Abstract
Poxviruses are an unusual family of large double-stranded (ds) DNA viruses that exhibit an incredible degree of self-sufficiency and complexity in their replication and immune evasion strategies. Indeed, amongst their ~200 open reading frames (ORFs), poxviruses encode ~100 immunomodulatory proteins to counter host responses along with complete DNA synthesis, transcription, mRNA processing and cytoplasmic redox systems that enable them to replicate exclusively in the cytoplasm of infected cells. However, like all other viruses poxviruses do not encode ribosomes and therefore remain completely dependent on gaining access to the host translational machinery in order to synthesize viral proteins. Early studies of these intriguing viruses helped discover the mRNA cap and polyA tail that we now know to be present on most eukaryotic messages and which play fundamental roles in mRNA translation, while more recent studies have begun to reveal the remarkable lengths poxviruses go to in order to control both host and viral protein synthesis. Here, we discuss some of the central strategies used by poxviruses and the broader battle that ensues with the host cell to control the translation system, the outcome of which ultimately dictates the fate of infection.
Graphical Abstract

Introduction
Poxviruses are large dsDNA viruses that encode all of the necessary components for DNA replication and transcription, as well as complete mRNA processing and cytoplasmic redox systems1. Indeed, poxviruses are so self-sufficient that they, along with the sole asfivirus, African Swine Fever Virus, are the only mammalian DNA viruses that replicate entirely in the cytoplasm of infected cells. Upon fusing into the cytosol, approximately 100 early mRNAs are transcribed and processed within the viral core that then exit into the cytoplasm where they are translated by host ribosomes (Figure 1). Synthesis of early viral proteins leads to core uncoating and entry into the post-replicative state. As detailed below, this involves the shut-down of host translation and formation of large cytoplasmic structures called Viral Factories (VFs). VFs are structurally complex and compartmentalized, serving as the sites of viral DNA replication and transcription, protein synthesis and virion assembly (Figure 1).
Figure 1. Overview of Poxvirus Infection and Effects on Translation.
After poxvirus entry into the cytosol, transcriptionally competent viral cores begin to produce early viral mRNAs that are translated in the cytoplasm amongst the broader pool of host transcripts. Synthesis of early proteins leads to core uncoating and entry into the post-replicative stage. This involves the degradation of a large portion of host mRNAs along with the formation of viral factories (VFs). VFs are structurally complex and compartmentalized to facilitate many processes including DNA synthesis, transcription of intermediate and late gene products, viral protein synthesis and virion assembly. Notably, as VFs mature cavities form that are the sub-VF sites where late viral mRNAs accumulate together with ribosomes for translation (inset).
Members of the poxvirus family include Variola Virus (VarV), which evolved from a rodent taterapox virus and as the causative agent of smallpox, killed more people than all other human pathogens combined2, 3. Smallpox was eradicated through a global vaccination campaign using another orthopoxvirus, Vaccinia Virus (VacV)4. Although the only remaining stocks of smallpox are now contained in two secure locations in the US and Russia, threats from its re-emergence as well as from zoonotic poxvirus infections are numerous. Such threats include recurrent outbreaks of monkeypox that exhibits smallpox-like symptoms and is adapting to human-human transmission in areas of Africa5. In addition, Molluscum Contagiosum Virus (MCV) is a relatively common infection in humans that is transmitted through contact with infected individuals or contaminated objects6. Although self-limiting and not life-threatening, pustules caused by MCV can cover large portions of the body and persist for many months but treatment options are limited. Tissue culture systems to study MCV have remained frustratingly elusive and there are no animal models (MCV only infects humans) meaning that we continue to have a poor understanding of MCV replication, much of which is inferred from studies using other poxviruses. However, beyond their negative effects on human health, poxviruses have also had significant positive impacts. Poxviruses were the first vaccines and now, due to their amenability to genetic manipulation and tolerance for large genetic inserts, are widely used as oncolytics and gene therapy vectors7. Furthermore, their use in basic research contributed to fundamental breakthroughs including the discovery of 5’ terminal “cap”, 2’O-Methylation and 3’ polyA structures that are present on the majority of eukaryotic mRNAs8–14.
A Beginner’s Guide to mRNA Translation
Eukaryotic mRNAs consist of an open reading frame (ORF) that is flanked by 5’ and 3’ untranslated regions (UTRs) (Figure 2). These UTRs are terminally modified, with the 5’UTR harboring a 7-Methyl-GTP cap (m7G-Cap) and the 3’UTR ending with a polyadenylated (polyA) tail. Both of these modifications play important roles in mRNA biogenesis, export, stability and translation. In terms of translation, the 5’ m7G-Cap is recognized by the eukaryotic initiation factor eIF4E, a subunit of the larger eIF4F complex15, 16. eIF4E binds the large scaffold protein, eIF4G that also binds the helicase, eIF4A. Together, these eIF4F subunits provide cap-recognition and helicase activities that facilitate the loading of 40S ribosome subunits onto the 5’ end of the mRNA. eIF4F does not directly load 40S subunits but does so through bridging interactions with the 13-subunit complex, eIF3. Ribosome recruitment is carefully regulated through the activity of various signaling pathways that respond to diverse environmental cues. In particular, the kinase mammalian Target of Rapamycin (mTOR) controls several aspects of eIF4F and eIF3 function to balance rates of translation with energy and nutrient availability, as well as in response to mitogenic signals (Figure 2)17, 18. mTOR resides in two distinct complexes, mTOCR1 and mTORC2. While both mTORCs share several common binding partners, each complex is distinguished by a unique regulator that controls their activity and substrate specificity; mTORC1 binds Raptor, while mTORC2 binds Rictor. mTORC2 regulates the actin cytoskeleton and stimulates Akt activity. Notably, mTORC1 is activated by PI3K-Akt signaling and so, to prevent formation of a feed-forward loop, specific mTORC1 substrates inhibit mTORC2 and PI3K activity. This feedback loop carefully balances mTORC1 and mTORC2 activity, as well as broader PI3K signaling17–19. Beyond upstream control by Akt, mTORC1 responds to energy and amino acid sensors, and directly controls protein synthesis. One of mTORC1’s primary effector targets is the translational repressor, eIF4E-binding protein 1 (4E-BP1). In its repressive, hypo-phosphorylated state 4E-BP1 competes with eIF4G for binding to eIF4E, thereby inhibiting eIF4F complex formation and translation initiation15, 17. Once activated, mTORC1 inactivates 4E-BP1 through multi-site phosphorylation that results in the release of eIF4E. Once released and assembled into an eIF4F complex, eIF4E can also be phosphorylated by the eIF4G-associated kinase, Mnk120. Mnk1 phosphorylation is stimulated by Extracellular Signal Regulated Kinase (ERK) and stress-related p38 Mitogen-Activated Protein Kinase (p38MAPK), further linking eIF4E phosphorylation and eIF4F activity to environmental cues. Although the mechanistic basis by which eIF4E phosphorylation functions remains unclear it preferentially regulates the translation of specific mRNAs, discussed further below.
Figure 2: Translational Control in the Host Cell.
