Abstract
Colistin is used as an alternative therapeutic for carbapenemase-producing Enterobacteriaceae (CPE) infections which are spreading at a very high rate due to the transfer of carbapenemase genes through mobile genetic elements. Due to the emergence of mcr-1, the plasmid-mediated colistin resistance gene, mcr-1-positive Enterobacteriaceae (MCRPEn) pose a high risk for the transfer of mcr-1-carrying plasmid to CPE, leading to a situation with no treatment alternatives for infections caused by Enterobacteriaceae possessing both mcr-1 and carbapenemase genes. Here, we report the application of PCR-dipstick-oriented surveillance strategy to control MCRPEn and CPE by conducting the PCR-dipstick technique for the detection of MCRPEn and CPE in a tertiary care hospital in Thailand and comparing its efficacy with conventional surveillance method. Our surveillance results showed a high MCRPEn (5.9%) and CPE (8.7%) carriage rate among the 219 rectal swab specimens examined. Three different CPE clones were determined by pulsed-field gel electrophoresis (PFGE) whereas only two MCRPEn isolates were found to be closely related as shown by single nucleotide polymorphism-based phylogenetic analysis. Whole genome sequencing (WGS) and plasmid analysis showed that MCRPEn carried mcr-1 in two plasmids types—IncX4 and IncI2 with ~99% identity to the previously reported mcr-1-carrying plasmids. The identification of both MCRPEn and CPE in the same specimen indicates the plausibility of plasmid-mediated transfer of mcr-1 genes leading to the emergence of colistin- and carbapenem-resistant Enterobacteriaceae. The rapidity (<2 h) and robust sensitivity (100%)/specificity (~99%) of PCR-dipstick show that this specimen-direct screening method could aid in implementing infection control measures at the earliest to control the dissemination of MCRPEn and CPE.
Keywords: mcr-1, carbapenemase, Enterobacteriaceae, PCR-dipstick, rapid detection
Introduction
Since carbapenems are regarded as the last line of defense for several multidrug-resistant bacterial infections, carbapenem-resistant organisms impede the effective treatment options and increase the mortality rate of afflicted patients (Kizny Gordon et al., 2017). Amongst different carbapenem-resistant organisms, carbapenemase-producing Enterobacteriaceae (CPE) are posing a global health threat due to their hasty dissemination, enabled through the horizontal transfer of carbapenemase genes (van Duin and Doi, 2017). Despite the adverse side-effects, colistin is used as a therapeutic alternative for multidrug-resistant organisms including CPE (Mansour et al., 2017; Poirel et al., 2017). However, due to the high usage of colistin in agriculture and livestock, colistin-resistant organisms have emerged as a serious threat to both the animals and humans (Caniaux et al., 2017; Wang et al., 2017b). Furthermore, clinical isolates of colistin-resistant Gram-negatives are being increasingly reported (Kontopidou et al., 2011; Thi Khanh Nhu et al., 2016). Several Enterobacteriaceae species are known to be intrinsically resistant to colistin, however, after the description of mcr-1, the plasmid-encoded colistin resistance gene, mcr-1-producing Enterobacteriaceae (MCRPEn) have been reported worldwide (Olaitan et al., 2014; Liu et al., 2016; Matamoros et al., 2017; Wang et al., 2018). Since multidrug-resistant CPE are well-known for their acquisition of plasmid carrying carbapenemase genes, it is suspected that they could also acquire the mcr-1-carrying plasmid from MCRPEn which might lead to the emergence of the superbug resistant to both colistin and carbapenem without alternate treatment regimens. Recent reports also show the sporadic identification of Enterobacteriaceae co-harboring carbapenemase and mcr-1 in plasmids (Beyrouthy et al., 2017; Pulss et al., 2017; Mendes et al., 2018).
Based on the currently available reports, MCRPEn are known for their prevalence in the environment from which they can spread to humans (Guenther et al., 2017; Kieffer et al., 2017). Few studies have reported on the epidemiology of clinical MCRPEn, while other reports screened for mcr-1 from the previous collections of clinical isolates (Chen et al., 2017; Saly et al., 2017; Terveer et al., 2017; Wang et al., 2017a; Zhong et al., 2018). In Thailand, there have been sporadic cases of mcr-1-positive Escherichia coli isolated from the clinical specimens of patients (Paveenkittiporn et al., 2017; Srijan et al., 2018). However, there have been no reports on the carriage of MCRPEn in hospitalized patients in the country. Similarly, the epidemiology of CPE carriage in Thai hospitals is still unknown. Thus, there is a lack of epidemiological data on the prevalence of MCRPEn and CPE to initiate efficient infection control measures.
To control their nosocomial transmission/infection, it is necessary to determine the fecal carriage rate of MCRPEn along with CPE in the hospitalized patients. In order to implement infection control measures instantly, a rapid detection system for the simultaneous detection of MCRPEn and CPE would be beneficial. Previously, we established the PCR-dipstick technique for the detection of CPE possessing blaNDM, blaKPC, blaIMP, and blaOXA−48 carbapenemase genes (Shanmugakani et al., 2017). Then, we included two more carbapenemase genes (blaVIM, blaGES) for CPE detection and mcr-1, the most prevalent mcr-1 gene variant for MCRPEn detection (Matamoros et al., 2017; Wang et al., 2018). Thus, the PCR-dipstick could be utilized for the rapid detection of MCRPEn and CPE directly from the clinical specimens by targeting mcr-1 and six carbapenemase (blaNDM, blaKPC, blaIMP, blaOXA−48, blaVIM, and blaGES) genes, respectively. Here, we attempted to develop a PCR-dipstick-oriented surveillance strategy for determining the MCRPEn and CPE fecal carriage and examined its efficacy by comparing with the conventional method in a tertiary care hospital in Thailand. Next, we analyzed the resistance profile and clonal relatedness of the MCRPEn and CPE isolates. Genetic characterization including multi-locus sequence typing and plasmid analysis of the MCRPEn isolates was carried out by whole genome sequencing (WGS) to determine the characteristics that mediate their mode of transmission. Furthermore, the clinical characteristics of the patients were analyzed to determine the risk factors that influence MCRPEn and CPE carriage in the hospital.
