Abstract
Aeromonas salmonicida (A. salmonicida) is a pathogenic bacterium that causes furunculosis and poses a significant global risk, particularly in economic activities such as Atlantic salmon (Salmo salar) farming. In a previous study, we identified proteins that are significantly upregulated in kidneys of Atlantic salmon challenged with A. salmonicida. Phosphoproteomic analyses were conducted to further clarify the dynamic changes in protein phosphorylation patterns triggered by bacterial infection. To our knowledge, this is the first study to characterize phosphorylation events in proteins from A. salmonicida-infected Atlantic salmon. Overall, we identified over 5635 phosphorylation sites in 3112 proteins, and 1502 up-regulated and 77 down-regulated proteins quantified as a 1.5-fold or greater change relative to control levels. Based on the combined data from proteomic and motif analyses, we hypothesize that five prospective novel kinases (VRK3, GAK, HCK, PKCδ and RSK6) with common functions in inflammatory processes and cellular pathways to regulate apoptosis and the cytoskeleton could serve as potential biomarkers against bacterial propagation in fish. Data from STRING-based functional network analyses indicate that fga is the most central protein. Our collective findings provide new insights into protein phosphorylation patterns, which may serve as effective indicators of A. salmonicida infection in Atlantic salmon.
Introduction
Aeromonas salmonicida subspecies salmonicida (A. salmonicida) is a gram-negative bacterium that triggers a major infectious and systematic disease affecting a wide range of marine and freshwater fish, especially salmonids1. The species can cause death in fish at any growth stage, with symptoms of hemorrhagic septicemia within as little as two or three days2,3. Previous studies have revealed symptoms, such as exophthalmia, skin hemorrhage, ulcers and necrosis, in muscle and different internal organs, mainly spleen and kidney4. As the major hematopoiesis organ in fish, the kidney has been well characterized in terms of immune responsiveness5. Despite its global incidence, the pathogenesis of this infectious disease in fish is currently unknown. Clarification of the mechanisms underlying the immune response to the pathogen infection process and identification of disease indicators should provide a good basis for understanding the bacterial infection resistance ability of Atlantic salmon.
Earlier proteomic analyses by our group using iTRAQ revealed 39 significantly regulated proteins between Atlantic salmon infected with A. salmonicida and healthy fish at different infection stages (7 and 14 days) and varying degrees of infection (low and high)6. However, the specific functions of these and other potential unidentified proteins are yet to be established. A number of studies have demonstrated that phosphorylation regulates pathogenesis. Therefore, to further clarify the mechanisms underlying the response of kidneys of Atlantic salmon to A. salmonicida infection, we selected the phosphoproteome as a tool to quantify and identify phosphoproteins. In recent years, mass spectrometry-based quantitative proteomic approaches have been extensively used to characterize the phosphoproteome7. Significant improvements in enrichment methodologies have been reported8. Protein phosphorylation represents a key post-translational modification in many cellular events that exerts regulatory effects on a range of essential biological processes, including metabolism, secretion, homeostasis, transcriptional and translational regulation, and cellular signaling9. Phosphoproteome analyses provide critical information on intracellular signaling events as well as the common post-translational modification, phosphorylation, without prior knowledge of function or distribution10. In this study, we employed mass spectrometry (MS)-based proteomics combined with phosphopeptide enrichment techniques to identify global protein phosphorylation responses in kidney of Atlantic salmon to infection with A. salmonicida. These collective findings not only facilitate the identification of phosphoproteins and phosphorylation sites of proteins in kidneys of Atlantic salmon activated in response to A. salmonicida infection, but also provide a further functional level to distinguish valuable biomarkers for disease diagnosis.
Materials and Methods
Fish and bacterial strain preparations
A. salmonicida strains (CGMCC No. 7335) used in our study were isolated as naturally occurring pathogens in Atlantic salmon cultured in Shandong Oriental Ocean Sci-Tech Co. (Yantai, Shandong Province, China). Affected fish displayed symptoms of furunculosis and mortality. Bacteria from glycerol stocks were incubated at 20 °C for 48–72 h and cultured in tryptone soy agar supplemented with 1% NaCl (w/v). Brain heart infusion (BHI) agar containing 1.5% NaCl was performed to verify that >95% of the bacteria were viable. Bacteria were adjusted to a final count of ~108 CFU/mL6.
