Abstract
Protein glycosylation, an important post-translational modification, plays an essential role in a wide range of biological processes such as immune response, intercellular signaling, inflammation, and host-pathogen interaction. Aberrant glycosylation has been correlated with various diseases. However, studying protein glycosylation remains challenging because of low abundance, microheterogeneities of glycosylation sites and poor ionization efficiency of glycopeptides. Therefore, the development of sensitive and accurate approaches to characterize protein glycosylation is crucial. The identification and characterization of protein glycosylation by mass spectrometry is referred to as the field of glycoproteomics. Methods such as enrichment, metabolic labeling, and derivatization of glycopeptides in conjunction with different mass spectrometry techniques and bioinformatics tools, have been developed to achieve an unequivocal quantitative and qualitative characterization of glycoproteins. This review summarizes the recent developments in the field of glycoproteomics over the past six years (2012 to 2018).
Keywords: Derivatization, Enrichment, Glycoproteins, Glycosylation, Metabolic labeling
1. Introduction
Post-translational modifications (PTMs) of proteins play significant roles in modulating protein functions [1–4]. Moreover, the biological impact of this PTMs on human health is critical. Protein glycosylation is a typical example of such PTMs. The two main types of glycosylated proteins are referred to as N-, and O-linked glycoproteins, where N-linked glycans are attached to asparagine residue while O-linked glycans are attached to serine or threonine residues [5–7]. Over half of all proteins are glycosylated, making glycosylation a valuable source of information for biofunction and disease studies. These glycan moieties and secreted glycoproteins modulate and control many biological functions, including cell signaling, adhesion, and communication [8–15]. Furthermore, protein folding, stability, and localization are dependent on protein glycosylation [2, 9]. Changes in glycan moieties of glycoproteins have been directly correlated with mammalian disease and hereditary disorders, such as immune deficiencies, cardiovascular disease, and cancer [1, 3, 16–19]. Since up to 50,000 proteins are encoded by the human genome, with over half being glycosylated, investigation of these protein modification sites has generated a demand for reliable qualitative and quantitative glycoproteomic methods [1–3, 20].
Mass spectrometry has become a prominent method for the unequivocal characterization of glycoproteins [5, 21]. Due to the surge in demand for accurate glycoprotein analysis in recent years, MS instrumentation and new techniques and strategies, facilitating effective analysis of glycoproteins, have been developed [5, 21–23]. MS methods utilized in glycoproteomics are similar to “bottom-up” and “top-down” proteomics [24]. Bottom-up glycoproteomics utilizes enzymes to digest glycoproteins and to characterize glycosylation at the glycopeptide level by LC-MS/MS [25]. The top-down method subject intact glycoproteins to LC-MS/MS analysis, thus permitting the identification of glycoprotein proteoforms with minimal sample preparation [26]. Analysis of glycoproteins is achieved using numerous analytical methods, including electrospray ionization-liquid chromatography-tandem mass spectrometry (LC-ESI-MS/MS) , capillary electrophoresis-mass spectrometry (CE-MS) [27], capillary zone electrophoresis-mass spectrometry (CZE-MS) [28], ion mobility-mass spectrometry (IM-MS) [29] and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) [30]. The use of these techniques to investigate aberrations in glycosylation associated with several diseases has already been demonstrated [19, 29, 31, 32], thus developing new sensitive and reliable techniques is of critical importance.
While advances in MS technologies and methods have propelled the progress of glycoproteomics, obstacles still currently exist. For example, glycoproteins in biological systems are present in low abundance, and MS analysis of glycopeptides is hampered by microheterogeneities and low ionization efficiencies in the presence of co-eluting peptides. Therefore, a comprehensive characterization of protein glycosylation microheterogeneity is challenging, and the identification and quantitation of isomeric glycan occupancies of potentially all glycosylation sites of proteins are a daunting task [33].
Although in-depth quantification of glycopeptides remains an overwhelming analytical task which has been facilitated because of recent progress in column chromatography, labeling, derivatization, enrichment methods and bioinformatics, and mass spectrometry [33–36]. As summarized in Figure 1, this review will provide an update on MS-based glycoproteomics, summarizing recent developments (i) in glycoprotein enrichment, (ii) in the derivatization of glycoproteins for enhanced detection and quantitation, (iii) in fragmentation methods for characterization of glycopeptides and glycoproteins, (iv) in ion mobility MS to characterize glycopeptides, (v) in metabolic labeling of glycoprotein to facilitate enrichment and quantitation, and (vi) in bioinformatics tools to facilitate glycopeptide and glycoprotein data processing. Also, this review includes rudimentary aspects of this field that helped lay the groundwork for recent advancements.
Figure 1.
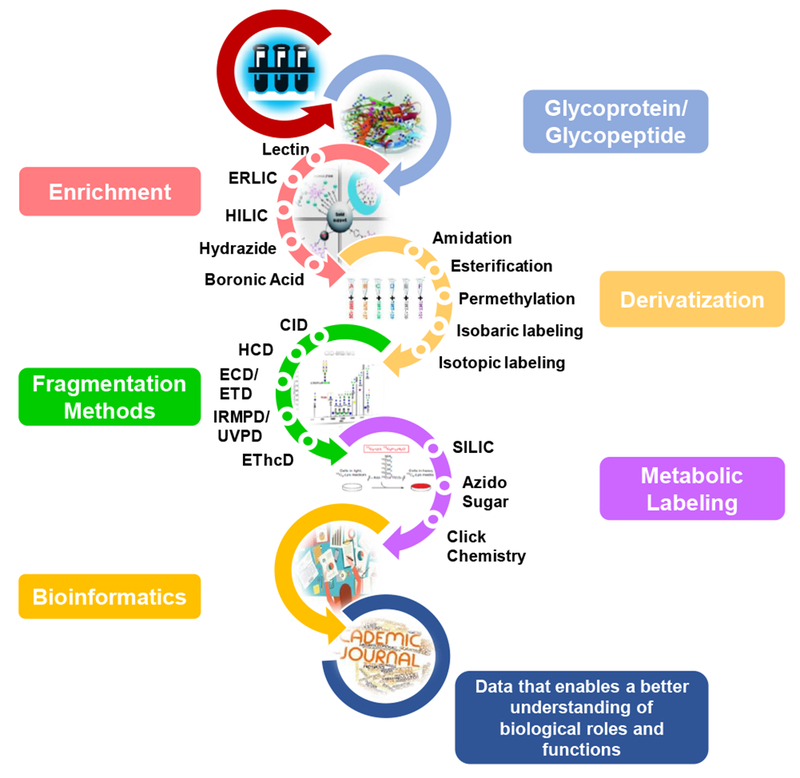
Workflow for recent developments on MS-based glycoproteomics.
2. Advances in glycoproteins and glycopeptides enrichment
Direct analysis of glycoproteins in complex samples by mass spectrometry-based strategies remains to be analytically challenging due to the low abundance of glycoproteins, microheterogeneity of protein glycosylation sites, the complexity of glycan structures and the low ionization efficiency of glycopeptides relative to unmodified peptides. Therefore, the selective enrichment and isolation of glycopeptides from biological samples are essential for glycoproteomics research. During the past decade, several enrichment methods have been developed and successfully utilized in glycoproteomics research.
2.1. Enrichment using lectin materials
Lectins have been widely used as affinity probes to efficiently detect glycans in complex biological fluids because of their selective recognition of specific glycan moieties of glycoconjugates. Immobilization of lectins on either a microarray, magnetic beads, or a resin material has been effectively employed for glycopeptide enrichment [37, 38]. The selectivity of a lectin towards a target originates from a combination of interactions, including hydrogen bonding, and van der Waals and/or hydrophobic interactions. Due to the distinct carbohydrate-binding specificities of lectins, multiple lectins with varying glycan-recognition specificities should be applied to achieve a comprehensive capture of glycosylated proteins from biological mixtures.
Recently, Samuelson and co-workers [39] reported selective isolation of N-linked glycopeptides from complex peptide mixtures or biological samples by improving the inherent properties of Fbs1 (aka Fbx2, FBXO2, NFB42) using protein engineering approach. Fbs1 is a eukaryotic lectin-like protein that generally functions in a ubiquitin-mediated process to eliminate misfolded N-glycosylated proteins. Mutant forms of Fbs1 were able to bind to diverse types of N-linked glycoproteins, whereas, wild-type Fbs1 preferentially binds high-mannose-containing glycans. Mutagenesis of serine to glycine (presenting smaller side chain) at position 155 resulted in a higher affinity to complex N-glycans, due to the removal of steric hindrance. Also, the conversion of phenylalanine to tyrosine at position 173, positively affected sugar ring binding, through the addition of a single hydroxyl group. The mutant type Fbs1 was successfully employed for the unbiased enrichment of N-linked glycopeptides from human serum. By using filter-aided sample preparation method, a total of 2142 N-glycopeptides were identified which was 2.2-folds more than the number of identified N-glycopeptide in the same sample papered by using a mixture of common lectins (concanavalin A, wheat germ agglutinin and Ricinus communis agglutinin).
More recently, Yang et al. [40] described a fabrication method of thermoresponsive porous polymer membrane reactors for online proteolysis and glycopeptide enrichment in an hour. The reactors were fabricated by coating polystyrene-maleic anhydride-N-isopropylacrylamide (PS-MAn-PNIPAm) on a Nylon sheet prior to packing in the precolumn filter units. The morphology of PNIPAm is dependent on temperature. PNIPAm forms micelle cavities at 37°C which incorporate trypsin or lectin (mixture of concanavalin A and wheat germ agglutinin) The thermoresponsive nature of PNIPAm improves the interactions between the enzyme or lectins and the proteins/peptides in the reactors. Increasing the temperature to 45°C resulted in the collapse of the micelle cavities and adversely affected the proteolysis and enrichment efficiencies. By using online proteolysis and glycopeptide enrichment at 37°C, a total of 262 N-glycopeptides were identified from 155 glycoproteins from the human plasma sample. This number was larger than the 115/84 glycopeptides/glycoproteins determined for the same sample prepared by off-line in-solution digestion and lectin enrichment [40]. Although the results were promising and demonstrated a fully automated protocol for online proteolysis and glycopeptide enrichment, the lifetime of the reactors was not adequately presented or discussed.
2.2. Enrichment using hydrophilic materials
Enrichment-based on hydrophilic interaction offers broad glycan specificity, unbiased recognition ability for glycopeptides, and excellent reproducibility and compatibility with MS analysis. Recently, various hydrophilic materials, such as metal–organic frameworks, polymer, and magnetic materials, have been used for glycopeptides enrichment.
2.2.1. Enrichment using hydrophilic metal-organic framework materials
Metal-organic frameworks (MOFs) have gained popularity during the past decade as a tool for the separation and enrichment of glycoproteins of biological samples because of their high surface area, adjustable pore size and the ease of functionalizing. MOFs which consist of metal ions and organic ligands have been employed extensively, in separation, gas adsorption, and drug delivery [41, 42].
