The complex genetic architecture of mutations in this gene, which includes large insertions and deletions, may obscure associations in studies using SNP platforms, which cannot directly query associations due to copy number variations.
Future directions
Although results from these three very large consortia have identified several new loci influencing smoking behavior, further studies are needed to replicate these findings and to resolve the candidate genes at each of these loci. Resequencing studies are also required to identify the functional variants at each locus as well to explore the roles of genetic factors in influencing each stage of smoking. In addition, analyses jointly modeling effects among multiple loci will help to characterize those combinations of variations that have large effects on smoking behavior. Finally, the current studies suggest possible pathways influencing smoking behavior that suggest promising areas for pharmacogenetic research.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturegenetics/.
References
- 1.Li MD Curr. Psychiatry Rep 8, 158–164 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Thorgeirsson TE et al. Nat. Genet 42, 448–453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu JZ et al. Nat. Genet 42, 436–440 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.TAG Consortium. Nat. Genet 42, 441–447 (2010).20418890 [Google Scholar]
- 5.Fanous S, Hammer RP Jr. & Nikulina EM Neuroscience 10.1016/j.neuroscience.2010.02.064 (3 March 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cazorla M et al. PLoS ONE 5, e9777 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vink JM et al. Am. J. Hum. Genet 84, 367–379 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bierut LJ et al. Hum. Mol. Genet 16, 24–35 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer EL, Yoshikami D & McIntosh JM J. Neurochem 105, 1761–1769 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klink R, de Kerchove, d’Exaerde A, Zoli M & Changeux JP J. Neurosci 21, 1452–1463 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swan GE et al. Pharmacogenet. Genomics 15, 115–125 (2005). [DOI] [PubMed] [Google Scholar]