Abstract
Recent studies have examined genetic correlations of single nucleotide polymorphism (SNP) effect sizes across pairs of populations to better understand the genetic architectures of complex traits. These studies have estimated ρg, the cross-population correlation of joint-fit effect sizes at genotyped SNPs. However, the value of ρg depends both on the cross-population correlation of true causal effect sizes (ρb) and on the similarity in linkage disequilibrium (LD) patterns in the two populations, which drive tagging effects. Here, we derive the value of the ratio ρg/ρb as a function of LD in each population. By applying existing methods to obtain estimates of ρg, we can use this ratio to estimate ρb. Our estimates of ρb were equal to 0.55 (s.e. 0.14) between Europeans and East Asians averaged across 9 traits in the Genetic Epidemiology Research on Adult Health and Aging (GERA) data set, 0.54 (s.e. 0.18) between Europeans and South Asians averaged across 13 traits in the UK Biobank data set, and 0.48 (s.e. 0.06) and 0.65 (s.e. 0.09) between Europeans and East Asians in summary statistic data sets for type 2 diabetes and rheumatoid arthritis, respectively. These results implicate substantially different causal genetic architectures across continental populations.
Keywords: genetic correlation, genetic architecture, multi-ethnic
Introduction
There has been substantial recent interest in comparing the genetic architecture of complex traits across world populations (de Candia et al., 2013, Mancuso et al., 2016, Brown et al., 2016). The global phenotypic distributions of complex traits can vary based on a combination of genetic and environmental factors (Robinson et al., 2015, Burt et al., 1995), and uncovering these factors is key to both understanding complex traits and ensuring that medical genetics research is globally equitable(Popejoy and Fullerton, 2016). Multi-ethnic studies have analyzed the replication rates of associations from genome-wide association studies (GWAS)(Marigorta and Navarro, 2013), improved fine-mapping resolution(Zaitlen et al., 2010, Kichaev and Pasaniuc, 2015), increased meta-analysis power(Mahajan et al., 2014, Coram et al., 2015, Morris, 2011), and assessed the global relationships between allelic effect sizes via the genetic correlations(de Candia et al., 2013, Mancuso et al., 2016, Brown et al., 2016). However, differences in joint-fit effect sizes are influenced both by differences in causal variant effect sizes and by differences in linkage disequilibrium (LD) patterns between the populations. In this study, we derive an approach for estimating genetic correlations of causal variant effect sizes across populations, leveraging data from densely genotyped reference panels to apply a correction factor to conventional estimates of genetic correlations of joint-fit effect sizes.
The cross-population genetic correlation of joint-fit effect sizes (ρg) is a scalar quantity that summarizes the similarity of joint-fit allelic effects between two populations(de Candia et al., 2013, Mancuso et al., 2016, Brown et al., 2016). It is defined as the correlation between the vectors of joint-fit effect sizes at single nucleotide polymorphisms (SNPs) shared between two populations (see Materials and Methods). It is closely related to the genetic correlation of two phenotypes in a single population, which is a scalar quantity that summarizes the shared genetic architecture between the traits (Lee et al., 2013, Bulik-Sullivan et al., 2015a). Rather than focusing on a limited number of GWAS associations, the cross-population genetic correlation provides a genome-wide estimate of the similarity in genetic effects between the two populations.
Several recent studies have estimated cross-population genetic correlations (de Candia et al., 2013, Mancuso et al., 2016, Brown et al., 2016) by extending previous methods to estimate cross-trait genetic correlations from either raw genotype/phenotype data(Lee et al., 2012a, Lee et al., 2012b, Lee et al., 2013) or summary association statistic data(Bulik-Sullivan et al., 2015a). These studies estimated the correlation of joint-fit effect sizes at genotyped SNPs that are shared between the populations. However, ρg may depend on patterns of LD between SNPs, which differ across populations (Lee et al., 2013). For example, consider the case of an untyped causal SNP u with the same effect size in two populations, and two SNPs t1 and t2 that are genotyped in both populations. If t1 perfectly tags u in population 1 (but not in population 2), and t2 perfectly tags u in population 2 (but not in population 1), then the ρg at those genotyped SNPs will be 0 despite identical causal effects in the two populations.
In contrast, our goal here is to estimate the cross-population correlation of causal effect sizes (ρb; see Materials and Methods). To accomplish this, we derive the value of the ratio ρg/ρb as a function of LD patterns in each population, which can be obtained from a reference panel such as 1000 Genomes(Auton et al., 2015). We first estimate ρg as in previous studies, and then divide this estimate by the value of the ratio ρg/ρb to obtain an estimate of ρb. We validate our method using simulations, and apply the method to raw genotype/phenotype and summary association statistic data sets with 46K-116K European samples and 2K-23K East Asian or South Asian samples.
Materials and Methods
Genotype-phenotype model
For population k, let gk,i denote the joint-fit effect size at SNP i in population k, so that gk is the vector of joint-fit effect sizes at genotyped SNPs. Similarly, let bk denote the vector of causal effect sizes at all SNPs (in practice, reference panel SNPs with minor allele frequency (MAF)>1%). We note that gk and bk are population-level parameters rather than estimates in a finite sample, but gk can be viewed as the value of joint-fit effect size estimates in the limit of infinite sample size. We also note that values of gk, but not bk, depend on the LD patterns in the population and on the set of genotyped SNPs. We use “genotyped SNPs” as shorthand to denote the set of SNPs for which raw genotype/phenotype data or summary association statistic data is available; in some cases this may include both genotyped and imputed SNPs.
Let the heritability at genotyped SNPs of the trait in populations 1 and 2 be and , respectively18. Likewise, let the heritability at causal SNPs in populations 1 and 2 be and , respectively. We assume the additive infinitesimal model for a quantitative phenotype,
where Yk is an Nk × 1 vector of phenotypes in Nk individuals from population k, XA,k is an Nk × M matrix of mean-centered genotypes at all M SNPs, is an M × 1 vector of causal effect sizes and is an Nk × 1 vector of environmental noise.