Xie et al. [43] synthesized a hydrophilic magnetic amino-functionalized MOF which permitted selective enrichment of glycopeptides. The sorbent was synthesized by initially preparing Fe3O4 nanoparticles at high temperature (265°C) in phenyl ether as a solvent. Fe3O4 nanoparticles were then functionalized with polydopamine (PDA). The functionalized nanoparticles were then dispersed in N, N-dimethylformamide, containing ZrCl4 and heated at 120°C for 45 min to produce Fe3O4@DA@MOF-NH2 MOF magnetic nanoparticles. The glycopeptides enriched on the surface by interaction with the amine groups present on the MOF surface. A total of 517 N-glycopeptides were identified from 151 glycoproteins in the human plasma sample.
Ma et al. [44] employed cysteine-functionalized MOF for the enrichment of N-linked glycopeptides derived from both model glycoprotein and HeLa cells. The sorbent material was synthesized by heating chromic nitrate and 2-aminoterephthalic acid as MOF precursors at 130°C for 24 h. The produced MOF-NH2 nanoparticles were then modified with gold nanoparticles (MOF@Au) and followed by the cysteine immobilization to produce MOF@Au-cysteine material. This MOF material exhibited excellent binding capacity, good selectivity, high recovery, and low detection limit, allowing the identification of 1069 N-glycopeptides corresponding to 614 N-glycoproteins in HeLa cells. Although both of the abovementioned studies [43, 44] suggested that functionalized MOFs can be used as a sorbent for glycopeptide enrichment, routine use of such material is limited by the fact that the synthesis of MOFs is time-consuming, tedious, and expensive.
2.2.2. Enrichment using hydrophilic polymers
Hydrophilic polymers have also been used for the enrichment of glycoproteins and glycopeptides in complex biological samples. Recently, L-cysteine functionalized polymers were synthesized by combining distillation-precipitation polymerization and “Thiol–ene” click chemistry [45]. The synthesized hydrophilic polymer was successfully employed for the enrichment of glycopeptides in human plasma. The functionalized polymer particles were utilized to identify 208 unique glycopeptides corresponding to 121 glycoproteins, which was higher than the number of glycopeptides (168 glycopeptides corresponding to 100 glycoproteins) that was detected using commercial ZIC@HILIC beads (SeQuant, Merck, Germany) [45].
Polyamidoamine dendrimer(PAMAM) was recently used for the synthesis of zwitterionically functionalized (ZICF) materials for glycopeptide enrichment [46]. Briefly, PAMAM was sulfonated with 1,3-propanesultone under inert atmosphere (18 h, 50°C) and then methylated with an excessive amount of iodomethane (12 h, 50°C) to produce the ZICF-PAMAM material. Glycopeptides were enriched through the interaction with amine and sulfite groups present on the surface of the sorbent. The multiple branched structures and good solubility of ZICF-PAMAM facilitated adequate interaction with glycopeptides. The ZICF-PAMAM combined with the filter-aided sample preparation strategy allowed the identification of 395 unique glycopeptides corresponding to 178 glycoproteins in 0.1 μl human serum. Also, the performance of this strategy for the enrichment of fetuin glycopeptides was compared to that using commercial ZIC@HILIC beads (SeQuant, Merck, Germany). 5 and 3 glycopeptides were identified by using ZICF-PAMAM and commercial ZIC@HILIC beads, respectively, suggesting the more superior performance of the ZICF-PAMAM over the commercial beads. Although the abovementioned studies [45, 46] suggested the potential use of polymerized materials in glycopeptide enrichment, the multistep fabrication of the material is still limiting a broader use of such material for enriching glycoproteins. This issue is easily overcome by making the material commercially available.
2.2.3. Enrichment using hydrophilic magnetic materials
Over the past decade, magnetic materials, especially Fe3O4 nanoparticles, have accumulated much attention in drug delivery, biochemistry, and bioseparation. This increase in awareness is due to the fast-magnetic response, biocompatibility, the ease of preparation, and flexible functionalization of these materials [47–50]. The combination of magnetic materials and covalently bound hydrophilic functional molecules permits the simultaneous enrichment of glycopeptides from complex peptide mixture.
Recently, Wan et al. [51] fabricated a dendrimer-assisted magnetic graphene–silica hydrophilic composite for the efficient and selective enrichment of glycopeptides from biological samples, such as mouse liver. The functionalization of magnetic graphene oxide with PAMAM dendrimer provided the composite with numerous active sites (hydroxyl and amine groups) which could be used for the addition of hydrophilic components. The synthesis process consisted of the oxidation of graphene to graphene oxide (GO), deposition of iron oxide (GO@ Fe3O4), encapsulation with silica shell (GO@Fe3O4@SiO2), addition of the dendrimer (GO@Fe3O4@SiO2@PAMAM) and graft of maltose (GO@Fe3O4@SiO2@PAMAM@maltose. The use of this material facilitated the identification of 1529 N-linked glycopeptides derived from 760 glycoproteins isolated from mouse liver [51]. Complex multistep synthesis of the sorbent makes the functionalized process time-consuming.
More recently, Sun et al. [52] synthesized hydrophilic magnetic mesoporous silica materials, presenting high selectivity, low detection limit and good binding capacity of glycopeptides. A total of 424 glycopeptides assigned to 140 glycoproteins were identified in the human serum sample using such hydrophilic magnetic mesoporous silica materials. The hydrophilic magnetic mesoporous silica materials exhibited a more superior performance compared to a variety of hydrophilic functionalized magnetic nanoparticles used for glycopeptide enrichment, including Fe3O4@maltose [53], Fe3O4@polyethylene glycol [54], Fe3O4@glucose-6-phosphate [55], and graphene@Fe3O4@glucose [56].
2.3. Enrichment using electrostatic repulsion hydrophilic interaction chromatography
Alpert [57] introduced electrostatic repulsion hydrophilic interaction chromatography (ERLIC), using a combination of interactive forces in hydrophilic interaction chromatography (HILIC) and ion-exchange chromatography. Due to the stronger interaction of glycopeptides compared to peptides by both the hydrophilic and electrostatic interactions, efficient isolation is permitted over the ion-exchange solid supports. Recently, an investigation of glycopeptides in six different breast cancer cell lines using HILIC and ERLIC enrichment methods was reported by Mechref and coworkers [58]. A total of 497 glycopeptides were characterized, of which 401 were found common (80.6% overlap) between the two enrichment techniques. Using HILIC and HILIC-ERLIC, 320 and 214 glycopeptides, respectively, were determined to be significantly over- or under-expressed in 231BR cell line relative to the other cell lines.
2.4. Enrichment using hydrazide functionalized materials
In 2003, Aebersold and coworkers [59] introduced a solid-phase extraction method for the adsorption of glycoproteins based on hydrazide chemistry which has been widely used for glycopeptide enrichment. The method involves oxidation of cis-diol groups of carbohydrate to aldehydes with periodate and then the formation of hydrazone, which allows covalent bond formation between glycopeptides and the support material. Compared with the lectin methods, hydrazide chemistry-based enrichment is less specific and could capture various glycopeptides. However, since the covalent bond between the glycopeptide and support is not reversible, the release of glycopeptides with intact glycan structures becomes impossible. Moreover, periodate oxidizes the saccharide chain at different positions, which impedes further analysis of intact glycopeptides or glycan structures [60, 61].
Recently, hydrazide functionalized PAMAM dendrimers were synthesized for the efficient and selective enrichment of N-linked glycopeptides from a human serum sample using FASP [62]. The sorbent was synthesized by adding a vinyl ester to PAMAM via the Michael addition method. Next, functionalization of the surface with hydrazide groups was achieved by hydrazinolysis. In total 158 unique glycopeptides corresponding to 60 different glycoproteins were identified in a human serum sample. Also, in comparison with commercial hydrazide beads (Bioclone, CA, USA), hydrazide functionalized PAMAM dendrimers, showed better performance for the enrichment of the glycopeptides derived from fetuin samples. 3 and 5 glycopeptides were identified by using commercial hydrazide beads and hydrazide functionalized PAMAM dendrimers, respectively.
In another study [63], a hydrazide functionalized core–shell magnetic nanocomposite was used for highly specific enrichment of N-glycopeptides. The nanocomposite was prepared by the functionalization of the magnetic core with poly methacrylic acid to obtain the Fe3O4@poly (methacrylic acid) prior to the modification of the surface with adipic acid dihydrazide to obtain Fe3O4@poly(methacrylic hydrazide). The abundant hydrazide groups showed highly specific enrichment of glycopeptides and the magnetic core made it suitable for large-scale, high-throughput, automated sample processing [63]. The nanocomposite utilized for the profiling of N-glycopeptides derived from colorectal cancer patient serum and allowed the identification of 175 unique glycopeptides corresponding to 63 unique glycoproteins. Moreover, in comparison with commercially hydrazide resin (Affi-Gel Hz resin, Bio-Rad, CA, USA), the nanocomposite improved the signal-to-noise ratios of fetuin glycopeptides by at least five times.
2.5. Enrichment using boronic acid functionalized materials
Since boronic acids form reversible cyclic boronate esters with cis-diol groups of glycans, it can be employed for the capture and isolation of glycopeptides [60]. The reversibility is the main difference between boronic acid and hydrazide chemistry. While boronic acid chemistry is easily reversible under acidic conditions without altering the glycan structure, hydrazide chemistry requires irreversible and destructive glycan oxidation to generate aldehyde groups.
Boronic acid-functionalized magnetic carbon nanotubes were synthesized with a large specific surface area and a high density of boronic acid groups [64]. This material was then used to enrich glycopeptides from horseradish peroxidase sample and permitted the identification of 10 glycopeptides. Commercial boronic acid-agarose (Sigma-Aldrich, USA) material was used for comparison and only permitted the identification of 7 glycopeptides. Additionally, 3-acrylaminophenylboronic acid functionalized Fe3O4 nanoparticles were synthesized and also used to enrich glycopeptides in horseradish peroxidase sample, resulting in the identification of 16 glycopeptides [65]. Although the results of these two studies [64, 65] show the applicability of the proposed sorbents in the enrichment of glycopeptides, their efficiencies in more complex samples such as the enrichment of glycopeptides from human serum samples were not demonstrated.
Recently, boronic acid-functionalized magnetic metal-organic framework composite was used for the enrichment of glycopeptides from human serum samples, allowing the identification of 209 N-glycosylation peptides derived from 89 unique glycoproteins [66]. The functionalization process is only achieved under harsh conditions (nonaqueous solvents and high temperatures) and involves multistep reactions. Also, the reusability of the sorbents and the reproducibility of the methods were not reported.
2.6. Challenges in using recently developed materials in the enrichment of glycoproteins
The recent advancement in the fields of glycoprotein enrichment based on using new functionalized materials was summarized. Although researchers have turned to the world of nanomaterials because of their superior physicochemical characteristics and variety of functionalized nanomaterials for the enrichment of glycopeptides have been reported [43–46, 51, 52, 54–56, 62–66], there are still challenges to utilize these materials in routine laboratories. Most of these functionalized materials (e.g. Fe3O4@DA@MOF [43], MOF@Au-cysteine [44], ZICF-PAMAM [46], and GO@Fe3O4@SiO2@PAMAM@maltose [51]) are not commercially available and suffers from time-consuming, tedious and multistep fabrication. Moreover, separation from the mixtures is the parameter that should be carefully considered when using nanomaterials for enrichment. Although functionalized magnetic nanomaterials can be easily separated from the mixture by using an external magnet, the other nanomaterials (e.g. MOF-, carbon-, and polymer-based nanomaterials) need an inconvenient and time-consuming centrifugation step. Also, more studies such as inter- and intra-laboratory comparisons, need to be performed to validate the analytical performance of such materials.