Results
Prevalence and Characteristics of MCRPEn and CPE
MCRPEn and CPE surveillance was conducted using both the PCR-dipstick-oriented method and conventional method to examine the efficacy of PCR-dipstick-oriented surveillance strategy (Figure 1). The surveillance results showed that out of the 219 rectal swab specimens collected from the hospitalized patients in a Thai hospital, 13 specimens (5.9%) carried MCRPEn with the mcr-1 gene and 19 specimens (8.7%) carried CPE with two different carbapenemase genes—blaNDM−1 (n = 7), blaOXA−232 (n = 10), or both (n = 2). Two different species of MCRPEn (E. coli and Klebsiella pneumoniae) and two different species of CPE (K. pneumoniae and Citrobacter farmeri) were identified (Table 1). Two MCRPEn-positive specimens carried both E. coli and K. pneumoniae with mcr-1 gene and thus, we found 15 MCRPEn isolates from 13 MCRPEn-positive specimens. Two specimens were found to carry both MCRPEn and CPE isolates whereas there were no isolates carrying both carbapenemase and mcr-1 genes. Of the two specimens with both MCRPEn and CPE isolates, one carried K. pneumoniae with blaNDM−1/blaOXA−232 and E. coli with mcr-1 and the other specimen carried K. pneumoniae with blaNDM−1 and E. coli with mcr-1. However, neither the MCRPEn were identified with carbapenemase genes nor were the CPE shown to possess the mcr-1 gene. Antimicrobial susceptibility testing of MCRPEn and CPE showed a varied resistance profile against diverse antibiotics including carbapenems and other β-lactams, fluoroquinolones, tetracyclines, aminoglycosides, sulphonamides, and colistin (Table S1). Notably, all the MCRPEn isolates (15/15) were resistant to colistin with a minimum inhibitory concentration of ≥4 μg/ml.
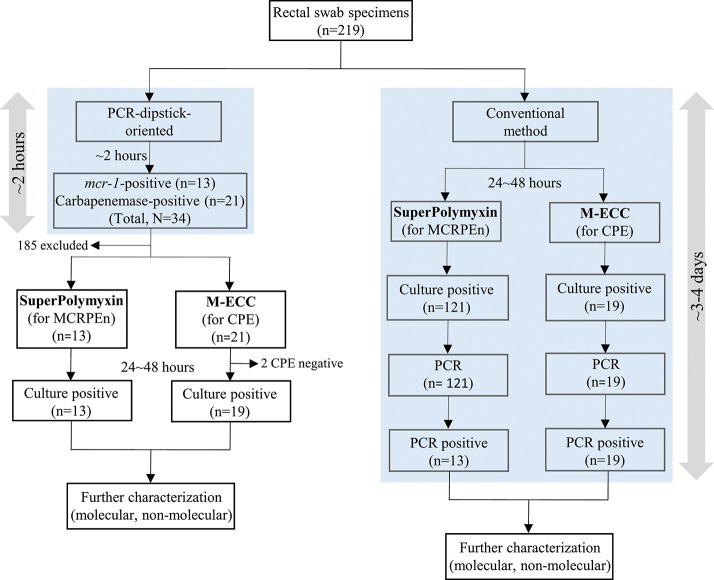
Figure 1.
Flow diagram of PCR-dipstick-oriented and conventional surveillance method. All the rectal swab specimens were subjected to PCR-dipstick-oriented and conventional surveillance method. PCR-dipstick detected the presence of resistance genes within 2 h and thus, only the specimens detected with resistance genes were cultured on selective media for the isolation of MCRPEn and CPE. In the conventional method, all of the specimens were cultured on the selective media and all the isolates grown (24~48 h) on them were screened for the resistance genes using PCR.
Table 1.
Bacterial species and the mcr-1/carbapenemsae genes of MCRPEn and CPE isolated from 219 rectal swab specimens.
| Isolate type | No. of positive specimens (%) | Species (no. of specimens) | Resistance genes | |
|---|---|---|---|---|
| Family | Variants | |||
| MCRPEn (n = 15) | 13 (5.9) | E. coli (11) | mcr-1 | mcr-1 |
| K. pneumoniae and E. coli (2) | mcr-1 | mcr-1 | ||
| CPE (n = 19) | 19 (8.7) | K. pneumoniae (6) | blaNDM | blaNDM−1 |
| C. farmeri (1) | blaNDM | blaNDM−1 | ||
| K. pneumoniae (10) | blaOXA−48 | blaOXA−232 | ||
| K. pneumoniae (2) | blaNDM/blaOXA−48 | blaNDM−1/blaOXA−232 | ||
Clonal Relatedness of MCRPEn and CPE Isolates
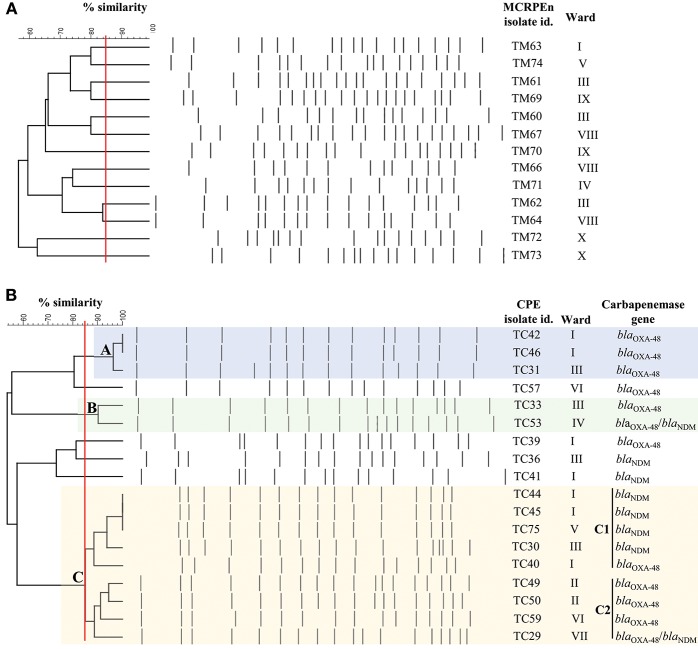
Pulsed-field gel electrophoresis (PFGE) was performed after XbaI digestion of the 12 MCRPEn (E. coli) and 18 CPE (K. pneumoniae) isolates to determine their clonal relatedness. It was found that none of the MCRPEn isolates belong to the same clone (≥85% Dice similarity) suggesting that there was no clonal spreading of MCRPEn in the hospital (Figure 2A). Among the 18 K. pneumoniae CPE isolates, we found three different clusters, namely A, B, and C (C1 and C2), of isolates with ≥85% Dice similarity (Figure 2B). The isolates in all the three clusters were isolated from the patients admitted not only in the same ward but also in different wards. The isolates in cluster A carry blaOXA−48 family, in B carry blaOXA−48 and blaOXA−48/blaNDM family, in C1 carry blaOXA−48 and blaNDM and in C2 carry blaOXA−48 and blaOXA−48/blaNDM as confirmed by PCR.
Figure 2.
PFGE analysis of XbaI-digested MCRPEn (E. coli) and CPE (K. pneumoniae) isolates. (A) Clonal relatedness of 13 mcr-1-producing E. coli isolates. (B) Clonal relatedness of 18 carbapenemase-producing K. pneumoniae isolates. A, B, C1, and C2 represents the cluster of isolates with ≥85% similarity (red line). Isolates with Dice similarity coefficient ≥85% fall under one cluster.
