Abstract
More than 50% of all known proteins are glycosylated, which is critical for many biological processes such as protein folding and signal transduction. Glycosylation has proven to be associated with different mammalian diseases such as breast and liver cancers. Therefore characterization of glycans is highly important to facilitate a better understanding of the development and progression of many human diseases. Although matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) offers several advantages such as ease of operation and short analysis times, however, due to the complexity of glycan structures and their low ionization efficiency, there are still challenges that need to be addressed to achieve sensitive glycan analysis. Here, magnetic carbon nanocomposites (CNPs@Fe3O4 NCs) were used as a new MALDI matrix or co-matrix for the analysis of glycans derived from different model glycoproteins and human blood serum samples. The addition of CNPs@Fe3O4 NCs to the matrix significantly enhanced glycan signal intensity by several orders of magnitude, and effectively controlled/reduced/eliminated in-source decay (ISD) fragmentation. The latter was attained by modulating CNPs@Fe3O4 NCs concentrations and allowed the simultaneous study of intact and fragmented glycans, and pseudo-MS3 analysis. Moreover, CNPs@Fe3O4 NCs was also effectively employed to desalt samples directly on MALDI plate, thus enabling direct MALDIMS analysis of unpurified permethylated glycans derived from both model glycoproteins and biological samples. On-plate desalting enhanced sensitivity by reducing sample loss.
Keywords: Glycomics, MALDI, Magnetic carbon nanoparticles, In-source decay fragmentation, Human blood serum
Graphical Abstract
1. Introduction
Glycosylation is an essential post-translational modification of proteins and plays a critical role in several biological functions. These functions include protein folding, cell division, tumor immunology, and signal transduction. Changes in glycosylation and glycan structure have also been linked to a variety of human diseases and illnesses, thus monitoring these changes can provide better diagnosis and outlook for such disorders [1–7]. Therefore, it is essential to identify and characterize glycan structures at high sensitivities to aid in better understanding glycan attributes the development and progression of diseases which in turn will facilitate reliable treatment strategies.
Mass spectrometry (MS) is currently considered one of the most prominent methods used in glycan analysis, and when coupled with different separation techniques, a powerful tool for glycan profiling can be achieved. Some of these techniques include ion mobility [8–10], capillary electrophoresis [11–13], and high-performance liquid chromatography [14–16]. On the other hand, MALDI analysis permits fast glycan analysis due to the simplicity of the operation, rapid analysis time, relatively small sample volumes and straightforward data interpretation [17–19]. Although several recent advances in instrumentation and technique in the field of glycomics have been reported, many challenges in MALDI-MS of glycans still exist due to the complex nature of glycan structures and their low abundance in biological systems. Utilizing novel matrix materials that improve the selective enrichment of glycans and ionization efficiency could overcome these obstacles and may assist in providing an accurate MS analysis of glycans by MALDI-MS.
In recent years, the superior physical and chemical characteristics of nanomaterials, especially carbon-based nanomaterials, have prompted a significant interest in exploring the advantage of using such materials as analytical tools. When applied as MALDI matrices, carbon nanomaterials like graphene nanosheets, carbon nanodots, and carbon nanotubes have shown to be quite effective because of their notable electrical properties, optical absorption, and minimal background interference [20–26]. In addition to carbon-based nanomaterials, some other nanomaterials have also been used as MALDI matrices. For example, silver nanoparticles (NPs) were used for profiling lipids in the brain [27], mesoporous silica NPs were used for analysis of peptides [28], and BaTiO3 NPs were applied as the matrix for MALDI analysis of phospholipids [29]. Glutathione-capped iron oxide NPs were also synthesized and used in analysis of simple glycans such as maltoheptaose and isomaltotriose [30]. Although abundant glycosidic and crossring cleavages confirmed that the matrix provided a strong energy transfer to the sample, the possibility of controlling the degree of fragmentation was not investigated.
Recently, we investigate the advantages of using graphene nanosheets (GNs) and carbon nanoparticles (CNPs) as MALDI matrices and co-matrices in glycan profiling [26]. The modulation of CNPs and GNs concentrations significantly enhanced glycan signal intensity by several orders of magnitude, and controlled the degree of in-source decay (ISD) fragmentation [26]. Generally, ISD in the MALDI source prompts the formation of product-ions which could be beneficial in glycan profiling and structural identification as we have recently shown [26].
Here, we investigated the performance of magnetic carbon nanocomposites (CNPs@Fe3O4 NCs) as a MALDI matrix and co-matrix in MALDI analysis of native and permethylated glycans. In comparison with CNPs and Fe3O4 NPs, not only the signal intensities were improved by using CNPs@ Fe3O4 NCs, but it also enabled desalting of the samples directly on MALDI plate resulting in direct analysis of unpurified permethylated glycans with minimal sample loss. Similar to CNPs and GNs, CNPs@ Fe3O4 NCs allowed controlling the degree of ISD fragmentation and simultaneous study of both intact and fragmented glycans.
2. Materials and Methods
2.1. Materials
Carbon nanoparticles (particle size less than 100 nm and specific surface area higher than 100 m2/g), ribonuclease B (RNase B) from bovine pancreas, fetuin from fetal bovine serum, human alpha-1 acid glycoprotein (AGP), human serum from human male AB plasma, 2,5dihydroxybenzoic acid (DHB), and iodomethane were purchased from Sigma-Aldrich (St. Louis, MO). Microspin columns were purchased from Harvard Apparatus (Holliston, MA) and PNGase F with 10×G7 reaction buffer (0.5 M sodium phosphate) was obtained from New England Biolabs (Ipswich, MA). HPLC grade ethanol, acetonitrile (ACN), and water were used for sample preparation and were purchased from Sigma-Aldrich (St. Louis, MO).