3. Advances in the derivatization of glycoproteins for enhanced detection and quantitation
Although low abundant glycoproteins can be enriched by different methods, enhancement of the ionization efficiency of glycopeptides is still needed [67]. Direct analysis of intact glycoproteins is needed to reflect the biological information of the protein glycosylation. Various chemical derivatization strategies have been used to modify the structures of glycopeptides to enhance MS ionization and stabilize acidic glycans. Also, stable isobaric-isotopic approaches, which are used to reduce bias caused by sample preparation and instrument instability, can simultaneously facilitate qualitative and quantitative analysis of glycopeptides.
3.1. Amidation of sialylated glycopeptides
Sialylation plays a role in the development and progression of diseases [18, 19, 68]. However, it is difficult to assess the sialylation of glycopeptides due to in-source and metastable decay of sialic acids. Amidation by dimethylamine and methylamine have been used to stabilize sialic acid [69, 70].
Recently, a highly reproducible derivatization method of sialic acid coupled with MALDI-TOF-MS analysis was introduced to enhance the detection of tryptic immunoglobulin G (IgG) glycopeptides [69]. Dimethylamidation with 1-ethyl-3-(3-dimethylamino) propyl carbodiimide (EDC) and the catalyst 1-hydroxybenzotriazole (HOBt) was used to selectively react with a carboxyl group in both glycan and peptide moieties. The differential derivatization method also resulted in a mass difference for α2,3- and α2,6-linked sialic acids linkages (Figure 2). The α2,3-linked sialic acids underwent lactonization resulting in an 18 Da loss, while α2,6-linkages were dimethylamidated, prompting a 27 Da gain. This mass difference facilitated the direct differentiation of sialic acid linkages of glycopeptides. This fast and relatively inexpensive approach stabilizes sialylated glycopeptides, thus facilitating MALDI-TOF-MS analysis of complex IgG samples. However, the dimethylamidation reaction only applied to α2,3-linked and α2,6-linked N-acetylneuraminic acid and N-glycolylneuraminic acid associated with IgG glycopeptides. This reaction suitability for α2,8-linked sialic acids was not assessed or investigated.
Figure 2.

MALDI-TOF-MS analyses of sialylated glycopeptides by dimethylamine reaction under EDC and HOBt at 60° C for 3 hours. Reprinted with permission from [69].
Nishikaze et al. [70] also presented an approach which used (7azabenzotriazol-1-yloxy) tripyrrolidinophosphonium hexafluorophosphate (PyAOP) as a condensing reagent to derivatize all carboxyl groups in both sialic acid and peptides through methylamidation. PyAOP completely converts both α2,3- and α2,6-linked sialic acids into methylamine forms with no byproducts [71]. Antenna-specific D ions from 6-antenna and E ions from 3-antenna of glycan structure were observed in negative-ion mode CID of the derivatized glycopeptides, which facilitated the in-depth characterization of glycan moieties. Also, methylamine derivatization of glycopeptide prohibited sialic acid loss during MS-based analysis. However, after amidation reaction, excess derivatization reagents remained in the sample, which would interfere with the detection of low abundant glycopeptides. Extra HILIC-based cleanup or enrichment steps were required after methylamine derivatization [70].
3.2. Esterification of sialylated glycopeptides
Beside amidation, esterification is a common derivatization technique used in glycomics field [72, 73], which neutralizes carboxyl groups in sialic acids and improves sialylation quantitation by stabilizing sialic acid moieties and enhancing ionization efficiencies. Esterification is also capable of distinguishing sialic acid linkage isomers without separation techniques.
Similar to the aforementioned amidation derivatizations, esterification also differentially reacts with α2,3 and α2,6-linked sialic acids, which creates a discrepancy in molecular mass by forming intramolecular lactone and esterification products. This strategy has been used for the characterization of the glycosylation of Fc fragment derived from IgG1 [74]. A synthesized peptide that has the same amino acid sequence was also used to study the peptide backbone modification under esterification conditions mentioned in this study. However, it is hard to achieve complete derivatization of α2,3 linked species because the reactivity of carboxyl group in α2,3 linked sialic acid is lower than that of α2,6-linked sialic acid [75]. Therefore, this approach is considered only appropriate for mono- and disialylated glycopeptides. Complex samples that contain tri- and tetra-antennary sialic acid linkages would introduce multiple products such as different level of lactone and esterification formations.
Recently, a solid phase derivatization method to sequentially perform esterification and amidation on immobilized sialoglycopeptides with specific sialic acid linkages was introduced by Yang et al. [76]. Ethyl esterification of α2,6-linked sialic acids was conducted with EDC and HOBt before the carbodiimide coupling reaction of α2,3-linkages sialic acids using ethylenediamine (EDA) and EDC under acidic conditions. The derivatized sialoglycopeptides were further enriched by HILIC solid phase extraction and analyzed by C18-LC-MS/MS. The α2,3 and α2,6-linked sialic acids can be easily distinguished by the mass difference (28.03 Da and 42.06 Da shift, respectively), which potentially provides a better understanding of the biological roles of sialic acid isomers. Moreover, introducing additional amine group by EDA enabled better glycopeptide HILIC enrichment and improved electrospray ionization. Combining both labeling approaches could efficiently stabilize sialic acids. Esterification would predominantly derivatize α2,6-linkages while EDA amidation would derivatize α2,3-linkages. Therefore, two different sialylation linkages are sequentially and completely derivatized with different mass tags. This method allows the analysis of complex sialoglycopeptides containing mono, di, tri- and tetra-sialylated glycan.
3.3. Permethylation of glycopeptides
Permethylation, which substitutes every free hydroxyl and amino groups of glycans and peptide backbone with methoxy groups, offers several advantages, including increased detection sensitivity and eliminiated/reduced in-source fragmentation. Recently, a comprehensive method that permethylated glycopeptides derived from protease digestion was reported [77]. High-resolution tandem-MSn experiments characterized glycopeptides from different standard glycoproteins. By isolating permethylated backbone fragment ions in MS2, the peptide sequences were characterized via MS3. Topological isomers of the permethylated Man5 glycopeptide (Figure 3(A)) and sialic acid linkages isomers (Figure 3(B)) were identified by diagnostic peaks in the MSn spectra. To achieve positive glycosylation site identification and straightforward data interpretation, this approach requires glycopeptide limited to 3-6 amino acid residues. Therefore, non-specific protease digestion using pronase is needed. However, this step introduces variability originating from the nonspecific pronase digestion that could potentially produce multiple glycopeptides representing the same glycosylation sites but with different peptide backbones. This multi-glycopeptide generation adversely influences the sensitivity of the approach.
Figure 3.

MSn analyses of structure and linkage isomers of permethylated glycopeptides using LTQ Orbitrap Fusion Tribrid mass spectrometer. (A) MSn identification of permethylated structural isomers among the high mannose glycoforms of RNase B. (B) MSn identification of permethylated sialylated α 2,3 and α 2,6-linkage isomers of same N-glycopeptide from fetuin sample. α 2,3-linkage of O-glycan was also verified. Reprinted with permission from [77].
3.4. Derivatization of glycopeptides using isobaric tags
Two commercially available isobaric stable isotope tags, namely isobaric tags for relative and absolute quantification (iTRAQ) and tandem mass tags (TMT), has been developed for relative quantification of proteins. However, those two tags could also be employed to tag glycopeptides. Peptides or glycopeptides are typically labeled with iTRAQ or TMT reagents to achieve quantification with advantages such as quantitative reliability, improving quantitation analytical efficiency, and time-saving.
iTRAQ is first reported by Shah et al. [78] to investigate the glycoprotein profiles that were related to protein abundance, glycosylation occupancy, and heterogeneity at specific glycosylation sites from two different cell lines. In this method, one aliquot of digested glycopeptides and peptides labeled with 4plex iTRAQ reagents were captured using solid-phase extraction of glycosylated peptides (SPEG) [59]. The other aliquot underwent basic reverse phase liquid chromatography separation and analysis of glycopeptides. LC-MS/MS analysis combined with 4plex iTRAQ labeling allows simultaneous identification and quantification of glycopeptides from up to 4 samples. Combining the “omics” approach with iTRAQ labeling allowed the detection of 1810 N-linked glycosylated peptides and 1145 iTRAQ labeled N-linked glycopeptides derived from 653 glycoproteins. However, extra purification steps are needed to remove excess iTRAQ reagents before LC-MS/MS analysis which could result in excessive sample loss, thus potentially leading to glycopeptides quantitation confidence decrease.
TMT is also employed in glycoproteomic studies. TMT labels the lysine residue, and the N-terminal of the glycopeptides can generate specific fragment ions in MS/MS that help to confirm the analytical reproducibility of this approach. Recently, Ye et al. [79] described an improved TMT labeling strategy for glycoproteomics. This strategy enabled a higher labeling efficiency without acetone precipitation and the direct glycopeptides characterization and quantitation coupled with different dissociation techniques. Moreover, 10plex TMT was used to evaluate changes in fucose-containing glycosylation patterns in prostate cancer cell lines. The core fucosylation glycopeptides were enriched using Lens culinaris agglutinin lectin and then labeled with 10plex TMT before separation on hydrophilic-based strong anion exchange column and MS/MS identification [80]. After enrichment, 973 fucosylated glycopeptides from 252 fucosylated proteins in total were identified and quantified by this approach. 10plex TMT labeling technique, which enables simultaneous quantification of up to 10 different samples, offers higher throughput than iTRAQ as shown above.
3.5. Derivatization of glycopeptides using isotope-coded affinity tags
A new method was reported to achieve simultaneous analysis and the direct combination of two glycopeptide profiles in a single MS scan by using differential isotope labeling for glycopeptides [81]. Succinic anhydride and its natural isotope form D413C4 (8 Da difference) labeled succinic anhydride can react with ε-amino groups of lysine of glycopeptide without introducing bias to the glycopeptide profile and glycan portion. Both light and heavy labeled glycopeptides were analyzed by MALDI-MS and nano-LC-ESI-MS. Bias from sample preparation, ionization processes, as well as MALDI spot crystallization, were significantly reduced. The isotope labeling method was used to assess fucosylated glycopeptides derived from human IgG1 concerning their galactosylation and sialylation changes. Galactosylation differences were observed in both MALDI-MS and nano-LC-ESI-MS, but slightly sialylation differences were only observed in nano-LC-ESI-MS maybe because of the sialylated glycans are not easily detected in positive ion-mode MALDI-MS analysis.
Kurogochi and Amano [82] developed an approach that used benzoic acid-d0 N-succinimidyl ester (BzOSu) and benzoic acid-d5 N-succinimidyl ester (d-BzOSu) as light and heavy isotope reagents for stable isotope labeling of glycopeptides combined with MALDI-TOF MS. The detection limit of glycopeptides derived from human IgG, egg yolk, and bovine ribonuclease B was approximately 2 fmol. Moreover, this technique distinguishes glycan branch isomers by combining specific endo-β-N-acetyl glucosaminidases with MS/MS profiles of x, y, a, and b-type fragmentations in both positive and negative modes. This approach was shown to enable the efficient characterization of IgG glycopeptides derived from human serum. Although BzOSu and d-BzOSu labeling tags can be used to analyze only two samples at a time, they are compatible with other isotopic reagents such as TMT for high sensitive multiplex quantification of glycopeptides.