Genomic Characteristics of MCRPEn Isolates
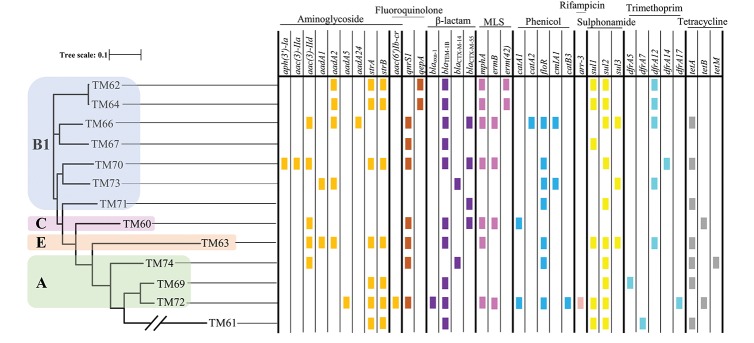
From the PFGE results, the CPE isolates were found to be clonally related; there are several previous reports describing their nosocomial transmission (Nordmann et al., 2011; Mataseje et al., 2016). However, all the MCRPEn isolates were clonally unrelated to each other as shown by PFGE; a high rate of MCRPEn carriage (5.9%) was also determined in this surveillance study. For a detailed understanding of the genomic characteristics of MCRPEn isolates, whole genome sequencing (WGS) was carried out. WGS of MCRPEn isolates showed that all MCRPEn isolates belonged to different individual sequence types except two E. coli isolates (TM62 and TM64) both of which belong to ST424 (Table 2). The phylogenetic analysis based on single nucleotide polymorphisms showed that the E. coli isolates (n = 13) of the MCRPEn are diverse and belong to four different phylotypes—A, B1, C, and E (Figure 3). One isolate (TM61) does not belong to any of the phylotypes. The phylogenetic analysis also showed that TM62 and TM64 were more closely related than any other MCRPEn isolates. In addition to the mcr-1 gene, the MCRPEn isolates were found to possess several resistance genes for a diverse range of antibiotics (Figure 3). S1 nuclease-PFGE followed by Southern hybridization showed that two E. coli MCRPEn isolates (TM63 and TM66) carried the mcr-1 gene in their chromosome. All other 13 MCRPEn possess the mcr-1 gene in the plasmids of two different sizes: ~33 and ~60 kb (Figure S1).
Table 2.
Description of mcr-1 gene location, plasmid analysis, and other characteristics of the MCRPEn isolates.
| Isolate id | Species | Sequence type | MIC of CST (μg/ml) | Location of mcr-1 | Plasmid replicon | Plasmid size (kb) | Query coverage (Identity) (%) | Reference accession no. |
|---|---|---|---|---|---|---|---|---|
| TM60 | E. coli | ST410 | 4 | Plasmid | IncX4 | ~33 | 98.3 (100) | CP02404.1 |
| TM61 | ST131 | 16 | Plasmid | IncX4 | ~33 | 99.1 (100) | CP02404.1 | |
| TM62 | ST424 | 16 | Plasmid | IncI2 | ~68 | 90.3 (99.8) | CP025679.1 | |
| TM63 | ST1011 | 16 | Chromosome | NA | NA | NA | NA | |
| TM64 | ST424 | 8 | Plasmid | IncI2 | ~68 | 89.9 (99.8) | CP025679.1 | |
| TM66 | ST4014 | 8 | Chromosome | NA | NA | NA | NA | |
| TM67 | ST156 | 8 | Plasmid | IncI2 | ~60 | 97.2 (99.9) | MF978388.1 | |
| TM69 | ST4741 | 4 | Plasmid | IncI2 | ~60 | 98.2 (99.8) | MF978388.1 | |
| TM70 | ST448 | 8 | Plasmid | IncX4 | ~33 | 98.4 (100) | CP02404.1 | |
| TM71 | ST1196 | 4 | Plasmid | IncI2 | ~60 | 98.9 (99.9) | MF978388.1 | |
| TM72 | ST744 | 8 | Plasmid | IncX4 | ~33 | 98.9 (100) | CP02404.1 | |
| TM73 | ST3695 | 8 | Plasmid | ND | ~60 | NA | NA | |
| TM74 | ST8335 | 8 | Plasmid | IncX4 | ~33 | 98.9 (100) | CP02404.1 | |
| TM65 | K. pneumoniae | ST394 | 16 | Plasmid | IncI2 | ~68 | 90.4 (99.8) | CP025679.1 |
| TM68 | ST709 | 8 | Plasmid | IncX4 | ~33 | 98.8 (99.9) | CP02404.1 |
CST, colistin; NA, not applicable; ND, not determined.
Figure 3.
Phylogenetic relationship and distribution of different resistance genes among MCRPEn isolates. Based on single nucleotide polymorphisms, the 13 E. coli MCRPEn isolates were classified into four different phylogroups (A, B1, C, and E) as represented by differently colored boxes over the phylogenetic tree. The presence of different resistance genes that confer resistance toward each of the antimicrobials determined by WGS are shown as small colored rectangles. The two diagonal bars across a branch of the phylogenetic tree represent scale break. MLS, macrolide, lincosamide, and streptogramin B.
Table 2 shows that out of the 13 isolates carrying mcr-1 in plasmids, two types of plasmids, IncX4 (n = 6) and IncI2 (n = 6) were found by WGS analysis. Along with mcr-1 gene, both the IncX4 and IncI2 plasmids carried a wide range of the different variants of other resistance genes for β-lactams, fluoroquinolones, aminoglycosides, macrolides, phenicol, sulphonamides, tetracyclines etc. (Figure 3). One of the specimens from which two MCRPEn (TM64-E. coli and TM65-K. pneumoniae) were isolated carries IncI2 plasmids whereas the other specimen with the two isolates, TM67- E. coli and TM68- K. pneumoniae carries IncI2 and IncX4, respectively. The mcr-1 genes in all six IncX4 plasmids (~33 kb) were found to possess a single point mutation (C27T) without causing any amino acid substitution. Interestingly, the same point mutation was observed in the previously reported plasmid carrying mcr-1 in K. pneumoniae (accession no. CP024041.1) isolated from Thailand. This reference plasmid also has both a high query coverage and identity of 99% with the IncX4 plasmids identified in our study. Among the six IncI2 plasmids, three (TM62, TM64, TM65) were similar to the IncI2 plasmid (~68 kb) carrying mcr-1 in an Escherichia albertii isolate from China (accession no. CP025679.1). Likewise, the other three IncI2 plasmids in TM67, TM69, and TM71 were similar to the mcr-1-carrying IncI2 plasmid (~60 kb) found in an E. coli isolate from China (accession no. MF978388.1). In one isolate (TM73), the contig with mcr-1 (~11 kb) showed high similarity to the previously reported IncI2 plasmids and S1 nuclease-PFGE showed that mcr-1 is located in a ~60 kb plasmid which is similar to the size of IncI2 plasmids identified in other MCRPEn isolates in our study. However, no IncI2 replicon was identified in TM63 by WGS and hence the type of plasmid carrying mcr-1 could not be determined.