In response to mitogenic signaling, PI3K activates Akt that in turn inactivates the Tuberous Sclerosis Complex (TSC1/2), a repressor of mTOR’s GTPase, RHEB1. This activates mTORC1, containing Raptor, resulting in phosphorylation and inactivation of 4E-BP1. mTORC2, containing Rictor, also activates Akt. Through complex pathways (dotted lines) that prevent a feed-forward loop from forming, mTORC1 substrates such as p70S6K or Grb10 suppress mTORC2 and/or PI3K activity to control Akt. Once 4E-BP1 has been inactivated, eIF4E is released to bind eIF4G and can be phosphorylated by the kinase, Mnk1 as part of the eIF4F complex. Through bridging interactions with eIF3, eIF4F recruit the 40S ribosome that is loaded with the eIF2-tRNAMet ternary complex. Once the AUG start codon is recognized, GTP hydrolysis and eIF2-GDP release occurs in a manner facilitated by other eIFs (dotted lines). eIF2-GDP is then recycled by eIF2B. Further details of these processes are provided in the main text.
Once loaded onto the 5’ end of the mRNA, the 40S ribosome subunit associated with a large number of eIFs in the form of a 48S pre-initiation complex scans the UTR in search of the start codon, which is often but not always an AUG (Figure 2)15. AUG recognition is mediated by a specialized methionine initiator tRNA (tRNAMet) bound to eIF2-GTP as a ternary complex. During initiation, GTP hydrolysis releases eIF2-GDP and tRNAMet, and the 60S ribosomal subunit joins to form a translationally competent 80S ribosome. The guanine nucleotide exchange factor eIF2B converts free eIF2-GDP back into eIF2-GTP to be reloaded with tRNAMet for a new round of initiation. Meanwhile, translation of the ORF proceeds until a stop codon is reached at which point the ribosome either disengages the mRNA, assisted by a termination and release factors, or initiates a new round of translation on the same mRNA21. Both initiation and re-initiation are facilitated by the 3’polyA tail (Figure 2); the polyA tail is bound by polyA-binding protein (PABP) that in turn binds the eIF4G scaffold subunit of eIF4F. These interactions circularize mRNAs and are thought to ensure efficient translation on properly processed, mature mRNAs.
Control of virtually every step in this complex translational control pathway plays a role in the outcome of infection16. Regulating gene expression at the level of translation enables the host to respond to infection by swiftly synthesizing new antiviral proteins, including interferon (IFN) stimulated genes that have direct antiviral activity and IFNs that signal to neighboring cells, alerting them to the presence of a pathogen. Additional host cell responses also center on preventing viral access to the ribosomes they need to make viral proteins. Viruses in turn have evolved means to wrangle control of the translation system from the host to make viral polypeptides and to counter antiviral responses. While many RNA viruses employ cap-independent modes of initiation that allow them to impair cap-dependent translation of host mRNAs16, DNA viruses including poxviruses generate capped and polyadenylated mRNAs and therefore require very different strategies.
Enter Poxviruses: Masters Manipulators of Translation Initiation
Poxviruses are not only transcriptionally self-sufficient they also encode their own capping and polyadenylation machinery1, 8, 11, 22, 23, which other DNA viruses usurp from their host by replicating in the nucleus24. Early studies found that poxviruses methylate a number of nucleotides around the 5’ end of their mRNAs and established the particular importance of the m7G-Cap for translation of viral transcripts using in vitro translation systems13. By contrast, 2’O-Methylation was not found to play a significant role in VacV mRNA translation using these cell-free systems. Interestingly, it would later be discovered that 2’O-Methylation is used by the host to mark “self” transcripts while unmodified “foreign” mRNAs are recognized by antiviral proteins and translationally repressed25. As such, while this 2’O-Methylation does not directly stimulate translation in cell extracts it likely enables poxvirus mRNAs to evade detection by their host.
VacV, the most widely studied prototype for poxvirus infection, stimulates both p38MAPK and ERK signaling pathways to increase eIF4E phosphorylation26. While this is thought to promote translation of capped viral mRNAs, eIF4E phosphorylation also selectively increases the synthesis of negative regulators of the IFN response27. As such, stimulating eIF4E phosphorylation may serve the dual purpose of enhancing viral protein synthesis and suppressing IFN responses, although this remains to be fully explored. VacV, like a number of other large DNA viruses, also stimulates the formation of eIF4F complexes16. This stimulation is achieved in part through the activation of mTORC1 to inactivate 4E-BP1 and through the concentration of eIF4F subunits within cytoplasmic VFs, where late viral protein synthesis occurs26, 28. The concentration of eIF4F subunits is achieved in part through binding of eIF4G by the viral I3 protein that localizes to VFs29. Although the m7G-cap plays an important role in VacV mRNA translation in cell extracts13 there continues to be debate over the degree to which poxvirus mRNAs actually require eIF4F. Early studies using viral proteases to cleave eIF4G reported minimal effects on VacV protein synthesis in infected cells30. However, more recent studies suggest a significant dependence of VacV translation on eIF4G in several cellular contexts31–33, as well as dependencies on eIF4E phosphorylation and mTORC1 activity in primary normal human cells26, 34–36. These different observations may reflect the type of approaches or transformed cell types used in specific studies. However, understanding the dependence of VacV translation on eIF4F and its subunits is further complicated by observations that poxvirus mRNAs have the capacity to utilize either cap-dependent or -independent modes of initiation37, 38, discussed below. Finally, microRNAs (miRNAs) can be deployed by host cells during infection, which repress eIF4F activity. To counter this VacV can impair the host miRNA processing machinery, in part through I7-mediated cleavage of Dicer as well as through miRNA polyadenylation by the viral polyA-polymerase that results in miRNA degradation39–42.
Impairing Host Translation
Despite capping their own transcripts and stimulating the host cap-dependent translation machinery, poxviruses encode two decapping enzymes, D9 and D10 that do not discriminate between host or viral mRNAs43–45. These enzymes destabilize host transcripts as part of the host shut-off strategy and also help to remove early viral mRNAs as infection transits into intermediate and late stages. Poxviruses also encode small non-translated polyadenylated mRNAs termed POLADS that appear to act as decoys for PABP to suppress translation46, 47. In addition, the VacV 169 protein resides in the cytoplasm and impairs translation initiation, but is not present at VFs where late viral protein synthesis occurs48. The localized activity of 169 protein likely offers a means to selectively impair host translation and antiviral responses. Indeed, poxviruses utilize complex multi-pronged strategies to control the overall translational landscape. In cells infected with either VacV or Rabbit poxvirus (RPV) the abundance of many host mRNAs dramatically declines yet specific transcripts are spared or even increased in abundance49–51. Studies using VacV suggest that those genes selectively increased in expression at early times likely represent host responses to infection50 while those increased at later times, such as the actin regulator WASP, are functionally important for VacV replication51. This selectivity suggests a certain degree of viral control over the host gene expression program despite replicating in the cytoplasm. Moreover, on a translational level, certain host mRNAs are also selectively retained on polysomes during VacV infection52. This selective translational control of host mRNAs also appears to have a purpose, at least to some extent, as some of these mRNAs encode components of metabolic pathways that benefit virus replication. How poxviruses accomplish this selectivity remains unknown, but this illustrates the mastery by which they take control of the host translation system.