4. Advances in fragmentation methods for characterization of glycopeptides and glycoproteins
Although advances in separation techniques and high-resolution mass analyzers facilitate differentiation of glycopeptides, the assignment of glycopeptides based on merely m/z values is ambiguous and can be insufficient for the analysis of complex biological samples [83]. In the attempt to improve the ability of profiling glycopeptides by MS, various fragmentation methods have been employed to reveal the protein site-specific glycosylation. In this section, recent development in tandem MS strategies is presented and discussed.
4.1. Collisional dissociation methods
Collisional dissociation is a traditional mean of ion dissociation, which is implemented in almost every type of commercial MS, such as triple quadrupole, quadrupole TOF (Q-TOF), Fourier transform-ion cyclotron resonance (FT-ICR) and linear ion trap-orbitrap (LTQ-Orbitrap). The advances of collisional dissociation in the field of glycoproteomics center on generating informative spectra for the characterization of the glycosylation sites.
4.1.1. Collision-induced dissociation
Collision-induced dissociation (CID) can be subdivided into ion trap collision-induced/activated dissociation (ion trap CID) and beam-type CID [84]. Ion trap CID requires relatively lower activation energy and longer activation time, while beam-type CID occurs in a shorter time with higher energy applied [84]. Both cases involve the collisions of selected precursor ions with neutral gas molecules to generate fragment ions [85]. In a typical CID spectrum, major peaks are B-type, and Y-ions are resulting from the cleavages of glycosidic bonds, while fragmentation of peptide backbones is rarely observed [86]. Herein, CID can readily determine the glycan structure information. An issue associated with CID is monosaccharide rearrangements [87, 88] since the less stable protonated species instead of glycopeptides carrying sodium adducts are selected for fragmentation [89]. So far, both fucose and hexose rearrangements have been observed in the MS/MS spectra of glycopeptides [89]. To prevent misinterpretation, this phenomenon is worth being considered during the interpretation of glycopeptide spectra. Although CID is a useful tool for structural elucidation of glycan moieties, this technique alone is not sufficient for the glycopeptide identification due to the lack of adequate peptide backbone fragmentation.
Nozzle-skimmer dissociation (NSD) [90], or in-source CID [91] is a particular type of CID taking place in the nozzle-skimmer region of the electrospray ionization (ESI) source. Although NSD is mainly applied for proteins, Mechref and coworkers [67] have successfully adopted NSD to glycopeptide identification, allowing the acquisition of peptide sequence and glycan composition in a single run. More recently, stepped CID/HCD where precursor ions are subjected to multiple collisional conditions within a single MS analysis are explored to improve the number of N- and O-linked glycopeptides identified [92, 93]. With elevated collisional energy, both glycan and peptide sequence information can be captured in a single spectrum [94].
4.1.2. Higher-energy collisional dissociation
Glycan oxonium ions (e.g., HexNAc at m/z 204.0867; NeuAc at m/z 292.1027, and HexNAc1Hex1 at m/z 366.1395) are fragments originated from glycans, therefore indicating the presence of glycopeptides. However, due to the 1/3 cut-off associated with an ion trap, glycan oxonium ions are not detected in CID spectra when m/z values of glycopeptide precursors are high. The limitation was vanquished by using higher-energy collisional dissociation (HCD) [58, 95], which is also known as higher-energy C-trap dissociation [95]. In the case of HCD, fragment ions are detected in orbitrap following dissociation of precursors in C-trap. Additionally, an acquisition strategy was developed based on the presence of the oxonium ions. The top intense precursor ions are subjected to the HCD fragmentation first. The subsequent MS/MS events (e.g., electron transfer dissociation, ETD) [96] are only triggered when the oxonium ions are detected in the HCD. The implementation of this strategy improves the dynamic range and duty cycle of the instrument [97].
4.2. Electron capture dissociation and electron transfer dissociation
Two electron-based dissociation techniques, namely electron capture dissociation (ECD) and electron transfer dissociation (ETD), are proven to be of great significance in glycoproteomics [83]. The formation of c- and z-ions from multiply charged proteins by capturing low-energy electrons were first observed by McLafferty and coworkers [98]. Dissociation based on the ion/ion reaction was extended by Hunt and coworkers [99] by introducing ETD. In ETD, the dissociation is prompted by the electron transferring from anions to the precursor ions [99]. Both ECD and ETD feature the fragmentation of peptide backbones, without significant loss of glycans or other modifications. Herein, ECD/ETD and CID/HCD are complementary techniques and are commonly employed in MS-based glycoproteomics.
Although ECD and ETD exhibit advantages for identification of glycosylation sites, their application is hampered by low fragmentation efficiency and limit useful m/z range of precursors [100]. Efforts have been made to master these issues. Zaia and coworkers [101] employed hot electron capture dissociation (hECD) to acquire more useful information for middle-down glycoproteomics by using increased electron energy. In ETD, a significant portion of precursors are not excited or undergo nondissociative electron transfer (ETnoD), which is caused by the fact that product-ions of peptide backbone dissociation are attached by noncovalent interactions [102, 103]. Coon and coworkers [102] adopted activated ion-electron transfer dissociation (AI-ETD) to improve the fragmentation efficiency by infrared photon activation of the precursors undergoing ETD. In more recent studies, they proved the ability of AI-ETD being used to characterize phosphorylation at both peptide level [104] and intact protein level [105]. They also highlighted the potential of AI-ETD being used in glycoproteomics study [104]. Costello and coworkers [86] studied the fragmentation patterns of a glycoprotein and its non-glycosylated analog. Individual vibrational (CID/IRMPD), radical (ECD/ETD) and combined activation techniques were compared. Among different fragmentation modes, CID pre-activated prior to ETD or ECD was observed to slightly increase the sequence coverage. Additionally, the limitation in fragmentation efficiency was also overcome by performing subsequent CID on charge reduced and unreacted species after ET excitation [106, 107].
4.3. Photon-induced dissociation
The internal energy required for dissociation of the glycopeptide precursors can be attained by absorbing photons. Infrared multiphoton dissociation (IRMPD) is a photon-based and vibrational dissociation technique, using a CO2 laser at a wavelength of 10.6 μm [85]. Given the low energy provided by each photon (hυ = 0.12 eV), multiple photons must be absorbed to accumulate sufficient internal energy for dissociation [108]. Similar to CID, labile glycosidic bonds rather than peptide bonds are preferentially cleaved in IRMPD [109]. Hence, IPMPD is also a useful tool for glycan structure elucidation. In addition to functioning as a complementary technique of other dissociation methods, infrared irritation was also used to assist ETD and ECD in a way where ions are activated by IR photons before ETD/ECD events to improve the peptide sequence coverage as described in section 4.2.
Ultraviolet photodissociation (UVPD) is a newly emerging method for the characterization of protein glycosylation. In contrast with IR photon, UV photons consist of high energies (e.g., 193 nm; 6.4 eV). Therefore, absorption of a single UV photon can excite the precursor ion to an electronic state that allows fragmentation via a higher energy pathway [108]. Reilly and Zhang [110] have employed 157 nm photodissociation to singly charged N-linked glycopeptide, obtaining informative spectra qualified for both glycan and peptide structure elucidation. More recently, the use of UVPD at 193 nm on O-linked glycopeptides (UVPD data shown in Figure 4) was shown by Brodbelt and coworkers [111]. UVPD generated a-/x- ions that contain intact glycans, allowing peptide sequencing. At the same time, fragment ions corresponding to glycosidic bond cleavages, as well as some cross-ring fragmentation, were observed, thus enabling glycan structural characterization [111]. They also presented using UVPD in a middle-down strategy to characterize a therapeutic monoclonal antibody [112]. In short, the ability to generate various types of fragment ions makes UVPD a new tool for glycosylation characterization. Although the UVPD technique enables intact glycoprotein analysis, the wide application requires the support of a bioinformatics platform to efficiently aid in deciphering the complex spectra from middle down and top-down analyses. Although several efforts are focused on developing such platforms, they are not yet fully developed.
Figure 4.

UVPD of a doubly deprotonated O-linked glycopeptide anion from kappa-casein: (A) UVPD spectrum; (B) zoom-in of low m/z regions of spectrum shown in A; (C) Zoom-in of high m/z range of UVPD spectrum shown in A. Reprinted with permission from [111].
4.4. Combined electron-transfer and higher-energy collision dissociation
As aforementioned, various combinations of tandem mass spectrometry techniques are widely employed in glycoproteomic studies. Different dissociation experiments can be conducted consecutively, resulting in separate spectra for corresponding fragmentation events. Another option is to combine different fragmentation techniques and generate a single spectrum for a given glycopeptide. One recently developed technique combined electron-transfer and higher-energy collision dissociation (EThcD) [113]. In EThcD, all ions including fragments, charge reduced and unreacted precursor ions after ET excitation are further subjected to HCD, resulting in b/y- and c/z-type fragment ions in a single spectrum [113]. EThcD was initially developed to achieve comprehensive peptide characterization, while the informative nature of EThcD allows it to be employed in glycoproteomic studies. More recently, a workflow highlighting redefined EThcD method was presented by Li and coworkers [114]. HCD was performed following ETD while the fragments were acquired in a single spectrum. Also, the effective speed was improved by reducing the ETD reaction time, and large-scale N-glycopeptide characterization was achieved as a result [114]. Additionally, Heck and coworkers [115] characterized recombinant therapeutic human erythropoietin at both intact glycoprotein and glycopeptide level by CID and EThcD. Moreover, the used of EThcD enabled the identification of O-linked glycopeptides associated with one or even multiple glycans, thus enabling O-glycoproteome profiling of human serum [116].
5. Characterization of glycopeptides using ion mobility spectroscopy
Ion mobility spectroscopy (IMS) separates analyte ions based on their mobilities, which are dictated by the shape and charge of the ions. In general, ionized molecules are introduced to a drift tube which consists of buffer gas in a weak electric field. The separation is driven by the electric field and interaction with the buffer gas [117]. Dodds and coworkers [106] reported using IMS to separate ions after extraction of N-linked glycopeptide ions, then resolved unreacted, and charge-reduced ions were subjected to CID and ETD, respectively. The addition of IM dimension not only simplified data interpretation by dispersing CID and ETD spectrum (data are shown in Figure 5), but also fully utilized the large fraction of precursor ions that fail to undergo ET reaction [106]. By combining IMS and CID techniques, Clemmer and coworkers [118] revealed the microheterogeneity of glycosylation sites of glycopeptides derived from chicken ovomucoid, which has 5 glycosylation sites. A significant outcome of this study is that the glycosylation pattern of a site can be observed from the IMS-MS data, allowing easier evaluation of the microheterogeneity and more confident glycan structural elucidation.
Figure 5.

Electron transfer MS analysis coupled with Ion mobility and vibrational activation (ET-IM-VA) of the N-glycopeptide derived from coral tree lectin. The IM-MS heat map is shown in (A). The CID spectrum is given in (B), while the ETD spectrum extracted is given in (C). Experimental sequence is summarized in insets in A-C. Fragmentation behaviors of the glycopeptide in ETD and CID are depicted in (D). Reprinted with permission from [106].