For a fixed set of MG genotyped SNPs, there also exists a vector of joint-fit effect sizes gk such that
where XG,k has dimension Nk × MG and eg,k is scaled such that the heritability explained by genotyped SNPs(Yang et al., 2010) is . Here, A denotes all SNPs and G denotes genotyped SNPs, so that XG,k represents a subset of the SNPs in XA,k. In the first model, where all SNPs are observed, the vector eb,k represents environmental noise. In the second model eg,k represents a combination of environmental noise and the remaining un-modeled SNP effects. Thus, Var(eg,k) ≥ Var(eb,k) and We can relate gk and bk via
where the last step follows from the law of large numbers and the fact that E[eb,k] = 0. Here, we introduce S as the M × M SNP cross-covariance matrix, which can be partitioned into genotyped and untyped SNPs (where G denotes genotyped SNPs, U denotes untyped SNPs and A denotes all SNPs):
The above model is based on genotypes X{G,A}, k that have been mean-centered but not normalized. It may also be of interest to consider mean-centered, normalized genotypes Wk. We can then define normalized causal effect sizes βk (instead of bk) and normalized joint-fit effect sizes γk (instead of gk), and relate γk and βk using a normalized SNP cross-correlation matrix Σk. We employ this approach when estimating ργ the cross-population correlation of normalized joint-fit effect sizes, and ρβ, the cross-population correlation of normalized causal effect sizes. We note that previous work has reported similar estimates of ργ and ρg, representing correlations of joint-fit effect sizes with or without normalization(Brown et al., 2016). We focus the derivations below on quantities without normalization (b, g, S, ρb, ρg), but all derivations are analogous when employing normalization (β, γ, Σ, ρβ, ργ).
Definition of ρg and ρb
We define the cross-population genetic correlation at genotyped SNPs as the correlation between g1 and g2,
Likewise, we define the cross-population genetic correlation at causal SNPs as the correlation between b1 and b2,
Based on these definitions, it follows that ρg (but not ρb) depends on the LD patterns in the two populations and on the set of genotyped SNPs.
The first step of our method for estimating ρb is to estimate ρg, the cross-population correlation of joint-fit effect sizes. When raw genotype/phenotype data is available, we use bivariate REML(Lee et al., 2013, Lee et al., 2012a, Lee et al., 2012b), as implemented in GCTA (see Web Resources). When only summary association statistic data is available, we use Popcorn(Brown et al., 2016) (see Web Resources), a maximum-likelihood based method that analyzes summary statistics and population-specific LD information from a reference panel.
Estimating ρg/ρb
The second step of our method for estimating ρb is to estimate the ratio ρg/ρb, which we derive as a function of LD in each population. We then divide our estimate of ρg by the value of ρg/ρb to obtain an estimate of ρb. In practice, this derivation requires that we estimate S, the cross-covariance LD matrix of all SNPs. We estimate S using an LD reference panel, because all SNP genotypes are unavailable in analyses of summary statistics and because SNP genotypes at untyped SNPs are unavailable in analyses of raw genotypes/phenotypes.
As noted above, the joint-fit effect sizes g (at genotyped SNPs) can be viewed as the value of joint-fit effect size estimates in the limit of infinite sample size:
where XA,k is the Nk × M matrix of mean-centered genotypes for all SNPs in population k, is the MG × MG cross-covariance sub-matrix between genotyped SNPs in population k and is that MG × M cross-covariance sub-matrix between genotyped SNPs and all SNPs in population k. It follows that and therefore that
where by Corr we refer to the scalar-valued correlation, rather than the matrix-valued cross-correlation. We now relate the right hand side of this equation to ρb. From the fact that E[bk] = 0M (the 0-vector) and properties of the trace, it follows that
so that
We define a function τ to simplify our notation:
so that
It similarly follows that
Combining the above equations, we have
so that
We note that the trace of the product of the LD matrices in the numerator and dominator of the τ function corresponds to the sum of the entries of the Hadamard product of the two matrices:
Thus, the denominator of the τ function contains the sums of LD scores, while the numerator contains the sum of a cross-population analog of LD scores(Brown et al., 2016). Since naïve estimates of squared correlations are upward biased, we adjust squared correlation estimates to remove this bias, as in previous work(Bulik-Sullivan et al., 2015b):
where Σ denotes the SNP cross-correlation matrix. We propagate this adjustment to squared covariance estimates:
We only consider LD within 1Mb windows, setting and to 0 if the distance between SNPs i and j is greater than 1Mb, similar to previous work(Bulik-Sullivan et al., 2015b, Kichaev and Pasaniuc, 2015).
Simulations with real genotypes and simulated phenotypes
To ensure realistic LD patterns, we performed simulations using real genotypes from the Genetic Epidemiology Research on Adult Health and Aging (GERA) data set and simulated phenotypes. The GERA data set contains 45,725 European ancestry (EUR) individuals, 3,357 East Asian ancestry (EAS) individuals, and 315,434 SNPs, after QC (see below). In each simulation, we sampled N1 EUR and N2 EAS samples, and restricted the simulation to all SNPs on chromosome 11 (we chose chromosome 11 because larger chromosomes tend to have higher LD, and smaller chromosomes tend to have lower LD). We selected a subset of MG SNPs that were considered as “genotyped” SNPs for the purpose of the simulation, and selected a subset of MC causal SNPs (from the set of all M SNPs) to simulate phenotypes. For each of the MC causal SNPs, we sampled per-allele causal effect sizes in the two populations from a bivariate normal distribution with variance N(0, Pb), where Pb is a 2 × 2 matrix with diagonal entries equal to 1 and off-diagonal entries equal to ρb, the cross-population correlation of causal effect sizes. In each population, we multiplied the matrix of real genotypes by the vector of causal effect sizes to construct simulated genetic values. We scaled the genetic values to have mean 0 and variance h2 and added environmental noise sampled from N(0, 1 – h2) to the genetic values to construct simulated phenotypes.
1000 Genomes data set
The 1000 Genomes data set(Auton et al., 2015) (see Web Resources) contains 503 individuals of European ancestry (EUR), 504 individuals of East Asian ancestry (EAS) and 489 individuals of South Asian ancestry (SAS). We performed QC in each population separately, retaining only bi-allelic SNPs in Hardy-Weinberg equilibrium (p>0.001) with MAF>0.1% and excluding SNPs with duplicate IDs, leaving 13,258,254 EUR SNPs, 12,285,372 EAS SNPs and 24,463,301 SAS SNPs. For each pair of populations analyzed (EUR-EAS and EUR-SAS), we restricted to SNPs with MAF>1% in each population (as in previous studies(Brown et al., 2016, de Candia et al., 2013, Mancuso et al., 2016)), resulting in 1,352,543 EUR-EAS SNPs and 2,115,911 EUR-SAS SNPs.