Host and Viral Transcripts: Similar, But Not Quite the Same
Although they harbor a 5’ m7G-cap and 3’ polyA-tail like their host counterparts, a unique feature of poxvirus mRNAs is the presence of 5’ polyA-leaders. While short polyA stretches can be found in the UTRs of a few early poxvirus transcripts, the UTRs of post-replicative (intermediate and late) mRNAs are unusual in that they consist almost entirely of polyA53–61. Post-replicative genes rarely encode a UTR directly but instead, the viral RNA polymerase slips on a unique “TTT” motif at the transcriptional start site (TSS) that immediately precedes the translation start codon62–65. Polymerase slippage results in the random reiteration of adenosine residues and is likely an efficient way to generate non-templated 5’ UTRs. Early studies of different late transcripts using a variety of approaches revealed that these leaders averaged 30–40 adenosines in length but could extent to hundreds or thousands of bases. Recent high-throughput sequencing confirmed the presence of polyA leaders on all post-replicative mRNAs, in line with the presence of the “TTT” motif at their TSS55. Notably, this study detected leaders as long as 51 nucleotides (nts), but medians were 8 (intermediate) and 11 (late) nts. However, Illumina sequencing is only capable of relatively short reads and homopolymeric adenosine runs pose problems for the polymerases used. As such, these median lengths may be underestimates given earlier size determinations and points below.
Recent work has begun to illuminate how these long-enigmatic leader elements contribute to poxvirus protein synthesis. Initial studies showed that polyA leaders conferred a reduced initiation factor dependency on mRNAs using cell free translation systems, and length comparisons showed that polyA leaders of 25nts functioned significantly more efficiently than those 12 or 5 nts long37. In line with this, subsequent studies showed that polyA-leader mRNAs could initiate in both cap-dependent and – independent manners in cells but determined that 12nts was optimal38. However, the longest leader tested in this study was 20nts. While differences in the leader lengths tested or the systems used may underlie these minor discrepancies, both studies conclusively show that poxvirus mRNAs can initiate translation with a lower dependence upon initiation factors than host transcripts. It would be of interest to determine if leaders of 30–40nt or greater that are found in infected cells function even more efficiently, and whether they also initiate in a cap-independent manner. Notably, these polyA leaders do not operate as internal ribosome entry sites that are employed by many RNA viruses16, 38. Although eIF4F activity is stimulated in primary cells and VacV replication is dependent on eIFs in a number of different cell types, the ability to utilize either cap-dependent or -independent ribosome loading strategies may allow viral mRNAs to outcompete host mRNAs or gain access to the translational machinery during conditions of cellular stress that impair cap-dependent initiation. The mechanistic basis by which poxvirus mRNAs initiate translation in a cap-independent manner, whether this only occurs for a subpopulation of mRNAs with shorter leaders, and why they do so when poxviruses do not inactivate eIFs as RNA viruses do will be of significant interest to determine.
PolyA-leaders also contribute to translation in other unusual ways. Despite differences in length estimates discussed above, polyA stretches ≥11nt cause ribosomes to undergo “phaseless wandering” or “sliding”, whereby they move bi-directionally37, 66, 67. This behavior may contribute to the ability of polyA leaders to initiate in a cap-independent manner37 but ribosome sliding is also central to mammalian ribosome quality control (RQC)68. RQC enables the host cell to detect aberrant mRNAs (e.g. lacking a stop codon or prematurely polyadenylated) or aberrant translation events (e.g. frameshifts and stop codon read-through). Decoding of the 3’ polyA tail stalls ribosomes, alerting the cell to the aberrant transcript or events resulting in destruction of the mRNA in question. Two key features of polyA sequences function in this process; ribosomes lose processivity on polyA tracts while reiteration of lysines encoded by AAA codons causes ribosome stalling. In addition, ribosome sliding on polyA likely exacerbates the number of AAA codons that are decoded even on shorter polyA tracts. For this reason, polyA stretches ≥11nt are selected against outside the 3’UTR in mammals, leaving the polyA tail to act as a sensor in RQC, highlighting the unusual nature of poxvirus post-replicative mRNAs. It is important to note that while initiating 40S ribosome subunits can slide37, lysine-induced stalling would not operate on 5’ polyA as it is not being decoded during scanning. Despite this, recent evidence suggests that poxviruses exploit several factors involved in RQC to promote viral protein synthesis. One such factor is Receptor for Activated C Kinase 1 (RACK1), a 40S subunit protein that also controls a form of ribosome slippage called frameshifting68–70. The poxvirus kinase, B1 phosphorylates RACK1 to stimulate translation of polyA-leader mRNAs71. Notably, unlike the VacV 169 protein that impairs host translation in the cytoplasm, RACK1 accumulates with other ribosomal proteins and the B1 kinase within cavity substructures of the VF where late viral mRNAs reside. More intriguingly, the RACK1 phosphorylation event appears to be unique to poxvirus infection and occurs in a flexible loop region, which mimics negative charge that is found in the loop of plant RACK1. Plants utilize A-rich leaders in their immune response mRNAs72 and some plant viruses utilize A-rich leader elements, such as the TMV “Omega Leader”73. As such, poxviruses not only exploit RQC factors but also appear to mimic aspects of translational control strategies that operate in plants where A-rich leaders function well as enhancers. Beyond RACK1, recent studies have shown that regulatory mono-ubiquitination of the small ribosomal proteins, RPS10 and RPS20 by the E3 ligase, ZNF598 regulate the early stages of ribosome stalling during RQC69, 74. While this represses translational read-through of polyA in their hosts, poxviruses coopt ZNF598 and RPS ubiquitination to promote viral protein synthesis75. These findings suggest that poxviruses have adapted to exploit several components of the host RQC pathway that revolves around polyA-based sensing, turning what are normally repressive factors into positive ones for virus replication.
Exploiting mTOR to Control Host Translation and Beyond
mTORC1 regulates the activity of several stages in the translation process by both directly and indirectly targeting initiation and elongation factors as well as ribosomal proteins. It also regulates a wider range of metabolic processes that are important to infection and is emerging as a key regulator of both innate and adaptive immunity. Several poxviruses stimulate upstream signaling pathways that control mTORC1, in particular PI3K-Akt. The rabbit poxvirus, myxomavirus (MYXV) encodes a protein M-T5 that binds and activates Akt76–78, but effects of M-T5 on mTORC1 or translation have not been examined. However, pharmacological modulation of mTOR activity has been found to influence MYXV tropism for human cells and shows potential for their use as oncolytics79. VacV also activates Akt and does so in a bi-phasic manner80. The first activation occurs early during entry when viral engagement of integrin receptors triggers PI3K-Akt signaling81. Studies have shown that Akt contributes to several aspects of infection ranging from early entry processes to late stage virion morphogenesis and cell survival80 as well as viral protein synthesis26, 34. However, recent studies have shown that the second phase of Akt-mTORC1 activation that occurs later in infection is quite unusual and facilitates virus replication in multiple ways. On a mechanistic level, the poxvirus late protein F17 binds and sequesters both Raptor and Rictor82. As Raptor and Rictor act as the regulatory substrate gatekeepers for each mTORC, their removal by F17 hyperactivates mTOR kinase activity to enhance protein synthesis. However, this strategy also serves to dysregulate mTORC1-mTORC2 crosstalk and thereby disrupt cytosolic sensing of poxvirus DNA by the cGAS-STING pathway82. As outlined earlier, mTORC1 negatively regulates mTORC2 to prevent formation of a feed-forward loop through Akt (Figure 2). This regulatory circuit also plays a critical role in host sensing and response pathways because active mTORC1 dampens STING activity82, 83. Yet, if the cell lowers mTORC1 activity to enable STING to operate the circuit will activate mTORC2, which in turn activates Akt that destabilizes cGAS82, 84, 85. As such, maintaining expression of the DNA sensor cGAS and mounting a cGAS-STING-mediated response necessitates that mTORC1 and mTORC2 activities are carefully balanced (Figure 2). By disrupting this mTORC1-mTORC2 regulatory circuit F17 causes degradation of cGAS and dampens STING signaling. While many viruses control mTORC1, they normally do so by acting on upstream components of the pathway16. This unusual strategy of directly targeting and dysregulating mTOR may be unique to cytoplasmically replicating poxviruses in order to evade sensing while retaining the metabolic benefits of mTOR.