Procedure of fish challenge and sampling
All fish in this study were handled in strict accordance with China’s legislation on scientific procedures on living animals. The protocol was approved by the ethics committee at the University of Chinese Academy of Science (Beijing, China). Breeding Atlantic salmon (body weight 113 ± 20 g) were obtained from Shandong Oriental Ocean Sci-Tech Co. Fish were transferred to cycle-filtered plastic tanks and reared on commercial dry pellets (42% crude protein and 22% crude lipid, Beijing Han Yeanye Science & Technology CO., LTD, Beijing, China) daily and acclimatized temporarily for two weeks (18 °C). Each tank was supplied with fresh water, and the oxygen concentration and temperature monitored daily. Thirty-six fish judged as healthy based on clinical parameters were randomly selected from the experimental group and divided into three groups (two infected and one control). Fish in the two infected groups were challenged with the bacterial suspension at final doses of 107 CFU/mL and 104 CFU/mL, respectively. Fish were challenged for 1 h in separate tanks. Under the same conditions, 12 fish in the control groups were placed in a bath with phosphate-buffered saline solution passed through a 0.22 μm membrane filter. Following the challenge, fish were transferred to their original tanks. No mortality was observed during the experimental period. Kidneys from four fish in infected and PBS incubation groups were collected at 0, 7 and 14 days.
Protein extraction and trypsin digestion
Total protein extraction and purification procedures were performed according to previously described methods6. Fish kidney samples were initially transferred to a 5 mL centrifuge tube and sonicated three times on ice using a high intensity ultrasonic processor (Scientz, Ningbo, China) in lysis buffer (8 M urea, 2 mM EDTA, 10 mM DTT and 1% Protease Inhibitor Cocktail). The remaining debris was removed by centrifugation at 20,000 g at 4 °C for 10 min and the supernatant transferred to new tubes. The protein concentration was determined with a 2-D Quant kit (GE, Boston, America) according to the manufacturer’s instructions.
Protein solution was reduced with 10 mM dithiothreitol (DTT) for 1 h at 37 °C and alkylated with 20 mM iodoacetamide for 45 min at room temperature in the dark. For trypsin digestion, the protein sample was diluted by adding 100 mM TEAB to urea at a concentration less than 2 M. Finally, trypsin was added at a trypsin-to-protein mass ratio of 1:50 for the first digestion overnight and 1:100 for a second 4 h digestion. Approximately 50 μg protein for each sample was digested with trypsin for subsequent experiments. Acquired peptides were dissolved in solvent A (0.1% FA in 2% ACN) and directly loaded onto a reversed-phase analytical column (Thermo, USA).
iTRAQ labeling of proteins for proteomic data analysis
The peptide mixture obtained was labeled using the iTRAQ Reagent-8plex Multiplex Kit (AB SCIEX, USA) according to the manufacturer’s protocol. The experimental groups, each including three biological replicates, were respectively labeled as 113, C-7 d, 114, C-14 d, 115, L-I-7 d, 116, H-I-7 d, 117, L-I-14 d and 118, H-I-14 (Control: C, Low-infected group: L-I, High-infected group: H-I). All labeled samples were separated with a gradient of 2–60% acetonitrile in 10 mM ammonium bicarbonate, pH 10, over 80 min into 12 fractions with an equal volumes collected via HPLC (Thermo DINOEX Ultimate 3000 BioRS, USA). Fractionated samples were analyzed via liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) performed on an AB SCIEX nanoLC-MS/MS (Thermo Q exactive Orbitrap, USA) system. Subsequent procedures were performed according to previous reports6.