2.2. Synthesis of magnetic carbon nanocomposites
CNPs@Fe3O4 NCs with 2:1 weight ratio of CNPs:Fe3O4 were synthesized using the modified coprecipitation method [31]. Briefly, 10 μl of CNPs (10 μg μl−1 in H2O) was dispersed in 1 ml aqueous solution containing 119.3 μg FeCl3.6H2O and 43.0 μg of FeCl2.4H2O at 50 0C under N2 atmosphere. After sonicating for 10 min, 300 μl of 25% ammonia solution was added for the formation of Fe3O4 NPs. The reaction was maintained at 50 0C for 30 min to complete the growth of Fe3O4 NPs on the surface of CNPs. After washing several times with water, the CNPs@Fe3O4 NCs were separated by using an external magnet. Finally, the nanocomposite was dried under vacuum. CNPs@Fe3O4 NCs with different weight ratios were also prepared by using the above procedure but with different amounts of precursors.
2.3. Sample Preparation Methods
N-Glycans were released from RNase B, fetuin, AGP and human blood serum according to a previously reported method, using PNGase F [32, 33]. A 5 μl aliquot of each model glycoproteins (10 μg μl−1) or 10 μl aliquot of human blood serum (stock solution) were mixed with 40 μl of diluted G7 buffer (50mM sodium phosphate buffer, pH 7.5, diluted 10 times with water). The mixtures were then denatured in a water bath at 800C for 30 min. After cooling to room temperature, 0.5 μl of PNGase F stock solution (500 units μl−1) was added, and the solutions were incubated in a 37 0C water bath for 18 hours to complete the enzymatic digestion. Released N-glycans were then purified by the addition of 90% ethanol and incubation at −20 0C for 30 min, to precipitate proteins. Purified glycans were then analyzed directly or permethylated using solid-phase permethylation method [32–35]. The permethylated glycans were then desalted using CNPs@Fe3O4 NCs (see section 2.4) or liquid-liquid extraction method [34, 35].
For matrix preparation, a 1 mg of CNPs@Fe3O4 NCs was dispersed in 1 ml of 50% ACN/H2O solution using sonication for 10 min prior to use. Different concentrations of CNPs@Fe3O4 NCs solutions were then prepared and mixed with an equal volume of DHB (20 μg μl–1, 50% ACN/H2O) to form binary matrices. A 0.5 μl aliquot of the resulting mixtures was added to the MALDI plate, followed by the addition of 0.5 μl of the sample solutions. The efficiencies of CNPs@Fe3O4 NCs and DHB as MALDI matrices were also examined separately by spotting 0.5 μl of each matrix solution to the MALDI plate.
2.4. Desalting of permethylated glycans on MALDI plate using CNPs@Fe3O4 NCs
A 1-μl of CNPs@Fe3O4 NCs suspension (0.2 μg μl−1 in 10% ACN) and 1 μl of the permethylated sample were carefully mixed in 500 μl centrifuge tube, using micropipette. 1 μl of resulting mixture were added to the MALDI plate and allowed to dry. The desired spot was then washed by using 1 μl of water for 3 times for desalting, followed by the addition of 0.5 μl of DHB (20 μg μl–1 in 50% ACN) prior the MALDI analysis.
2.5. Instrumentation
The morphologies of Fe3O4 NPs, CNPs, and CNPs@Fe3O4 NCs were characterized by using transmission electron microscopy (TEM, Hitachi H-8100). UV-Vis spectra were recorded using Shimadzu UV-1800 spectrophotometer. Powder XRD analyses were performed on a Rigaku Ultima III diffractometer operating with a Cu-Ka radiation source filtered with a graphite monochromator (λ = 1.5406 Å). A 4800 MALDI TOF/TOF (AB SCIEX) equipped with a pulsed Nd:YAG laser at an excitation wavelength of 355 nm, was used for the analyses. For each MS spectrum, 1250 laser shots were fired with a total of 50 sub-spectra and 25 shots per subspectrum. Data Explorer 4.9 software (AB SCIEX) and GlycoWorkbench software were used for MS data interpretation and glycan analyses.
3. Results and discussion
3.1. Characterization of matrix materials
The crystalline nature and phase purity of the synthesized nanomaterials were characterized by using XRD. Fig. S1a (see Electronic Supplementary Material, ESM) depicts the XRD patterns of CNPs, Fe3O4 NPs, and CNPs@Fe3O4 NCs. Accordinlgy, the CNPs have a graphite-like structure, with a broad diraction peak around 23.54° and interlayer spacing of 0.36 nm [36]. Six characteristic peaks of Fe3O4 NPs (2θ = 30.2, 35.6, 43.4, 53.7, 57.3 and 62.8) were also observed in both XRD spectra of Fe3O4 NPs and CNPs@Fe3O4 NCs, confirming the formation of Fe3O4 NPs on the surface of CNPs [37, 38]. The UV−Vis absorption spectra (ESM Fig. S1b) indicated that CNPs@Fe3O4 NCs have a broad absorption peak from 250 nm to 375 nm which corresponds to a π−π* transition of aromatic sp2 carbon domains. As it was shown in our previous work [26], Raman spectra also confirmed π-conjugation structures of CNPs. Comparing CNPs and Fe3O4 NPs, the UV−Vis data verified higher molar absorptivity of CNPs@Fe3O4 NCs, at the MALDI laser radiation wavelength (355 nm). The morphology of CNPs and CNPs@Fe3O4 NCs were characterized by TEM analysis. The TEM images show that the CNPs are monodisperse with an average size of 57 ± 3 nm (ESM Fig. S1c), whereas immobilized Fe3O4 NPs possess a spherical structure with an average size of 4.1 ± 0.7 nm (ESM Fig. S1d). Moreover, the TEM image of CNPs@Fe3O4 NCs (ESM Fig. S1d), clearly displays the formation of Fe3O4 NPs on the surface of CNPs.