Efficacy of PCR-Dipstick in Detecting MCRPEn and CPE as Evaluated During This Surveillance Study
Evaluation of the sensitivity and specificity of PCR-dipstick compared to conventional PCR, showed that PCR-dipstick had both 100% sensitivity and specificity in detecting the MCRPEn directly from the 219 rectal swab specimens examined in this study (Table 3). Likewise, it showed 100% sensitivity and 98.9% specificity in detecting CPE directly from clinical specimens (Table 3). The conventional method requires all the 219 specimens to be screened for the presence of MCRPEn and CPE isolates followed by confirmation of the presence of resistance genes which takes ~3–4 days (Figure 1). In contrast, PCR-dipstick extensively reduced the number of specimens which needed to be subjected for the isolation of MCRPEn and CPE from 219 to 34. Furthermore, PCR-dipstick was found to be rapid in detecting the presence of resistance genes thereby identifying the MCRPEn- and CPE-positive specimens in <2 h.
Table 3.
Sensitivity and specificity of PCR-dipstick vs. conventional PCR for 219 rectal swab specimens.
| Conventional PCR | Sensitivity (95% CI) | Specificity (95% CI) | |||
|---|---|---|---|---|---|
| Positive | Negative | ||||
| PCR-dipstick (MCRPEn) | Positive | 13 | 0 | 100 (75.3–100) | 100 (98.2–100) |
| Negative | 0 | 206 | |||
| PCR-dipstick (CPE) | Positive | 19 | 2 | 100 (82.4–100) | 98.9 (96.4–99.9) |
| Negative | 0 | 198 | |||
CI, confidence interval.
Risk Factors Associated With MCRPEn and CPE Carriage
The univariate analysis showed that chronic kidney disease (p = 0.02) alone was significantly associated with MCRPEn carriage (Table 4). On the other hand, diaper usage (p = 0.02), health complications such as chronic respiratory diseases (p = 0.009) and chronic kidney diseases (p = 0.02), invasive procedures of mechanical ventilation (p = 0.02), and tracheostomy (p = 0.003), history of multidrug-resistant organisms (p = 0.002) and prior exposure to antimicrobials (p = 0.05) were significantly associated with CPE carriage (Table 4).
Table 4.
Risk factors associated with MCRPEn and CPE carriage.
| Characteristics | MCRPEn | CPE | ||
|---|---|---|---|---|
| OR (95% CI) | p-value | OR (95% CI) | p-value | |
| Diaper use | 0.61 (0.16–2.28) | 0.56 | 3.21 (1.23–8.38) | 0.02 |
| Health complications | ||||
| Chronic respiratory diseases | 0.60 (0.08–4.84) | 1.00 | 4.15 (1.42–12.13) | 0.009 |
| Chronic kidney diseases | 3.95 (1.26–12.32) | 0.02 | 3.10 (1.19–8.09) | 0.02 |
| Invasive procedures | ||||
| Mechanical ventilation | 1.84 (0.54–6.30) | 0.30 | 3.31 (1.24–8.82) | 0.02 |
| Tracheostomy | 0.73 (0.10–5.93) | 1.00 | 5.31 (1.78–15.85) | 0.003 |
| Carriage of MDROsa (<3 months) | 1.28 (0.38–4.34) | 0.75 | 4.60 (1.75–12.12) | 0.002 |
| Exposure to antimicrobialsb (<3 months) | 2.29 (0.29-18.21) | 0.70 | ** | 0.05 |
OR, odds ratio; CI, confidence of interval; MDROs, multidrug-resistant organisms.
MDROs include methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococci, multidrug-resistant Pseudomonas aeruginosa, multidrug-resistant Acinetobacter baumannii, and extended-spectrum β-lactamase producers.
Antimicrobials include β-lactams, β-lactamase inhibitors, quinolones, tetracyclines, glycopeptides, colistin, sulfamethoxazole-trimethoprim, clindamycin, and metronidazole.
All 19 cases of CPE were exposed to antimicrobials.
Discussion
Due to their resistance toward the drugs of last resort, colistin/carbapenem, mcr-1, and carbapenemase co-producing Enterobacteriaceae have been reported from different parts of the world (Pulss et al., 2017; Mendes et al., 2018). Since there are no alternate therapeutics available, their rapid spreading has become an alarming threat which needs to be controlled. It is essential to understand the prevalence of both in healthcare settings to prevent their transmission. Recently, Jin et al. reported the simultaneous screening of MCRPEn and CPE from sewage water in China (Jin et al., 2018). However, there were no reports of the screening of MCRPEn together with CPE in hospitalized patients. We conducted a surveillance of MCRPEn and CPE together using the direct clinical specimens and divulged the MCRPEn (5.9%) and CPE (8.7%) carriage rate in a tertiary care hospital in Thailand by applying the PCR-dipstick-oriented specimen-direct surveillance strategy. Moreover, we demonstrated that the underlying medical conditions of MCRPEn and CPE carriage differ in that hospital.
CPE isolated in this study were found to carry only two types of the six carbapenemase genes tested—blaNDM−1, blaOXA−232, or both and they showed high level of resistance to most of the antibiotics tested. From the PFGE pattern, the clonal relatedness of the CPE isolates suggested that they have been spreading in the hospital within and between the wards. Thus, it becomes obvious that there is nosocomial transmission of CPE. These results are in line with the previous reports on the nosocomial transmission of CPE in clinical settings (Onori et al., 2015; Sotgiu et al., 2018). Furthermore, the currently determined high carriage rate (8.7%) of CPE is suspected to increase in the next few years if left uncontrolled.
