Abstract
Concrete is susceptible to cracking owing to drying shrinkage, freeze-thaw cycles, delayed ettringite formation, reinforcement corrosion, creep and fatigue, etc. Continuous inspection and maintenance of concrete infrastructure require onerous labor and high costs. If the damaging cracks can heal by themselves without any human interference or intervention, that could be of great attraction. In this study, a novel self-healing approach is investigated, in which fungi are applied to heal cracks in concrete by promoting calcium carbonate precipitation. The goal of this investigation is to discover the most appropriate species of fungi for the application of biogenic crack repair. Our results showed that, despite the significant pH increase owing to the leaching of calcium hydroxide from concrete, Aspergillus nidulans (MAD1445), a pH regulatory mutant, could grow on concrete plates and promote calcium carbonate precipitation.
Introduction
Sustainable infrastructure is the key to creating a sustainable community, due to its significant impact on energy consumption, land use, and global economy. However, many countries are facing the downfall of progressively aging infrastructure that needs rehabilitation. In particular, concrete infrastructure suffers from serious deterioration owing to the effect of various physical and chemical phenomena, such as drying shrinkage, freeze-thaw cycles, reinforcement corrosion, creep and fatigue, and delayed ettringite formation, all of which could lead to concrete cracking.
Cracks themselves may not significantly reduce the load-carrying capacity of concrete in the short run, but they considerably weaken the durability of concrete structures, as they channel in water, oxygen, and carbon dioxide, which could potentially corrode the steel reinforcement. Moreover, cracking may promote severe degradation of the non-mechanical properties of concrete, such as the radiation-shielding properties of concrete elements used in nuclear applications. Nowadays, concrete has been the key construction material for reactor containment and biological shielding structures, which are essential components of the nuclear reactors in service worldwide for power generation1. In addition, cementitious grouts, mortars, and concrete are also often used to provide shielding and encapsulation of various radioactive waste materials from military, research, and power generation applications. Some waste isotopes as well as their decay products will become a serious radiation hazard for hundreds of thousands of years, which requests exceptionally durable storage.
In view of the remarkable social significance of concrete infrastructures and their exceptional service demands, the maintenance and inspection for concrete structures have come into focus. However, continuous inspection and maintenance usually require onerous labor and high investments, which presents a huge and costly challenge. Fortunately, inspired by the amazing capability of the human body to fix broken bones through mineralization, researchers have conducted numerous investigations to equip concrete structures with self-healing properties and generated many innovative solutions2–23.
So far, concrete can fix its own cracks mainly through the following three mechanisms: autogenous healing, embedment of polymeric material, and bacteria-mediated CaCO3 precipitation, which have been summarized in a comprehensive review provided by Seifan et al.20. Among them, the biotechnological approach by using mineral-producing microorganisms is most desirable. It has been demonstrated that in the environment of concrete, CaCO3 can be precipitated by certain species of bacteria through biologically induced mineralization processes2–21. Owing to its sufficient compatibility with concrete compositions, CaCO3 is often regarded as the most appropriate filler for concrete cracks.
Bacteria can precipitate CaCO3 mainly through three different pathways, as summarized below. (1) Certain species of ureolytic bacteria have been explored based on their ability to indirectly cause the formation of calcium precipitates by increasing the local pH via catalyzing the hydrolysis of urea to ammonium2–5. (2) Microbiologists at Delft University of Technology in the Netherlands11–13 tested the second pathway, in which aerobic oxidation of organic acids results in production of CO2, leading to CaCO3 production in the environment of concrete. However, in the case of low oxygen concentration, such as for most underground structures, the efficiency of this approach can be limited. (3) The third pathway is known as dissimilatory nitrate reduction16. This approach is less efficient than the other two in terms of the amount of produced CaCO3, but it can be applied in anaerobic zones.
Fungi-Mediated Self-Healing Concrete
Although the research on bacteria-mediated self-healing concrete indeed achieved a certain level of success, it still suffers from serious limitations. So far, the viability of bacterial spores embedded in concrete is generally less than six months13, which is far from being practical considering the fact that the lifetime of concrete infrastructure can easily be fifty years or even a century. The detrimental environment of concrete, such as very high pH values, tiny pores, serious moisture deficit, varied temperatures, and limited nutrient availability, dramatically influences the microbial metabolic activities and makes bacteria and their spores susceptible to death. In addition, owing to the limited ability of bacteria to produce large amounts of CaCO3, bacteria can only heal small cracks with crack widths less than 0.8 mm10–20.
To address the above-mentioned problems, further investigation on alternative microorganisms becomes potentially essential. In fact, microorganisms are very diverse, and besides bacteria, they also include Archaea, protists, and fungi. Among them, fungi form a species-rich group of eukaryotic organisms, which exhibits vast biodiversity with more than 100 K described species and approximately 1.5 M unknown species. Common fungi include yeasts, lichen-forming fungi, molds, and mushrooms, etc. Recent studies in geomycology demonstrated that certain species of fungi can play a central role in CaCO3 precipitation24,25, but those species have never been tested in the environment of concrete. Currently, the investigation of fungi has been mostly focusing on their importance in the degradation of organic matter, and their connection with inorganic constituents is limited to acquisition of mineral nutrition by mycorrhizal fungi as well as mineral weathering of lichens and ectomycorrhizae.
The cells of filamentous fungi grow as threadlike structures called hyphae, which possess multiple nuclei and grow apically with new apices emerging from the formation of lateral branches, creating an intertwined 3D network called mycelium. In fact, the existing literature suggests that filamentous fungi have unique features to be used in various applications of biomineralization-based technologies26–30. For example, compared with other microbial groups, filamentous fungi exhibit higher surface-to-volume ratios, and therefore possess a larger fraction of organic substrates available for mineral precipitation.
The calcification of fungal filaments is a complex process, and it remains incompletely understood29–36. However, it has been concluded that there are two critical factors determining the amount of CaCO3 production, i.e., carbonate alkalinity and Ca2+ concentration24. The metabolic activities of filamentous fungi that can increase carbonate alkalinity typically include water consumption, physicochemical degassing of fungal respired CO2, oxidation of organic acids, nitrate assimilation, and urea mineralization24. In addition to carbonate alkalinity, fungal metabolic activities can also affect calcium concentration. Ca2+ concentration within fungal cells should be strictly controlled, i.e., most of the calcium ions need to be gathered at the apex for apical growth and sharply reduced in subapical regions24. To maintain this sharp gradient, fungi need to effectively regulate Ca2+. Ca2+ in the cytoplasm is kept at sufficiently low concentrations by deliberately pumping it out of the cell or by binding it onto cytoplasmic proteins24.
In addition, fungi can also influence calcium concentration out of self-protection. Metals that are required for fungal growth and metabolism may become toxic if their concentrations are too high. However, unlike other microbial group, many species of fungi can survive and prosper in regions that are seriously polluted by metals. Precipitation of metal minerals onto hyphae was considered as one of the most important mechanisms to explain the superior metal tolerance in fungi. Concrete is a calcium-rich environment, which represents a stress for fungal cells owing to calcium cytotoxicity and subsequent osmotic pressure24. The production of calcium oxalates has been regarded as a method to decrease their internal Ca2+ concentration24. Precipitation of CaCO3 may be due to a similar passive mechanism to immobilize excessive Ca2+. Excessive alkalinity could also pose a source of stress and precipitation of CaCO3 may be due to intracellular protection24.