GERA data set
The Genetic Epidemiology Research on Adult Health and Aging (GERA) data set(Banda et al., 2015) (see Web Resources) includes 62,318 individuals of European ancestry (EUR) and 5,188 individuals of East Asian ancestry (EAS) genotyped on population-specific microarrays containing 657,184 and 694,877 SNPs, respectively. We performed QC in each population separately, retaining only bi-allelic SNPs with MAF>1% (as in previous studies(Brown et al., 2016, de Candia et al., 2013, Mancuso et al., 2016)) and missing genotype rate less than 2%. Only SNPs that passed QC in both populations were retained, resulting in 351,421 SNPs. This SNP set was further intersected with the 1000 Genomes EUR-EAS SNPs, resulting in 315,434 EUR-EAS SNPs. Related individuals and individuals with a greater than 2% missing data rate were also excluded from the study, resulting in 45,725 EUR and 3,357 EAS samples. We analyzed 9 traits that were previously analyzed in(Loh et al., 2015): allergic rhinitis, asthma, cardiovascular disease, type 2 diabetes, dyslipidemia, hypertension, macular degeneration, osteoarthritis and osteoporosis.
UK Biobank data set
The UK Biobank data set(Sudlow et al., 2015) (see Web Resources) includes 120,286 individuals of British ancestry QC-ed for GWAS analysis (EUR) and 1,784 individuals of South Asian ancestry (SAS) genotyped at 847,131 SNPs. We performed QC as with the GERA data set, resulting in 392,598 EUR-SAS SNPs, 116,478 EUR samples and 1,706 SAS samples. We analyzed 13 traits: bone-densitometry of heel, height, weight-height ratio, diastolic blood pressure, systolic blood pressure, college education, smoking status, eczema, asthma, hypertension, FEV1, FEV1-FCV ratio and age at menarche.
RA and T2D summary statistic data sets
We analyzed rheumatoid arthritis (RA) and type 2 diabetes (T2D) summary statistic data sets that were used to estimate ρg between Europeans and East Asians in a previous study(Brown et al., 2016). The RA data set included summary statistics from 58,284 European ancestry individuals(Okada et al., 2014) and summary statistics from 22,515 East Asian ancestry individuals(Okada et al., 2014), each computed at 2,539,629 genotyped or imputed SNPs. The T2D data set included summary statistics from 69,033 European ancestry individuals(Morris et al., 2012) and summary statistics from 18,817 East Asian ancestry individuals(Cho et al., 2011), each computed at 1,054,079 genotyped or imputed SNPs. For both RA and T2D, we used the estimates of ρg from the previous study(Brown et al., 2016), so that we only directly analyzed 1000 Genomes data (informed by the set of genotyped/imputed SNPs in the summary statistic data sets). As noted in the previous study(Brown et al., 2016), estimates of in these data sets were incorrectly scaled due to genomic control correction, which does not affect estimates of ρg, but were greater than 0 with very high statistical significance.
Results
Simulations with real genotypes and simulated phenotypes
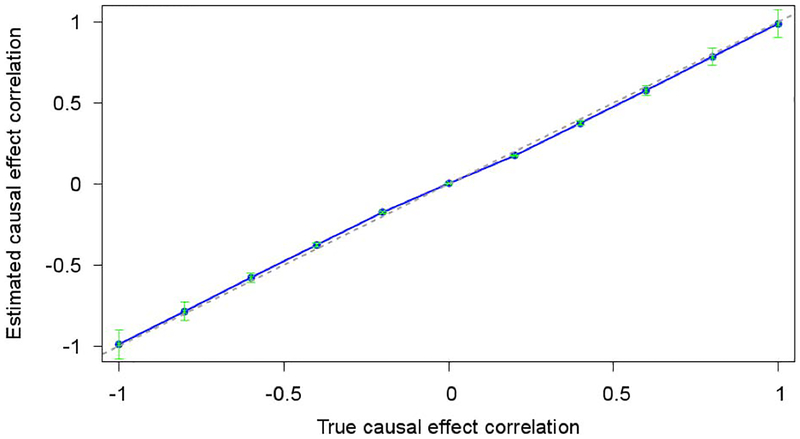
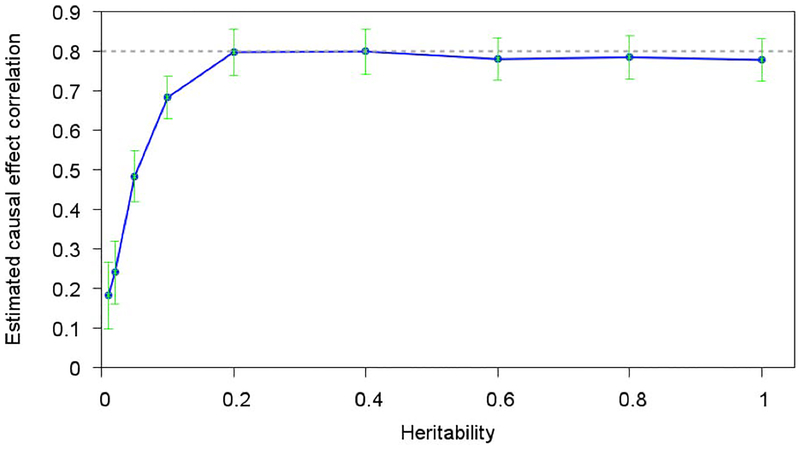
We first evaluated our method using simulations in which the true value of ρb is known. To ensure realistic LD patterns, we used real genotypes from chromosome 11 of the GERA data set and simulated causal effect sizes and phenotypes in EUR and EAS samples (see Materials and Methods). We included NEUR=2K EUR samples, NEAS=2K EAS samples, MT=5,000 SNPs that were considered as “genotyped” SNPs (used to estimate ρg) and MC=100 causal SNPs with nonzero causal effect sizes (selected from set of all MT SNPs). We estimated ρg using bivariate REML, and transformed this into an estimate of ρb using our derivation of ρg/ρb (see Materials and Methods). We first fixed h2=0.8 and varied ρb. We determined that our method produced accurate estimates of ρb across all values of ρb (Figure 1). We then fixed h2=0.8 and ρb =0.8 and varied MT, MC, NEUR=NEAS, and NEUR only, respectively. In each case, our method continued to produce accurate estimates of ρb (Figure S1). However, our results are subject to two caveats. First, we noted that regularizing LD estimates by restricting to 1Mb windows reduced slight biases (Figure S2). Second, we varied h2 and determined that estimates of ρb were downward biased at very low values of h2 (less than 0.2; Figure S3); this cannot be a limitation of our derivation of ρg/ρb, which does not depend on h2 (or on any phenotypic values), and must instead be a limitation of estimation of ρg at very low values of h2 (and , although the true value of in these simulations is unknown). Thus, efforts to estimate either ρg or ρb should avoid traits with very low values of h2.