Additional Defenses Through RNA Sensing and “Expanded” Viral Counter-Strategies
Beyond sensors for viral DNA, a central line of defense against a wide range of both RNA and DNA viruses centers on detection of dsRNA that is produced at some point in the replication cycle of virtually all viruses. dsRNA activates two sensors that subsequently impair translation; 2’−5’ Oligoadenylate Synthetase (OAS), which degrades ribosomal mRNAs, and Protein Kinase R (PKR)16. Production of IFN by infected cells also signals to neighboring cells to increase PKR levels, preparing them to fight the impending infection. When PKR binds dsRNA it dimerizes and auto-phosphorylates, activating its kinase activity to phosphorylate eIF2α. Phosphorylated eIF2 tightly binds and sequesters the limited amounts of the critical GTP recycling factor, eIF2B resulting in a potent shutdown of translation. While eIF2 inactivation drastically impairs translation and kills the host cell it very effectively limits viral protein synthesis and the spread of infection, in a form of altruistic cellular suicide for the greater good of the organism. Perhaps unsurprisingly, viruses have evolved an array of strategies to counteract PKR and OAS activity.
In the case of poxviruses, transcriptional read-through of early and intermediate genes generates transcripts that overlap and form dsRNA species that can be detected by the cell63, 86–88. To counter this, the viral decapping enzymes D9 and D10 function together with the endonuclease XRN1 to reduce the accumulation of dsRNA89, 90. The inability of D9/D10 mutants to clear dsRNA limits their replication in vivo and is now being explored as a potential oncolytic virotherapy45, 90, 91. Interestingly, the levels of dsRNA production vary between different poxviruses and may contribute to the relatively greater resistance of monkeypox over VacV to the antiviral, isatin-β-thiosemicarbazone (IBT)92. Beyond limiting dsRNA accumulation, the VacV K3 protein (or its related proteins, C8 in Swine Pox or M156 in MYXV) acts as an eIF2α pseudosubstrate and decoy for PKR, while the VacV E3 protein prevents PKR activation both by sequestering dsRNA and through additional mechanisms independent of its dsRNA-binding activity93–109. Although PKR is a major effector of host restriction toward E3 mutants, E3 also counters several PKR-independent host responses108, 110–112. Due to its importance in virus replication, VacV E3L (the gene encoding the E3 protein) mutants are also being studied for their potential use as vaccines113. Similarly, M029, a truncated relative of E3 encoded by MYXV blocks antiviral responses through both PKR-dependent and – independent mechanisms114, 115, highlighting the multi-functionality of many viral proteins. In addition to this, the host range proteins K1 and C7 of VacV, along with their counterparts in other poxviruses, also function to block PKR activity in part through binding dsRNA87, 116, 117. However, their functions in this regard are less well understood than E3 or K3, and is complicated by their roles in countering broader antiviral responses118–121. Preventing eIF2α phosphorylation also serves to prevent the formation of stress granules, which are sites of mRNA sequestration into translationally inactive structures that localize around VFs and have antiviral activity122, 123. These “antiviral granules” form through eIF2α phosphorylation but may also have eIF2-independent antiviral activity through factors such as TIA-1122.
As the crux of host antiviral defense, PKR can evolutionarily adapt through subtle changes to evade viral antagonists124–127. This in turn forces viral counter-adaptation. A remarkable example of this is seen when VacV deficient in E3, the more potent PKR antagonist, is challenged to replicate in human cells using its weaker PKR antagonist, K3. Unlike RNA viruses that adapt through high mutation rates, DNA viruses have a relatively low mutation rate. During serial passage in culture however, E3L-null poxviruses amplify multiple copies of K3L in what is termed a “gene accordion”128. This allows the virus to retain an original copy of this weak antagonist while other K3L copies gradually acquire mutations and explore fitness benefits. Once a K3L copy acquires a mutation that results in a better PKR antogonist the “gene accordion” can collapse again to retain the newly adapted K3L gene128–130. In further exploring the role of gene accordions VacV lacking either E3L, or VacV lacking both E3L and K3L but expressing a weak Cytomegalovirus antagonist TRS1, unexpectedly revealed additional adaptive mutations that can be acquired in to enable PKR evasion in diverse ways131, 132. This includes distinct mutations in A24R, which encodes the viral RNA polymerase, and changes in its functionality may influence the levels of dsRNA accumulation during infection131, 132. Interestingly, mutations in A24R also conferred greater resistance to IBT131, similar to monkeypox that produces less dsRNA, above. The consequences of this continuous coevolution and adaptation that revolves around PKR has also been observed in the wild. MYXV was released in Australia to control the explosive spread of European rabbits, with devastating effect. However, eventual attenuation occurred and this was recently mapped to the acquisition of specific loss-of-function mutations in M156, the MYXV-encoded antagonist of rabbit PKR133. This highlights the ongoing and continually adapting war that poxviruses, and many other viruses, wage with their host to control the translation system through PKR.
Concluding Remarks
Poxviruses have arguably had a more profound impact on human health, in both positive and negative ways, than any other pathogen. As DNA viruses, they are also quite remarkable in their cytoplasmic replication and self-sufficiency. Studies of poxviruses provided fundamental insights into mRNA processing and stability, unusual modes of translation such as cap-independent initiation through phase-less wandering and plant ribosome mimicry, as well as extraordinary adaptation to host translation-based defenses through gene accordions. As they have done repeatedly in the past, future studies of poxviruses will undoubtedly continue to reveal new and fascinating insights into host-pathogen interactions as well as more fundamental aspects of translational control.