While bacteria promote mineral precipitation only through induced biomineralization processes, fungi can do it through both induced biomineralization and organomineralization processes. The cell walls of many fungi contain chitin, a polymer of N-acetylglucosamine37. Chitin is known for its ability to bind Ca2+, as it forms a substrate that could considerably reduce the required activation energies for nuclei formation so that the interfacial energy between the fungus and the mineral crystal becomes significantly lower than the one between the mineral crystal and the solution38–42. Thus, both living and dead fungal biomass can bind ions onto their cell walls, resulting in nucleation and deposition of mineral phases31. Bound Ca2+ can then interact with the dissolved carbonate, resulting in CaCO3 precipitation on the fungal hyphae.
Therefore, thanks to their ability to directly and indirectly promote CaCO3 production, fungi could be used as self-healing agents. The goal of this investigation is to discover the most appropriate species of fungi for the application of biogenic crack repair. To make fungi-mediated self-healing concrete, the fungal spores alongside their nutrients, will be blended into the concrete before the curing process starts. When cracks appear and water trickles into the concrete, the dormant fungal spores will wake up, grow, consume the nutrient soup, and promote CaCO3 precipitates to fix the cracks in situ. After the cracks are finally healed, the bacteria or fungi will make spores and go dormant once more – ready to start a new cycle of self-healing when cracks form again. For existing concrete infrastructures with cracks, the fungal spores and their nutrients can be injected or sprayed into the cracks.
Besides wild-type fungal strains, genetically engineered fungi are also important candidates for self-healing concrete. Since the extremely high pH of the concrete environment can be handled by only a few fungi, pH regulatory mutants were the focus here. It is well known that many microorganisms that are capable of growing over a relatively wide pH range can adjust their gene expression according to the ambient pH levels43,44. Fungi make responses to the extracellular pH by means of activation of a dedicated transcription factor, PacC45–50. Mutants in the pacC regulatory gene are extremely heterogeneous in phenotype43. A major class of mutations are gain-of-function pacCc created by truncating the C-terminal region44. The pacCc mutations bypassing the need for the extracellular pH signal result in permanent activation of alkaline genes and superrepression of acidic genes, which leads to alkalinity mimicry47. Irrespective of ambient pH, fungi exhibiting alkalinity-mimicking mutations believe that they are always at alkaline pH and trigger a gene expression pattern similar to that of the wild-type grown at high pH values, which is exactly what is needed for self-healing concrete.
Materials and Methods
In our previous investigation51, we have found that the spores of a wild-type Trichoderma reesei (ATCC13631) germinated into hyphal mycelium on concrete plates and grew well. However, there exist only a few studies on the pH regulation of T. reesei. In filamentous fungi, the best characterized member for gene regulation by ambient pH is Aspergillus nidulans44. Therefore, gene manipulations are easily achievable in A. nidulans. In this study, three different types of alkalinity-mimicking mutants of A. nidulans, i.e., MAD1445, MAD0305, and MAD0306, will be tested.
The following five different wild-type strains were purchased from American Type Culture Collection (ATCC): Rhizopus oryzae (ATCC22961), Phanerochaete chrysosporium (ATCC24725), A. nidulans (ATCC38163), A. terreus (ATCC1012), and A. oryzae (ATCC1011). The genomes of these fungal strains have been sequenced and annotated and are publicly available52–60. In addition, these species do not exhibit any toxicity and belong to Biosafety Level 1 (BSL-1), which is the lowest risk level.
The above-mentioned fungi are all filamentous fungi. In addition to filamentous fungi, there are also single-celled fungi, i.e., yeasts, that do not form hyphae. One of the most well-known species of yeast is Saccharomyces cerevisiae, also called baker’s yeast, as it has been instrumental to winemaking, baking, and brewing since ancient times. Recently, a series of publications were released on the success of designing synthetic set of S. cerevisiae chromosomes, which could allow S. cerevisiae to rapidly evolve and rearrange its genome, optimizing itself for certain applications61–63. Thus, in this study, S. cerevisiae will also be tested.
To characterize the fungal precipitates, X-ray diffraction (XRD), scanning electron microscope (SEM), and transmission electron microscope (TEM) were applied in this study. XRD is an established technique to identify unknown crystalline phases64,65. SEM has been used to visualize fungal precipitates66,67, and TEM has been used to study fungal-biotite interfaces and weathered alkali feldspars68,69. SEM will be applied to analyze the morphology and composition of the fungal precipitates, which will complement the very local characterization by TEM.
In this section, the experimental procedures of gene replacement to obtain alkalinity-mimicking mutations, survival test of fungi on concrete plates, as well as phase identification and microscopic characterization of fungal precipitates will be presented. The procedures of the yeast strain identification and preparation of concrete specimens are described in the Supplementary Information.
Gene Replacement Procedure to Obtain Alkalinity-Mimicking Mutations
The full genotypes of the strains investigated in the current work are given in Table 1. The detailed about the parental strains and the selection procedures of isolating the pacC mutations were previously published70–74.
Table 1.
The full genotypes of the strains.
Briefly, the gene replacement procedure to obtain alleles pacC14 and pacC202 can be described as follows. The parental strains with genotype pabaA1 gatA2 palF15 fwA1 (p-aminobenzoate-requiring, lacking ω-amino acid transaminase, mimicking growth at acidic pH, and having fawn conidial color) were mutagenized with ultraviolet treatment followed by growth selection on media supplemented with 5 mM γ-amino-n-butyrate in presence of 10 mM ammonium tartrate as nitrogen source75–77. The pacC14 mutant strain was selected as it mimics the growth at alkaline pH values, downregulates the γ-amino-n-butyrate permease, decreases the γ-amino-n-butyrate level in cells, and thus relieves the γ-amino-n-butyrate-induced cytotoxicity. The pacC202 strain was a spontaneous mutant strain derived from the parental strains with genotype pabaA1 sasA60 (p-aminobenzoate-requiring and semialdehyde sensitive), selected as it relieves the toxicity of 10 mM γ-amino-n-butyrate. The pacC genomic sequences in pacC14 and pacC202 strains were then obtained by Polymerase Chain Reaction (PCR) amplification.
The procedure to obtain MYC3-tagged wild-type pacC allele, designated as pacC900, as well as the engineered allele pacC14900 can be described as follows. A null allele of pacC was first constructed by transforming the orthologous pyr4 (orotidine 5′-phosphate decarboxylase) gene of Neurospora crassa flanked by 750 bp of upstream and 700 bp of downstream pacC genomic sequences into a pyrG89 (resulting in pyrimidine auxotrophy) pacC14 strain. Transformation was carried out using linearized fragments containing the pacC moiety of the plasmid. The long flanking sequence ensures high efficiency of homologous recombination-mediated gene replacement. The transformants displaying pyrimidine prototrophy were then selected and tested by Southern blot analyses to confirm that pacC14 was replaced by pyr4. The pacC null strain was then used as recipient strain and transformed with linearized MYC3-tagged pacC carrying C2437A mutation flanked with genomic sequence 825 bp upstream of pacC start codon and 925 bp downstream of stop codon78. Transformants displaying pacC colony morphology were tested for pyrimidine auxotrophy. The gene replacement was again confirmed using Southern blot analyses. The genotypes in the resulting pacC14900 strain were further confirmed by PCR sequencing. The protein expression was confirmed by Western blot analyses using anti-MYC antibody.