Figure 1. Estimates of ρb are accurate in simulations at various values of ρb.
We report estimates of ρb for each value of true ρb. Dashed line is y=x.
Application to 9 traits from GERA data set
We applied our method for estimate ρb to 9 traits from the GERA data set, which includes data from 45,725 Europeans (EUR) and 3,357 East Asians (EAS) at 315,434 genotyped SNPs (see Materials and Methods). We first computed a value of 0.93 for the ratio ρg/ρb, for this set of genotyped SNPs relative to 1000 Genomes reference SNPs. We then used bivariate REML to estimate ρg for each trait (restricting the computation to 10K EUR and all EAS samples), and divided by 0.93 to obtain estimates of ρb. Estimates of ρg and ρb are reported in Table 1. The inverse-variance weighted average of was 0.51 with standard error 0.13, and the inverse-variance weighted average of was 0.55 with standard error 0.14. Estimates of cross-population correlations of normalized effect sizes ( and ) were slightly lower, with inverse-variance weighted averages of 0.41 (SE=0.13) and 0.44 (SE=0.14) respectively.
Table 1. Estimates of cross-population genetic correlations for 9 GERA traits.
We report estimates of ρg, ργ, ρb and ρβ for each trait. (Adjusting estimates of ρg that are close to 1 by dividing by estimates of ρg/ρb can result in estimates of ρb that are greater than 1; although the true value of ρb cannot be greater than 1, it is possible for estimates of ρb to be greater than 1.) Numbers in parentheses denote standard errors. Standard errors of (resp. ) are estimated by GCTA using bi-variate REML, while standard errors of (resp. ) are estimated by correcting the standard error estimated by GCTA by the estimated correction factor ρg/ρb (resp. ργ/ρβ).
| Phenotype | ||||
|---|---|---|---|---|
| Allergic rhinitis | 1.00 (1.06) | 1.00 (1.00) | 1.08 (1.14) | 1.08 (1.08) |
| Asthma | −0.04 (0.40) | −0.34 (0.33) | −0.04 (0.43) | −0.37 (0.35) |
| Cardiovascular Disease | 0.48 (0.35) | 0.30 (0.32) | 0.52 (0.38) | 0.32 (0.34) |
| Type 2 Diabetes | 0.35 (0.27) | 0.29 (0.27) | 0.38 (0.29) | 0.31 (0.29) |
| Dyslipidemia | 0.52 (0.21) | 0.47 (0.21) | 0.56 (0.23) | 0.51 (0.23) |
| Hypertension | 0.27 (0.19) | 0.24 (0.19) | 0.29 (0.20) | 0.26 (0.20) |
| Macular Degeneration | 1.00 (2.09) | 1.00 (2.08) | 1.08 (2.25) | 1.08 (2.24) |
| Osteoarthritis | 0.53 (0.35) | 0.42 (0.32) | 0.57 (0.38) | 0.45 (0.34) |
| Osteoporosis | −0.07 (0.47) | −0.12 (0.53) | −0.08 (0.51) | −0.13 (0.57) |
Application to 13 traits from UK Biobank data set
We next applied our method for estimating ρb to 13 traits from the UK Biobank data set, which includes data from 116,478 Europeans (EUR) and 1,706 South Asians (SAS) at 392,598 genotyped SNPs (see Materials and Methods). We first computed a value of 0.98 for the ratio ρg/ρb, for this set of genotyped SNPs relative to the 1000 Genomes reference SNPs. The larger value of ρg/ρb between Europeans and South Asians than between Europeans and East Asians (despite similar numbers of genotyped SNPs) is expected because Europeans and South Asians are more recently diverged than Europeans and East Asians(Sved et al., 2008). We then used bivariate REML to estimateρg for each trait (restricting the computation to 10K EUR and all SAS samples), and divided by 0.98 to obtain estimates of ρb. Estimates of ρg and ρb are reported in Table 2. The inverse-variance weighted average of was 0.53 with standard error 0.17, and the inverse-variance weighted average of was 0.54 with standard error 0.18. Estimates of cross-population correlations of normalized effect sizes ( and ) were slightly lower, with inverse-variance weighted averages of 0.50 (SE=0.17) and 0.51 (SE=0.17) respectively.
Table 2. Estimates of cross-population genetic correlations for 13 UK Biobank traits.
We report estimates of ρg, ργ, ρb and ρβ for each trait. (Adjusting estimates of ρg that are close to 1 by dividing by estimates of ρg/ρb can result in estimates of ρb that are greater than 1; although the true value of ρb cannot be greater than 1, it is possible for estimates of ρb to be greater than 1.) Numbers in parentheses denote standard errors. Standard errors of (resp. ) are estimated by GCTA using bi-variate REML, while standard errors of (resp. ) are estimated by correcting the standard error estimated by GCTA by the estimated correction factor ρg/ρb (resp. ργ/ρβ).