The original MS/MS file data were submitted to MaxQuant with the integrated Andromeda search engine (v.1.5.2.8) for data analysis. Raw data files were processed and quantified using Maxquant v1.5.7.4 (21) and searched against the Atlantic salmon database in the NCBI website (97,714 items, updated on December 2015) concatenated with the reverse decoy database. Trypsin/P was specified as the cleavage enzyme allowing up to 2 missing cleavages. Mass error was set to 10 ppm for precursor ions and 0.02 Da for fragment ions. Carbamidomethylation on Cys was specified as fixed modification and oxidation on Met. False discovery rate (FDR) thresholds for protein, peptide and modification sites were specified as 1%. Minimum peptide length was set at 7. We set a 1.5-fold change (which reduced the false positive to a better extent than 2.0-fold change) and p-value < 0.05 as the threshold to distinguish significant changes in data.
Tandem mass tag (TMT) labeling of proteins for phosphoproteomic data analysis
To identify the protein phosphorylation events in kidney of Atlantic salmon, we applied multiplexed TMT and LC/LC-MS/MS approaches to quantify the phosphoproteomes of healthy fish and those infected with A. salmonicida for 7 d with biological replicates. The three biological replicates were labeled as 126, K-C-1, 127, K-C-2, 128, K-C-3, 129, K-I-2 and 130, K-I-3 (Control: C, High-infected group: I). TMT-6 plex was selected for quantification. Briefly, one unit of TMT reagent (defined as the amount of reagent required to label 100 μg protein) was thawed and reconstituted in 24 μL ACN. Peptide mixtures were incubated for 2 h at room temperature, pooled, desalted and dried via vacuum centrifugation.
The sample was fractionated using high pH reverse-phase HPLC on an Agilent 300 Extend C18 column (5 μm particles, 4.6 mm ID, 250 mm length). The method used was the same as that described in section 2.4. After combining into 8 overall fractions, peptides were dried via vacuum centrifugation.
Peptide mixtures were initially incubated with an IMAC microsphere suspension with vibration. IMAC microspheres with enriched phosphopeptides were collected via centrifugation and the supernatant removed11. To eliminate nonspecifically adsorbed peptides, IMAC microspheres were sequentially washed with 50% ACN/6% TFA and 30% ACN/0.1% TFA. To elute enriched phosphopeptides from IMAC microspheres, elution buffer containing 10% NH4OH was added and elution performed with vibration. The supernatant containing phosphopeptides was collected and lyophilized for LC-MS/MS analysis.
The resulting MS/MS data on the phosphoproteome were processed using the same search engine as that for proteomic data analysis, except for the processes of phosphorylation at Ser, Thr, Tyr and acetylation at the protein N-terminus, which were specified as variable modifications. The site localization probability set as >0.75.
Proteome and phosphoproteome analyses of kidneys of atlantic salmon infected with A. salmonicida
Gene ontology (GO) analysis was performed using STRING version 10.5 (https://string-db.org)12. Proteins and phosphoproteins were classified as regulated based on inclusion of at least one regulated phosphosite. The Encyclopedia of Genes and Genomes (KEGG) database was used to identify enriched pathways. For each category, a two-tailed Fisher’s exact test was employed to establish enrichment of differentially expressed against all identified proteins. Correction for multiple hypothesis testing was carried out using standard false discovery rate control methods. GO and pathways with corrected p-values < 0.05 were considered significant. All the pathways identified were classified into hierarchical categories according to the KEGG website.
Network analysis
Proteins were filtered if they were not sampled in at least two of the replicates. All common protein and phosphoprotein candidates were searched against STRING database version 10.5 for protein-protein interactions. We followed these initial filtering steps with STRING analysis13. Only interactions between proteins belonging to the searched dataset were selected, thereby excluding external candidates. STRING defines a metric designated “confidence score” to classify interaction confidence. We fetched all interactions with confidence scores ≥0.7, representing high confidence. The interaction network from STRING was visualized in Cytoscape, a graph-theoretical clustering algorithm.