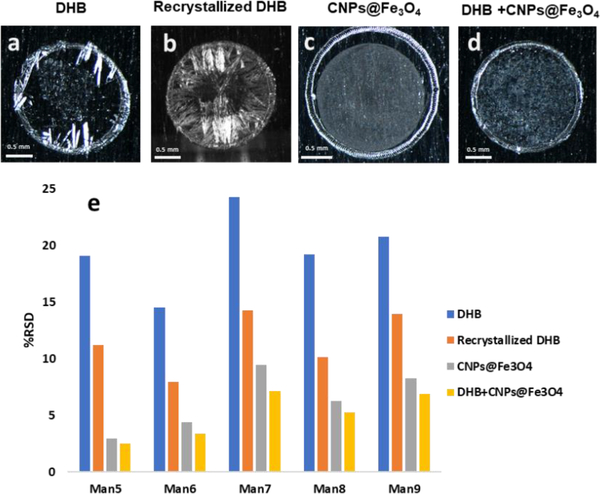
In general, DHB crystallizes heterogeneously and forms large needle-shaped crystals near the rim on the MALDI plate (Fig. 1a), resulting in poor shot-to-shot reproducibility in MALDI analyses [26, 39, 40]. Although crystallization could be improved via recrystallization using methanol, still small crystals formed near the rim (Fig. 1b). On the other hand, more homogeneous layers were formed when CNPs@Fe3O4 NCs as a matrix (Fig. 1c) or co-matrix (Fig. 1d). To evaluate the effect of CNPs@Fe3O4 NCs on measurement reproducibility, N-glycans derived from RNase B were analyzed using the different matrices studied. The relative standard deviations (RSD) of the peak intensities for the five N-glycans derived from RNase B are shown in Fig. 1e, which clearly demonstrates an improvement in reproducibility in the presence of CNPs@Fe3O4 NCs.
Fig. 1.
Optical images of the MALDI spots (a) 10 μg DHB (b) 10 μg DHB after recrystallization (c) 0.1 μg CNPs@Fe3O4 NCs and (d) 10 μg DHB+0.025 μg CNPs@Fe3O4 NCs. (e) the relative standard deviations of the peak intensities for glycans derived from RNase B, acquired three times by automatic measurements on three different spots, using different matrices. DHB (blue), recrystallized DHB (orange), CNPs@Fe3O4 NCs (gray), and DHB+CNPs@Fe3O4 NCs (yellow).
3.2. MALDI-TOF-MS spectra of N-glycans derived from RNase B using different matrices
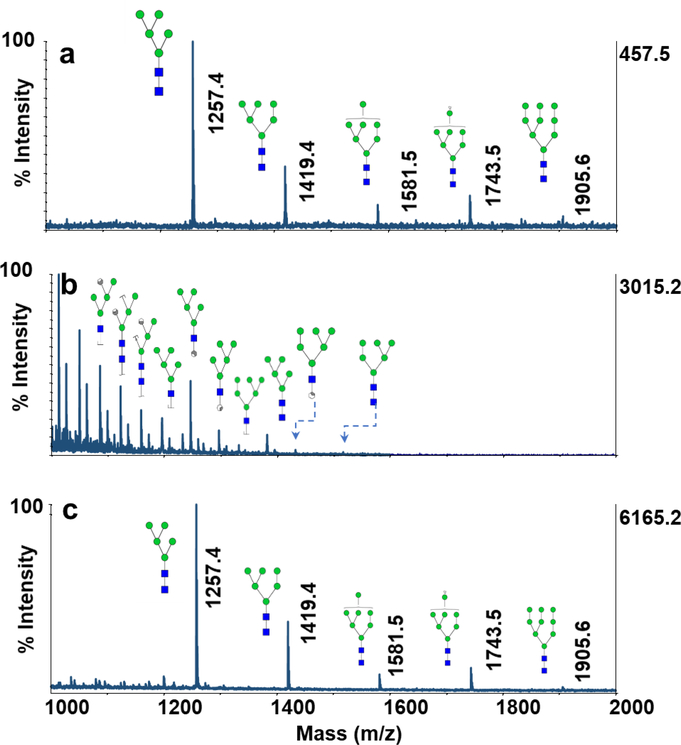
The performance of CNPs@Fe3O4 NCs as a MALDI matrix for analysis of glycans was first evaluated using N-glycans derived from 100 ng of RNase B, as a representative sample. Fig. 2a,b shows MALDI-TOF-MS spectra obtained using DHB, commonly used for MALDI analysis of glycans, and CNPs@Fe3O4 NCs. As shown in Fig. 2a, all five high mannose glycans were detected with relatively low intensities when using DHB as a matrix. Although the signal intensities were enhanced by using CNPs@Fe3O4 NCs (Fig. 2b), abundant ISD glycosidic and cross-ring cleavages were also observed. The matrix appears to provid strong energy transfer to glycans, thus prompting fragmentation. Similar results were obtained by using CNPs (ESM Fig. S2a) and Fe3O4 NPs (ESM Fig. S2b), with lower signal intensities, confirming higher laser absorption and transfer efficiency of CNPs@Fe3O4 NCs. Also, CNPs@Fe3O4 NCs showed interference-free backgrounds between m/z 700–5000, the mass range where most glycans are usually observed.