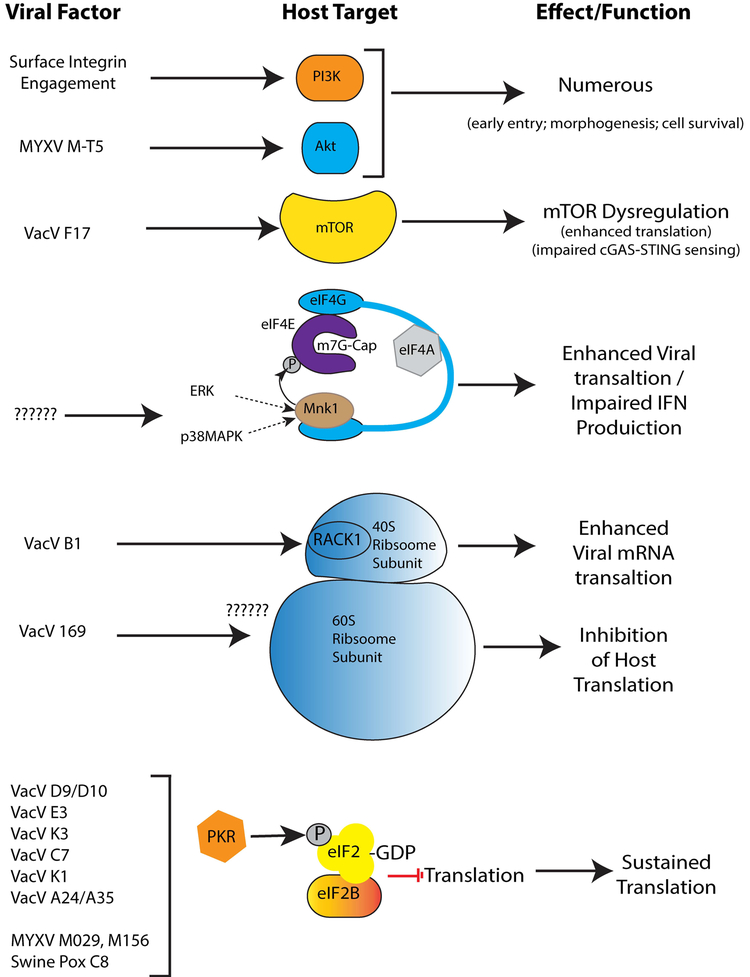
Figure 3: Poxvirus Factors that Regulate Translation during Infection.
Examples of poxvirus proteins that control various aspects of translation, as discussed in the main text. “?????” point to some interesting gaps in our current knowledge; how poxviruses activate ERK/p38MAPK signaling and the target (ribosomal or non-ribosomal?) of the 169 protein that results in the formation of inactive 80S ribosomes in the cytoplasm remains unknown. Poxviruses such as VacV encode a wide array of proteins that counter the function of PKR in phosphorylating eIF2α, which sequesters eIF2B and potently inhibits translation.
References
- 1.Moss B Poxviridae: the viruses and their replication. In: DMKaPM Howley, ed. Fields virology. Philadelphia, PA: Lippincott Williams & Wilkins; 2007, 2849–2883. [Google Scholar]
- 2.McFadden G Poxvirus tropism. Nat Rev Microbiol 2005, 3:201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Carroll DS, Gardner SN, Walsh MC, Vitalis EA, Damon IK. On the origin of smallpox: correlating variola phylogenics with historical smallpox records. Proc Natl Acad Sci U S A 2007, 104:15787–15792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moss B Smallpox vaccines: targets of protective immunity. Immunol Rev 2011, 239:8–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shchelkunov SN. An increasing danger of zoonotic orthopoxvirus infections. PLoS Pathog 2013, 9:e1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shisler JL. Immune evasion strategies of molluscum contagiosum virus. Adv Virus Res 2015, 92:201–252. [DOI] [PubMed] [Google Scholar]
- 7.Chan WM, McFadden G. Oncolytic Poxviruses. Annu Rev Virol 2014, 1:119–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urushibara T, Furuichi Y, Nishimura C, Miura K. A modified structure at the 5’-terminus of mRNA of vaccinia virus. FEBS Lett 1975, 49:385–389. [DOI] [PubMed] [Google Scholar]
- 9.Ensinger MJ, Martin SA, Paoletti E, Moss B. Modification of the 5’-terminus of mRNA by soluble guanylyl and methyl transferases from vaccinia virus. Proc Natl Acad Sci U S A 1975, 72:2525–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kates J, Beeson J. Ribonucleic acid synthesis in vaccinia virus. II. Synthesis of polyriboadenylic acid. J Mol Biol 1970, 50:19–33. [DOI] [PubMed] [Google Scholar]
- 11.Wei C, Moss B. 5’-Terminal capping of RNA by guanylyltransferase from HeLa cell nuclei. Proc Natl Acad Sci U S A 1977, 74:3758–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbosa E, Moss B. mRNA(nucleoside-2’-)-methyltransferase from vaccinia virus. Characteristics and substrate specificity. J Biol Chem 1978, 253:7698–7702. [PubMed] [Google Scholar]
- 13.Muthukrishnan S, Moss B, Cooper JA, Maxwell ES. Influence of 5’-terminal cap structure on the initiation of translation of vaccinia virus mRNA. J Biol Chem 1978, 253:1710–1715. [PubMed] [Google Scholar]
- 14.Shuman S, Moss B. Vaccinia virus poly(A) polymerase. Specificity for nucleotides and nucleotide analogs. J Biol Chem 1988, 263:8405–8412. [PubMed] [Google Scholar]
- 15.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 2014, 83:779–812. [DOI] [PubMed] [Google Scholar]
- 16.Jan E, Mohr I, Walsh D. A Cap-to-Tail Guide to mRNA Translation Strategies in Virus-Infected Cells. Annu Rev Virol 2016, 3:283–307. [DOI] [PubMed] [Google Scholar]
- 17.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 169:361–371. [DOI] [PubMed] [Google Scholar]
- 18.Kim SG, Buel GR, Blenis J. Nutrient regulation of the mTOR complex 1 signaling pathway. Mol Cells 2013, 35:463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011, 332:1322–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proud CG. Mnks, eIF4E phosphorylation and cancer. Biochim Biophys Acta 2015, 1849:766–773. [DOI] [PubMed] [Google Scholar]
- 21.Schuller AP, Green R. Roadblocks and resolutions in eukaryotic translation. Nat Rev Mol Cell Biol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei CM, Moss B. Methylated nucleotides block 5’-terminus of vaccinia virus messenger RNA. Proc Natl Acad Sci U S A 1975, 72:318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei C, Gershowitz A, Moss B. N6, O2’-dimethyladenosine a novel methylated ribonucleoside next to the 5’ terminal of animal cell and virus mRNAs. Nature 1975, 257:251–253. [DOI] [PubMed] [Google Scholar]
- 24.Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 2012, 10:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, et al. 2’-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 2010, 468:452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walsh D, Arias C, Perez C, Halladin D, Escandon M, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Mohr I. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol Cell Biol 2008, 28:2648–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herdy B, Jaramillo M, Svitkin YV, Rosenfeld AB, Kobayashi M, Walsh D, Alain T, Sean P, Robichaud N, Topisirovic I, et al. Translational control of the activation of transcription factor NF-kappaB and production of type I interferon by phosphorylation of the translation factor eIF4E . Nat Immunol 2012, 13:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsafanas GC, Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2007, 2:221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaborowska I, Kellner K, Henry M, Meleady P, Walsh D. Recruitment of host translation initiation factor eIF4G by the Vaccinia Virus ssDNA-binding protein I3. Virology 2012, 425:11–22. [DOI] [PubMed] [Google Scholar]
- 30.Mulder J, Robertson ME, Seamons RA, Belsham GJ. Vaccinia virus protein synthesis has a low requirement for the intact translation initiation factor eIF4F, the cap-binding complex, within infected cells. J Virol 1998, 72:8813–8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barco A, Feduchi E, Carrasco L. A stable HeLa cell line that inducibly expresses poliovirus 2A(pro): effects on cellular and viral gene expression. J Virol 2000, 74:2383–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welnowska E, Castelló A, Moral P, Carrasco L. Translation of mRNAs from vesicular stomatitis virus and vaccinia virus is differentially blocked in cells with depletion of eIF4GI and/or eIF4GII. J Mol Biol 2009, 394:506–521. [DOI] [PubMed] [Google Scholar]
- 33.Marcet-Palacios M, Duggan BL, Shostak I, Barry M, Geskes T, Wilkins JA, Yanagiya A, Sonenberg N, Bleackley RC. Granzyme B inhibits vaccinia virus production through proteolytic cleavage of eukaryotic initiation factor 4 gamma 3. PLoS Pathog 2011, 7:e1002447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaborowska I, Walsh D. PI3K signaling regulates rapamycin-insensitive translation initiation complex formation in vaccinia virus-infected cells. J Virol 2009, 83:3988–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahon R, Zaborowska I, Walsh D. Noncytotoxic inhibition of viral infection through eIF4F-independent suppression of translation by 4EGi-1. J Virol 2011, 85:853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh D, Mohr I. Coupling 40S ribosome recruitment to modification of a cap-binding initiation factor by eIF3 subunit e. Genes Dev 2014, 28:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shirokikh NE, Spirin AS. Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors. Proc Natl Acad Sci U S A 2008, 105:10738–10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dhungel P, Cao S, Yang Z. The 5’-poly(A) leader of poxvirus mRNA confers a translational advantage that can be achieved in cells with impaired cap-dependent translation. Plos Pathogens 2017, 13:e1006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Backes S, Shapiro JS, Sabin LR, Pham AM, Reyes I, Moss B, Cherry S, tenOever BR. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe 2012, 12:200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grinberg M, Gilad S, Meiri E, Levy A, Isakov O, Ronen R, Shomron N, Bentwich Z, Shemer-Avni Y. Vaccinia virus infection suppresses the cell microRNA machinery. Arch Virol 2012, 157:1719–1727. [DOI] [PubMed] [Google Scholar]