Identification of over-represented kinase substrate motifs
The software tool Motif-x was used to analyze the models of sequences constituted with amino acids in specific positions of modify-21-mers (10 residues upstream and downstream of the site) in all protein sequences14. All database protein sequences were used as the background parameter, with other parameters set as default. To further determine whether the identified phosphoproteins activate additional kinases, we investigated the influence of amino acids in the immediate vicinity of each phosphorylation site on phosphorylation of kinase substrates using the Group-based Prediction System (GPS)15.
Results
Selection of fish infected with A. salmonicida at different doses and times
We sampled fish at two stages, specifically, 7 d and 14 d after infection. The two infection times were selected to determine the potential proteins involved in the early and late stages of infection. Based on LD50 (104 CFU/mL), we selected 107 and 104 CFU/mL as high and low infection doses for challenge. Kidney is the major hematopoiesis organ in fish that performs key roles in immune responsiveness and clearing bacteria from blood16,17. Accordingly, kidneys of normal and infected fish (both low and high doses) were isolated at 7 d and 14 d for iTRAQ analysis. We only determined the phosphorylation state of proteins in kidney samples challenged with the high bacterial dose (107 CFU/mL) at 7 d, with a view to improving analysis and identifying the molecules involved in the early stages of infection.
Multiplexed quantitative analysis of the whole proteome and phosphoproteome
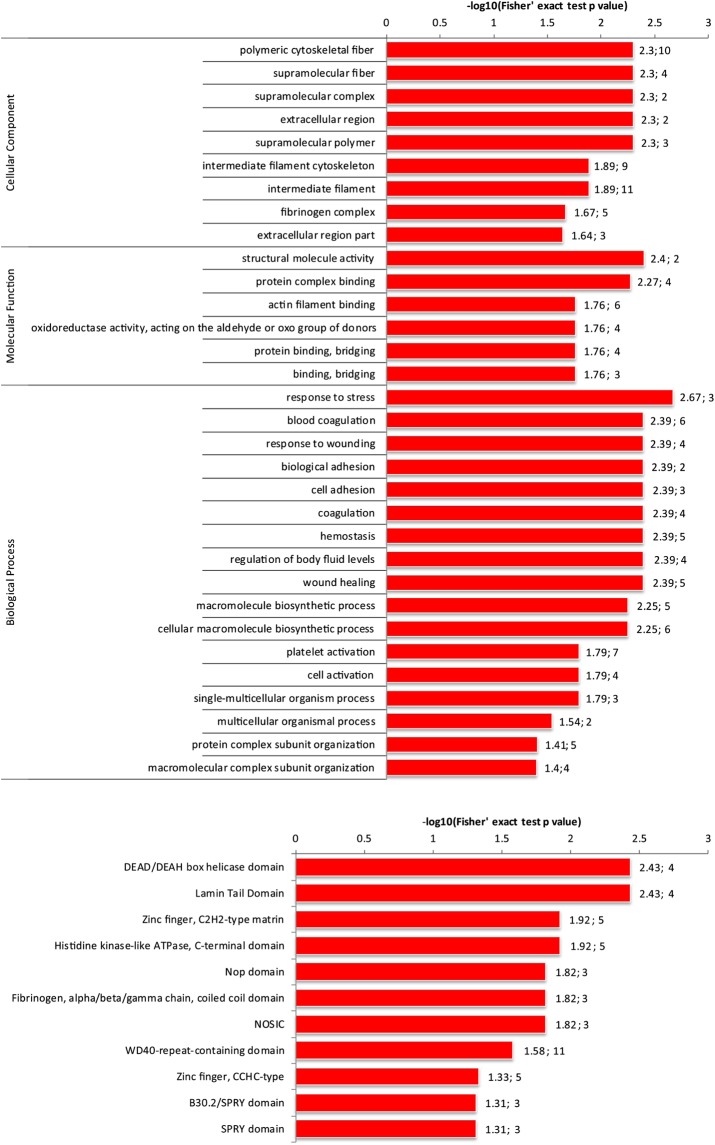
In a previous study, we established a fish model infected with A. salmonicida6. MS-based iTRAQ quantitative proteomics in combination with phosphopeptide enrichment was employed to identify phosphopeptides and phosphoproteins and quantify their expression levels. In total, 5635 phosphorylation sites in 3112 proteins were detected in kidneys of fish, and 1502 and 77 proteins were quantitatively increased and decreased by ≥1.5-fold, respectively (Supplemental Table 1). For proteins possessing multiple phosphorylation sites, GO was used to analyze total phosphorylation in the high-infection group, compared to controls at 7 d. As a result, differentially phosphorylated proteins were sub-categorized into 26 hierarchically structured GO classifications, including 9 biological processes, 5 cellular components and 6 molecular functions (Fig. 1A–C). In particular, “metabolic process” (17%) and “cellular process” (25%) were highly represented in “biological process”, “cell” (37%) and “organelle” (27%) in “cellular component”, and “catalytic activity” (20%) and “binding” (67%) in “molecular function” branches of ontology.