| Phenotype | ||||
|---|---|---|---|---|
| Bone-densitometry of heel | 0.60 (0.18) | 0.49 (0.19) | 0.62 (0.18) | 0.50 (0.20) |
| Height | 0.77 (0.26) | 0.63 (0.24) | 0.78 (0.26) | 0.64 (0.24) |
| Weight-height ratio | 1.00 (2.19) | 1.00 (2.64) | 1.02 (2.24) | 1.02 (2.69) |
| Diastolic blood pressure | 1.00 (0.56) | 0.73 (0.35) | 1.02 (0.57) | 0.74 (0.36) |
| Systolic blood pressure | 1.00 (0.91) | 0.76 (0.59) | 1.02 (0.93) | 0.77 (0.60) |
| College education | 0.36 (0.22) | 0.38 (0.22) | 0.37 (0.23) | 0.39 (0.22) |
| Smoking status | 0.37 (0.39) | 0.22 (0.37) | 0.38 (0.40) | 0.22 (0.38) |
| Eczema | −0.19 (0.62) | 0.14 (0.59) | −0.19 (0.63) | 0.15 (0.60) |
| Asthma | 0.92 (1.49) | 0.65 (0.72) | 0.94 (1.52) | 0.66 (0.73) |
| Hypertension | 0.32 (0.32) | 0.38 (0.32) | 0.32 (0.33) | 0.38 (0.33) |
| FEV1 | 0.57 (0.27) | 0.50 (0.29) | 0.58 (0.28) | 0.51 (0.30) |
| FEV1-FCV ratio | 0.39 (0.29) | 0.58 (0.41) | 0.40 (0.30) | 0.59 (0.42) |
| Age at menarche | 0.70 (1.07) | 0.59 (1.00) | 0.71 (1.09) | 0.60 (1.02) |
Application to RA and T2D summary statistics
We next applied our method for estimating ρb to summary statistic data sets for RA (58,284 EUR and 22,151 EAS samples, 2,539,629 genotyped/imputed SNPs) and T2D (69,033 EUR and 18,817 EAS samples, 1,054,079 genotyped/imputed SNPs) that were used to estimate ρg in a previous study(Brown et al., 2016), which reported estimates of ρg of 0.463 (s.e. 0.058) for RA and 0.621 (s.e. 0.088) for T2D. We computed a value of 0.96 for the ratio ρg/ρb for the RA genotyped/imputed SNPs relative to the 1000 Genomes reference SNPs, and a value of 0.97 for the ratio ρg/ρb for the T2D genotyped/imputed SNPs relative to the 1000 Genomes reference SNPs. The larger values of ρg/ρb between Europeans and East Asians for these SNP sets, compared to 0.93 for the GERA genotyped SNP set, is expected due to the larger numbers of genotyped/imputed SNPs. We divided the previously reported estimates of ρg by the values of ρg/ρb to obtain estimates of ρb. The resulting estimates of were 0.48 (s.e. 0.06) for RA and 0.65 (s.e. 0.09) for T2D, which are still significantly less than 1.
Discussion
Recent work comparing the genetic architecture of complex traits across continental populations has established that GWAS results do not always transfer across populations(Brown et al., 2016, Mahajan et al., 2014), however, this finding may be explained by the fact that continental populations have different LD patterns. Our results demonstrate for the first time that causal genetic architectures differ between continental populations, and therefore that differences in GWAS results across populations cannot be explained by differences in LD patterns alone. We introduced a new method for estimating ρb, the cross-population correlation of causal effect sizes: we first estimate ρg (the cross-population correlation of joint-fit effect sizes) using existing methods(Brown et al., 2016, Lee et al., 2013, Lee et al., 2012a, Lee et al., 2012b), and then divide by the value of ρg/ρb that we obtain from 1000 Genomes reference data (as a function of the set of genotyped SNPs used to define ρg) using a new derivation. We applied our method to estimate ρb in GERA and UK Biobank data sets for which ρg and ρb had not previously been estimated, and to RA and T2D summary statistic data sets for which ρg (but not ρb) had previously been estimated. For each of the genotyped SNP sets and population pairs that we analyzed, ρg/ρb was only modestly smaller than 1, so that ρb was only modestly larger than ρg, and remained significantly smaller than 1. This could for example be explained by gene-gene or gene-environment interaction. Importantly, we have only analyzed data from European and Asian populations, which are known to have relatively similar LD patterns(Lonjou et al., 2003); our findings may not generalize to African-ancestry populations, which have more divergent LD patterns.
Our method is subject to several limitations. First, our method relies on LD information from a reference panel, and restricts to SNPs that are present in the reference panel. Methods that use LD information from a reference panel to analyze summary statistic data(Pasaniuc and Price, 2017) rely on the assumption that the LD in each study population is well approximated by the LD in the respective reference populations(Ni et al., 2018). In addition, due to complexities of admixture-LD, such methods may not work well in admixed populations(Bulik-Sullivan et al., 2015b, Brown et al., 2016), and thus our method is not currently applicable to populations such as African and Latin Americans that often provide the most practical route to assaying African and Native American genetic variation(Seldin et al., 2011). Second, limitations in existing methods for estimating ρg will carry over to our estimates of ρb; this is a particular concern for traits with very low heritability. Third, our method assumes that selection of the set of genotyped SNPs is independent of LD. This may not be strictly true in instances where the set of genotyped SNPs is selected based on their tagging efficiency, but restricting our analyses to SNPs that are genotyped in both populations minimizes the impact of SNPs with population-specific tagging efficiency. Finally, we restricted our analyses to SNPs with MAF>1% in both populations, as in previous studies(de Candia et al., 2013, Mancuso et al., 2016, Brown et al., 2016). Thus, many SNPs that have MAF<1% in either population are excluded (see Materials and Methods). It is possible that ρg/ρb would be smaller (i.e. ρb would be larger) when including the effects of rare (MAF<1%) causal variants. It will be possible to formally assess this when larger multi-ethnic reference panels become available, but we anticipate that the impact on our results will be small. This is because most common variation is shared across populations and because emerging research suggests that rare and low-frequency causal variants contribute only modestly to the heritability of complex traits(Yang et al., 2015, Zeng et al., 2018, Schoech et al., 2017) Despite these limitations, our method provides a promising way to assess cross-population correlations of causal effect sizes.
Supplementary Material
Figure S1. Estimates of ρb are accurate in simulations at various parameter settings. We report estimates of ρb for various values of MT, the number of “genotyped” SNPs; MC, the number of causal SNPs; NEUR=NEAS; and NEUR (with NEAS fixed at 2K). The dashed line is the true value ρb =0.8.
Figure S2. Regularizing LD estimates is necessary to minimize biases. We report the value of the correction factor ρg/ρb (resp. ργ/ρβ) as a function of the LD window used. We report estimates using only an LD window (dashed curve), as well as estimates using both an LD window and the regularization method of Wen & Stephens 2010 Ann Appl Stat (solid curve). The horizontal dashed line is the true correction factor, that is, the value of (resp. ). The results support the use of an LD window, and indicate than the regularization method of Wen & Stephens does not impact results when an LD window is used. Standardized and mean-centered approaches tend to give similar results in practice, but they assume a different trait model and therefore can give different results, especially if causal alleles have very different alleles frequencies between the two populations.
Figure S3. Estimates of ρb are downward biased at very low values of h2. We report estimates of ρb for various values of h2. The dashed line is the true value ρb =0.8.
Figure 2. Estimates of ρb are downward biased at very low values of h2.