Liquid chromatography (LC) and capillary electrophoresis (CE) are frequently used separation techniques in conjunction with MS; however, separation of isomeric glycopeptides remains a challenge in current workflows. IMS has been of great interest due to its potential in separating isomeric glycopeptides. Examples include separation of O-glycopeptides that differs in monosaccharides which occupy the sites (GlcNAc or GalNAc) [119] and sites of glycosylation [34]. For N-glycopeptides, separation of linkage isomers using IMS was proved in several recent studies [118–120]. Overall, IMS adds additional structural information to the traditional MS platform, and thus holds potentials for the characterization of glycopeptides. The broad application of IMS in separating isomeric glycopeptides is limited by difficulties originating from the lack of standards and the presence of various endogenous isomers associated with the inherent complexity of glycopeptide structures.
6. Metabolic labeling of the glycoprotein to facilitate enrichment and quantitation
More accurate and sensitive quantitation of glycoproteins or glycopeptides has always been challenging in the field of glycoproteomics. Chemical derivatization techniques have been utilized successfully in glycoproteomic quantitation. However, these strategies were performed on the glycoprotein level or after cell/tissue lysis, which introduces the deviations during sample preparation. Also, chemical labeling cannot reduce the variation caused by individual differences among biological samples [121]. Thus, metabolic labeling has been widely used in cell line glycoproteomics research. These metabolic labels can be coupled to any techniques such as enrichment, derivatization, or characterization strategies, as previously discussed, to acquire a better glycoproteomic quantitation in living systems.
6.1. Stable isotope labeling by amino acids in cell culture
A common metabolic labeling method is stable isotope labeling by amino acids in cell culture (SILAC). SILAC was first introduced by Mann and coworkers [122] in 2002 using deuterated leucine (Leu-D3). Although the first use of SILAC was for a proteomic purpose, it has been expanded to glycoproteomic researches [123–125]. However, labeling in only one amino acid (such as Leu-D3) cannot ensure that all glycopeptides have been labeled after enzymatic digestion (e.g., tryptic digestion), which may decrease the labeling coverage. Recently, a better SILAC method was introduced, in which 13C-15N-lysine and 13C-15N-arginine were used instead of Leu-D3 [126].
Recently, Mann and coworkers [124] reported the successful use of SILAC in the investigation of glycopeptides derived from diffused large B-cell lymphoma subtypes in conjunction with Lectin enrichment. Furthermore, Ji et al. [127] applied SILAC to investigate the glycoproteomic alterations in doxorubicin sensitive and resistant ovarian cancer cells for a better understanding of multidrug resistance of ovarian cancer. 13C6-15N4-arginine and 13C6-15N2-lysine were used to achieve “heavy” labeling of glycoproteins. LC-MS/MS allowed the identification of 1525 unique N-glycopeptides from 740 N-glycoproteins, and 253 N-glycopeptides exhibited significant alterations in the drug-resistant cells. Through comparing with proteomics results, 14 different occupancy and expression pattern changes of glycopeptides were observed. Although the labeling of SILAC is efficient and straightforward, this strategy does not allow subsequent bioorthogonal reactions, thus inhibiting the improvements of glycoprotein enrichment. Therefore, it is not widely applied in glycoproteomic research.
6.2. Metabolic labeling of glycans
Besides SILAC, another approach for glycoproteomic labeling is associated with unnatural monosaccharide incorporation. The conjunction of modified monosaccharides allows a bioorthogonal moiety that can be utilized for highly effective glycoprotein enrichment strategy [128]. Bertozzi and coworkers [129] first reported the incorporation of unnatural N-acetyl-mannosamine (ManNAc) in the cell culture, which eventually converted to sialic acid and incorporated ketone group in the cell surface through sialic acid biosynthesis pathway.
6.2.1. Metabolic labeling of glycoprotein using azido-sugar
During the development of glycan metabolic labeling, azide is considered to be a better reagent than ketones [130]. The limit of early sugar analog is their low cell membrane permeability which prevents the high efficiency of metabolic labeling. The use of these unnatural sugar analogs was then improved by per-acetylation with an acetyl group to increase their cellular uptake [131–133]. Also, other azido sugar analogs, including GalNAz [134, 135], FucNAz [136, 137] and GlcNAz [138, 139], have been utilized to incorporate azido groups to different positions on the glycan structure, not only for sialic acid but also for other specific glycan studies. The favorable properties of azide-based cell metabolic labeling were not fully realized until a satisfactory functional partner – Staudinger ligation - was developed by Saxon and Bertozzi in 2000 [132]. However, the main disadvantage of this method is still the relatively low reaction efficiency due to the poor solubility of the functional group of the Staudinger ligation reagent. Also, the reagent is susceptible to oxidation, thus limiting the reagent shelf-life.
6.2.2. Metabolic labeling of glycoprotein using click chemistry
An alternative efficient bioorthogonal approach coupled with azido-glycan metabolic labeling is known as click chemistry. In 2003, Finn and coworkers [140] reported the potential of using copper-catalyzed cycloaddition reaction as a general bioconjugation method. This method resulted in a rapid and reliable covalent attachment of azides and alkynes, which is now known as copper-catalyzed azide-alkyne 1,3-dipolar cycloaddition (CuAAC). Since then, CuAAC was commonly used in the glycoproteomic field.
Recently, Bai and coworkers [141] reported the metabolic labeling using AC4ManNAz coupled to CuAAC on HeLa, A549, and SW1990 cells, and identified 56 cell surface glycoproteins. Moreover, Bertozzi and coworkers [142] introduced a novel isotope targeted glycoproteomics (IsoTaG) strategy for N- and O-sialoglycopeptide analysis. The authors described the design of an IsoTaG-compatible azide probe which could achieve analysis of alkyne-labeled glycopeptides. Two pairs of azide-alkynyl reagents were introduced. Either of them could be used for IsoTaG strategy. This strategy was performed on PC-3 cells using LC-MS/MS and allowed the identification of 699 glycopeptides from 192 glycoproteins, including 8 sialylated glycan structures across 126 N-glycopeptides and 576 O-glycopeptides. However, the use of CuAAC in a cell or living organism glycoproteomics were hampered due to the toxicity of Cu(I), which is necessary for this reaction, against both bacteria and mammalian cells [130, 143].
6.2.3. Metabolic labeling of glycoprotein using copper-free click chemistry
One solution of CuAAC for living organisms is copper-free click chemistry. Bertozzi and coworkers [144] reported the synthesis of a biotin-conjugated cyclooctyne reagent, which was well known as “Strain-Promoted [3 + 2] Azide-Alkyne Cycloaddition”. The reported cycloaddition between cyclooctynes and azides could be performed under physiological conditions without copper. This method was then investigated using Jurkat cells and no apparent toxicity was observed. Since no copper is needed in this kind of method, it is called “copper-free click chemistry” [145]. In the following studies, Bertozzi and coworkers [146–148] reported several modifications of the first-generation strain-promoted cycloaddition to improve its reaction efficiency. Boons and coworkers [149] introduced the novel 4-Dibenzocyclooctynol derivatives as a copper-free click chemistry reagent.
According to these early works, copper-free click chemistry coupled to azide metabolic labeling has become a powerful tool for glycoproteomic studies in living organisms. New reagents for this purpose have been commercialized and can be purchased through many vendors such as Thermo Fisher Scientific and Sigma-Aldrich (detailed information can be accessed via their website). These commercialized reagents or even click chemistry kits significantly prompted the development of glycoproteomic research.
Recently, Drake and coworkers [150] investigated glycoproteins in WPMY-1 and HS5 prostate stromal cell secretome. AC4ManNAz were used to label secretome, glycoproteins and Click-iT™ Protein Analysis Detection Kit (Thermo Fisher Scientific, #C33372) was used to achieve copper-free click chemistry. Enriched glycoproteins were experienced on-beads-tryptic digestion and analyzed using tandem MS, allowing the identification of over 100 secreted glycoproteins. In the same study, GalNAz labeling was also estimated using the same strategy and proved to be comparable with ManNAz labeling.
Wu and coworkers [151] introduced a strategy for cell surface sialoglycoproteins study. The principle of this strategy is summarized in Figure 6 (A). DBCO-sulfo-biotin was used for copper-free click chemistry. The fact that DBCO-sulfo-biotin could not pass through cell member resulted in the labeling of only cell surface glycoproteins. This site-specific sialoglycoproteomics strategy was initially tested on HEK293T cells and then applied to breast cancer cell line MDA-MB-231 and MCF-7 to investigate the correlation between cell surface sialoglycoprotein and breast cancer invasion. Figure 6B depicts the strategy applied to the two cancer cell lines. The cell surface N-sialoglycosylation site-specific strategy allowed the identification of 439 and 237 glycosylation sites for MDA-MB-231 and MCF-7, respectively (Figure 6C), corresponding to 274 N-sialoglycoproteins identified (Figure 6D). Among the 274 glycoproteins, more than 50% were type I membrane proteins. The transmembrane domain analysis suggested that all the glycosylation sites identified were in the extracellular space, as shown in Figure 6E.
Figure 6.

(A) The principle of the site-specific identification of the cell surface N sialoglycoproteome by integrating metabolic labeling, copper-free click chemistry and MS-based proteomics techniques. (B) Experimental procedure of cell surface N-sialoglycoproteome site-specific identification on breast cancer cell line MDA-MB-231 and MCF-7. (C) Comparison of cell surface N-sialoglycosylation sites between two cell lines. (D) Cell surface N-sialoglycoproteins identified in two cell lines. (E) Glycosylation site location of the type I and II N-sialoglycoproteins. Reprinted with permission from [151].
Recently, azido-based metabolic labeling coupled to copper-free click chemistry has become the most common cell surface labeling strategy due to its high labeling efficiency and low toxicity to living systems. The typical protocol of this strategy involves (i) the use of per-acetylated unnatural sugar analogs such as AC4ManNAz and AC4GlcNAz to incorporate azido group into glycans; (ii) the cycloaddition reaction between azido-glycans and modified cyclooctyne reagent that usually contains bio-functional group such as biotin; and (iii) specific enrichment based on corresponding bio-functional group (e.g., use avidin beads to enrich biotin ligated glycans). Despite the advantages of this strategy, metabolic labeling can only be achieved in living systems such as cell lines and mice, which limits its use on clinical, biological specimens such as serum and tissue. Moreover, the incorporation efficiency of unnatural sugars is still inadequate, and the metabolism influence caused by unnatural sugars is not effffectively addressed.
7. Bioinformatics development in glycoproteomics data processing
Due to the time consuming and tedious interpretation of MS output data related to glycan structure and glycosylation site identification, there is a demand for the development of bioinformatics tools that would facilitate a rapid and accurate interpretation of glycoproteomics data [152–153]. Although commercial software such as SimGlycan® (SCIEX), MassyLynx™ (Waters) and Bionyc™ (Protein Metrics) have been available for glycoproteomics analysis, highly competitive and legitimate open-source software is still being developed since manual data validation is still needed as most commercial tools are returning high false discovery rates. Even though there have been several types of bioinformatics software, specifically in glycoproteomics [109, 154, 155], we provide here a brief overview of the recent efforts devoted to the development of open-source glycobioinformatics tools. These recently developed bioinformatics tools are summarized in Table 1.