Figure 1.
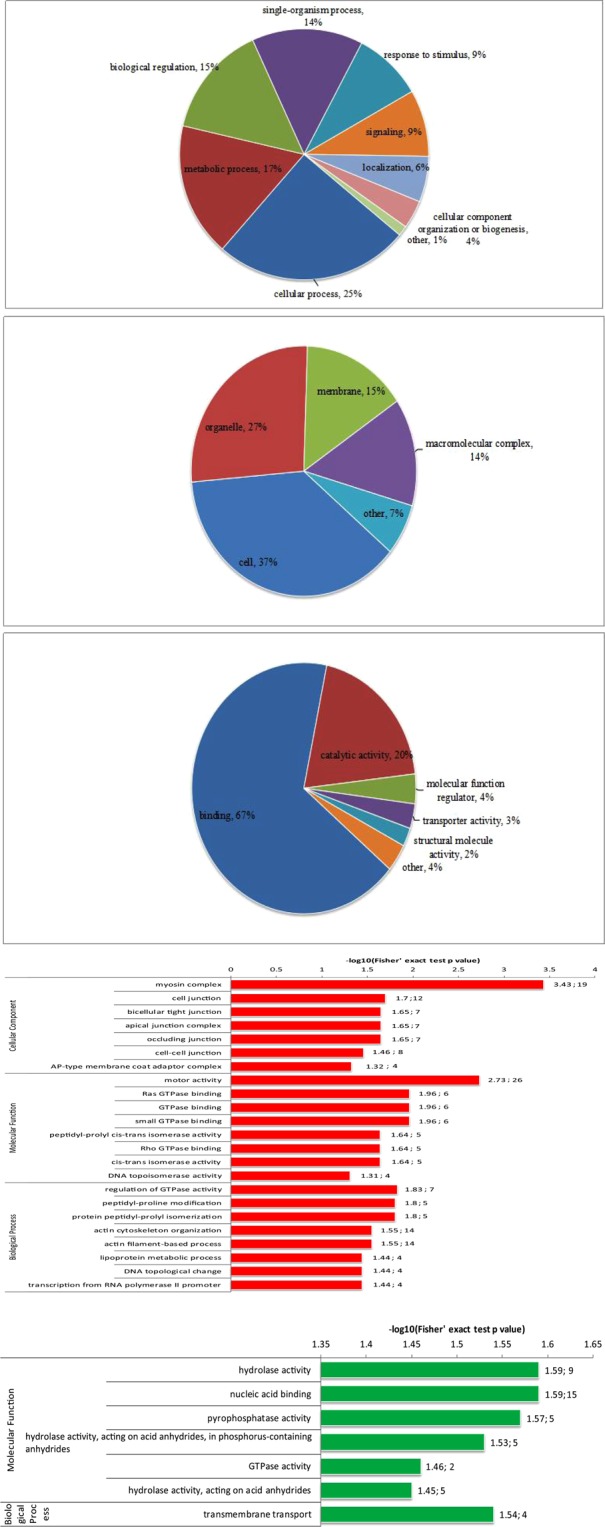
Gene Ontology classification of proteins between high-infected and control groups at 7 d, including (A) biological processes, (B) cellular components and (C) molecular functions. (D) GO-based enrichment analysis of up-regulated proteins between high-infected and control groups at 7 d. (E) GO-based enrichment analysis of down-regulated proteins between high-infected and control groups at 7 d.