Regarding MCRPEn, all of the MCRPEn (100%) isolated in this study were found to be colistin-resistant. MCRPEn identified in this study were all unrelated clones, except two E. coli isolates, which indicates that the MCRPEn isolates were acquired from outside the hospital. MCRPEn isolates could also have been acquired from other wards of the hospital which were not included in this surveillance study. WGS showed that the MCRPEn isolates carry two types of mcr-1-carrying plasmids, IncX4, and IncI2, which are also known to be two of the major plasmid types that mediate the spread of mcr-1 gene (Cui et al., 2017; Li et al., 2017). The high identity of IncX4 plasmids with the previously reported IncX4 plasmid along with the same single point mutation detected in the mcr-1 gene, suggests that IncX4 carrying mcr-1 might be spreading in Thailand, thereby mediating MCRPEn dissemination. Similarly, the mcr-1-carrying IncI2 plasmids were highly identical to the previously reported IncI2 plasmids identified from food sources in China. Thus, the mcr-1-bearing IncI2 plasmids might have been imported from China; however, this association between the plasmids needs to be further elucidated. On the other hand, plasmid mediated spread of mcr-1 gene between isolates would also have occurred in the intestinal flora of the patients and/or the isolates residing in the hospital environment. Furthermore, ST1011 and ST4014 E. coli isolates carrying mcr-1 in the chromosome were epidemiologically unrelated to the previously reported mcr-1-positive E. coli isolates with the same sequence types (Guenther et al., 2017; Wang et al., 2017a). Altogether, these results suggest that MCRPEn would have been acquired from the community.
The identification of single specimens carrying two MCRPEn isolates with same or different types of plasmids shows the plausibility of the transfer of mcr-1 gene from MCRPEn to naïve Enterobacteriaceae (Wang et al., 2018). In addition, the presence of both MCRPEn and CPE in the same specimen suggests that there is a risk of plasmid-mediated transfer of the mcr-1 gene from MCRPEn to CPE or carbapenemase genes from CPE to MCRPEn, thereby leading to the emergence of carbapenem- and colistin-resistant Enterobacteriaceae without any therapeutic options (Beyrouthy et al., 2017; Pulss et al., 2017; Mendes et al., 2018). We showed a very high rate of MCRPEn (5.9%) even without any duplicated specimens whereas other studies showed a low rate of clinical MCRPEn (Yu et al., 2016; Chan et al., 2018). This might be due to the high sensitivity of PCR-dipstick in detecting the MCRPEn directly from clinical specimens, whereas the other methods, such as screening with selective media, are less sensitive.
Our risk factor analysis shows that only the chronic kidney disease is associated with the carriage of MCRPEn in this hospital. In contrast, the patients colonized with CPE have health complications such as chronic respiratory and kidney diseases, used diaper frequently, underwent invasive procedures and had prior history of infection with multidrug-resistant organisms and antibiotics exposure. These backgrounds in patients with CPE carriage are highly consistent with the previous reports (Han et al., 2017; McConville et al., 2017; Yamamoto et al., 2017a).
The main advantage of using PCR-dipstick is its rapidity to detect the presence of these multidrug-resistant pathogens by targeting the resistance genes directly from the clinical specimens within 2 h. We compared PCR-dipstick-oriented surveillance with the conventional method for the surveillance of MCRPEn and CPE and found that the conventional method is more time-consuming and laborious than PCR-dipstick-oriented surveillance. For instance, the rectal swab specimens examined in this study showed the growth of a mixture of several Gram-negatives including mcr-1-negative isolates in the selective media leading to the screening of all the suspected isolates. However, the specimen-direct detection of resistance genes by PCR-dipstick aids in the culturing of only the specimens that were found to be positive for the resistance genes thereby reducing the number of specimens for screening of MCRPEn and CPE (Figure 1). Since PCR-dipstick could simultaneously identify MCRPEn and CPE from direct clinical specimens by targeting the resistance genes within 2 h, it could aid in the faster implementation of infection control measures such as patient cohorting to prevent their dissemination.
The limitation of applying PCR-dipstick-oriented surveillance is that the Enterobacteriaceae that exhibit carbapenem and colistin resistance through mechanisms other than carbapenemase and mcr-1 genes could not be detected unlike the detection systems which target the genes that mediate other modes of resistance. However, the risk of dissemination of carbapenem and colistin resistance resides in the plasmid-mediated transfer of carbapenemase and mcr-1 genes, respectively (van Duin and Doi, 2017; MacNair et al., 2018). Considering this aspect of MCRPEn and CPE, the currently proposed and examined surveillance strategy will be indispensable for both the clinical and environmental screening of these superbugs. As it is feasible to design the PCR-dipstick technique for any other newly reported variants of carbapenemase and mcr genes by designing the primers and evaluating the optimal hybridization conditions, CPE and MCRPEn harboring other resistance genes could be detected with a customized dipstick (Shanmugakani et al., 2017). Two specimens that were identified as blaIMP-positive by PCR-dipstick did not show the growth of any CPE isolates on the selective media which might be due the blaIMP carried by non-fermenters such as Acinetobacter baumannii, Pseudomonas aeruginosa, for example. Due to fewer number of MCRPEn- and CPE-positive cases, we could not apply a multivariate approach for the statistical analysis. Thus, a multiplicity could not be adjusted appropriately, influencing the interpretation of putative statistically significant findings. To determine the risk factors for MCRPEn and CPE in a hospital setting, a prospective, large-scaled study is warranted.
In summary, the MCRPEn isolates are undetectably existing in the hospital community due to the lack of a proper detection system. Our PCR-dipstick-oriented surveillance strategy is highly efficient in detecting MCRPEn directly from clinical specimens with a short turnaround time. The shockingly high prevalence of MCRPEn and CPE in the Thai hospital as determined in our surveillance study will be very helpful in undertaking potent infection control measures to control these deadly pathogens. Furthermore, a simple and cost-effective PCR-dipstick-oriented specimen-direct surveillance strategy for MCRPEn and CPE was developed and examined for its utility in clinical settings.
Materials and Methods
Ethics Statement and Specimen Collection
A total of 219 non-duplicated rectal swab specimens were collected using eSwab (COPAN Italia S.p.A., Brescia, Italy) from hospitalized patients (121 male and 98 female) with a mean age of 61 years in the surgical, medical, intensive care, intermediate care, cardiac care, and geriatric wards in Ramathibodi Hospital, Mahidol University in Bangkok, Thailand, during Jan–Feb 2018 as part of the Infection Control Committee cross-sectional surveillance activity. The rectal swabs collected were transported to the research laboratory and processed on the same day. Ethical approval was obtained from the Ethics Committee of Osaka University Graduate School of Medicine, Osaka, Japan and Ethical Clearance Committee on Human Rights Related to Research Involving Human Subjects, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand. Since rectal swab specimens were collected as a component of antimicrobial resistance surveillance in the hospital, the need for informed consent was waived by the Institutional Review Boards.
Surveillance of MCRPEn and CPE Using PCR-Dipstick
PCR-dipstick was used for the direct detection of six carbapenemase genes (blaNDM, blaKPC, blaIMP, blaOXA−48, blaVIM, blaGES) and the mcr-1 gene from the rectal swab specimens. We previously reported the development of this technique for the detection of four carbapenemase genes—blaNDM, blaKPC, blaIMP, and blaOXA−48, from direct clinical specimens (Shanmugakani et al., 2017). Here, we constructed another dipstick that could detect two additional carbapenemase genes, blaVIM, blaGES, and the mcr-1 gene (Supplementary Material). Thus, two dipsticks were used for the detection of MCRPEn and CPE directly from rectal swab specimens (Supplementary Material and Figure S2).