Survival Test of Fungi on Concrete Plates
In this study, potato dextrose agar (PDA) was chosen as growth medium. To investigate the impact of the extracellular pH on fungal growth, the inert pH buffer 3-(N-morpholino)propanesulfonic acid (MOPS, 20 mM, pH 7.0) was added to some of the growth media. Sterilization of concrete plate was carried out in a steam autoclave at 121 °C under 15 psi of pressure for 30 min. A 5 mm diameter mycelial disc of each fungal strain removed from 7-day-old cultures was aseptically placed at the center of each 60 mm Petri dish containing 10 ml growth medium. In some Petri dishes, there exists a concrete plate of 3 mm thickness at the bottom. Sterile PDA plugs were used as the negative inoculum control. The Petri dishes were incubated at 22 °C and 30 °C, respectively, for 21 days.
To summarize, as shown in Fig. S1 in the Supplementary Information, for each type of fungal strain, eight different cases were investigated including PDA30, PDA22, MPDA30, MPDA22, CPDA30, CPDA22, CMPDA30, and CMPDA22. All the cases were investigated independently in triplicates.
Radial growth was measured based on two perpendicular diameters. The fungal samples prepared by the tape touch method79 were studied using an optical microscope. pH values were recorded after 21 days of incubation by five independent measurements using an Orion 9110DJWP double junction pH electrode, which is a semi-micro combination pH electrode.
Phase Identification of Fungal Precipitates
Phase identification of the fungal precipitates were performed on a Siemens-Bruker D5000 powder diffractometer with Cu-Kα radiation in the θ/θ configuration operating at 40 kV and 30 mA. The samples were analyzed over the range of 10° to 80° 2θ at a scan rate of 1°/min with an increment of 0.02° 2θ. Before the measurements, the solid precipitates were isolated by dissolving the fungal sample in a vat of dilute bleach (NaOCl less than 3% by weight) at room temperature and washing it repeatedly in methanol following the published protocol65.
Microscopic Characterization of Fungal Precipitates
Scanning Electron Microscope (SEM)
The fungal precipitates were studied by a Zeiss Supra 55 VP Field Emission SEM with an EDAX Genesis energy-dispersive X-ray spectrometer (EDS) at accelerating voltages of 5 kV to 20 kV. The collected solids were placed in an oven at 50 °C for two days, and then were sputter-coated with carbon by an Emitech K950X Carbon Vacuum Evaporator in order to ensure electrical conductivity. The chemical microanalysis technique EDS was applied in conjunction with the SEM analysis.
Transmission Electron Microscope (TEM)
TEM analysis was performed by a JEOL JEM-2100F TEM operating at 200 kV. The precipitates were collected by centrifugation (6000 × g, 10 min) and washed multiple times in methanol to get rid of the growth medium. The particles were dried at room temperature then ground using a set of agate mortar and pestle into fine powders. The powders were then collected on copper grids overlain with a carbon-coated collodion film prior to the examination. Bright-field imaging (BFI), selected area electron diffraction (SAED), and high-resolution transmission electron microscopy (HRTEM) were applied to analyze the calcite phases.
Pore Size Distribution Inside Cement Paste Specimen
High-resolution X-ray computed microtomography (μCT) was applied to characterize the pore size distribution inside the cement paste specimen. Images were acquired using UltraTom X-ray μ-CT system equipped with a LaB6 X-ray source and a CCD camera with resolution of 4000 × 2624 pixels. Small cubic samples of 0.4 cm × 0.4 cm × 0.4 cm were used, for which the voxel size of the obtained images will be about 1.2 μm. Samples were firmly fixed on a cylindrical holder mounted on a rotating table using an adhesive paste. Acquisition parameters were set as follows: source 230 kV, voltage 90 kV, intensity 200–300 μA, filament LaB6, frame rate 0.4, and 8 averaging images. To impose small angular increments to cover 360° along the rotation axis, a total of 1664 phase contrast images were collected. The acquisition duration was about 30 min for each sample. Based on the raw data measured by μCT, the reconstruction of inner microstructure was carried out by using the Octopus Imaging software80. Images were post-processed by using the ImageJ software81.
Results and Discussion
Mutant Sequence Changes and Resulting Proteins
The mutant sequence changes in pacC and the resulting proteins are summarized in Table 2. Note that c denotes constitutive phenotype, which bypasses the requirement for the pH signal and mimics alkaline growth conditions; and an asterisk denotes a stop codon. It can be seen that the pacC14 genotype carries a C2437A allele, causing an early stop codon that results in a C-terminal truncation of PacC protein at 492 amino acid. The pacC202 genotype has a deletion from 2353 bp to 2555 bp, resulting in an open reading frame shift after amino acid 464 and an early stop codon. The resulting pacC14900 strain has a MYC3-tagged pacC carrying the pacC14 mutation at the pacC locus.
Table 2.
Mutant sequence changes in pacC and resulting proteins.
Identification of the Yeast Strain
The yeast strain has 99% ITS sequence similarity to the S. cerevisiae ex-type culture CBS 1171 (NR_111007) and has the same morphological features of S. cerevisiae. Therefore, it is confirmed as the baker’s yeast, i.e., S. cerevisiae.
Fungal Growth on Concrete Plates
The measurement results show that, because of the leaching of calcium hydroxide from concrete, the pH values increased significantly from 6.5 to 13.0 (Table 3). On the concrete plates, only one type of pH regulatory mutants of A. nidulans, i.e., MAD1445, has been found to be able to grow well (Table 4). At 30 °C, its growth rate was 3.2 mm/day in the case of CMPDA30. Plentiful conidia were found on the concrete plates, which possess similar morphology with those found in the Petri dishes without concrete (Fig. 1). However, A. nidulans (MAD1445) did not grow in the cases of CPDA22 or CPDA30. The other eight species grew well in all the cases without concrete but did not grow on any concrete plate. The fungal growth in each case and the associated optical microscopic images are shown in Fig. S1, Fig. S2, and Fig. S3 in the Supplementary Information.