All phosphoproteins involved in biological processes, cellular components and molecular functions were differentially phosphorylated as log10 transformations relative to their average Con expression levels during infection in fish kidney (Fig. 1D,E). Most of the upregulated proteins were associated with myosin complex, motor activity, Ras GTPase binding, GTPase binding, small GTPase binding and regulation of GTPase activity while downregulated proteins were predominantly linked with hydrolase, nucleic acid binding and pyrophosphatase activity.
Samples at 7 and 14 d after A. salmonicida treatment exhibited alterations in expression in 189 (L-I-7), 244 (H-I-7), 161 (L-I-14) and 135 (H-I-14) proteins by at least 1.5-fold with statistical significance (p < 0.05), compared with the Control group. Interestingly, 21, 39, 30 and 25 proteins were exclusively identified as phosphoproteins in the four treatment groups respectively (Fig. 2). Enrichment analysis further revealed that these proteins are mainly involved in the apoptosis pathway and display characteristic structural features, such as DEAD/DEAH box helicase domain, Lamin Tail Domain, Zinc finger, C2H2-type matrin, Histidine kinase-like ATPase, C-terminal domain, Nop domain and Fibrinogen, alpha/beta/gamma chain, and coiled coil domain (Fig. 3A,B).
Figure 2.
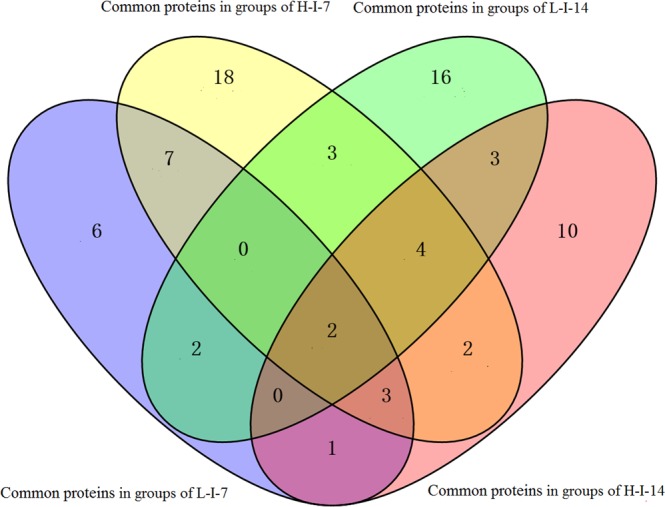
Venn diagram of the numbers of proteins from both whole proteome and phosphoproteome in groups of L-I-7, H-I-7, L-I-14 and H-I-14 significantly expressed proteins at least 1.5 fold with statistical significance (q < 0.05) compared with those in controls respectively.
Figure 3.
Enrichment of common proteins in both the whole proteome and phosphoproteome. (A) Gene Ontology of common proteins; (B) Domain enrichment of common proteins.
To further determine the differentially phosphorylated and coordinative proteins, we selected significantly altered phosphoproteins with different trends in expression at 7 d (listed in Table 1). Overall, 8 and 11 proteins were identified in the high-dose and low-dose infection groups at 7 d, respectively. Five of the proteins (keratin, type II cytoskeletal cochleal-like, prothrombin-like, membrane-associated progesterone receptor component 1, fibrinogen alpha chain-like, and apolipoprotein A-I precursor) were common between the two groups. Notably, only five (epoxide hydrolase 1, LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like, matrix metalloproteinase-9 precursor, stanniocalcin and lymphocyte-specific protein 1-like) and three (keratin, type I cytoskeletal 18-like, phosphoenolpyruvate carboxykinase, cytosolic [GTP] and hematopoietic cell-specific Lyn substrate 1 isoform X1) proteins were specifically induced in the H-I-7 and L-I-7 treatment groups, respectively, which could potentially serve as indicators of resistance against A. salmonicida infection.