Fig. 2.
MALDI-TOF-MS spectra of N-glycans derived from 100 ng of RNase B using (a) 10 μg DHB (b) 0.1 μg CNPs@Fe3O4 NCs (c) 10 μg DHB+0.025 μg CNPs@Fe3O4 NCs. Symbols:  , N-acetylglucosamine;
, N-acetylglucosamine;  , Galactose;
, Galactose; , Fucose;
, Fucose;  , Mannose;
, Mannose; , N-acetylneuraminic acid.
, N-acetylneuraminic acid.
Our previous report [26] on CNPs and GNs, demonstrated that the degree of ISD could be controlled when when such material are used as co-matrices. In order to evaluate the performance of CNPs@Fe3O4 NCs as a co-matrix, MALDI-TOF-MS spectrum of RNase B glycans was obtained using DHB+CNPs@Fe3O4 NCs (Fig. 2c). The signal intensities were improved clearly, with almost no visible ISD products. The obtained data also demonstrated the superior performance of CNPs@Fe3O4 NCs over the CNPs (ESM Fig. S2c) and Fe3O4 NPs (ESM Fig. S2d) as co-matrices.
3.3. Effect the ratio CNPs to Fe3O4 on MALDI-TOF-MS of glycans
Next, CNPs@Fe3O4 NCs were prepared with different weight ratios of CNPs to Fe3O4 and used as a co-matrix with DHB to assess the effect of CNPs:Fe3O4 ratios on MALDI-TOFMS of glycans (Fig. 3a-c). According to the peak intensities, the best performance was obtained using 2:1 weight ratio of CNPs:Fe3O4. Higher ratios of CNPs:Fe3O4 (4:1, 8:1, ESM Fig. S3) were also tested however, the nanocomposites hardly showed any magnetic property at such high concentrations. The magnetic property of the nanocomposites is important for the purification process described below (section 3.6). The corresponding TEM images are also shown in Fig. 3d-f, which illustrate full coverage of CNPs with Fe3O4 NPs at a 1:2 weight ratio of CNPs Fe3O4.
Fig. 3.
MALDI-TOF-MS spectra of N-glycans derived from 100 ng of RNase B using 10 μg DHB+0.025 μg CNPs@Fe3O4 NCs, synthesized with different weight ratios of CNPs:Fe3O4 (a) 2:1 (b) 1:1 (c) 1:2. TEM images of CNPs@Fe3O4 NCs synthesized with different weight ratios of CNPs:Fe3O4 (d) 2:1 (e) 1:1 (f) 1:2. The arrows indicate some example of the Fe3O4 NPs.
3.4. Effect of CNPs@Fe3O4 concentration on MALDI-TOF-MS of glycans
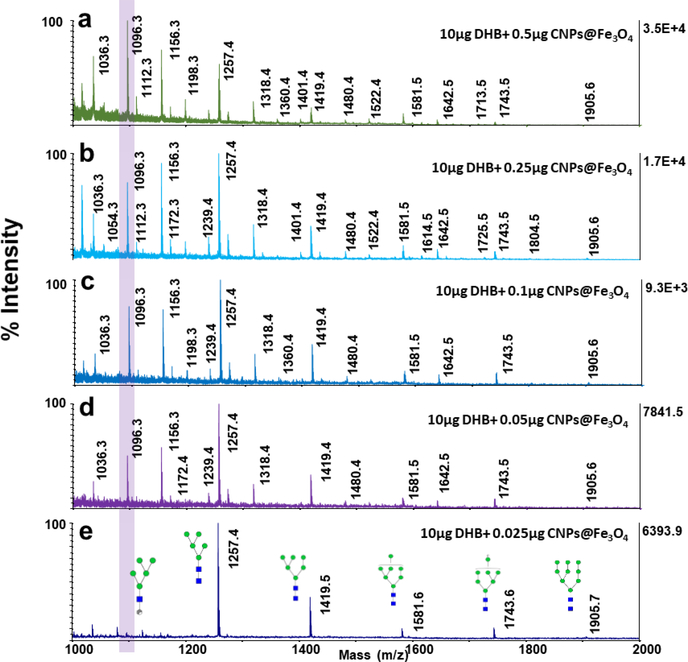
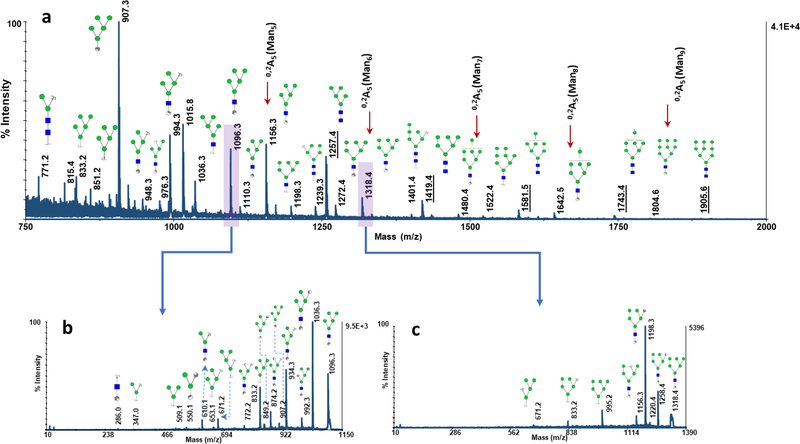
The effect of CNPs@Fe3O4 NCs concentrations on glycan profiling, when used as comatrix, was also investigated (Fig. 4). At higher amounts of nanomaterials, not only the signal intensities were improved, but the degree of ISD was also increased, generating extensive glycosidic and cross-ring cleavages. For instance, variation in the intensity of the peak at m/z 1096.3 (one of the cross-ring fragment product of Man5) is shown in Fig. 4a-e, as a representative example. The degree of ISD fragmentation could be controlled by adjusting the concentration of CNPs@Fe3O4 NCs added to the matrix. This could be employed to generate pseudo-MS3 spectra which can provide valuable data for structural identification. Fig. 5a depicts the MALDI-TOF-MS spectrum of N-glycans derived from 100 ng of RNase B, using 0.5 μg of CNPs@Fe3O4 as a co-matrix. Abundant cross-ring and glycosidic bond fragmentations were produced, containing frequently observed Y-, B-ions as well as A- and X- and C-ions. As it is observed, cross-ring fragmented ions are more abundant, compared to those resulting from the cleavage of glycosidic bonds, which can be very informative in linkage study. Pseudo-MS3 spectra were acquired for the peaks at m/z 1096.3 (Fig. 5b) and 1318.4 (Fig. 5c), as representative ISD fragment ions. Several fragmented ions with relatively high signal to noise ratios were observed, which can be beneficial for further structural analysis.