Table 3.
pH measurement results after 12 days of inoculation.
| PDA | MPDA | CPDA | CMPDA | |
|---|---|---|---|---|
| Control | 6.13 ± 0.12 | 6.83 ± 0.14 | 13.14 ± 0.14 | 12.02 ± 0.11 |
| Aspergillus nidulans (ATCC38163) | 6.50 ± 0.12 | 7.22 ± 0.12 | 13.04 ± 0.13 | 11.60 ± 0.10 |
| Aspergillus nidulans (MAD1445) | 6.42 ± 0.09 | 7.12 ± 0.15 | 13.22 ± 0.16 | 11.21 ± 0.10 |
| Aspergillus nidulans (MAD0305) | 6.31 ± 0.12 | 7.08 ± 0.13 | 13.01 ± 0.10 | 12.11 ± 0.10 |
| Aspergillus nidulans (MAD0306) | 6.19 ± 0.19 | 7.11 ± 0.19 | 13.11 ± 0.06 | 11.32 ± 0.08 |
| Aspergillus oryzae (ATCC1011) | 6.34 ± 0.13 | 7.14 ± 0.20 | 13.63 ± 0.13 | 12.19 ± 0.19 |
| Aspergillus terreus (ATCC1012) | 6.33 ± 0.22 | 7.03 ± 0.15 | 13.14 ± 0.20 | 13.04 ± 0.08 |
| Rhizopus oryzae (ATCC22961) | 6.71 ± 0.20 | 7.02 ± 0.06 | 13.24 ± 0.19 | 12.84 ± 0.12 |
| Phanerochaete chrysosporium (ATCC24725) | 6.81 ± 0.20 | 7.00 ± 0.05 | 13.31 ± 0.13 | 12.11 ± 0.13 |
| Saccharomyces cerevisiae (Yeast) | 6.25 ± 0.13 | 7.04 ± 0.19 | 13.01 ± 0.06 | 12.81 ± 0.15 |
Table 4.
Average growth rates (mm/day) after 21 days of inoculation (n = 6). Note that [mean ± standard deviation] followed by different letters are remarkably different according to Tukey’s HSD test (p = 0.05).
| PDA30 | PDA22 | MPDA30 | MPDA22 | CPDA30 | CPDA22 | CMPDA30 | CMPDA22 | |
|---|---|---|---|---|---|---|---|---|
| Aspergillus nidulans (ATCC38163) | 2.6 ± 0.07a | 2.6 ± 0.25a | 0.5 ± 0.12b | 0.7 ± 0.23b | 0 | 0 | 0 | 0 |
| Aspergillus nidulans (MAD1445) | 3.9 ± 0.18a | 1.9 ± 0.14c | 1.2 ± 0.15d | 1.2 ± 0.17d | 0 | 0 | 3.2 ± 0.52b | 0.9 ± 0.23e |
| Aspergillus nidulans (MAD0305) | 3.2 ± 0.17a | 2.1 ± 0.06b | 1.6 ± 0.08c | 1.5 ± 0.21c | 0 | 0 | 0 | 0 |
| Aspergillus nidulans (MAD0306) | 2.1 ± 0.20a | 1.4 ± 0.33b | 0.3 ± 0.06c | 0.3 ± 0.12c | 0 | 0 | 0 | 0 |
| Aspergillus oryzae (ATCC1011) | 4.3 ± 0.06a | 2.8 ± 0.14c | 3.3 ± 0.09b | 2.3 ± 0.13d | 0 | 0 | 0 | 0 |
| Aspergillus terreus (ATCC1012) | 4.6 ± 0.46a | 2.8 ± 0.35b | 1.8 ± 0.06c | 1.7 ± 0.22c | 0 | 0 | 0 | 0 |
| Rhizopus oryzae (ATCC22961) | 7.8 ± 0.07a | 7.8 ± 0.15a | 7.8 ± 0.06a | 7.8 ± 0.12a | 0 | 0 | 0 | 0 |
| Phanerochaete chrysosporium (ATCC24725) | 7.8 ± 0.06a | 7.8 ± 0.07a | 7.8 ± 0.15a | 4.2 ± 0.07b | 0 | 0 | 0 | 0 |
| Saccharomyces cerevisiae (Yeast) | 1.9 ± 0.09a | 1.9 ± 0.23a | 1.9 ± 0.29a | 1.5 ± 0.11b | 0 | 0 | 0 | 0 |
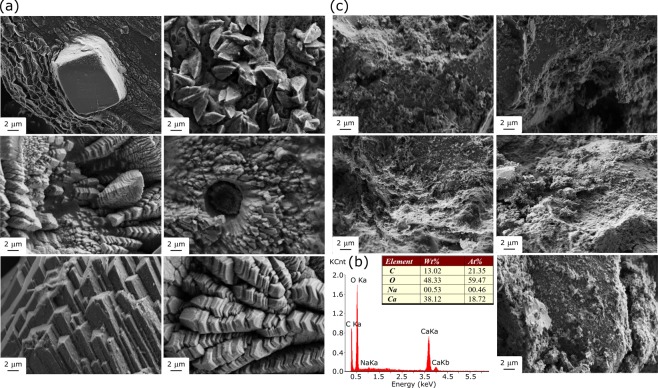
Figure 1.
For the case of A. nidulans (MAD1445), plentiful conidia were found in the case of CMPDA 30. The diameter of A. nidulans spores is typically larger than 3 μm. More images are shown in Fig. S3 in the Supplementary Information.
Note that of all these three strains, which carry pacC constitutive mutations and otherwise possess very similar phenotypes, only MAD1445 grows well on the concrete plates in the presence of MOPS. This could be due to the fact that other unknown genetic factors could lead to increased survival against high pH values. The three strains were obtained through either mutagenesis or homologous recombination. Although the pacC locus was sequence-verified, it is possible that mutations happened at other undesired loci of the MAD1445 strain resulted in increased survival in the alkaline environment.
Characteristics of the Fungal Precipitates
The XRD analysis shows that the precipitates promoted by fungi are indeed CaCO3 crystals (Fig. 2). The sharp peak at approximately 30° 2θ indicates highly crystalline phases of calcite. In comparison, the mortar specimens obtained from the control medium were composed of both quartz and calcite.
Figure 2.

XRD results for the solids collected from A. nidulans (MAD1445)-inoculated media and fungus-free media.
Figure 3(a) shows massive mineral crystals in the A. nidulans (MAD1445)-inoculated media: CMPDA22 and CMPDA30. The particles had regular shapes with well-defined crystalline faces, indicating high crystallinity of the precipitates. The mineral crystals exhibited a range of crystalline morphologies from a single pure bulk crystal to plate-like crystals stacked upon each other. Besides, the crystals confirmed fungal involvement, as many cylindrical holes were found in the crystals, which probably occurred in the space inhabited by fungi. The holes also suggested that fungal filaments functioned as nucleation sites during the biomineralization process. As shown in Fig. 3(b), EDS results show that the crystals are composed of calcium, carbon, and oxygen and their atomic percentage approximately matches that of CaCO3. Note that EDS data may carry error if the sample is not homogeneous or flat. In our test, EDS data are obtained from several relatively flat sample areas, and the results are consistent. In comparison, the amount of calcium precipitation in the control medium was significantly reduced, as evidenced by the absence of faceted particles (Fig. 3(c)).
Figure 3.
SEM and EDS results of the collected solids: (a) A. nidulans (MAD1445)-inoculated media; (b) EDS spectra; and (c) fungus-free medium.