Table 1.
Recent developed bioinformatic tools for glycoproteomics analysis
| Software Name | Software Validation Samples | Functionality | FDR Available | Ref. |
|---|---|---|---|---|
| ArMone 2.0 | Proteins from HEK293T, Fetuin from fetal calf serum | Qualitative | No | [156] |
| GlycoPep Evaluator | Bovine fetuin, Ribonuclease B, Human IgG, Transferrin, Alpha-1-acid glycoprotein, HIV envelope protein | Qualitative | Yes | [157] |
| GlycoMaster DB | Ribonuclease B, Human IgG, Human urine | Qualitative | Yes | [158] |
| MAGIC | Horseradish peroxidase, Fetuin, Chicken Ovalbumin, Lactotransferrin, Serotransferrin | Qualitative | Yes | [159] |
| MassyTools | IgG1-derived monoclonal antibody, Human plasmas | Qualitative/Quantitative | No | [167] |
| GPQuest | Prostate tumor LNCaP cells | Qualitative | Yes | [160] |
| GlycoMID | Bovine collagen α-(II) chain protein | Qualitative | Yes | [165] |
| GlycoSeq | Breast cancer cell lines | Qualitative | Yes | [161] |
| SweetNET | Human urine, Human cerebrospinal fluid | Qualitative | Yes | [166] |
| GlycoProteome Analyzer | Human plasmas, α1-acid glycoprotein | Qualitative/Quantitative | Yes | [162] |
| GlycoPAT | Fetuin, Fibronectin, Ribonuclease B, Human α1-acid glycoprotein | Qualitative | Yes | [163] |
| GlycoPep MassList | Fetuin from fetal bovine serum, Avidin from chicken egg white, Human IgG | Qualitative | No | [164] |
| MoFi | Ado-trastuzumab emtansine, Rituximab, Human erythropoietin | Qualitative | Yes | [168] |
Over the past few years, several bioinformatics tools were introduced and developed for the identification of N-glycopeptides and structures of attached N-glycan from proteomics samples, including ArMone 2.0 [156], GlycoPep Evaluator [157] , GlycoMaster DB [158], MAGIC [159], GPQuest [160], GlycoSeq [161], Integrated GlycoProteome Analyzer (I-GPA) [162], GlycoPAT [163], and GlycoPep MassList [164]. GlycoMID [165] was developed for the identification of O-linked glycosylation sites. On the other hand, SweetNET [166] bioinformatics workflow allows the analysis of both N- and O-linked glycosylation sites and also CS-glycopeptide data.
On the other hand, GlycoPAT [163] and GlycoPep MassList [164] utilize three fragmentation modes, including CID, HCD and ETD, to generate inclusion lists for targeted analysis of glycoproteins. However, GlycoMaster DB [158] is able to search peptide sequences and glycans database by using a combination of glycopeptide HCD and ETD tandem mass spectra. MAGIC [159] and GlycoSeq [161] have the ability to identify peptide sequences and glycan compositions from CID spectra. On the other hand, GlycoPep Evaluator [157] can generate decoy glycopeptide de novo for every target glycopeptide by scoring against the ETD data while GPQuest [160] can construct a library of deglycosylated peptides and classify glycopeptide by using HCD tandem mass spectra on the presence of oxonium ions.
While integrated GlycoProteome Analyzer (I-GPA) software [162], which was developed for qualification and quantitation of N-glycoproteomes, MassyTools [167] was developed to process glycan and glycopeptide data for quality control and spectra calibration as well as quantitation. This software performed data analysis and calibration significantly better and faster than the commercial flexAnalysis software. More recently, MoFi [168] was developed to characterize glycoproteins and changes in their proteome profiles. This software can be used for both top-down proteomics and biopharma characterization, providing a solution to the challenges of intact proteins MS and glycan analysis.
8. Concluding Remarks
This review surveys the recent MS-based methods facilitating the profiling of glycoproteins in biological samples. In the glycoproteomics study, low abundance glycopeptides are effectively enriched by the developed enrichment methods. The glycopeptide ionization efficiency and sensitivity of the glycopeptide profiling have been improved by chemical derivatization to stabilize glycopeptides and reduce variations for further MS/MS analyses. Moreover, different metabolic labeling techniques that are conjunct with other advanced enrichment and derivatization strategies facilitate accurate glycopeptide quantification. Indeed, these chemical-related advancements of glycoproteomics, the development on various fragmentation modes of mass spectrometry and the software tools that aim to obtain high throughput data acquisition efficiently address the difficulties existing in the glycopeptide analysis. The MS-based glycoproteomic study is expected to benefit from ongoing advancements in enrichment method, chemical derivatization, metabolic labeling in cell study, tandem MS fragmentation techniques and bioinformatics.
Acknowledgment
This work was supported by the grant from National Institutes of Health, NIH (1R01GM112490-04 and 1U01CA225753-01) and Cancer Prevention and Research Institute of Texas, CPRIT (RP130624).
Abbreviations:
- AI-ETD
activated ion-electron transfer dissociation
- BzOSu
benzoic acid-d0 N-succinimidyl ester
- CuAAC
copper-catalyzed azide–alkyne 1,3-dipolar cycloaddition
- d-BzOSu
benzoic acid-d5 N-succinimidyl ester
- ECD
electron capture dissociation
- EDA
ethylenediamine
- EDC
1-ethyl-3-(3-dimethylamino) propyl) carbodiimide
- ERLIC
electrostatic repulsion hydrophilic interaction chromatography
- ETD
electron transfer dissociation
- EThcD
electron-transfer/higher-energy collision dissociation
- ETnoD
nondissociative electron transfer
- FDR
false discovery rate
- HCD
high collision dissociation
- hECD
hot electron capture dissociation
- HOBt
1-hydroxybenzotriazole
- IM-MS
ion mobility-mass spectrometry
- IMS
Ion mobility spectroscopy
- IRMPD
Infrared multiphoton dissociation
- IsoTaG
isotope targeted glycoproteomics
- iTRAQ
isobaric tags for relative and absolute quantification
- MOFs
metal-organic frameworks
- PAMAM
polyamidoamine
- PyAOP
(7azabenzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate
- SILAC
stable isotope labeling by amino acids in cell culture
- SPEG
glycosite-containing peptides
- TMT
tandem mass tags
- UVPD
ultraviolet photodissociation
Footnotes
Reference
- [1].Dennis JW, Granovsky M, Warren CE, Bioessays 1999, 21, 412–421. [DOI] [PubMed] [Google Scholar]
- [2].Dwek RA, Chem. Rev 1996, 96, 683–720. [DOI] [PubMed] [Google Scholar]
- [3].Lowe JB, Marth JD, Annu. Rev. Biochem 2003, 72, 643–691. [DOI] [PubMed] [Google Scholar]
- [4].Banazadeh A, Peng W, Veillon L, Mechref Y, J. Am. Soc. Mass Spectrom 2018, doi: 10.1007/s13361-13018-11985-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Morelle W, Michalski J-C, Nat. Protoc 2007, 2, 1585–1602. [DOI] [PubMed] [Google Scholar]
- [6].Varki A, Gagneux P, Essentials of Glycobiology (Eds.: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, Seeberger PH), Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2015–2017. [PubMed] [Google Scholar]
- [7].Zhu R, Zhou S, Peng W, Huang Y, Mirzaei P, Donohoo K, Mechref Y, J. Proteome Res 2018, doi: 10.1021/acs.jproteome.1028b00127. [DOI] [PubMed] [Google Scholar]
- [8].Hart GW, Copeland RJ, Cell 2010, 143, 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Helenius A, Aebi M, Science 2001, 291, 2364–2369. [DOI] [PubMed] [Google Scholar]
- [10].Moremen KW, Tiemeyer M, Nairn AV, Nat. Rev. Mol. Cell Biol 2012, 13, 448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rudd PM, Woods RJ, Wormald MR, Opdenakker G, Downing AK, Campbell ID, Dwek RA, Biochim. Biophys. Acta 1995, 1248, 1–10. [DOI] [PubMed] [Google Scholar]
- [12].Rudd PM, Wormald MR, Stanfield RL, Huang M, Mattsson N, Speir JA, DiGennaro JA, Fetrow JS, Dwek RA, Wilson IA, J. Mol. Biol 1999, 293, 351–366. [DOI] [PubMed] [Google Scholar]
- [13].Varki A, Glycobiology 1993, 3, 97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Veillon L, Huang Y, Peng W, Dong X, Cho BG, Mechref Y, Electrophoresis 2017, 38, 2100–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wooding KM, Peng W, Mechref Y, Curr. Pharm. Biotechnol 2016, 17, 788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xia B, Royall JA, Damera G, Sachdev GP, Cummings RD, Glycobiology 2005, 15, 747–775. [DOI] [PubMed] [Google Scholar]
- [17].Chace KV, Flux M, Sachdev GP, Biochemistry 1985, 24, 7334–7341. [DOI] [PubMed] [Google Scholar]
- [18].Davril M, Degroote S, Humbert P, Galabert C, Dumur V, Lafitte J-J, Lamblin G, Roussel P, Glycobiology 1999, 9, 311–321. [DOI] [PubMed] [Google Scholar]
- [19].Mechref Y, Hu Y, Garcia A, Hussein A, Electrophoresis 2012, 33, 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fukui S, Feizi T, Galustian C, Lawson AM, Chai W, Nat. Biotechnol 2002, 20, 1011–1017. [DOI] [PubMed] [Google Scholar]
- [21].North SJ, Hitchen PG, Haslam SM, Dell A, Curr. Opin. Struct. Biol 2009, 19, 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yang Y, Franc V, Heck AJ, Trends Biotechnol 2017, 35, 598–609. [DOI] [PubMed] [Google Scholar]
- [23].Hitchen PG, Twigger K, Valiente E, Langdon RH, Wren BW, Dell A, Biochem. Soc. Trans 2010, 38, 1307–1313. [DOI] [PubMed] [Google Scholar]
- [24].Tholey A, Becker A, Biochim Biophys Acta. 2017, 1864, 2191–2199. [DOI] [PubMed] [Google Scholar]