Fig. 4.
MALDI-TOF-MS spectra of N-glycans derived from 100 ng of RNase B using 10 μg DHB mixed with different amounts of CNPs@Fe3O4 NCs (a) 0.5 μg (b) 0.25 μg (c) 0.1 μg (d) 0.05 μg (e) 0.025 μg. Highlight shows the variations in the intensity of the peak at m/z=1096.3, with changing the amount of CNPs@Fe3O4 NCs in the matrix. Symbols as in Fig. 2.
Fig. 5.
(a) MALDI-TOF-MS spectrum of N-glycans derived from 100 ng of RNase B using 10 μg DHB+ 0.5 μg CNPs@Fe3O4 NCs. Underlines indicate the five intact high mannose structures and the arrows designate the fragmented 0,2A5 ions corresponding to the five high mannose structures. (b) MS/MS spectrum of the ISD fragmented ion at m/z=1096.3 and (c) MS/MS spectrum of the ISD fragmented ion at m/z=1318.4. Symbols as in Fig. 2.
3.5. MALDI-MS of permethylated glycans using CNPs@Fe3O4 NCs
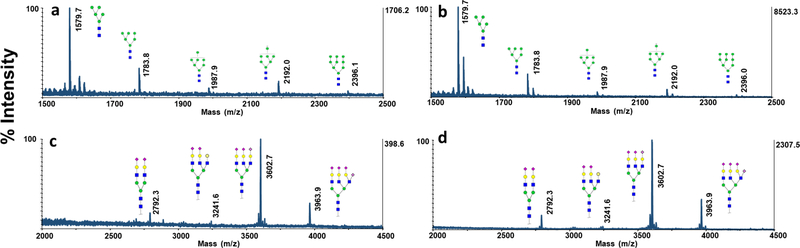
It has been previously demonstrated that the ionization efficiency of glycans significantly increase upon permethylation [33, 41, 42]. Moreover, permethylation allows comprehensive mapping of both sialylated and neutral glycans, by stabilization of sialic acid residues [17–19]., CNPs@Fe3O4 NCs were utilized as MALDI matrix for the analysis of permethylated glycans derived from 100 ng of fetuin and RNase B to assess the performance of co-matrix (Fig. 6). Relative to DHB (Fig. 6a,c), the presence of CNPs@Fe3O4 NCs as a co-matrix significantly increased the signal intensities of both high mannose (Fig. 6b) and sialylated glycans (Fig. 6d) by several folds. Moreover, the relative abundances of RNase B and fetuin glycans were comparable to those previously reported by NMR (ESM Fig. S4) [43, 44], confirming the usability of the proposed matrix with no bias.
Fig. 6.
MALDI-TOF-MS spectra of permethylated N-glycans derived from 100 ng of RNase B, using (a) 10 μg DHB (b) 10 μg DHB+ 0.025 μg CNPs@Fe3O4 NCs. MALDI-TOFMS spectra of permethylated N-glycans derived from 100 ng of bovine fetuin, using (c) 10 μg DHB (d) 10 μg DHB+ 0.025μg CNPs@Fe3O4 NCs. Symbols as in Fig. 2.
3.6. Use of CNPs@Fe3O4 NCs enables desalting on MALDI plate
MALDI analysis depends on the amount of salts present in samples. High concentration of salts can affect ionization efficiency and suppress signal intensities. Biological samples usually contain noticeable amounts of both endogenous salts or salts originating from sample preparation, thus purification and desalting of biological samples are necessary prior to MALDI analyses. Moreover, during the permethylation process, a high amount of salts are introduced to samples, thus sharply decreasing ionization efficiency. Carbon materials have been used as excellent sorbents for both native and permethylated glycans. They can adsorb glycans via hydrophobic and polar interactions [15, 45, 46]. Recently, we compared the efficiency of porous graphitic carbon, charcoal, graphene nanosheets and CNPs for purification of permethylated glycans derived from standard glycoproteins and human sera in different esophageal disease stages, demonstrating higher purification efficiency of CNPs relative to the other shapes of carbons [47].