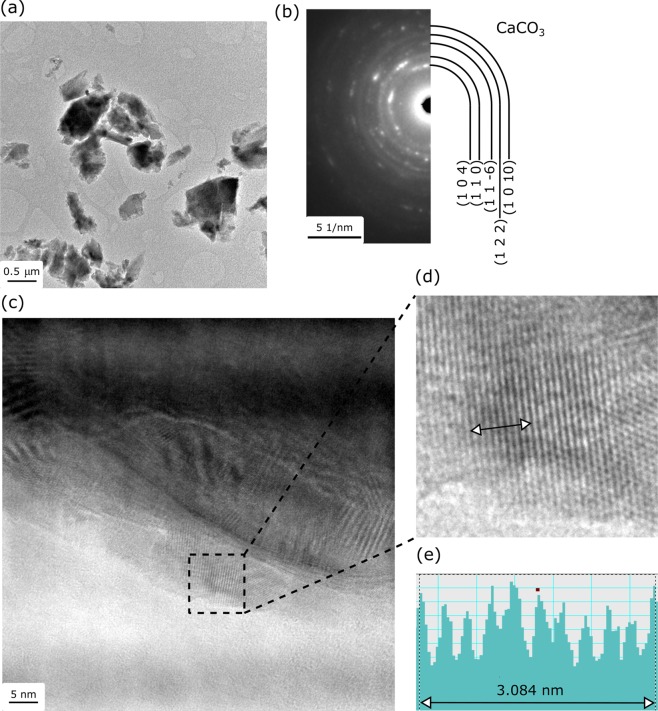
TEM analyses, as shown in Fig. 4, further confirm the crystalline nature of the CaCO3. Figure 4(a) shows a bright-field TEM image of the fragments. Figure 4(b) is a SAED pattern from the collection of the particles shown in Fig. 4(a), which demonstrates the crystalline nature of the precipitated particles, as evidenced by the presence of well resolved diffraction rings of (104), (110), (11–6), (122), and (1010) planes, matching well with the calcite phase. Figure 4(c) is a higher magnification TEM image of the precipitate particle, which shows the presence of crystalline lattice (moiré fringes are also visible due to the overlapping of particles). Figure 4(d) is a HRTEM image obtained from the area as marked with the black dashed square in Fig. 4(c). As shown in Fig. 4(e), the interplanar spacing of the crystalline planes in Fig. 4(d) is measured to be 0.308 nm, which matches with the interplanar spacing of (104) of the hexagonal crystal of calcite with the lattice parameters of a = b = 4.989 Å, c = 17.062 Å, α = β = 90°, and γ = 120°.
Figure 4.
TEM analysis of the fragments of precipitated particles: (a) Bright-field TEM image of the fragments of crushed precipitated calcite; (b) SAED obtained from the crushed precipitate shown in (a), the indexing of the diffraction rings matches well with the crystal structure of the calcite phase; (c) Higher-magnification TEM image of the precipitated particles, showing the presence of crystalline lattice fringes; (d) HRTEM image from the area marked with the black dashed square in (c); (e) Measurement of the interplanar spacing by averaging ten lattice fringes marked in (d).
The experiments presented in this study have been repeated three times, confirming that the results are reproducible. However, it is still not clear what are the optimal conditions for fungal growth and CaCO3 precipitation. Along with our previous work51, we found CaCO3 precipitates in both A. nidulans (MAD1445)-inoculated and T. reesei (ATCC13631)-inoculated media, but the amount of mineral crystals was rather limited. As our future endeavors, the effects of the multiple factors promoting fungal growth and CaCO3 precipitation will be tested, such as concentrations of calcium ions and dissolved inorganic carbon, temperature, exposure to ultraviolet light, growth medium composition, fungal spore concentration, and availability of nucleation sites, etc. For examples, so far, our tests have confirmed that adding 4.0 µg/ml aminobenzoic acid and 0.02 µg/ml biotin to the growth medium significantly promotes the growth of A. nidulans (MAD1445); and adding 5.0 mM CaCl2 to the growth medium can promote calcium carbonate precipitation. In addition, a systematic investigation will be conducted to assess the fungal spore germination. According to Wang et al., whether or not the fungal spores in/on the concrete crack germinate can be measured by the oxygen consumption on the crack surface82. Furthermore, we are also testing urease-positive fungi Neurospora crassa83,84 and Myrothecium gramineum85 for this specific application.
Embedment of Fungal Healing Agents in Concrete
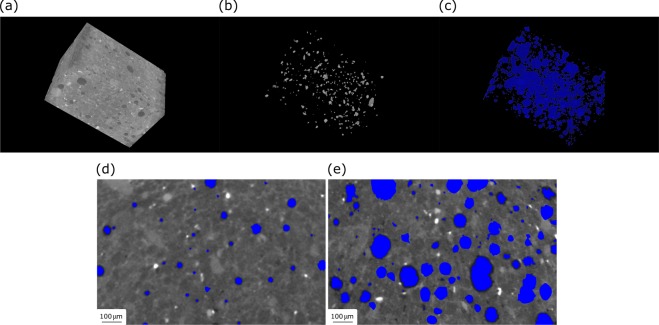
If the fungal spores are larger than the pore sizes in concrete, if they are directly put into cement paste, most spores will be crushed and lost viability during the hydration process. Our results on μCT characterization of concrete microstructure have been shown in Fig. 5. The 3D renderings, as shown in Fig. 5(a) to Fig. 5(c), are particularly useful for qualitative assessment of phase distribution, which is complementary to higher resolution 2D microscopy imaging. From a typical 2D slice as shown in Fig. 5(d), it shows that the matrix pore sizes after 28 days of curing are much smaller than the typical diameters of fungal spores, which are usually larger than 3 μm (Fig. 1). The matrix pore sizes in air-entrained specimens after 28 days of curing are shown in Fig. 5(e). It shows that air-entraining agents could be applied to generate plentiful additional tiny air bubbles in concrete matrix to provide the housing for the healing agents. Although the specimen without air-entraining agents may also contain a small number of inhomogeneous air voids, the specimen containing air-entraining agents usually shows a large number of homogeneous air voids with appropriate size, which is more beneficial for both workability and durability of concrete.
Figure 5.
A 3D rendering of a grayscale volume is shown in (a), the corresponding high-density particles and pores are shown in (b,c), respectively. (d) Pore size distribution inside cement paste specimens after 28 days of curing measured by μCT. (e) Effect of air-entraining agent on pore size distribution measured by μCT.
Concluding Remarks
Concrete is considered as an extreme environment for fungi mainly due to its high pH values. To use fungi to heal cracks in concrete, the first step, which is also the most critical step, is to find the fungi strains which can survive the harsh environment of concrete and can promote calcium carbonate precipitation. The experimental results of this study showed that, despite the significant pH rise caused by the leaching of calcium hydroxide from concrete, one type of pH regulatory mutants of A. nidulans (MAD1445) could grow on concrete plates. In comparison, A. nidulans (ATCC38163), A. nidulans (MAD0305), A. nidulans (MAD0306), R. oryzae (ATCC22961), P. chrysosporium (ATCC24725), A. terreus (ATCC1012), and A. oryzae (ATCC1011) could not grow in the alkali solution caused by the leaching of calcium hydroxide from concrete. The characterization by XRD, SEM, and TEM confirmed that the precipitates promoted by fungal activities were mainly composed of calcite.
It is important to note that according to the existing literature, A. nidulans is usually considered relatively harmless to healthy human beings. However, according to the investigation conducted by Bignnell et al.86 on the role of the PacC-mediated pH adaptation in the pathogenesis of pulmonary aspergillosis, elimination of the PacC protein increases virulence in a murine neutropenic model, indicating that activities under PacC control may be crucial for A. nidulans pathogenicity. Therefore, a complete assessment needs to be performed to examine the possible short-term and long-term adverse outcomes of pacCc mutation of A. nidulans on both environment and human beings before it is used in concrete structures.