- 41.Buck AH, Ivens A, Gordon K, Craig N, Houzelle A, Roche A, Turnbull N, Beard PM. Quantitative Analysis of MicroRNAs in Vaccinia virus Infection Reveals Diversity in Their Susceptibility to Modification and Suppression. PLoS One 2015, 10:e0131787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen JS, Li HC, Lin SI, Yang CH, Chien WY, Syu CL, Lo SY. Cleavage of Dicer protein by I7 protease during vaccinia virus infection. PLoS One 2015, 10:e0120390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parrish S, Resch W, Moss B. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc Natl Acad Sci U S A 2007, 104:2139–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parrish S, Moss B. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J Virol 2007, 81:12973–12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu SW, Wyatt LS, Orandle MS, Minai M, Moss B. The D10 decapping enzyme of vaccinia virus contributes to decay of cellular and viral mRNAs and to virulence in mice. J Virol 2014, 88:202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cacoullos N, Bablanian R. Polyadenylated RNA sequences produced in vaccinia virus-infected cells under aberrant conditions inhibit protein synthesis in vitro. Virology 1991, 184:747–751. [DOI] [PubMed] [Google Scholar]
- 47.Lu C, Bablanian R. Characterization of small nontranslated polyadenylylated RNAs in vaccinia virus-infected cells. Proc Natl Acad Sci U S A 1996, 93:2037–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strnadova P, Ren H, Valentine R, Mazzon M, Sweeney TR, Brierley I, Smith GL. Inhibition of Translation Initiation by Protein 169: A Vaccinia Virus Strategy to Suppress Innate and Adaptive Immunity and Alter Virus Virulence. PLoS Pathog 2015, 11:e1005151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brum LM, Lopez MC, Varela JC, Baker HV, Moyer RW. Microarray analysis of A549 cells infected with rabbitpox virus (RPV): a comparison of wild-type RPV and RPV deleted for the host range gene, SPI-1. Virology 2003, 315:322–334. [DOI] [PubMed] [Google Scholar]
- 50.Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc Natl Acad Sci U S A 2010, 107:11513–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guerra S, Lopez-Fernandez LA, Pascual-Montano A, Munoz M, Harshman K, Esteban M. Cellular gene expression survey of vaccinia virus infection of human HeLa cells. J Virol 2003, 77:6493–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai A, Cao S, Dhungel P, Luan Y, Liu Y, Xie Z, Yang Z. Ribosome Profiling Reveals Translational Upregulation of Cellular Oxidative Phosphorylation mRNAs during Vaccinia Virus-Induced Host Shutoff. J Virol 2017, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahn BY, Jones EV, Moss B. Identification of the vaccinia virus gene encoding an 18-kilodalton subunit of RNA polymerase and demonstration of a 5’ poly(A) leader on its early transcript. J Virol 1990, 64:3019–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel DD, Pickup DJ. Messenger RNAs of a strongly-expressed late gene of cowpox virus contain 5’-terminal poly(A) sequences. EMBO J 1987, 6:3787–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. Genome-wide analysis of the 5’ and 3’ ends of vaccinia virus early mRNAs delineates regulatory sequences of annotated and anomalous transcripts. J Virol 2011, 85:5897–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wright CF, Moss B. In vitro synthesis of vaccinia virus late mRNA containing a 5’ poly(A) leader sequence. Proc Natl Acad Sci U S A 1987, 84:8883–8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwer B, Visca P, Vos JC, Stunnenberg HG. Discontinuous transcription or RNA processing of vaccinia virus late messengers results in a 5’ poly(A) leader. Cell 1987, 50:163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bertholet C, Van Meir E, ten Heggeler-Bordier B, Wittek R. Vaccinia virus produces late mRNAs by discontinuous synthesis. Cell 1987, 50:153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baldick CJ Jr., Moss B. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate-stage genes. J Virol 1993, 67:3515–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee-Chen GJ, Niles EG. Map positions of the 5’ ends of eight mRNAs synthesized from the late genes in the vaccinia virus HindIII D fragment. Virology 1988, 163:80–92. [DOI] [PubMed] [Google Scholar]
- 61.Ahn BY, Moss B. Capped poly(A) leaders of variable lengths at the 5’ ends of vaccinia virus late mRNAs. J Virol 1989, 63:226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davison AJ, Moss B. Structure of vaccinia virus late promoters. J Mol Biol 1989, 210:771–784. [DOI] [PubMed] [Google Scholar]
- 63.Rosel JL, Earl PL, Weir JP, Moss B. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J Virol 1986, 60:436–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwer B, Stunnenberg HG. Vaccinia virus late transcripts generated in vitro have a poly(A) head. EMBO J 1988, 7:1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Magistris L, Stunnenberg HG. Cis-acting sequences affecting the length of the poly(A) head of vaccinia virus late transcripts. Nucleic Acids Res 1988, 16:3141–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arthur L, Pavlovic-Djuranovic S, Smith-Koutmou K, Green R, Szczesny P, Djuranovic S. Translational control by lysine-encoding A-rich sequences. Sci Adv 2015, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, Green R. Ribosomes slide on lysine-encoding homopolymeric A stretches. Elife 2015, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joazeiro CAP. Ribosomal Stalling During Translation: Providing Substrates for Ribosome-Associated Protein Quality Control. Annu Rev Cell Dev Biol 2017, 33:343–368. [DOI] [PubMed] [Google Scholar]