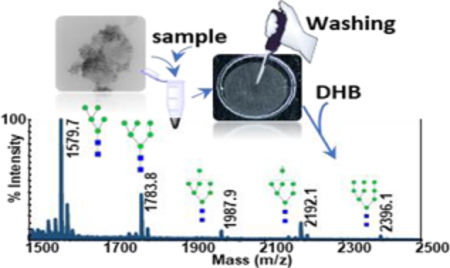
Here, we investigated the performance of CNPs@Fe3O4 NCs for purification of permethylated glycans. The main advantages of CNPs@Fe3O4 NCs over the CNPs, is their magnetic property, which helps to keep CNPs@Fe3O4 NCs attached to MALDI plate, even after mildly washing the spots with water. Fig. S5a,b (see ESM) shows optical microscopic images of the MALDI spots covered with a thin layer of CNPs@Fe3O4 NCs, before (ESM Fig. S5a) and after (ESM Fig. S5b) washing with 5μl of water. No clear changes were observed, which confirms the high stability of CNPs@Fe3O4 NCs on the MALDI plate. This property and the unique interaction of carbon materials with permethylated glycans enabled desalting directly on MALDI plate. Permethylated glycans derived from RNase B were desalted on-plate according to the procedure explained in section 2.4 using CNPs@Fe3O4 NCs. As shown in ESM Fig. S5c, the sample exhibited very low signals with four of the glycan structures peaks missing before washing. However, after washing of the spot with 3μl of water, all the five high mannose glycans were detected with high intensities (ESM Fig. S5d).
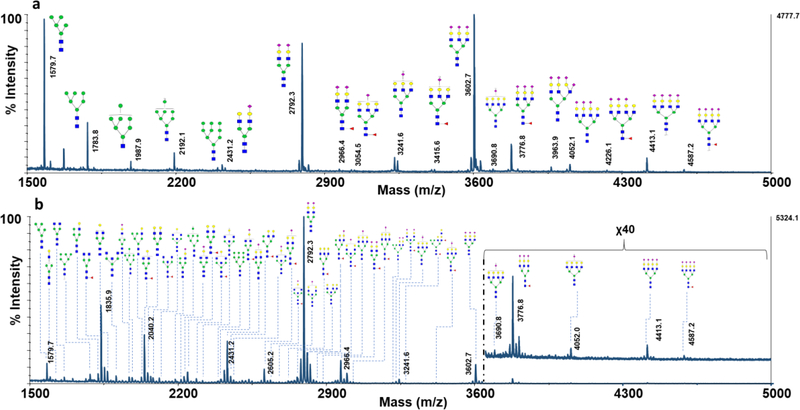
The utility of CNPs@Fe3O4 NCs was also investigated for the direct analysis of permethylated glycans in more complex sample, containing glycans derived from RNase B, fetuin and AGP (Fig. 7a). Although before the washing, the spectra obtained had very weak intensities with missing glycans peaks (ESM Fig. S6a), after the washing step all the high mannose, sialylated and fucosylated glycans were detected with excellent signal to noise ratios (Fig. 7a). Also, the performance of the CNPs@Fe3O4 NCs was examined for the direct analysis of permethylated glycans derived from human blood serum. MALDI-MS analysis of the spot before washing only allowed the detection of few permethylated glycan structures (ESM Fig. S6b); however, 56 glycan structures were detected when the same spot was washed with water as described in the Experimental section (Fig. 7b). This number of detected glycans is comparable to previously reported results [26, 48]. Accordingly, CNPs@Fe3O4 NCs permitted efficient desalting of samples on MALDI plate, and provided an effective and fast technique for analysis of glycans, via simplifying the process of sample pretreatment and isolation.
Fig. 7.
MALDI-TOF-MS spectra of permethylated glycans desalted directly on MALDI plate, using CNPs@Fe3O4 NCs, spotting the equivalent of glycans derived from (a) a mixture containing 100ng of RNase B, 200 ng of fetuin, and 500 ng of AGP (b) 0.5μl of human blood serum. The total amount of sample prepared is detailed in the Experimental section. Symbols as in Fig. 2.
Conclusion
In this work, we studied the performance of CNPs@Fe3O4 NCs as a MALDI matrix or co-matrix for glycan profiling of three model glycoproteins and human blood serum. The material exhibited excellent adsorption and transfer of laser energy to glycans, resulting in improved signal intensities by several orders of magnitude. Also, in contrast to CNPs, the magnetic property of CNPs@Fe3O4 NCs enabled the effective use of the material to achieve desalting directly on MALDI plate, thus permitting direct analysis of unpurified permethylated glycans with minimal sample loss. Moreover, similar to CNPs, the level of ISD fragmentation can be adjusted easily by modulating CNPs@Fe3O4 NCs concentration, allowing simultaneous study of intact and fragmented glycans in full MS scan, as well as pseudo-MS3.
Supplementary Material
Acknowledgment
This work was supported by an NIH grant (1R01GM112490–04).