Supplementary information
Acknowledgements
Congrui Jin was funded by the Research Foundation for the State University of New York (TAE-16083068). Congrui Jin was also supported by the Small Scale Systems Integration and Packaging (S3IP) Center of Excellence. Dr. Miguel Penalva at the Biological Research Center of the Spanish National Research Council was thanked for kindly providing three different alkalinity-mimicking mutants of A. nidulans, i.e., MAD1445, MAD0306, and MAD0305. Dr. Joan Tilburn at Department of Microbiology of Imperial College London, Dr. Daniel Lucena-Agell at the Biological Research Center of the Spanish National Research Council, and Dr. America Hervas-Aguilar at the Division of Biomedical Sciences of University of Warwick were thanked for useful discussion.
Author Contributions
C.J. and N.Z. conceived and initiated the project. R.R.M., J.L., H.Z. and X.C. designed and performed the experimental work. G.Z., N.Z. and C.J. supervised the research tasks. R.R.M. and C.J. wrote the initial draft of the manuscript. R.R.M., J.L., X.C., Z.L., G.Z., N.Z. and C.J. co-edited the manuscript. All authors reviewed the manuscript before submission.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ning Zhang, Email: ningz@rutgers.edu.
Congrui Jin, Email: cjin@binghamton.edu.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39156-8.
References
- 1.Kurtis K, et al. Can we design concrete to survive nuclear environments? Concr. Int. 2017;39:29–35. [Google Scholar]
- 2.Ramachandran SK, Ramakrishnan V, Bang SS. Remediation of concrete using micro-organisms. ACI Mater. J. 2001;98:3–9. [Google Scholar]
- 3.Bang SS, Galinat JK, Ramakrishnan V. Calcite precipitation induced by polyurethane-immobilized Bacillus pasteurii. Enzyme Microb. Technol. 2001;28:404–409. doi: 10.1016/S0141-0229(00)00348-3. [DOI] [PubMed] [Google Scholar]
- 4.Dick J, et al. Bio-deposition of a calcium carbonate layer on degraded limestone by Bacillus species. Biodegradation. 2006;17:357–367. doi: 10.1007/s10532-005-9006-x. [DOI] [PubMed] [Google Scholar]
- 5.Ramakrishnan, V. Performance characteristics of bacterial concrete: a smart biomaterial. In: Proceedings of the 1st International Conference on Recent Advances in Concrete Technology, Washington, D.C., 67-78 (2007).
- 6.De Muynck W, Debrouwer D, De Belie N, Verstraete W. Bacterial carbonate precipitation improves the durability of cementitious materials. Cement Concrete Res. 2008;38:1005–1014. doi: 10.1016/j.cemconres.2008.03.005. [DOI] [Google Scholar]
- 7.Van Tittelboom K, De Belie N, Van Loo D, Jacobs P. Self-healing efficiency of cementitious materials containing tubular capsules filled with healing agent. Cement Concrete Composites. 2011;33:497–505. doi: 10.1016/j.cemconcomp.2011.01.004. [DOI] [Google Scholar]
- 8.De Muynck W, De Belie N, Verstraete W. Microbial carbonate precipitation improves the durability of cementitious materials: a review. Ecol. Eng. 2010;36:118–36. doi: 10.1016/j.ecoleng.2009.02.006. [DOI] [Google Scholar]
- 9.Van Tittelboom K, De Belie N, De Muynck W, Verstraete W. Use of bacteria to repair cracks in concrete. Cem. Concr. Res. 2010;40:157–66. doi: 10.1016/j.cemconres.2009.08.025. [DOI] [Google Scholar]
- 10.Wang J, Van Tittelboom K, De Belie N, Verstraete W. Use of silica gel or polyurethane immobilized bacteria for self-healing concrete. Constr. Build. Mater. 2012;26:532–540. doi: 10.1016/j.conbuildmat.2011.06.054. [DOI] [Google Scholar]
- 11.Jonkers, H. M., Schlangen, E. A two component bacteria-based self-healing concrete. In: Alexander, M. G., Beushausen, H. D., Dehn, F. & Moyo, P. editors Concrete Repair, Rehabilitation and Retrofitting II. Boca Raton, CRC Press, Taylor and Francis Group (2009).
- 12.Jonkers HM, Thijssen A, Muyzer G, Copuroglu O, Schlangen E. Application of bacteria as self-healing agent for the development of sustainable concrete. Ecol. Eng. 2010;36:230–235. doi: 10.1016/j.ecoleng.2008.12.036. [DOI] [Google Scholar]
- 13.Jonkers HM. Bacteria-based self-healing concrete. Heron. 2011;56:1–12. [Google Scholar]
- 14.Wiktor V, Jonkers HM. Quantification of crack-healing in novel bacteria-based self-healing concrete. Cement Concrete Composites. 2011;33:763–770. doi: 10.1016/j.cemconcomp.2011.03.012. [DOI] [Google Scholar]
- 15.Bang SS, Lippert JJ, Yerra U, Mulukutla S, Ramakrishnan V. Microbial calcite, a bio-based smart nanomaterial in concrete remediation. Int. J. Smart Nano Mater. 2010;1:28–39. doi: 10.1080/19475411003593451. [DOI] [Google Scholar]
- 16.Ersan YC, De Belie N, Boon N. Microbially induced CaCO3 precipitation through denitrification: an optimization study in minimal nutrient environment. Biochem. Eng. J. 2015;101:108–118. doi: 10.1016/j.bej.2015.05.006. [DOI] [Google Scholar]
- 17.Wang JY, De Belie N, Verstraete W. Diatomaceous earth as a protective vehicle for bacteria applied for self-healing concrete. J. Ind. Microbiol. Biotechnol. 2012;39:567–577. doi: 10.1007/s10295-011-1037-1. [DOI] [PubMed] [Google Scholar]
- 18.Wang JY, Soens H, Verstraete W, De Belie N. Self-healing concrete by use of microencapsulated bacterial spores. Cement Concrete Res. 2014;56:139–152. doi: 10.1016/j.cemconres.2013.11.009. [DOI] [Google Scholar]
- 19.Wang JY, Snoeck D, Van Vlierberghe S, Verstraete W, De Belie N. Application of hydrogel encapsulated carbonate precipitating bacteria for approaching a realistic self-healing in concrete. Constr. Build. Mater. 2014;68:110–119. doi: 10.1016/j.conbuildmat.2014.06.018. [DOI] [Google Scholar]
- 20.Seifan M, Samani A, Berenjian A. Bioconcrete: next generation of self-healing concrete. Appl. Microbiol. Biotechnol. 2016;100:2591–2602. doi: 10.1007/s00253-016-7316-z. [DOI] [PubMed] [Google Scholar]
- 21.Fortin D, Ferris FG, Beveridge TJ. Surface-mediated mineral development by bacteria. Rev. Mineral. 1997;35:161–180. [Google Scholar]
- 22.Dry C. Matrix cracking repair and filling using active and passive modes for smart timed release of chemicals from fibers into cement matrices. Smart Mater. Struct. 1994;3:118–123. doi: 10.1088/0964-1726/3/2/006. [DOI] [Google Scholar]
- 23.Sangadji S, Schlangen E. Mimicking bone healing process to self-repair concrete structure novel approach using porous network concrete. Procedia Engineering. 2013;54:315–326. doi: 10.1016/j.proeng.2013.03.029. [DOI] [Google Scholar]
- 24.Bindschedler S, Cailleau G, Verrecchia E. Role of fungi in the biomineralization of calcite. Minerals. 2016;6:1–19. doi: 10.3390/min6020041. [DOI] [Google Scholar]
- 25.Zhu T, Dittrich M. Carbonate precipitation through microbial activities in natural environment, and their potential in biotechnology: a review. Frontiers in Bioengineering and Biotechnology. 2016;4:1–21. doi: 10.3389/fbioe.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volesky B, Holan ZR. Biosorption of heavy metals. Biotechnology Progress. 1995;11:235–250. doi: 10.1021/bp00033a001. [DOI] [PubMed] [Google Scholar]
- 27.Dias MA, Lacerda ICA, Pimentel PF, De Castro HF, Rosa CA. Removal of heavy metals by an Aspergillus terreus strain immobilized in a polyurethane matrix. Lett. Appl. Microbiol. 2002;34:46–50. doi: 10.1046/j.1472-765x.2002.01040.x. [DOI] [PubMed] [Google Scholar]
- 28.Phanjom P, Ahmed G. Biosynthesis of silver nanoparticles by Aspergillus oryzae (MTCC No. 1846) and its characterizations. Nanosci. Nanotech. 2015;5:14–21. [Google Scholar]
- 29.Gadd, G. M. Fungi and yeasts for metal accumulation. In: Ehrilich, H. L. & Brierley, C. editors Microbial Mineral Recovery. New York, McGraw-Hill (1990).