- 69.Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ. ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol Cell 2017, 65:751–760 e754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolf AS, Grayhack EJ. Asc1, homolog of human RACK1, prevents frameshifting in yeast by ribosomes stalled at CGA codon repeats. RNA 2015, 21:935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jha S, Rollins MG, Fuchs G, Procter DJ, Hall EA, Cozzolino K, Sarnow P, Savas JN, Walsh D. Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature 2017, 546:651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu G, Greene GH, Yoo H, Liu L, Marques J, Motley J, Dong X. Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 2017, 545:487–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gallie DR, Sleat DE, Watts JW, Turner PC, Wilson TM. The 5’-leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo. Nucleic Acids Res 1987, 15:3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, Amiri M, Shapiro M, Molina H, Tuschl T, et al. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat Commun 2017, 8:16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DiGiuseppe S, Rollins MG, Bartom ET, Walsh D. ZNF598 Plays Distinct Roles in Interferon-Stimulated Gene Expression and Poxvirus Protein Synthesis. Cell Rep 2018, 23:1249–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Werden SJ, Barrett JW, Wang G, Stanford MM, McFadden G. M-T5, the ankyrin repeat, host range protein of myxoma virus, activates Akt and can be functionally replaced by cellular PIKE-A. J Virol 2007, 81:2340–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Werden SJ, McFadden G. The role of cell signaling in poxvirus tropism: the case of the M-T5 host range protein of myxoma virus. Biochim Biophys Acta 2008, 1784:228–237. [DOI] [PubMed] [Google Scholar]
- 78.Werden SJ, Lanchbury J, Shattuck D, Neff C, Dufford M, McFadden G. The myxoma virus m-t5 ankyrin repeat host range protein is a novel adaptor that coordinately links the cellular signaling pathways mediated by Akt and Skp1 in virus-infected cells. J Virol 2009, 83:12068–12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stanford MM, Barrett JW, Nazarian SH, Werden S, McFadden G. Oncolytic virotherapy synergism with signaling inhibitors: Rapamycin increases myxoma virus tropism for human tumor cells. J Virol 2007, 81:1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soares JA, Leite FG, Andrade LG, Torres AA, De Sousa LP, Barcelos LS, Teixeira MM, Ferreira PC, Kroon EG, Souto-Padron T, et al. Activation of the PI3K/Akt pathway early during vaccinia and cowpox virus infections is required for both host survival and viral replication. J Virol 2009, 83:6883–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Izmailyan R, Hsao JC, Chung CS, Chen CH, Hsu PW, Liao CL, Chang W. Integrin beta1 mediates vaccinia virus entry through activation of PI3K/Akt signaling. J Virol 2012, 86:6677–6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meade N, Furey C, Li H, Verma R, Chai Q, Rollins MG, DiGiuseppe S, Naghavi MH, Walsh D. Poxviruses Evade Cytosolic Sensing through Disruption of an mTORC1-mTORC2 Regulatory Circuit. Cell 2018, 174:1143–1157 e1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moretti J, Roy S, Bozec D, Martinez J, Chapman JR, Ueberheide B, Lamming DW, Chen ZJ, Horng T, Yeretssian G, et al. STING Senses Microbial Viability to Orchestrate Stress-Mediated Autophagy of the Endoplasmic Reticulum. Cell 2017, 171:809–823 e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 2016, 17:1142–1149. [DOI] [PubMed] [Google Scholar]
- 85.Seo GJ, Yang A, Tan B, Kim S, Liang Q, Choi Y, Yuan W, Feng P, Park HS, Jung JU. Akt Kinase-Mediated Checkpoint of cGAS DNA Sensing Pathway. Cell Rep 2015, 13:440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Myskiw C, Arsenio J, Booy EP, Hammett C, Deschambault Y, Gibson SB, Cao J. RNA species generated in vaccinia virus infected cells activate cell type-specific MDA5 or RIG-I dependent interferon gene transcription and PKR dependent apoptosis. Virology 2011, 413:183–193. [DOI] [PubMed] [Google Scholar]
- 87.Willis KL, Langland JO, Shisler JL. Viral double-stranded RNAs from vaccinia virus early or intermediate gene transcripts possess PKR activating function, resulting in NF-kappaB activation, when the K1 protein is absent or mutated. J Biol Chem 2011, 286:7765–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang Z, Cao S, Martens CA, Porcella SF, Xie Z, Ma M, Shen B, Moss B. Deciphering poxvirus gene expression by RNA sequencing and ribosome profiling. J Virol 2015, 89:6874–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burgess HM, Mohr I. Cellular 5’−3’ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 2015, 17:332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu SW, Katsafanas GC, Liu R, Wyatt LS, Moss B. Poxvirus decapping enzymes enhance virulence by preventing the accumulation of dsRNA and the induction of innate antiviral responses. Cell Host Microbe 2015, 17:320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burgess HM, Pourchet A, Hajdu CH, Chiriboga L, Frey AB, Mohr I. Targeting Poxvirus Decapping Enzymes and mRNA Decay to Generate an Effective Oncolytic Virus. Mol Ther Oncolytics 2018, 8:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arndt WD, White SD, Johnson BP, Huynh T, Liao J, Harrington H, Cotsmire S, Kibler KV, Langland J, Jacobs BL. Monkeypox virus induces the synthesis of less dsRNA than vaccinia virus, and is more resistant to the anti-poxvirus drug, IBT, than vaccinia virus. Virology 2016, 497:125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu R, Moss B. Opposing Roles of Double-Stranded RNA Effector Pathways and Viral Defense Proteins Revealed with CRISPR-Cas9 Knockout Cell Lines and Vaccinia Virus Mutants. J Virol 2016, 90:7864–7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang HW, Watson JC, Jacobs BL. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci U S A 1992, 89:4825–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Davies MV, Elroy-Stein O, Jagus R, Moss B, Kaufman RJ. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J Virol 1992, 66:1943-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carroll K, Elroy-Stein O, Moss B, Jagus R. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2 alpha-specific protein kinase. J Biol Chem 1993, 268:12837–12842. [PubMed] [Google Scholar]
- 97.Davies MV, Chang HW, Jacobs BL, Kaufman RJ. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J Virol 1993, 67:1688–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Craig AW, Cosentino GP, Donze O, Sonenberg N. The kinase insert domain of interferon-induced protein kinase PKR is required for activity but not for interaction with the pseudosubstrate K3L. J Biol Chem 1996, 271:24526–24533. [DOI] [PubMed] [Google Scholar]
- 99.Gale M Jr., Tan SL, Wambach M, Katze MG. Interaction of the interferon-induced PKR protein kinase with inhibitory proteins P58IPK and vaccinia virus K3L is mediated by unique domains: implications for kinase regulation. Mol Cell Biol 1996, 16:4172–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kawagishi-Kobayashi M, Silverman JB, Ung TL, Dever TE. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2alpha. Mol Cell Biol 1997, 17:4146–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kawagishi-Kobayashi M, Cao C, Lu J, Ozato K, Dever TE. Pseudosubstrate inhibition of protein kinase PKR by swine pox virus C8L gene product. Virology 2000, 276:424–434. [DOI] [PubMed] [Google Scholar]
- 102.Dar AC, Sicheri F. X-ray crystal structure and functional analysis of vaccinia virus K3L reveals molecular determinants for PKR subversion and substrate recognition. Mol Cell 2002, 10:295–305. [DOI] [PubMed] [Google Scholar]