Footnotes
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Drake RR, Powers TW, Jones EE, Bruner E, Mehta AS, Angel PM. MALDI Mass Spectrometry Imaging of N-Linked Glycans in Cancer Tissues. Adv Cancer Res. 2017;134:85–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kailemia MJ, Park D, Lebrilla CB. Glycans and glycoproteins as specific biomarkers for cancer. Anal Bioanal Chem. 2017;409(2):395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veillon L, Huang Y, Peng W, Dong X, Cho BG, Mechref Y. Characterization of isomeric glycan structures by LC-MS/MS. Electrophoresis. 2017;38(17):2100–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veillon L, Zhou S, Mechref Y. Quantitative Glycomics: A Combined Analytical and Bioinformatics Approach. Methods Enzymol. 2017;585:431–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou S, Wooding KM, Mechref Y. Analysis of Permethylated Glycan by Liquid Chromatography (LC) and Mass Spectrometry (MS). Methods Mol Biol. 2017;1503:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kailemia MJ, Xu G, Wong M, Li Q, Goonatilleke E, Leon F et al. Recent Advances in the Mass Spectrometry Methods for Glycomics and Cancer. Anal Chem. 2018;90(1):208–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruhaak LR, Xu G, Li Q, Goonatilleke E, Lebrilla CB. Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses. Chem Rev. 2018:DOI: 10.1021/acs.chemrev.7b00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaye MM, Ding T, Shion H, Hussein A, Hu Y, Zhou S et al. Delineation of disease phenotypes associated with esophageal adenocarcinoma by MALDI-IMS-MS analysis of serum N-linked glycans. Analyst. 2017;142(9):1525–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmann J, Pagel K. Glycan Analysis by Ion Mobility-Mass Spectrometry. Angew Chem Int Ed Engl. 2017;56(29):8342–9. [DOI] [PubMed] [Google Scholar]
- 10.Manz C, Pagel K. Glycan analysis by ion mobility-mass spectrometry and gas-phase spectroscopy. Curr Opin Chem Biol. 2018;42:16–24. [DOI] [PubMed] [Google Scholar]
- 11.Mechref Y Analysis of glycans derived from glycoconjugates by capillary electrophoresismass spectrometry. Electrophoresis. 2011;32(24):3467–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snyder CM, Zhou X, Karty JA, Fonslow BR, Novotny MV, Jacobson SC. Capillary electrophoresis-mass spectrometry for direct structural identification of serum N-glycans. J Chromatogr A. 2017;1523:127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu G, Crihfield CL, Gattu S, Veltri LM, Holland LA. Capillary Electrophoresis Separations of Glycans. Chem Rev. 2018:doi: 10.1021/acs.chemrev.7b00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuhrer M, de Boer AR, Deelder AM. Structural glycomics using hydrophilic interaction chromatography (HILIC) with mass spectrometry. Mass Spectrom Rev. 2009;28(2):192–206. [DOI] [PubMed] [Google Scholar]
- 15.West C, Elfakir C, Lafosse M. Porous graphitic carbon: a versatile stationary phase for liquid chromatography. J Chromatogr A. 2010;1217(19):3201–16. [DOI] [PubMed] [Google Scholar]
- 16.Vreeker GC, Wuhrer M. Reversed-phase separation methods for glycan analysis. Anal Bioanal Chem. 2017;409(2):359–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harvey DJ. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: an update for 2009–2010. Mass Spectrom Rev. 2015;34(3):268–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harvey DJ. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update for 2011–2012. Mass Spectrom Rev. 2017;36(3):255–422. [DOI] [PubMed] [Google Scholar]
- 19.Harvey DJ. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update for 2013–2014. Mass Spectrom Rev. 2018;37(4):353–491. [DOI] [PubMed] [Google Scholar]
- 20.Li XS, Wu JH, Xu LD, Zhao Q, Luo YB, Yuan BF et al. A magnetite/oxidized carbon nanotube composite used as an adsorbent and a matrix of MALDI-TOF-MS for the determination of benzo[a]pyrene. Chem Commun (Camb). 2011;47(35):9816–8. [DOI] [PubMed] [Google Scholar]
- 21.Min Q, Zhang X, Chen X, Li S, Zhu JJ. N-doped graphene: an alternative carbon-based matrix for highly efficient detection of small molecules by negative ion MALDI-TOF MS. Anal Chem. 2014;86(18):9122–30. [DOI] [PubMed] [Google Scholar]
- 22.Friesen WL, Schultz BJ, Destino JF, Alivio TE, Steet JR, Banerjee S et al. Two-dimensional graphene as a matrix for MALDI imaging mass spectrometry. J Am Soc Mass Spectrom. 2015;26(11):1963–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez CE, Palacios J, Fajardo I, Urdiales JL, Le Guevel X, Lozano J et al. Conventional Matrices Loaded Onto a Graphene Layer Enhances MALDI-TOF/TOF Signal: Its Application to Improve Detection of Phosphorylated Peptides. J Am Soc Mass Spectrom. 2016;27(2):366–9. [DOI] [PubMed] [Google Scholar]