- [25].Sun B, Ranish JA, Utleg AG, White JT, Yan X, Lin B, Hood L, Mol. Cell. Proteomics 2007, 6, 141–149. [DOI] [PubMed] [Google Scholar]
- [26].Resing KA, Ahn NG, FEBS Lett 2005, 579, 885–889. [DOI] [PubMed] [Google Scholar]
- [27].Stadlmann J, Pabst M, Kolarich D, Kunert R, Altmann F, Proteomics 2008, 8, 2858–2871. [DOI] [PubMed] [Google Scholar]
- [28].Belov AM, Viner R, Santos MR, Horn DM, Bern M, Karger BL, Ivanov AR, J. Am. Soc. Mass Spectrom 2017, 28, 2614–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Glaskin RS, Khatri K, Wang Q, Zaia J, Costello CE, Anal. Chem 2017, 89, 4452–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Selman MH, Hoffmann M, Zauner G, McDonnell LA, Balog CI, Rapp E, Deelder AM, Wuhrer M, Proteomics 2012, 12, 1337–1348. [DOI] [PubMed] [Google Scholar]
- [31].Whelan SA, Lu M, He J, Yan W, Saxton RE, Faull KF, Whitelegge JP, Chang HR, J. Proteome Res 2009, 8, 4151–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wada Y, Kadoya M, Okamoto N, Glycobiology 2012, 22, 1140–1144. [DOI] [PubMed] [Google Scholar]
- [33].Hua S, Nwosu CC, Strum JS, Seipert RR, An HJ, Zivkovic AM, German JB, Lebrilla CB, Anal. Bioanal. Chem 2012, 403, 1291–1302. [DOI] [PubMed] [Google Scholar]
- [34].Creese AJ, Cooper HJ, Anal. Chem 2012, 84, 2597–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li H, Bendiak B, Siems WF, Gang DR, Hill HH, Int. J. Ion. Mobil. Spectrom 2013, 16, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Takegawa Y, Deguchi K, Keira T, Ito H, Nakagawa H, Nishimura S-I, J. Chromatogr 2006, 1113, 177–181. [DOI] [PubMed] [Google Scholar]
- [37].Abbott KL, Pierce JM, Methods Enzymol 2010, 480, 461–476. [DOI] [PubMed] [Google Scholar]
- [38].Mechref Y, Madera M, Novotny MV, Methods Mol. Biol 2008, 424, 373–396. [DOI] [PubMed] [Google Scholar]
- [39].Chen M, Shi X, Duke RM, Ruse CI, Dai N, Taron CH, Samuelson JC, Nat. Commun 2017, 8, doi: 10.1038/ncomms15487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yang JS, Qiao J, Kim JY, Zhao L, Qi L, Moon MH, Anal. Chem 2018, 90, 3124–3131. [DOI] [PubMed] [Google Scholar]
- [41].Wen C, Cheng-Chao C, Gang L, J. WY-X, Small 2015, 11, 4806–4822.26193176 [Google Scholar]
- [42].Wu MX, Yang YW, Adv. Mater 2017, 29, doi: 10.1002/adma.201606134. [DOI] [PubMed] [Google Scholar]
- [43].Xie Y, Deng C, Sci. Rep 2017, 7, doi: 10.1038/s41598-41017-01341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ma W, Xu L, Li X, Shen S, Wu M, Bai Y, Liu H, ACS Appl. Mater. Interfaces 2017, 9, 19562–19568. [DOI] [PubMed] [Google Scholar]
- [45].Liu J, Yang K, Shao W, Li S, Wu Q, Zhang S, Qu Y, Zhang L, Zhang Y, ACS Appl. Mater. Interfaces 2016, 8, 22018–22024. [DOI] [PubMed] [Google Scholar]
- [46].Cao W, Huang J, Jiang B, Gao X, Yang P, Sci. Rep 2016, 6, doi: 10.1038/srep29776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cardoso VF, Francesko A, Ribeiro C, Banobre-Lopez M, Martins P, Lanceros-Mendez S, Adv. Healthc. Mater 2018, 7, doi: 10.1002/adhm.201700845. [DOI] [PubMed] [Google Scholar]
- [48].Gutierrez AM, Dziubla TD, Hilt JZ, Rev. Environ. Health 2017, 32, 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mehta RV, Mater. Sci. Eng. C Mater. Biol. Appl 2017, 79, 901–916. [DOI] [PubMed] [Google Scholar]
- [50].Wong J, Prout J, Seifalian A, Curr. Pharm. Des 2017, 23, 2908–2917. [DOI] [PubMed] [Google Scholar]
- [51].Wan H, Huang J, Liu Z, Li J, Zhang W, Zou H, Chem. Commun 2015, 51, 9391–9394. [DOI] [PubMed] [Google Scholar]
- [52].Sun N, Wang J, Yao J, Deng C, Anal. Chem 2017, 89, 1764–1771. [DOI] [PubMed] [Google Scholar]
- [53].Bi C, Zhao Y, Shen L, Zhang K, He X, Chen L, Zhang Y, ACS Appl. Mater. Interfaces 2015, 7, 24670–24678. [DOI] [PubMed] [Google Scholar]
- [54].Jiang B, Wu Q, Deng N, Chen Y, Zhang L, Liang Z, Zhang Y, Nanoscale 2016, 8, 4894–4897. [DOI] [PubMed] [Google Scholar]
- [55].Li Y, Wang J, Sun N, Deng CH, Anal. Chem 2017, 89, 11151–11158. [DOI] [PubMed] [Google Scholar]
- [56].Feng X, Deng C, Gao M, Yan G, Zhang X, Talanta 2018, 179, 377–385. [DOI] [PubMed] [Google Scholar]
- [57].Alpert AJ, Anal. Chem 2008, 80, 62–76. [DOI] [PubMed] [Google Scholar]
- [58].Zacharias LG, Hartmann AK, Song E, Zhao J, Zhu R, Mirzaei P, Mechref Y, J. Proteome Res 2016, 15, 3624–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhang H, Li XJ, Martin DB, Aebersold R, Nat. Biotechnol 2003, 21, 660–606. [DOI] [PubMed] [Google Scholar]
- [60].Chen CC, Su WC, Huang BY, Chen YJ, Tai HC, Obena RP, Analyst 2014, 139, 688–704. [DOI] [PubMed] [Google Scholar]
- [61].Ongay S, Boichenko A, Govorukhina N, Bischoff R, J. Sep. Sci 2012, 35, 2341–2372. [DOI] [PubMed] [Google Scholar]
- [62].Zhang L, Jiang H, Yao J, Wang Y, Fang C, Yang P, Lu H, Chem. Commun 2014, 50, 1027–1029. [DOI] [PubMed] [Google Scholar]
- [63].Liu L, Yu M, Zhang Y, Wang C, Lu H, ACS Appl Mater Interfaces. 2014, 6, 7823–7832. [DOI] [PubMed] [Google Scholar]
- [64].Ma R, Hu J, Cai Z, Ju H, Nanoscale 2014, 6, 3150–3156. [DOI] [PubMed] [Google Scholar]
- [65].Zhang X, Wang J, He X, Chen L, Zhang Y, ACS Appl. Mater. Interfaces 2015, 7, 24576–24584. [DOI] [PubMed] [Google Scholar]
- [66].Xie Y, Liu Q, Li Y, Deng C, J. Chromatogr. A 2018, 1540, 87–93. [DOI] [PubMed] [Google Scholar]
- [67].Zhao J, Song E, Zhu R, Mechref Y, Electrophoresis 2016, 37, 1420–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Varki A, Trends Mol. Med 2008, 14, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].de Haan N, Reiding KR, Haberger M, Reusch D, Falck D, Wuhrer M, Anal. Chem 2015, 87, 8284–8291. [DOI] [PubMed] [Google Scholar]
- [70].Nishikaze T, Kawabata S. i., Tanaka K, Anal. Chem 2014, 86, 5360–5369. [DOI] [PubMed] [Google Scholar]
- [71].Liu X, Qiu H, Lee RK, Chen W, Li J, Anal. Chem 2010, 82, 8300–8306. [DOI] [PubMed] [Google Scholar]
- [72].Wheeler SF, Domann P, Harvey DJ, Rapid Commun. Mass Spectrom 2009, 23, 303–312. [DOI] [PubMed] [Google Scholar]
- [73].Hansson GC, Bouhours JF, Karlsson H, Carlstedt I, Carbohydr. Res 1991, 221, 179–189. [DOI] [PubMed] [Google Scholar]
- [74].Gomes de Oliveira AG, Roy R, Raymond C, Bodnar ED, Tayi VS, Butler M, Durocher Y, Perreault H, Rapid Commun. Mass Spectrom 2015, 29, 1817–1826. [DOI] [PubMed] [Google Scholar]
- [75].Toyoda M, Ito H, Matsuno YK, Narimatsu H, Kameyama A, Anal. Chem 2008, 80, 5211–5218. [DOI] [PubMed] [Google Scholar]
- [76].Yang S, Wu WW, Shen R-F, Bern M, Cipollo J, J. Am. Soc. Mass Spectrom 2018, 29, 1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Shajahan A, Supekar NT, Heiss C, Ishihara M, Azadi P, Anal. Chem 2017, 89, 10734–10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Shah P, Wang X, Yang W, Eshghi ST, Sun S, Hoti N, Chen L, Yang S, Pasay J, Rubin A, Mol. Cell. Proteomics 2015, 14, 2753–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ye H, Boyne MT, Buhse LF, Hill J, Anal. Chem 2013, 85, 1531–1539. [DOI] [PubMed] [Google Scholar]
- [80].Zhou J, Yang W, Hu Y, Hoti N, Liu Y, Shah P, Sun S, Clark D, Thomas S, Zhang H, Anal. Chem 2017, 89, 7623–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Pabst M, Benesšovaá I, Fagerer SR, Jacobsen M, Eyer K, Schmidt G, Steinhoff R, Krismer J, Wahl F, Preisler J, J. Proteome Res 2015, 15, 326–331. [DOI] [PubMed] [Google Scholar]
- [82].Kurogochi M, Amano J, Molecules 2014, 19, 9944–9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Zhu ZK, Desaire H, Annu. Rev. Anal. Chem 2015, 8, 463–483. [DOI] [PubMed] [Google Scholar]
- [84].Frese CK, Altelaar AF, Hennrich ML, Nolting D, Zeller M, Griep-Raming J, Heck AJ, Mohammed S, J. Proteome Res 2011, 10, 2377–2388. [DOI] [PubMed] [Google Scholar]
- [85].Dodds ED, Mass Spectrom. Rev 2012, 31, 666–682. [DOI] [PubMed] [Google Scholar]
- [86].Bourgoin-Voillard S, Leymarie N, Costello CE, Proteomics 2014, 14, 1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wuhrer M, Deelder AM, van der Burgt YEM, Mass Spectrom. Rev 2011, 30, 664–680. [DOI] [PubMed] [Google Scholar]
- [88].Ruhaak LR, Xu G, Li Q, Goonatilleke E, Lebrilla CB, Chem. Rev 2018, doi: 10.1021/acs.chemrev.1027b00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wuhrer M, Koeleman CAM, Deelder AM, Anal. Chem 2009, 81, 4422–4432. [DOI] [PubMed] [Google Scholar]
- [90].Loo JA, Udseth HR, Smith RD, Futrell J, Rapid Commun. Mass Spectrom 1988, 2, 207–210. [Google Scholar]
- [91].Gabelica V, De Pauw E, Mass Spectrom. Rev 2005, 24, 566–587. [DOI] [PubMed] [Google Scholar]
- [92].Hinneburg H, Stavenhagen K, Schweiger-Hufnagel U, Pengelley S, Jabs W, Seeberger PH, Silva DV, Wuhrer M, Kolarich D, J. Am. Soc. Mass Spectrom 2016, 27, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Cao QC, Zhao XY, Zhao Q, Lv XD, Ma C, Li XY, Zhao Y, Peng B, Ying WT, Qian XH, Anal. Chem 2014, 86, 6804–6811. [DOI] [PubMed] [Google Scholar]
- [94].Kolli V, Dodds ED, Analyst 2014, 139, 2144–2153. [DOI] [PubMed] [Google Scholar]
- [95].Segu ZM, Mechref Y, Rapid Commun. Mass Spectrom 2010, 24, 1217–1225. [DOI] [PubMed] [Google Scholar]
- [96].Singh C, Zampronio CG, Creese AJ, Cooper HJ, J. Proteome Res 2012, 11, 4517–4525. [DOI] [PubMed] [Google Scholar]
- [97].Saba J, Dutta S, Hemenway E, Viner R, J. Proteomics 2012, doi: 10.1155/2012/560391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zubarev RA, Kelleher NL, McLafferty FW, J. Am. Chem. Soc 1998, 120, 3265–3266. [Google Scholar]