- 30.Gadd GM. Interactions of fungi with toxic metals. New Phytol. 1993;124:25–60. doi: 10.1111/j.1469-8137.1993.tb03796.x. [DOI] [Google Scholar]
- 31.Takey M, Shaikh T, Mane N, Majumder DR. Bioremediation of xenobiotics: use of dead fungal biomass as biosorbent. International Journal of Research in Engineering and Technology. 2013;3:565–570. [Google Scholar]
- 32.Roncero C. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 2002;41:367–378. doi: 10.1007/s00294-002-0318-7. [DOI] [PubMed] [Google Scholar]
- 33.Manoli F, Koutsopoulos E, Dalas E. Crystallization of calcite on chitin. J. Cryst. Growth. 1997;182:116–124. doi: 10.1016/S0022-0248(97)00318-7. [DOI] [Google Scholar]
- 34.Sayer JA, Kierans M, Gadd GM. Solubilisation of some naturally occurring metal-bearing minerals, limescale and lead phosphate by Aspergillus niger. FEMS Microbiol. Lett. 1997;154:29–35. doi: 10.1111/j.1574-6968.1997.tb12620.x. [DOI] [PubMed] [Google Scholar]
- 35.Gharieb MM, Sayer JA, Gadd GM. Solubilization of natural gypsum (CaSO4·2H2O) and formation of calcium oxalate by Aspergillus niger and Serpula himantioides. Mycol. Res. 1998;102:825–830. doi: 10.1017/S0953756297005510. [DOI] [Google Scholar]
- 36.Verrecchia, E. P., Braissant, O. & Cailleau, G. The oxalate-carbonate pathway in soil carbon storage: the role of fungi and oxalotrophic bacteria. In: Gadd, G. M. editor. Fungi in Biogeochemical Cycles. Cambridge, Cambridge University Press (2006).
- 37.Blumenthal HJ, Roseman S. Quantitative estimation of chitin in fungi. J. Bacteriol. 1957;74:222–224. doi: 10.1128/jb.74.2.222-224.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ehrlich H. Chitin and collagen as universal and alternative templates in biomineralization. International Geology Review. 2010;35:661–699. doi: 10.1080/00206811003679521. [DOI] [Google Scholar]
- 39.Wysokowski M, et al. Poriferan chitin as a versatile template for extreme biomimetics. Polymers. 2015;7:235–265. doi: 10.3390/polym7020235. [DOI] [Google Scholar]
- 40.Brunner E, et al. Chitin-based organic networks: an integral part of cell wall biosilicain the diatom Thalassiosira pseudonana. Angew. Chem. Int. Ed. 2009;48:9724–9727. doi: 10.1002/anie.200905028. [DOI] [PubMed] [Google Scholar]
- 41.Al-Sawalmih A, et al. Microtexture and chitin/calcite orientation relationship in the mineralized exoskeleton of the American lobster. Adv. Funct. Mater. 2008;18:3307–3314. doi: 10.1002/adfm.200800520. [DOI] [Google Scholar]
- 42.Wysokowski M, et al. Preparation of chitin-silica composites by in vitro silicification of two-dimensional Ianthella basta demosponge chitinous scaffolds under modified Stöber conditions. Mater. Sci. Eng. C. 2013;33:3935–3941. doi: 10.1016/j.msec.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 43.Penalva MA, Arst HN., Jr. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 2002;66:426–446. doi: 10.1128/MMBR.66.3.426-446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Penalva MA, Arst HN., Jr. Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 2004;58:425–451. doi: 10.1146/annurev.micro.58.030603.123715. [DOI] [PubMed] [Google Scholar]
- 45.Lamb TM, Xu W, Diamond A, Mitchell AP. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 2001;276:1850–1856. doi: 10.1074/jbc.M008381200. [DOI] [PubMed] [Google Scholar]
- 46.Nobile CJ, et al. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol. 2008;10:2180–2196. doi: 10.1111/j.1462-5822.2008.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Penalva MA, Tilburn J, Bignell E, Arst HN., Jr. Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 2008;16:291–300. doi: 10.1016/j.tim.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 48.Caracuel Z, et al. The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum. Mol. Microbiol. 2003;48:765–779. doi: 10.1046/j.1365-2958.2003.03465.x. [DOI] [PubMed] [Google Scholar]
- 49.Caracuel Z, Casanova C, Roncero MI, Di Pietro A, Ramos J. pH response transcription factor PacC controls salt stress tolerance and expression of the P-Type Na+ −ATPase Ena1 in Fusarium oxysporum. Eukaryot. Cell. 2003;2:1246–1252. doi: 10.1128/EC.2.6.1246-1252.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hervas-Aguilar A, Galindo A, Penalva MA. Receptor-independent Ambient pH signaling by ubiquitin attachment to fungal arrestin-like PalF. J. Biol. Chem. 2010;285:18095–18102. doi: 10.1074/jbc.M110.114371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo J, et al. Interactions of fungi with concrete: significant importance for bio-based self-healing concrete. Construction and Building Materials. 2018;164:275–285. doi: 10.1016/j.conbuildmat.2017.12.233. [DOI] [Google Scholar]
- 52.Machida M, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 53.Vanden Wymelenberg A, et al. The Phanerochaete chrysosporium secretome: Database predictions and initial mass spectrometry peptide identifications in cellulose-grown medium. J. Biotechnol. 2005;118:17–34. doi: 10.1016/j.jbiotec.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 54.Ma LJ, et al. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 2009;5:e1000549. doi: 10.1371/journal.pgen.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savitha J, Bhargavi SD, Praveen VK. Complete genome sequence of soil fungus Aspergillus terreus (KM017963), a potent lovastatin producer. Genome Announc. 2016;4:9–10. doi: 10.1128/genomeA.00491-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Powell MD, Amott HJ. Calcium oxalate crystal production in two members of the mucorales. Scanning Electron Microscopy. 1985;1:183. [Google Scholar]
- 57.Hojo M, et al. Unexpected link between polyketide synthase and calcium carbonate biomineralization. Zoological Letters. 2015;1:1–16. doi: 10.1186/s40851-014-0001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pinzari F, Zotti M, De Mico A, Calvini P. Biodegradation of inorganic components in paper documents: formation of calcium oxalate crystals as a consequence of Aspergillus terreus Thom growth. Int. Biodeterior. Biodegradation. 2010;64:499–505. doi: 10.1016/j.ibiod.2010.06.001. [DOI] [Google Scholar]