We report estimates of ρb for various values of h2. The dashed line is the true value ρb =0.8.
Acknowledgements
This research was funded by NIH grant R01 HG006399. This research was conducted using the UK Biobank Resource under Application #16549.
Footnotes
Web Resources
GCTA: http://cnsgenomics.com/software/gcta
Popcorn: https://github.com/brielin/popcorn
1000 Genomes: http://www.internationalgenome.org
GERA: http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000674.v1.p1
UK Biobank: http://www.ukbiobank.ac.uk
References
- AUTON A, BROOKS LD, DURBIN RM, GARRISON EP, KANG HM, KORBEL JO, MARCHINI JL, MCCARTHY S, MCVEAN GA & ABECASIS GR 2015. A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BANDA Y, KVALE MN, HOFFMANN TJ, HESSELSON SE, RANATUNGA D, TANG H, SABATTI C, CROEN LA, DISPENSA BP, HENDERSON M, IRIBARREN C, JORGENSON E, KUSHI LH, LUDWIG D, OLBERG D, QUESENBERRY CP JR., ROWELL S, SADLER M, SAKODA LC, SCIORTINO S, SHEN L, SMETHURST D, SOMKIN CP, VAN DEN EEDEN SK, WALTER L, WHITMER RA, KWOK PY, SCHAEFER C & RISCH N 2015. Characterizing Race/Ethnicity and Genetic Ancestry for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics, 200, 1285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BROWN BC, YE CJ, PRICE AL & ZAITLEN N 2016. Transethnic Genetic-Correlation Estimates from Summary Statistics. Am J Hum Genet, 99, 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BULIK-SULLIVAN B, FINUCANE HK, ANTTILA V, GUSEV A, DAY FR, LOH PR, DUNCAN L, PERRY JR, PATTERSON N, ROBINSON EB, DALY MJ, PRICE AL & NEALE BM 2015a. An atlas of genetic correlations across human diseases and traits. Nat Genet, 47, 1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BULIK-SULLIVAN BK, LOH PR, FINUCANE HK, RIPKE S, YANG J, PATTERSON N, DALY MJ, PRICE AL & NEALE BM 2015b. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet, 47, 291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURT VL, WHELTON P, ROCCELLA EJ, BROWN C, CUTLER JA, HIGGINS M, HORAN MJ & LABARTHE D 1995. Prevalence of hypertension in the US adult population. Results from the Third National Health and Nutrition Examination Survey, 1988–1991. Hypertension, 25, 305–13. [DOI] [PubMed] [Google Scholar]
- CHO YS, CHEN CH, HU C, LONG J, ONG RT, SIM X, TAKEUCHI F, WU Y, GO MJ, YAMAUCHI T, CHANG YC, KWAK SH, MA RC, YAMAMOTO K, ADAIR LS, AUNG T, CAI Q, CHANG LC, CHEN YT, GAO Y, HU FB, KIM HL, KIM S, KIM YJ, LEE JJ, LEE NR, LI Y, LIU JJ, LU W, NAKAMURA J, NAKASHIMA E, NG DP, TAY WT, TSAI FJ, WONG TY, YOKOTA M, ZHENG W, ZHANG R, WANG C, SO WY, OHNAKA K, IKEGAMI H, HARA K, CHO YM, CHO NH, CHANG TJ, BAO Y, HEDMAN AK, MORRIS AP, MCCARTHY MI, TAKAYANAGI R, PARK KS, JIA W, CHUANG LM, CHAN JC, MAEDA S, KADOWAKI T, LEE JY, WU JY, TEO YY, TAI ES, SHU XO, MOHLKE KL, KATO N, HAN BG & SEIELSTAD M 2011. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet, 44, 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CORAM MA, CANDILLE SI, DUAN Q, CHAN KH, LI Y, KOOPERBERG C, REINER AP & TANG H 2015. Leveraging Multi-ethnic Evidence for Mapping Complex Traits in Minority Populations: An Empirical Bayes Approach. Am J Hum Genet, 96, 740–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE CANDIA TR, LEE SH, YANG J, BROWNING BL, GEJMAN PV, LEVINSON DF, MOWRY BJ, HEWITT JK, GODDARD ME, O’DONOVAN MC, PURCELL SM, POSTHUMA D, VISSCHER PM, WRAY NR & KELLER MC 2013. Additive genetic variation in schizophrenia risk is shared by populations of African and European descent. Am J Hum Genet, 93, 463–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KICHAEV G & PASANIUC B 2015. Leveraging Functional-Annotation Data in Trans-ethnic Fine-Mapping Studies. Am J Hum Genet, 97, 260–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE SH, DECANDIA TR, RIPKE S, YANG J, SULLIVAN PF, GODDARD ME, KELLER MC, VISSCHER PM & WRAY NR 2012a. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet, 44, 247–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE SH, RIPKE S, NEALE BM, FARAONE SV, PURCELL SM, PERLIS RH, MOWRY BJ, THAPAR A, GODDARD ME, WITTE JS, ABSHER D, AGARTZ I, AKIL H, AMIN F, ANDREASSEN OA, ANJORIN A, ANNEY R, ANTTILA V, ARKING DE, ASHERSON P, AZEVEDO MH, BACKLUND L, BADNER JA, BAILEY AJ, BANASCHEWSKI T, BARCHAS JD, BARNES MR, BARRETT TB, BASS N, BATTAGLIA A, BAUER M, BAYES M, BELLIVIER F, BERGEN SE, BERRETTINI W, BETANCUR C, BETTECKEN T, BIEDERMAN J, BINDER EB, BLACK DW, BLACKWOOD DH, BLOSS CS, BOEHNKE M, BOOMSMA DI, BREEN G, BREUER R, BRUGGEMAN R, CORMICAN P, BUCCOLA NG, BUITELAAR JK, BUNNEY WE, BUXBAUM JD, BYERLEY WF, BYRNE EM, CAESAR S, CAHN W, CANTOR RM, CASAS M, CHAKRAVARTI A, CHAMBERT K, CHOUDHURY K, CICHON S, CLONINGER CR, COLLIER DA, COOK EH, COON H, CORMAND B, CORVIN A, CORYELL WH, CRAIG DW, CRAIG IW, CROSBIE J, CUCCARO ML, CURTIS D, CZAMARA D, DATTA S, DAWSON G, DAY R, DE GEUS EJ, DEGENHARDT F, DJUROVIC S, DONOHOE GJ, DOYLE AE, DUAN J, DUDBRIDGE F, DUKETIS E, EBSTEIN RP, EDENBERG HJ, ELIA J, ENNIS S, ETAIN B, FANOUS A, FARMER AE, FERRIER IN, FLICKINGER M, FOMBONNE E, FOROUD T, FRANK J, FRANKE B, FRASER C, et al. 2013. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet, 45, 984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE SH, YANG J, GODDARD ME, VISSCHER PM & WRAY NR 2012b. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics, 28, 2540–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOH PR, BHATIA G, GUSEV A, FINUCANE HK, BULIK-SULLIVAN BK, POLLACK SJ, DE CANDIA TR, LEE SH, WRAY NR, KENDLER KS, O’DONOVAN MC, NEALE BM, PATTERSON N & PRICE AL 2015. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat Genet, 47, 1385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LONJOU C, ZHANG W, COLLINS A, TAPPER WJ, ELAHI E, MANIATIS N & MORTON NE 2003. Linkage disequilibrium in human populations. Proc Natl Acad Sci U S A, 100, 6069–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAHAJAN A, GO MJ, ZHANG W, BELOW JE, GAULTON KJ, FERREIRA T, HORIKOSHI M, JOHNSON AD, NG MC, PROKOPENKO I, SALEHEEN D, WANG X, ZEGGINI E, ABECASIS GR, ADAIR LS, ALMGREN P, ATALAY M, AUNG T, BALDASSARRE D, BALKAU B, BAO Y, BARNETT AH, BARROSO I, BASIT A, BEEN LF, BEILBY J, BELL GI, BENEDIKTSSON R, BERGMAN RN, BOEHM BO, BOERWINKLE E, BONNYCASTLE LL, BURTT N, CAI Q, CAMPBELL H, CAREY J, CAUCHI S, CAULFIELD M, CHAN JC, CHANG LC, CHANG TJ, CHANG YC, CHARPENTIER G, CHEN CH, CHEN H, CHEN YT, CHIA KS, CHIDAMBARAM M, CHINES PS, CHO NH, CHO YM, CHUANG LM, COLLINS FS, CORNELIS MC, COUPER DJ, CRENSHAW AT, VAN DAM RM, DANESH J, DAS D, DE FAIRE U, DEDOUSSIS G, DELOUKAS P, DIMAS AS, DINA C, DONEY AS, DONNELLY PJ, DORKHAN M, VAN DUIJN C, DUPUIS J, EDKINS S, ELLIOTT P, EMILSSON V, ERBEL R, ERIKSSON JG, ESCOBEDO J, ESKO T, EURY E, FLOREZ JC, FONTANILLAS P, FOROUHI NG, FORSEN T, FOX C, FRASER RM, FRAYLING TM, FROGUEL P, FROSSARD P, GAO Y, GERTOW K, GIEGER C, GIGANTE B, GRALLERT H, GRANT GB, GRROP LC, GROVES CJ, GRUNDBERG E, GUIDUCCI C, HAMSTEN A, HAN BG, HARA K, HASSANALI N, et al. 2014. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet, 46, 234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANCUSO N, ROHLAND N, RAND KA, TANDON A, ALLEN A, QUINQUE D, MALLICK S, LI H, STRAM A, SHENG X, KOTE-JARAI Z, EASTON DF, EELES RA, LE MARCHAND L, LUBWAMA A, STRAM D, WATYA S, CONTI DV, HENDERSON B, HAIMAN CA, PASANIUC B & REICH D 2016. The contribution of rare variation to prostate cancer heritability. Nat Genet, 48, 30–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARIGORTA UM & NAVARRO A 2013. High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genet, 9, e1003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORRIS AP 2011. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol, 35, 809–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORRIS AP, VOIGHT BF, TESLOVICH TM, FERREIRA T, SEGRE AV, STEINTHORSDOTTIR V, STRAWBRIDGE RJ, KHAN H, GRALLERT H, MAHAJAN A, PROKOPENKO I, KANG HM, DINA C, ESKO T, FRASER RM, KANONI S, KUMAR A, LAGOU V, LANGENBERG C, LUAN J, LINDGREN CM, MULLER-NURASYID M, PECHLIVANIS S, RAYNER NW, SCOTT LJ, WILTSHIRE S, YENGO L, KINNUNEN L, ROSSIN EJ, RAYCHAUDHURI S, JOHNSON AD, DIMAS AS, LOOS RJ, VEDANTAM S, CHEN H, FLOREZ JC, FOX C, LIU CT, RYBIN D, COUPER DJ, KAO WH, LI M, CORNELIS MC, KRAFT P, SUN Q, VAN DAM RM, STRINGHAM HM, CHINES PS, FISCHER K, FONTANILLAS P, HOLMEN OL, HUNT SE, JACKSON AU, KONG A, LAWRENCE R, MEYER J, PERRY JR, PLATOU CG, POTTER S, REHNBERG E, ROBERTSON N, SIVAPALARATNAM S, STANCAKOVA A, STIRRUPS K, THORLEIFSSON G, TIKKANEN E, WOOD AR, ALMGREN P, ATALAY M, BENEDIKTSSON R, BONNYCASTLE LL, BURTT N, CAREY J, CHARPENTIER G, CRENSHAW AT, DONEY AS, DORKHAN M, EDKINS S, EMILSSON V, EURY E, FORSEN T, GERTOW K, GIGANTE B, GRANT GB, GROVES CJ, GUIDUCCI C, HERDER C, HREIDARSSON AB, HUI J, JAMES A, JONSSON A, RATHMANN W, KLOPP N, KRAVIC J, KRJUTSKOV K, LANGFORD C, LEANDER K, LINDHOLM E, LOBBENS S, MANNISTO S, et al. 2012. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet, 44, 981–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NI G, MOSER G, WRAY NR & LEE SH 2018. Estimation of Genetic Correlation via Linkage Disequilibrium Score Regression and Genomic Restricted Maximum Likelihood. Am J Hum Genet, 102, 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKADA Y, WU D, TRYNKA G, RAJ T, TERAO C, IKARI K, KOCHI Y, OHMURA K, SUZUKI A, YOSHIDA S, GRAHAM RR, MANOHARAN A, ORTMANN W, BHANGALE T, DENNY JC, CARROLL RJ, EYLER AE, GREENBERG JD, KREMER JM, PAPPAS DA, JIANG L, YIN J, YE L, SU DF, YANG J, XIE G, KEYSTONE E, WESTRA HJ, ESKO T, METSPALU A, ZHOU X, GUPTA N, MIREL D, STAHL EA, DIOGO D, CUI J, LIAO K, GUO MH, MYOUZEN K, KAWAGUCHI T, COENEN MJ, VAN RIEL PL, VAN DE LAAR MA, GUCHELAAR HJ, HUIZINGA TW, DIEUDE P, MARIETTE X, BRIDGES SL JR., ZHERNAKOVA A, TOES RE, TAK PP, MICELI-RICHARD C, BANG SY, LEE HS, MARTIN J, GONZALEZ-GAY MA, RODRIGUEZ-RODRIGUEZ L, RANTAPAA-DAHLQVIST S, ARLESTIG L, CHOI HK, KAMATANI Y, GALAN P, LATHROP M, EYRE S, BOWES J, BARTON A, DE VRIES N, MORELAND LW, CRISWELL LA, KARLSON EW, TANIGUCHI A, YAMADA R, KUBO M, LIU JS, BAE SC, WORTHINGTON J, PADYUKOV L, KLARESKOG L, GREGERSEN PK, RAYCHAUDHURI S, STRANGER BE, DE JAGER PL, FRANKE L, VISSCHER PM, BROWN MA, YAMANAKA H, MIMORI T, TAKAHASHI A, XU H, BEHRENS TW, SIMINOVITCH KA, MOMOHARA S, MATSUDA F, YAMAMOTO K & PLENGE RM 2014. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature, 506, 376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PASANIUC B & PRICE AL 2017. Dissecting the genetics of complex traits using summary association statistics. Nat Rev Genet, 18, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POPEJOY AB & FULLERTON SM 2016. Genomics is failing on diversity. Nature, 538, 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINSON MR, HEMANI G, MEDINA-GOMEZ C, MEZZAVILLA M, ESKO T, SHAKHBAZOV K, POWELL JE, VINKHUYZEN A, BERNDT SI, GUSTAFSSON S, JUSTICE AE, KAHALI B, LOCKE AE, PERS TH, VEDANTAM S, WOOD AR, VAN RHEENEN W, ANDREASSEN OA, GASPARINI P, METSPALU A, BERG LH, VELDINK JH, RIVADENEIRA F, WERGE TM, ABECASIS GR, BOOMSMA DI, CHASMAN DI, DE GEUS EJ, FRAYLING TM, HIRSCHHORN JN, HOTTENGA JJ, INGELSSON E, LOOS RJ, MAGNUSSON PK, MARTIN NG, MONTGOMERY GW, NORTH KE, PEDERSEN NL, SPECTOR TD, SPELIOTES EK, GODDARD ME, YANG J & VISSCHER PM 2015. Population genetic differentiation of height and body mass index across Europe. Nat Genet, 47, 1357–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHOECH A, JORDAN D, LOH P-R, GAZAL S, O’CONNOR L, BALICK DJ, PALAMARA PF, FINUCANE H, SUNYAEV SR & PRICE AL 2017. Quantification of frequency-dependent genetic architectures and action of negative selection in 25 UK Biobank traits. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SELDIN MF, PASANIUC B & PRICE AL 2011. New approaches to disease mapping in admixed populations. Nat Rev Genet, 12, 523–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUDLOW C, GALLACHER J, ALLEN N, BERAL V, BURTON P, DANESH J, DOWNEY P, ELLIOTT P, GREEN J, LANDRAY M, LIU B, MATTHEWS P, ONG G, PELL J, SILMAN A, YOUNG A, SPROSEN T, PEAKMAN T & COLLINS R 2015. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med, 12, e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SVED JA, MCRAE AF & VISSCHER PM 2008. Divergence between human populations estimated from linkage disequilibrium. Am J Hum Genet, 83, 737–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG J, BAKSHI A, ZHU Z, HEMANI G, VINKHUYZEN AA, LEE SH, ROBINSON MR, PERRY JR, NOLTE IM, VAN VLIET-OSTAPTCHOUK JV, SNIEDER H, ESKO T, MILANI L, MAGI R, METSPALU A, HAMSTEN A, MAGNUSSON PK, PEDERSEN NL, INGELSSON E, SORANZO N, KELLER MC, WRAY NR, GODDARD ME & VISSCHER PM 2015. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet, 47, 1114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG J, BENYAMIN B, MCEVOY BP, GORDON S, HENDERS AK, NYHOLT DR, MADDEN PA, HEATH AC, MARTIN NG, MONTGOMERY GW, GODDARD ME & VISSCHER PM 2010. Common SNPs explain a large proportion of the heritability for human height. Nat Genet, 42, 565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZAITLEN N, PASANIUC B, GUR T, ZIV E & HALPERIN E 2010. Leveraging genetic variability across populations for the identification of causal variants. Am J Hum Genet, 86, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZENG J, DE VLAMING R, WU Y, ROBINSON MR, LLOYD-JONES LR, YENGO L, YAP CX, XUE A, SIDORENKO J, MCRAE AF, POWELL JE, MONTGOMERY GW, METSPALU A, ESKO T, GIBSON G, WRAY NR, VISSCHER PM & YANG J 2018. Signatures of negative selection in the genetic architecture of human complex traits. Nat Genet, 50, 746–753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Estimates of ρb are accurate in simulations at various parameter settings. We report estimates of ρb for various values of MT, the number of “genotyped” SNPs; MC, the number of causal SNPs; NEUR=NEAS; and NEUR (with NEAS fixed at 2K). The dashed line is the true value ρb =0.8.
Figure S2. Regularizing LD estimates is necessary to minimize biases. We report the value of the correction factor ρg/ρb (resp. ργ/ρβ) as a function of the LD window used. We report estimates using only an LD window (dashed curve), as well as estimates using both an LD window and the regularization method of Wen & Stephens 2010 Ann Appl Stat (solid curve). The horizontal dashed line is the true correction factor, that is, the value of (resp. ). The results support the use of an LD window, and indicate than the regularization method of Wen & Stephens does not impact results when an LD window is used. Standardized and mean-centered approaches tend to give similar results in practice, but they assume a different trait model and therefore can give different results, especially if causal alleles have very different alleles frequencies between the two populations.
Figure S3. Estimates of ρb are downward biased at very low values of h2. We report estimates of ρb for various values of h2. The dashed line is the true value ρb =0.8.