- [99].Coon JJ, Syka JEP, Schwartz JC, Shabanowitz J, Hunt DF, Int. J. Mass spectrom 2004, 236, 33–42. [Google Scholar]
- [100].Mechref Y, Curr. Protoc. Protein. Sci 2012, 68, 12.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Khatri K, Pu Y, Klein JA, Wei J, Costello CE, Lin C, Zaia J, J. Am. Soc. Mass Spectrom 2018, 29, 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Ledvina AR, McAlister GC, Gardner MW, Smith SI, Madsen JA, Schwartz JC, Stafford GC Jr., Syka JE, Brodbelt JS, Coon JJ, Angew. Chem. Int. Ed. Engl 2009, 48, 8526–8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Riley NM, Westphall MS, Coon JJ, J. Proteome Res 2017, 16, 2653–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Ledvina AR, Rose CM, McAlister GC, Syka JEP, Westphall MS, Griep-Raming J, Schwartz JC, Coon JJ, J. Am. Soc. Mass Spectrom 2013, 24, 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Riley NM, Hebert AS, Durnberger G, Stanek F, Mechtler K, Westphall MS, Coon JJ, Anal. Chem 2017, 89, 6367–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kolli V, Schumacher KN, Dodds ED, Analyst 2017, 142, 4691–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JEP, Coon JJ, Anal. Chem 2007, 79, 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Brodbelt JS, Chem. Soc. Rev 2014, 43, 2757–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Hu H, Khatri K, Zaia J, Mass Spectrom. Rev 2017, 36, 475–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Zhang L, Reilly JP, J. Proteome Res 2009, 8, 734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Madsen JA, Ko BJ, Xu H, Iwashkiw JA, Robotham SA, Shaw JB, Feldman MF, Brodbelt JS, Anal. Chem 2013, 85, 9253–9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Cotham VC, Brodbelt JS, Anal. Chem 2016, 88, 4004–4013. [DOI] [PubMed] [Google Scholar]
- [113].Frese CK, Altelaar AFM, van den Toorn H, Nolting D, Griep-Raming J, Heck AJR, Mohammed S, Anal. Chem 2012, 84, 9668–9673. [DOI] [PubMed] [Google Scholar]
- [114].Yu Q, Wang BW, Chen ZW, Urabe G, Glover MS, Shi XD, Guo LW, Kent KC, Li LJ, J. Am. Soc. Mass Spectrom 2017, 28, 1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Yang Y, Liu F, Franc V, Halim LA, Schellekens H, Heck AJR, Nat. Commun 2016, 7, doi: 10.1038/ncomms13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zhang Y, Xie X, Zhao X, Tian F, Lv J, Ying W, Qian X, J. Proteomics 2018, 170, 14–27. [DOI] [PubMed] [Google Scholar]
- [117].Bohrer BC, Mererbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE, Annu. Rev. Anal. Chem 2008, 1, 293–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Zhu FF, Trinidad JC, Clemmer DE, J. Am. Soc. Mass Spectrom 2015, 26, 1092–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Both P, Green AP, Gray CJ, Sardzik R, Voglmeir J, Fontana C, Austeri M, Rejzek M, Richardson D, Field RA, Widmalm G, Flitsch SL, Eyers CE, Nat. Chem 2014, 6, 368–368. [DOI] [PubMed] [Google Scholar]
- [120].Guttman M, Lee KK, Anal. Chem 2016, 88, 5212–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Walker SH, Taylor AD, Muddiman DC, J. Am. Soc. Mass Spectrom 2013, 24, 1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M, Mol. Cell. Proteomics 2002, 1, 376–386. [DOI] [PubMed] [Google Scholar]
- [123].Clark DJ, Mei Y, Sun S, Zhang H, Yang AJ, Mao L, Theranostics 2016, 6, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Deeb SJ, Cox J, Schmidt-Supprian M, Mann M, Mol. Cell. Proteomics 2014, 13, 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Taga Y, Kusubata M, Ogawa-Goto K, Hattori S, Proteome Res J. 2013, 12, 2225–2232. [DOI] [PubMed] [Google Scholar]
- [126].Ong SE, Mann M, Nat. Protoc 2006, 1, 2650–2660. [DOI] [PubMed] [Google Scholar]
- [127].Ji Y, Wei S, Hou J, Zhang C, Xue P, Wang J, Chen X, Guo X, Yang F, Oncotarget 2017, 8, 13413–13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Palaniappan KK, Bertozzi CR, Chem. Rev 2016, 116, 14277–14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Mahal LK, Yarema KJ, Bertozzi CR, Science 1997, 276, 1125–1128. [DOI] [PubMed] [Google Scholar]
- [130].Sletten EM, Bertozzi CR, Angew. Chem. Int. Ed 2009, 48, 6974–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Sarkar AK, Fritz TA, Taylor WH, Esko JD, Proc. Natl. Acad. Sci. U. S. A 1995, 92, 3323–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Saxon E, Bertozzi CR, Science 2000, 287, 2007–2010. [DOI] [PubMed] [Google Scholar]
- [133].Saxon E, Luchansky SJ, Hang HC, Yu C, Lee SC, Bertozzi CR, J. Am. Chem. Soc 2002, 124, 14893–14902. [DOI] [PubMed] [Google Scholar]
- [134].Hang HC, Yu C, Kato DL, Bertozzi CR, Proc. Natl. Acad. Sci. U. S. A 2003, 100, 14846–14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR, Proc. Natl. Acad. Sci. U. S. A 2011, 108, 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Rabuka D, Hubbard SC, Laughlin ST, Argade SP, Bertozzi CR, J. Am. Chem. Soc 2006, 128, 12078–12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Sawa M, Hsu TL, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong CH, Proc. Natl. Acad. Sci. U. S. A 2006, 103, 12371–12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR, Proc. Natl. Acad. Sci. U. S. A 2003, 100, 9116–9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Gurcel C, Vercoutter-Edouart AS, Fonbonne C, Mortuaire M, Salvador A, Michalski JC, Lemoine J, Anal. Bioanal. Chem 2008, 390, 2089–2097. [DOI] [PubMed] [Google Scholar]
- [140].Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG, J. Am. Chem. Soc 2003, 125, 3192–3193. [DOI] [PubMed] [Google Scholar]
- [141].Pan PW, Zhang Q, Hou J, Liu Z, Bai F, Cao MR, Sun T, Bai G, Anal. Bioanal. Chem 2012, 403, 1661–1670. [DOI] [PubMed] [Google Scholar]
- [142].Woo CM, Felix A, Zhang L, Elias JE, Bertozzi CR, Anal. Bioanal. Chem 2017, 409, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Link AJ, Tirrell DA, J. Am. Chem. Soc 2003, 125, 11164–11165. [DOI] [PubMed] [Google Scholar]
- [144].Agard NJ, Prescher JA, Bertozzi CR, J. Am. Chem. Soc 2004, 126, 15046–15047. [DOI] [PubMed] [Google Scholar]
- [145].Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR, Proc. Natl. Acad. Sci. U. S. A 2010, 107, 1821–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR, ACS Chem. Biol 2006, 1, 644–648. [DOI] [PubMed] [Google Scholar]
- [147].Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR, Proc. Natl. Acad. Sci. U. S. A 2007, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Sletten EM, Bertozzi CR, Org. Lett 2008, 10, 3097–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Ning X, Guo J, Wolfert MA, Boons GJ, Angew. Chem. Int. Ed 2008, 47, 2253–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Roper SM, Zemskova M, Neely BA, Martin A, Gao P, Jones EE, Kraft AS, Drake RR, Proteomics Clin. Appl 2013, 7, 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Chen W, Smeekens JM, Wu R, Chem. Sci 2015, 6, 4681–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Li F, Glinskii OV, Glinsky VV, Proteomics 2013, 13, 341–354. [DOI] [PubMed] [Google Scholar]
- [153].Banazadeh A, Veillon L, Wooding KM, Zabet-moghaddam M, Mechref Y, Electrophoresis 2017, 38, 162–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Tsai PL, Chen SF, Mass Spectrom (Tokyo) 2017, 6, doi: 10.5702/massspectrometry.S0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Dallas DC, Martin WF, Hua S, German JB, Brief Bioinform 2013, 14, 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Cheng K, Chen R, Seebun D, Ye M, Figeys D, Zou H, J. Proteomics 2014, 110, 145–154. [DOI] [PubMed] [Google Scholar]
- [157].Zhu Z, Su X, Go EP, Desaire H, Anal. Chem 2014, 86, 9212–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].He L, Xin L, Shan B, Lajoie GA, Ma B, J. Proteome Res 2014, 13, 3881–3895. [DOI] [PubMed] [Google Scholar]
- [159].Lynn KS, Chen CC, Lih TM, Cheng CW, Su WC, Chang CH, Cheng CY, Hsu WL, Chen YJ, Sung TY, Anal. Chem 2015, 87, 2466–2473. [DOI] [PubMed] [Google Scholar]
- [160].Toghi Eshghi S, Shah P, Yang W, Li X, Zhang H, Anal. Chem 2015, 87, 5181–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Yu CY, Mayampurath A, Zhu R, Zacharias L, Song E, Wang L, Mechref Y, Tang H, Anal. Chem 2016, 88, 5725–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Park GW, Kim JY, Hwang H, Lee JY, Ahn YH, Lee HK, Ji ES, Kim KH, Jeong HK, Yun KN, Kim YS, Ko JH, An HJ, Kim JH, Paik YK, Yoo JS, Sci. Rep 2016, 6, doi: 10.1038/srep21175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Liu G, Cheng K, Lo CY, Li J, Qu J, Neelamegham S, Mol. Cell. Proteomics 2017, 16, 2032–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Hu W, Su X, Zhu Z, Go EP, Desaire H, Anal. Bioanal. Chem 2017, 409, 561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Zhang Y, Yu CY, Song E, Li SC, Mechref Y, Tang H, Liu X, J. Proteome Res 2015, 14, 5099–5108. [DOI] [PubMed] [Google Scholar]
- [166].Nasir W, Toledo AG, Noborn F, Nilsson J, Wang M, Bandeira N, Larson G, J. Proteome Res 2016, 15, 2826–2840. [DOI] [PubMed] [Google Scholar]
- [167].Jansen BC, Reiding KR, Bondt A, Hipgrave Ederveen AL, Palmblad M, Falck D, Wuhrer M, J. Proteome Res 2015, 14, 5088–5098. [DOI] [PubMed] [Google Scholar]
- [168].Skala W, Wohlschlager T, Senn S, Huber GE, Huber CG, Anal. Chem 2018, 90, 5728–5736. [DOI] [PubMed] [Google Scholar]