For the isolation of MCRPEn and CPE isolates from the PCR-dipstick-positive specimens, an aliquot of 100 μl each of the rectal swab specimens was inoculated onto SuperPolymyxin and M-ECC media, respectively (Nordmann et al., 2016; Yamamoto et al., 2017b). The plates were then incubated at 37°C for 24~48 h and the suspected colonies were subjected to species identification and antimicrobial susceptibility testing for carbapenems and other β-lactams, fluoroquinolones, tetracyclines, aminoglycosides, and sulphonamides using MicroScan WalkAway 96 Plus (Beckman Coulter, CA, USA) with a Neg Combo NF1J panel. The resistance profile of the isolates were then characterized according to the guideline M100, 28th edition of Clinical and Laboratory Standards Institute (CLSI) (Wayne, PA, USA). For colistin, antibiotic susceptibility testing was performed by broth microdilution using cation-adjusted Mueller Hinton II Broth according to CLSI document M07-A10, 2015 and the MIC breakpoints (≤2 susceptible, >2 resistant) given by European Committee on Antimicrobial Susceptibility Testing (http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.1_Breakpoint_Tables.pdf) was followed for result interpretation (Chew et al., 2017; Simar et al., 2017). The presence of carbapenemase/mcr-1 genes in the MCRPEn and CPE isolates was confirmed by conventional PCR and sequencing (Supplementary Material and Table S2). To analyze whether the MCRPEn and CPE isolates were clonally spreading in the hospital, they were subjected to PFGE using a previously reported protocol with slight modifications (Supplementary Material) (Ohno et al., 2017).
Genomic Characterization of MCRPEn Isolates
To determine the location of the mcr-1 gene in the MCRPEn isolates, S1 nuclease-PFGE followed by Southern hybridization with mcr-1 probe was performed (Supplementary Material). For WGS, all the MCRPEn isolates were grown overnight at 37°C in brain heart infusion broth (BD Bacto, Franklin Lakes, NJ, USA) and their genomic DNA was isolated using DNeasy Powersoil Kit (QIAGEN, Valencia, CA, USA). From the isolated genomic DNA, the libraries were prepared with MiSeq v2 500 cycle kit (Illumina, San Diego, CA, USA) and subjected to the MiSeq system (Illumina) (Miyamoto et al., 2014). De novo assembly of the sequence reads was performed using CLC Genomics Workbench 11.0.1 (CLC Bio, Aarhus, Denmark). The assembled sequences were submitted to MLST 2.0, PlasmidFinder 1.3, and ResFinder 2.1 to determine the sequence types, plasmid replicons and presence of different resistance genes, respectively (Larsen et al., 2012; Zankari et al., 2012; Carattoli et al., 2014). The mcr-1-carrying contigs were extracted and searched for similar sequences using BLAST in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Then, the sequence identified with the highest query coverage/identity was taken as the reference sequence for further analysis. The contigs were mapped with the reference sequence on CLC Genomics Workbench and the resultant mapped sequences were compared with the reference sequence to determine their similarity. With the E. coli str. K-12 substr. MG1655 (accession no: NC_00913.3) as the reference genome, phylogenetic relationships among the MCRPEn (E. coli) isolates were determined based on single nucleotide polymorphisms using CSI Phylogeny 1.3 (Kaas et al., 2014). Their phylotypes were determined based on the previously described phylo-typing method (Clermont et al., 2013).
Sequence Accession Numbers
The whole genome sequences of the 15 MCRPEn isolates identified in this study had been deposited to the DDBJ/ENA/GenBank under the accession numbers: DRX131210 (TM60), DRX131211 (TM61), DRX131212 (TM62), DRX131213 (TM63), DRX131214 (TM64), DRX131215 (TM65), DRX131216 (TM66), DRX131217 (TM67), DRX131218 (TM68), DRX131219 (TM69), DRX131220 (TM70), DRX131221 (TM71), DRX131222 (TM72), DRX131223 (TM73), and DRX131224 (TM74).
Evaluation of the Efficacy of PCR-Dipstick-Oriented Surveillance
During the PCR-dipstick-oriented surveillance of MCRPEn and CPE, conventional method of surveillance was carried out simultaneously (Figure 1). For the conventional method, initially all the rectal swab specimens were inoculated into SuperPolymyxin and M-ECC media. Then, the suspected MCRPEn and CPE isolates were isolated and subjected to genomic DNA isolation followed by conventional PCR for carbapenemase and mcr-1 genes (Supplementary Material). The efficiency of PCR-dipstick in detecting the MCRPEn and CPE directly in rectal swab specimens was determined by comparing the results of PCR-dipstick with a reference comparator in which the conventional PCR was performed for the genomic DNA of MCRPEn and CPE isolated from rectal swab specimens (Supplementary Material). The sensitivity and specificity of PCR-dipstick was calculated using GraphPad Prism version 6.05 (GraphPad Software, La Jolla, CA, USA). To evaluate the utility of PCR-dipstick-oriented surveillance, the work flow of PCR-dipstick-oriented method was compared with the conventional surveillance method (Figure 1).
Risk Factor Analysis
Clinical characteristics of the patients were analyzed for the risk factors associated with MCRPEn and CPE carriage separately. Clinical characteristics such as age, sex, length of hospitalization, invasive procedures, history of hospitalization (< 90 days), and prior antibiotics exposure (<90 days) were included for risk factor analysis. Logistic regression, Chi-square test, Fisher's exact test were performed, as appropriate, to assess the carriage rate of MCRPEn and CPE with each factor. The variables with two-tailed p ≤ 0.05 were considered statistically significant. The statistical analysis of the risk factors was performed with StataMP 15 (Stata Corp., College Station, TX, USA).
Author Contributions
YA and KT conceived and designed the study. RS, WL, NC, SS, NY, IN, HY, DT, NS, and PS contributed to the specimen collection, isolation, and the phenotypic/genotypic characterization of clinical isolates. RS and YS contributed to the whole genome sequencing. RS, HH, DM, and YF performed the statistical analysis. RS, YA, and KT drafted the manuscript drafting. YA, KM, PS, KT, and SH performed the critical revision of the manuscript for intellectual content. All the authors read, edited, and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Takuya Kodera and Dr. Mitsuo Kawase from Tohoku Bio-Array, Sendai, Japan for their support in manufacturing of the dipsticks and related reagents. We acknowledge Dr. Ryuji Kawahara from Osaka Prefectural Institute of Public Health, Osaka, Japan for providing the blaVIM and blaGES positive control isolates. We appreciate Dr. Daisuke Motooka from Osaka University, Osaka, Japan for his support to perform whole genome sequencing.