- 24.Wu BS, Gopal J, Hua PY, Wu HF. Graphene nanosheet mediated MALDI-MS (GN-MALDIMS) for rapid, in situ detection of intact incipient biofilm on material surfaces. Mater Sci Eng C Mater Biol Appl. 2016;66:285–96. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Cai Y, Wang Y, Zhou X, Zhang Y, Lu H. Improved MALDI imaging MS analysis of phospholipids using graphene oxide as new matrix. Sci Rep. 2017;7:doi: 10.1038/srep44466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banazadeh A, Peng W, Veillon L, Mechref Y. Carbon Nanoparticles and Graphene Nanosheets as MALDI Matrices in Glycomics: a New Approach to Improve Glycan Profiling in Biological Samples. J Am Soc Mass Spectrom. 2018:doi: 10.1007/s13361-018-1985-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan M, Zhang Z, Li S, Liu J, Liu L, Yang H et al. Silver nanoparticles as matrix for MALDI FTICR MS profiling and imaging of diverse lipids in brain. Talanta. 2018;179:62431. [DOI] [PubMed] [Google Scholar]
- 28.Jun C, Tianxi H, Xuemei F. A novel matrix additive, MCM-41-type mesoporous silica nanoparticles, used for analysis of peptides by MALDI-FT/ICRMS. Talanta. 2012;100:41924. [DOI] [PubMed] [Google Scholar]
- 29.Kailasa SK, Wu HF. Surface modified BaTiO3 nanoparticles as the matrix for phospholipids and as extracting probes for LLME of hydrophobic proteins in Escherichia coli by MALDIMS. Talanta. 2013;114:283–90. [DOI] [PubMed] [Google Scholar]
- 30.Liang Q, Macher T, Xu Y, Bao Y, Cassady CJ. MALDI MS in-source decay of glycans using a glutathione-capped iron oxide nanoparticle matrix. Anal Chem. 2014;86(16):8496–503. [DOI] [PubMed] [Google Scholar]
- 31.Wang C, Feng C, Gao Y, Ma X, Wu Q, Wang Z. Preparation of a graphene-based magnetic nanocomposite for the removal of an organic dye from aqueous solution. Chem Eng J. 2011;173(1):92–7. [Google Scholar]
- 32.Zhou S, Hu Y, DeSantos-Garcia JL, Mechref Y. Quantitation of permethylated N-glycans through multiple-reaction monitoring (MRM) LC-MS/MS. J Am Soc Mass Spectrom. 2015;26(4):596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou S, Dong X, Veillon L, Huang Y, Mechref Y. LC-MS/MS analysis of permethylated N-glycans facilitating isomeric characterization. Anal Bioanal Chem. 2017;409(2):453–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang P, Mechref Y, Klouckova I, Novotny MV. Solid-phase permethylation of glycans for mass spectrometric analysis. Rapid Commun Mass Spectrom. 2005;19(23):3421–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang P, Mechref Y, Novotny MV. High-throughput solid-phase permethylation of glycans prior to mass spectrometry. Rapid Commun Mass Spectrom. 2008;22(5):721–34. [DOI] [PubMed] [Google Scholar]
- 36.Krishnamoorthy K, Veerapandian M, Yun K, Kim S-J. The chemical and structural analysis of graphene oxide with different degrees of oxidation. Carbon. 2013;53:38–49. [Google Scholar]
- 37.Atacan K, Cakiroglu B, Ozacar M. Efficient protein digestion using immobilized trypsin onto tannin modified Fe3O4 magnetic nanoparticles. Colloids Surf B Biointerfaces. 2017;156:9–18. [DOI] [PubMed] [Google Scholar]
- 38.Chen F, Xie S, Huang X, Qiu X. Ionothermal synthesis of Fe3O4 magnetic nanoparticles as efficient heterogeneous Fenton-like catalysts for degradation of organic pollutants with H2O2. J Hazard Mater. 2017;322(Pt A):152–62. [DOI] [PubMed] [Google Scholar]
- 39.Cohen SL, Chait BT. Influence of matrix solution conditions on the MALDI-MS analysis of peptides and proteins. Anal Chem. 1996;68(1):31–7. [DOI] [PubMed] [Google Scholar]
- 40.Shin D, Kim I, Paek J, Kim J. A Novel “Freeze Vacuum Drying” Crystallization Method Toward Quantitative MALDI MS. Bull Korean Chem Soc. 2017;38(1):133–5. [Google Scholar]
- 41.Zhou S, Huang Y, Dong X, Peng W, Veillon L, Kitagawa DA et al. Isomeric separation of permethylated glycans by porous graphitic carbon (PGC)-LC-MS/MS at high temperatures. Anal chem. 2017;89(12):6590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou S, Wooding KM, Mechref Y. Analysis of Permethylated glycan by liquid chromatography (LC) and mass spectrometry (MS) High-Throughput Glycomics and Glycoproteomics. Springer; 2017. p. 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green ED, Adelt G, Baenziger JU, Wilson S, Van Halbeek H. The asparagine-linked oligosaccharides on bovine fetuin. Structural analysis of N-glycanase-released oligosaccharides by 500-megahertz 1H NMR spectroscopy. J Biol Chem. 1988;263(34):18253–68. [PubMed] [Google Scholar]
- 44.Fu D, Chen L, O’Neill RA. A detailed structural characterization of ribonuclease B oligosaccharides by 1H NMR spectroscopy and mass spectrometry. Carbohydr Res. 1994;261(2):173–86. [DOI] [PubMed] [Google Scholar]
- 45.Melmer M, Stangler T, Premstaller A, Lindner W. Comparison of hydrophilic-interaction, reversed-phase and porous graphitic carbon chromatography for glycan analysis. J Chromatogr A. 2011;1218(1):118–23. [DOI] [PubMed] [Google Scholar]
- 46.Stavenhagen K, Kolarich D, Wuhrer M. Clinical Glycomics Employing Graphitized Carbon Liquid Chromatography-Mass Spectrometry. Chromatographia. 2015;78(5–6):307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong J, Banazadeh A, Wenjing P, Mechref Y. A carbon nanoparticles-based solid-phase purification method for permethylated N-glycans. Submitted to Electrophoresis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu Y, Mechref Y. Comparing MALDI-MS, RP-LC-MALDI-MS and RP-LC-ESI-MS glycomic profiles of permethylated N-glycans derived from model glycoproteins and human blood serum. Electrophoresis. 2012;33(12):1768–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.