Table 1.
List of proteins identified and quantified in kidneys of Atlantic salmon infected with A. salmonicida from phosphoproteome.
| NCBI Accession | Protein description | Protein level | Phosphorylation level | Site | Fold change |
|---|---|---|---|---|---|
| Identified at 7 day of low-concentration infected group | |||||
| NP_001133921.1 | phosphoenolpyruvate carboxykinase, cytosolic [GTP] [Salmo salar] | down | up | Ser119 | 1.965 |
| NP_001134612.1 | Apolipoprotein A-I precursor [Salmo salar] | down | up | Ser50 | 2.567 |
| NP_001139831.1 | Membrane-associated progesterone receptor component 1 [Salmo salar] | down | up | Thr63 | 2.062 |
| XP_013981919.1 | PREDICTED: prothrombin-like [Salmo salar] | down | up | Ser319 | 1.816 |
| XP_014002674.1 | PREDICTED: keratin, type II cytoskeletal cochleal-like [Salmo salar] | down | up | Ser273 | 1.579 |
| XP_014002674.1 | PREDICTED: keratin, type II cytoskeletal cochleal-like [Salmo salar] | down | up | Ser31 | 1.882 |
| XP_014012273.1 | PREDICTED: fibrinogen alpha chain-like [Salmo salar] | down | up | Ser443 | 2.294 |
| XP_014012273.1 | PREDICTED: fibrinogen alpha chain-like [Salmo salar] | down | up | Ser457 | 2.367 |
| XP_014012273.1 | PREDICTED: fibrinogen alpha chain-like [Salmo salar] | down | up | Ser433 | 4.259 |
| XP_014023450.1 | PREDICTED: keratin, type I cytoskeletal 18-like [Salmo salar] | down | up | Ser152 | 1.547 |
| XP_014023450.1 | PREDICTED: keratin, type I cytoskeletal 18-like [Salmo salar] | down | up | Ser25 | 1.875 |
| XP_014063638.1 | PREDICTED: hematopoietic cell-specific Lyn substrate 1 isoform X1 [Salmo salar] | down | up | Ser148 | 1.981 |
| XP_014063638.1 | PREDICTED: hematopoietic cell-specific Lyn substrate 1 isoform X1 [Salmo salar] | down | up | Ser11 | 3.147 |
| NP_001133929.1 | Matrix metalloproteinase-9 precursor [Salmo salar] | down | up | Ser27 | 2.015 |
| NP_001134612.1 | Apolipoprotein A-I precursor [Salmo salar] | down | up | Ser50 | 2.567 |
| NP_001134917.1 | Epoxide hydrolase 1 [Salmo salar] | up | down | Ser117 | 0.642 |
| NP_001139831.1 | Membrane-associated progesterone receptor component 1 [Salmo salar] | down | up | Thr63 | 2.062 |
| XP_013981919.1 | PREDICTED: prothrombin-like [Salmo salar] | down | up | Ser319 | 1.816 |
| XP_013983369.1 | PREDICTED: lymphocyte-specific protein 1-like [Salmo salar] | down | up | Ser15 | 5.749 |
| XP_013993347.1 | PREDICTED: stanniocalcin [Salmo salar] | down | up | Ser247 | 2.08 |
| XP_013993347.1 | PREDICTED: stanniocalcin [Salmo salar] | down | up | Ser226 | 2.584 |
| XP_014002674.1 | PREDICTED: keratin, type II cytoskeletal cochleal-like [Salmo salar] | down | up | Ser273 | 1.579 |
| XP_014002674.1 | PREDICTED: keratin, type II cytoskeletal cochleal-like [Salmo salar] | down | up | Ser31 | 1.882 |
| XP_014002674.1 | PREDICTED: keratin, type II cytoskeletal cochleal-like [Salmo salar] | down | up | Ser959 | 2.429 |
| XP_014012273.1 | PREDICTED: fibrinogen alpha chain-like [Salmo salar] | down | up | Ser443 | 2.294 |
| XP_014012273.1 | PREDICTED: fibrinogen alpha chain-like [Salmo salar] | down | up | Ser457 | 2.367 |
| XP_014012273.1 | PREDICTED: fibrinogen alpha chain-like [Salmo salar] | down | up | Ser433 | 4.259 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser123 | 1.628 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser2649 | 1.659 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser2621 | 1.757 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser2651 | 1.816 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser130 | 1.842 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser1228 | 1.876 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser2678 | 2.774 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser1437 | 2.79 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser704 | 3.184 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser514 | 3.47 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser2733 | 3.626 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser598 | 4.952 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser2739 | 5.71 |