Footnotes
Funding. This work was supported by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) from Ministry of Education, Culture, Sport, Science, and Technology in Japan, and the Japan Agency for Medical Research and Development (AMED) under the Grant Number JP18fm0108003.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00149/full#supplementary-material
References
- 1.Beyrouthy R., Robin F., Lessene A., Lacombat I., Dortet L., Naas T., et al. (2017). Mcr-1 and oxa-48 in vivo acquisition in kpc-producing Escherichia coli after colistin treatment. Antimicrob. Agents Chemother. 61:e02540–16. 10.1128/AAC.02540-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caniaux I., van Belkum A., Zambardi G., Poirel L., Gros M. F. (2017). Mcr: modern colistin resistance. Eur. J. Clin. Microbiol. Infect. Dis. 36, 415–420. 10.1007/s10096-016-2846-y [DOI] [PubMed] [Google Scholar]
- 3.Carattoli A., Zankari E., Garcia-Fernandez A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan W. S., Au C. H., Ho D. N., Chan T. L., Ma E. S., Tang B. S. (2018). Prospective study on human fecal carriage of Enterobacteriaceae possessing mcr-1 and mcr-2 genes in a regional hospital in Hong Kong. BMC Infect. Dis. 18:81. 10.1186/s12879-018-2987-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X., Zhao X., Che J., Xiong Y., Xu Y., Zhang L., et al. (2017). Detection and dissemination of the colistin resistance gene, mcr-1, from isolates and faecal samples in china. J. Med. Microbiol. 66, 119–125. 10.1099/jmm.0.000425 [DOI] [PubMed] [Google Scholar]
- 6.Chew K. L., La M. V., Lin R. T. P., Teo J. W. P. (2017). Colistin and polymyxin b susceptibility testing for carbapenem-resistant and mcr-positive enterobacteriaceae: comparison of sensititre, microscan, vitek 2, and etest with broth microdilution. J. Clin. Microbiol. 55, 2609–2616. 10.1128/JCM.00268-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clermont O., Christenson J. K., Denamur E., Gordon D. M. (2013). The clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. 10.1111/1758-2229.12019 [DOI] [PubMed] [Google Scholar]
- 8.Cui M., Zhang J., Gu Z., Li R., Chan E. W., Yan M., et al. (2017). Prevalence and molecular characterization of mcr-1-positive Salmonella strains recovered from clinical specimens in china. Antimicrob. Agents Chemother. 61:e02471–16. 10.1128/AAC.02471-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guenther S., Falgenhauer L., Semmler T., Imirzalioglu C., Chakraborty T., Roesler U., et al. (2017). Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from german swine farms. J. Antimicrob. Chemother. 72, 1289–1292. 10.1093/jac/dkw585 [DOI] [PubMed] [Google Scholar]
- 10.Han J. H., Goldstein E. J., Wise J., Bilker W. B., Tolomeo P., Lautenbach E. (2017). Epidemiology of carbapenem-resistant Klebsiella pneumoniae in a network of long-term acute care hospitals. Clin. Infect. Dis. 64, 839–844. 10.1093/cid/ciw856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin L., Wang R., Wang X., Wang Q., Zhang Y., Yin Y., et al. (2018). Emergence of mcr-1 and carbapenemase genes in hospital sewage water in Beijing, China. J. Antimicrob. Chemother. 73, 84–87. 10.1093/jac/dkx355 [DOI] [PubMed] [Google Scholar]
- 12.Kaas R. S., Leekitcharoenphon P., Aarestrup F. M., Lund O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 9:e104984. 10.1371/journal.pone.0104984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kieffer N., Aires-de-Sousa M., Nordmann P., Poirel L. (2017). High rate of mcr-1-producing Escherichia coli and Klebsiella pneumoniae among pigs, Portugal. Emerg. Infect. Dis. 23, 2023–2029. 10.3201/eid2312.170883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kizny Gordon A. E., Mathers A. J., Cheong E. Y. L., Gottlieb T., Kotay S., Walker A. S., et al. (2017). The hospital water environment as a reservoir for carbapenem-resistant organisms causing hospital-acquired infections-a systematic review of the literature. Clin. Infect. Dis. 64, 1435–1444. 10.1093/cid/cix132 [DOI] [PubMed] [Google Scholar]
- 15.Kontopidou F., Plachouras D., Papadomichelakis E., Koukos G., Galani I., Poulakou G., et al. (2011). Colonization and infection by colistin-resistant Gram-negative bacteria in a cohort of critically ill patients. Clin. Microbiol. Infect. 17, E9–E11. 10.1111/j.1469-0691.2011.03649.x [DOI] [PubMed] [Google Scholar]
- 16.Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li R., Xie M., Zhang J., Yang Z., Liu L., Liu X., et al. (2017). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 72, 393–401. 10.1093/jac/dkw411 [DOI] [PubMed] [Google Scholar]
- 18.Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 19.MacNair C. R., Stokes J. M., Carfrae L. A., Fiebig-Comyn A. A., Coombes B. K., Mulvey M. R., et al. (2018). Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun. 9:458. 10.1038/s41467-018-02875-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mansour W., Haenni M., Saras E., Grami R., Mani Y., Ben Haj Khalifa A., et al. (2017). Outbreak of colistin-resistant carbapenemase-producing Klebsiella pneumoniae in Tunisia. J. Glob. Antimicrob. Resist. 10, 88–94. 10.1016/j.jgar.2017.03.017 [DOI] [PubMed] [Google Scholar]
- 21.Matamoros S., van Hattem J. M., Arcilla M. S., Willemse N., Melles D. C., Penders J., et al. (2017). Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 7:15364. 10.1038/s41598-017-15539-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mataseje L. F., Abdesselam K., Vachon J., Mitchel R., Bryce E., Roscoe D., et al. (2016). Results from the canadian nosocomial infection surveillance program on carbapenemase-producing Enterobacteriaceae, 2010 to 2014. Antimicrob. Agents Chemother. 60, 6787–6794. 10.1128/AAC.01359-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McConville T. H., Sullivan S. B., Gomez-Simmonds A., Whittier S., Uhlemann A. C. (2017). Carbapenem-resistant Enterobacteriaceae colonization (CRE) and subsequent risk of infection and 90-day mortality in critically ill patients, an observational study. PLoS ONE 12:e0186195. 10.1371/journal.pone.0186195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendes A. C., Novais A., Campos J., Rodrigues C., Santos C., Antunes P., et al. (2018). Mcr-1 in carbapenemase-producing Klebsiella pneumoniae with hospitalized patients, Portugal, 2016-2017. Emerg. Infect. Dis. 24, 762–766. 10.3201/eid2404.171787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamoto M., Motooka D., Gotoh K., Imai T., Yoshitake K., Goto N., et al. (2014). Performance comparison of second- and third-generation sequencers using a bacterial genome with two chromosomes. BMC Genomics 15:699. 10.1186/1471-2164-15-699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nordmann P., Jayol A., Poirel L. (2016). A universal culture medium for screening polymyxin-resistant Gram-negative isolates. J. Clin. Microbiol. 54, 1395–1399. 10.1128/JCM.00446-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordmann P., Naas T., Poirel L. (2011). Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohno Y., Nakamura A., Hashimoto E., Matsutani H., Abe N., Fukuda S., et al. (2017). Molecular epidemiology of carbapenemase-producing Enterobacteriaceae in a primary care hospital in Japan, 2010-2013. J. Infect. Chemother. 23, 224–229. 10.1016/j.jiac.2016.12.013 [DOI] [PubMed] [Google Scholar]