- 59.Gharieb MM, Gadd GM. Influence of nitrogen source on the solubilization of natural gypsum (CaSO4 2H2O) and the formation of calcium oxalate by different oxalic and citric acid-producing fungi. Mycol. Res. 1999;103:473–481. doi: 10.1017/S0953756298007382. [DOI] [Google Scholar]
- 60.Martinez D, et al. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 61.Richardson SM, et al. Design of a synthetic yeast genome. Science. 2017;355:1040–1044. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- 62.Xie Z-X, et al. “Perfect” designer chromosome V and behavior of a ring derivative. Science. 2017;355:eaaf4704. doi: 10.1126/science.aaf4704. [DOI] [PubMed] [Google Scholar]
- 63.Mitchell LA, et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science. 2017;355:eaaf4831. doi: 10.1126/science.aaf4831. [DOI] [PubMed] [Google Scholar]
- 64.Adamo P, Violante P. Weathering of rocks and neogenesis of minerals associated with lichen activity. Appl. Clay Sci. 2000;16:229–256. doi: 10.1016/S0169-1317(99)00056-3. [DOI] [Google Scholar]
- 65.Tuason MMS, Arocena JM. Calcium oxalate biomineralization by Piloderma fallax in response to various levels of calcium and phosphorus. Appl. Environ. Microbiol. 2009;75:7079–7085. doi: 10.1128/AEM.00325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosling A, Suttle KB, Johansson E, Van Hees PAW, Banfield JF. Phosphorous availability influences the dissolution of apatite by soil fungi. Geobiology. 2007;5:265–280. doi: 10.1111/j.1472-4669.2007.00107.x. [DOI] [Google Scholar]
- 67.Gleeson DB, Clipson N, Melville K, Gadd GM, McDermott FP. Characterization of fungal community structure on a weathered pegmatitic granite. Microb. Ecol. 2005;50:360–368. doi: 10.1007/s00248-005-0198-8. [DOI] [PubMed] [Google Scholar]
- 68.Bonneville S, et al. Plant-driven fungal weathering: early stages of mineral alteration at the nanometer scale. Geology. 2009;37:615–618. doi: 10.1130/G25699A.1. [DOI] [Google Scholar]
- 69.Lee MR, et al. Characterization of mineral surfaces using FIB and TEM: a case study of naturally weathered alkali feldspars. Am. Mineral. 2007;92:1383–1394. doi: 10.2138/am.2007.2453. [DOI] [Google Scholar]
- 70.Penas MM, et al. Further characterization of the signaling proteolysis step in the Aspergillus nidulans pH signal transduction pathway. Eukaryot Cell. 2007;6:960–970. doi: 10.1128/EC.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hervas-Aguilar A, Rodriguez JM, Tilburn J, Arst HN, Jr., Penalva MA. Evidence for the direct involvement of the proteasome in the proteolytic processing of the Aspergillus nidulans zinc finger transcription factor PacC. J. Biol. Chem. 2007;282:34735–47. doi: 10.1074/jbc.M706723200. [DOI] [PubMed] [Google Scholar]
- 72.Tilburn J, et al. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acidand alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mingot J-M, et al. Specificity determinants of proteolytic processing of Aspergillus PacC transcription factor are remote from the processing site and processing occurs in yeast if pH signaling is bypassed. Mol. Cell. Biol. 1999;19:1390–1400. doi: 10.1128/MCB.19.2.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caddick MX, Brownlee AG, Arst HN., Jr. Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol. Gen. Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 75.Arst HN., Jr. Integrator gene in Aspergillus nidulans. Nature. 1976;262:231–234. doi: 10.1038/262231a0. [DOI] [PubMed] [Google Scholar]
- 76.Arst HN, Jr., Penfold HA, Bailey CR. Lactam utilization in Aspergillus nidulans: evidence for a fourth gene under the control of the integrator gene. intA. Molecular Genetics and Genomics. 1978;166:321–327. doi: 10.1007/BF00267625. [DOI] [PubMed] [Google Scholar]
- 77.Arst HN, Jr., Bailey CR, Penfold HA. A possible role for acid phosphatase in γ-amino-n-butyrate uptake in Aspergillus nidulans. Archives of Microbiology. 1980;125:153–158. doi: 10.1007/BF00403213. [DOI] [PubMed] [Google Scholar]
- 78.Diez E, et al. Activation of the Aspergillus PacC zinc-finger transcription factor requires two proteolytic steps. EMBO J. 2002;21:1350–1359. doi: 10.1093/emboj/21.6.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harris JL. Safe, low-distortion tape touch method for fungal slide mounts. Journal of Clinical Microbiology. 2000;38:4683–4684. doi: 10.1128/jcm.38.12.4683-4684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Octopus Imaging software. http://octopusimaging.eu Accessed on September 29, 2017.
- 81.ImageJ software. http://imagej.nih.gov/ij/ Accessed on September 22, 2017.
- 82.Wang JY, et al. Application of modified-alginate encapsulated carbonate producing bacteria in concrete: A promising strategy for crack self-healing. Front. Microbiol. 2015;33:1088. doi: 10.3389/fmicb.2015.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Q, Gadd GM. Fungal nanoscale metal carbonates and production of electrochemical materials. Microb. Biotechnol. 2017;10:1131–1136. doi: 10.1111/1751-7915.12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Q, Csetenyi L, Gadd GM. Biomineralization of metal carbonates by Neurospora crassa. Environmental Science and Technology. 2014;48:14409–14416. doi: 10.1021/es5042546. [DOI] [PubMed] [Google Scholar]
- 85.Li Q, Csetenyi L, Paton GI, Gadd GM. CaCO3 and SrCO3 bioprecipitation by fungi isolated from calcareous soil. Environmental Microbiology. 2015;17:3082–3097. doi: 10.1111/1462-2920.12954. [DOI] [PubMed] [Google Scholar]
- 86.Bignell E, et al. The Aspergillus pH-responsive transcription factor PacC regulates virulence. Molecular Microbiology. 2005;55:1072–1084. doi: 10.1111/j.1365-2958.2004.04472.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.