- 103.Langland JO, Jacobs BL. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 2002, 299:133–141. [DOI] [PubMed] [Google Scholar]
- 104.Ramelot TA, Cort JR, Yee AA, Liu F, Goshe MB, Edwards AM, Smith RD, Arrowsmith CH, Dever TE, Kennedy MA. Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2alpha. J Mol Biol 2002, 322:943–954. [DOI] [PubMed] [Google Scholar]
- 105.Ludwig H, Suezer Y, Waibler Z, Kalinke U, Schnierle BS, Sutter G. Double-stranded RNA-binding protein E3 controls translation of viral intermediate RNA, marking an essential step in the life cycle of modified vaccinia virus Ankara. J Gen Virol 2006, 87:1145–1155. [DOI] [PubMed] [Google Scholar]
- 106.Arsenio J, Deschambault Y, Cao J. Antagonizing activity of vaccinia virus E3L against human interferons in Huh7 cells. Virology 2008, 377:124–132. [DOI] [PubMed] [Google Scholar]
- 107.Zhang P, Jacobs BL, Samuel CE. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. J Virol 2008, 82:840–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rice AD, Turner PC, Embury JE, Moldawer LL, Baker HV, Moyer RW. Roles of vaccinia virus genes E3L and K3L and host genes PKR and RNase L during intratracheal infection of C57BL/6 mice. J Virol 2011, 85:550–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dueck KJ, Hu YS, Chen P, Deschambault Y, Lee J, Varga J, Cao J. Mutational analysis of vaccinia virus E3 protein: the biological functions do not correlate with its biochemical capacity to bind double-stranded RNA. J Virol 2015, 89:5382–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Langland JO, Jacobs BL. Inhibition of PKR by vaccinia virus: role of the N- and C-terminal domains of E3 L. Virology 2004, 324:419–429. [DOI] [PubMed] [Google Scholar]
- 111.Myskiw C, Arsenio J, van Bruggen R, Deschambault Y, Cao J. Vaccinia virus E3 suppresses expression of diverse cytokines through inhibition of the PKR, NF-kappaB, and IRF3 pathways. J Virol 2009, 83:6757–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang P, Langland JO, Jacobs BL, Samuel CE. Protein kinase PKR-dependent activation of mitogen-activated protein kinases occurs through mitochondrial adapter IPS-1 and is antagonized by vaccinia virus E3L. J Virol 2009, 83:5718–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jentarra GM, Heck MC, Youn JW, Kibler K, Langland JO, Baskin CR, Ananieva O, Chang Y, Jacobs BL. Vaccinia viruses with mutations in the E3L gene as potential replication-competent, attenuated vaccines: scarification vaccination. Vaccine 2008, 26:2860–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rahman MM, McFadden G. Myxoma Virus dsRNA Binding Protein M029 Inhibits the Type I IFN-Induced Antiviral State in a Highly Species-Specific Fashion. Viruses 2017, 9. [Google Scholar]
- 115.Rahman MM, Liu J, Chan WM, Rothenburg S, McFadden G. Myxoma virus protein M029 is a dual function immunomodulator that inhibits PKR and also conscripts RHA/DHX9 to promote expanded host tropism and viral replication. PLoS Pathog 2013, 9:e1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Meng X, Chao J, Xiang Y. Identification from diverse mammalian poxviruses of host-range regulatory genes functioning equivalently to vaccinia virus C7L. Virology 2008, 372:372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Willis KL, Patel S, Xiang Y, Shisler JL. The effect of the vaccinia K1 protein on the PKR-eIF2alpha pathway in RK13 and HeLa cells. Virology 2009, 394:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Meng X, Jiang C, Arsenio J, Dick K, Cao J, Xiang Y. Vaccinia virus K1L and C7L inhibit antiviral activities induced by type I interferons. J Virol 2009, 83:10627–10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Backes S, Sperling KM, Zwilling J, Gasteiger G, Ludwig H, Kremmer E, Schwantes A, Staib C, Sutter G. Viral host-range factor C7 or K1 is essential for modified vaccinia virus Ankara late gene expression in human and murine cells, irrespective of their capacity to inhibit protein kinase R-mediated phosphorylation of eukaryotic translation initiation factor 2alpha. J Gen Virol 2010, 91:470–482. [DOI] [PubMed] [Google Scholar]
- 120.Liu J, McFadden G. SAMD9 is an innate antiviral host factor with stress response properties that can be antagonized by poxviruses. J Virol 2015, 89:1925–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bravo Cruz AG, Shisler JL. Vaccinia virus K1 ankyrin repeat protein inhibits NF-kappaB activation by preventing RelA acetylation. J Gen Virol 2016, 97:2691–2702. [DOI] [PubMed] [Google Scholar]
- 122.Simpson-Holley M, Kedersha N, Dower K, Rubins KH, Anderson P, Hensley LE, Connor JH. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J Virol 2011, 85:1581–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rozelle DK, Filone CM, Kedersha N, Connor JH. Activation of stress response pathways promotes formation of antiviral granules and restricts virus replication. Mol Cell Biol 2014, 34:2003–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carpentier KS, Esparo NM, Child SJ, Geballe AP. A Single Amino Acid Dictates Protein Kinase R Susceptibility to Unrelated Viral Antagonists. PLoS Pathog 2016, 12:e1005966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature 2009, 457:485–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Seo EJ, Liu F, Kawagishi-Kobayashi M, Ung TL, Cao C, Dar AC, Sicheri F, Dever TE. Protein kinase PKR mutants resistant to the poxvirus pseudosubstrate K3L protein. Proc Natl Acad Sci U S A 2008, 105:16894–16899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rothenburg S, Seo EJ, Gibbs JS, Dever TE, Dittmar K. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat Struct Mol Biol 2009, 16:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Elde NC, Child SJ, Eickbush MT, Kitzman JO, Rogers KS, Shendure J, Geballe AP, Malik HS. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell 2012, 150:831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Brennan G, Kitzman JO, Rothenburg S, Shendure J, Geballe AP. Adaptive gene amplification as an intermediate step in the expansion of virus host range. PLoS Pathog 2014, 10:e1004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sasani TA, Cone KR, Quinlan AR, Elde NC. Long read sequencing reveals poxvirus evolution through rapid homogenization of gene arrays. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Brennan G, Kitzman JO, Shendure J, Geballe AP. Experimental Evolution Identifies Vaccinia Virus Mutations in A24R and A35R That Antagonize the Protein Kinase R Pathway and Accompany Collapse of an Extragenic Gene Amplification. J Virol 2015, 89:9986–9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cone KR, Kronenberg ZN, Yandell M, Elde NC. Emergence of a Viral RNA Polymerase Variant during Gene Copy Number Amplification Promotes Rapid Evolution of Vaccinia Virus. J Virol 2017, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Peng C, Haller SL, Rahman MM, McFadden G, Rothenburg S. Myxoma virus M156 is a specific inhibitor of rabbit PKR but contains a loss-of-function mutation in Australian virus isolates. Proc Natl Acad Sci U S A 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]