- 29.Olaitan A. O., Morand S., Rolain J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. 10.3389/fmicb.2014.00643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onori R., Gaiarsa S., Comandatore F., Pongolini S., Brisse S., Colombo A., et al. (2015). Tracking nosocomial Klebsiella pneumoniae infections and outbreaks by whole-genome analysis: small-scale italian scenario within a single hospital. J. Clin. Microbiol. 53, 2861–2868. 10.1128/JCM.00545-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paveenkittiporn W., Kerdsin A., Chokngam S., Bunthi C., Sangkitporn S., Gregory C. J. (2017). Emergence of plasmid-mediated colistin resistance and New Delhi metallo-beta-lactamase genes in extensively drug-resistant Escherichia coli isolated from a patient in thailand. Diagn. Microbiol. Infect. Dis. 87, 157–159. 10.1016/j.diagmicrobio.2016.11.005 [DOI] [PubMed] [Google Scholar]
- 32.Poirel L., Jayol A., Nordmann P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. 10.1128/CMR.00064-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulss S., Semmler T., Prenger-Berninghoff E., Bauerfeind R., Ewers C. (2017). First report of an Escherichia coli strain from swine carrying an oxa-181 carbapenemase and the colistin resistance determinant mcr-1. Int. J. Antimicrob. Agents 50, 232–236. 10.1016/j.ijantimicag.2017.03.014 [DOI] [PubMed] [Google Scholar]
- 34.Saly M., Jayol A., Poirel L., Megraud F., Nordmann P., Dubois V. (2017). Prevalence of faecal carriage of colistin-resistant Gram-negative rods in a university hospital in western France, 2016. J. Med. Microbiol. 66, 842–843. 10.1099/jmm.0.000497 [DOI] [PubMed] [Google Scholar]
- 35.Shanmugakani R. K., Akeda Y., Yamamoto N., Sakamoto N., Hagiya H., Yoshida H., et al. (2017). Pcr-dipstick chromatography for differential detection of carbapenemase genes directly in stool specimens. Antimicrob. Agents Chemother. 61:e00067–17. 10.1128/AAC.00067-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simar S., Sibley D., Ashcraft D., Pankey G. (2017). Colistin and polymyxin b minimal inhibitory concentrations determined by etest found unreliable for gram-negative bacilli. Ochsner J. 17, 239–242. 10.1043/1524-5012-17.3.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sotgiu G., Are B. M., Pesapane L., Palmieri A., Muresu N., Cossu A., et al. (2018). Nosocomial transmission of carbapenem-resistant Klebsiella pneumoniae in an italian university hospital: a molecular epidemiological study. J. Hosp. Infect. 99, 413–418. 10.1016/j.jhin.2018.03.033 [DOI] [PubMed] [Google Scholar]
- 38.Srijan A., Margulieux K. R., Ruekit S., Snesrud E., Maybank R., Serichantalergs O., et al. (2018). Genomic characterization of nonclonal mcr-1-positive multidrug-resistant Klebsiella pneumoniae from clinical samples in thailand. Microb. Drug Resist. 24, 403–410. 10.1089/mdr.2017.0400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terveer E. M., Nijhuis R. H. T., Crobach M. J. T., Knetsch C. W., Veldkamp K. E., Gooskens J., et al. (2017). Prevalence of colistin resistance gene (mcr-1) containing Enterobacteriaceae in feces of patients attending a tertiary care hospital and detection of a mcr-1 containing, colistin susceptible E. coli. PLoS ONE 12:e0178598. 10.1371/journal.pone.0178598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thi Khanh Nhu N., Riordan D. W., Do Hoang Nhu T., Thanh D. P., Thwaites G., Huong Lan N. P., et al. (2016). The induction and identification of novel colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 6:28291. 10.1038/srep28291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Duin D., Doi Y. (2017). The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8, 460–469. 10.1080/21505594.2016.1222343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang R., van Dorp L., Shaw L. P., Bradley P., Wang Q., Wang X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. 10.1038/s41467-018-03205-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Tian G. B., Zhang R., Shen Y., Tyrrell J. M., Huang X., et al. (2017a). Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from china: an epidemiological and clinical study. Lancet Infect. Dis. 17, 390–399. 10.1016/S1473-3099(16)30527-8 [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Zhang R., Li J., Wu Z., Yin W., Schwarz S., et al. (2017b). Comprehensive resistome analysis reveals the prevalence of ndm and mcr-1 in chinese poultry production. Nat. Microbiol. 2:16260. 10.1038/nmicrobiol.2016.260 [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto N., Asada R., Kawahara R., Hagiya H., Akeda Y., Shanmugakani R. K., et al. (2017a). Prevalence of, and risk factors for, carriage of carbapenem-resistant Enterobacteriaceae among hospitalized patients in Japan. J. Hosp. Infect. 97, 212–217. 10.1016/j.jhin.2017.07.015 [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto N., Kawahara R., Akeda Y., Shanmugakani R. K., Yoshida H., Hagiya H., et al. (2017b). Development of selective medium for imp-type carbapenemase-producing Enterobacteriaceae in stool specimens. BMC Infect. Dis. 17:229. 10.1186/s12879-017-2312-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu H., Qu F., Shan B., Huang B., Jia W., Chen C., et al. (2016). Detection of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae from different hospitals in china. Antimicrob. Agents Chemother. 60, 5033–5035. 10.1128/AAC.00440-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong L. L., Phan H. T. T., Shen C., Vihta K. D., Sheppard A. E., Huang X., et al. (2018). High rates of human fecal carriage of mcr-1-positive multidrug-resistant Enterobacteriaceae emerge in china in association with successful plasmid families. Clin. Infect. Dis. 66, 676–685. 10.1093/cid/cix885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.