| XP_014054044.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser1554 | 9.949 |
| XP_014068765.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser4497 | 1.677 |
| XP_014068765.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser356 | 1.957 |
| XP_014068765.1 | PREDICTED: LOW QUALITY PROTEIN: neuroblast differentiation-associated protein AHNAK-like [Salmo salar] | down | up | Ser4533 | 1.984 |
Numbers on the bar chart represent−log10(p) and GO-terms level.
Motif analysis and regulation of protein kinases and phosphatases
Application of motif-x to determine the amino acid sequence patterns resulted in the identification of 22 highly enriched motifs including Ser and Tyr among all the phosphoproteins (Fig. 4A,B). Proteins were assigned as typical kinases based on an online search of the kinase database (http://www.kinase.com/human/kinome/), mainly including members of STE (17), CAMK (20), AGC (19), CK1(4), CMGC (13), TK (22), TKL (15), RGC (1) and other families (Supplemental Table 2). Additionally, the motif-x tool revealed significant overrepresentation of eight motifs (SxDx, xGSx, RxSx, KxxS, RxxS, RxxSL, RKxS and LXRxxS) among the upregulated A. salmonicida-responsive phosphorylation sites. These clustered proteins are presented in Supplemental Fig. 1. Combined with the protein data presented in Table 1, five proteins, specifically, inactive serine/threonine-protein kinase VRK3-like, cyclin-G-associated kinase isoform X2, tyrosine-protein kinase HCK isoform X1, protein kinase C delta type-like and ribosomal protein S6 kinase alpha-3-like, were identified as potential biomarkers matched three motifs including SxDx, RxSx and KxxS respectively were shown in Table 2. SxDx corresponds to the consensus motif of inactive serine/threonine-protein kinase, VRK3-like, with Ser118 and Ser151 phosphorylation sites, and cyclin-G-associated kinase isoform, X2, with a Ser1049 phosphorylation site and RxSx to the consensus motif for tyrosine-protein kinase HCK isoform X1 with a Ser236 phosphorylation site. The activities of protein kinase C delta type-like with Ser658, Ser297 and Ser344 phosphorylation sites and ribosomal protein S6 kinase alpha-3-like with Ser363 and Ser221 phosphorylation sites are regulated by the KxxS motif. The phosphorylation status of these kinase proteins may serve as an effective biomarker of A. salmonicida infection in Atlantic salmon.
Figure 4.

Search for putative kinase substrate motifs within up- (A) and downregulated (B) A. salmonicida responsive phosphorylation sites. Representative phosphorylation motifs identified from proteins at Ser (F) and Tyr (E) and centered on phosphorylated serine are shown for each top-scoring motif.
Table 2.
Kinases identified in A.
| NCBI Accession | Protein description | Kinase Name | Kinase Group | Kinase Family | Site | Motif |
|---|---|---|---|---|---|---|
| XP_014031257.1 | PREDICTED: inactive serine/threonine-protein kinase VRK3-like [Salmo salar] | VRK3 | CK1 | VRK | Ser118 | ……S.D…. |
| XP_014031257.1 | PREDICTED: inactive serine/threonine-protein kinase VRK3-like [Salmo salar] | VRK3 | CK1 | VRK | Ser151 | ……S.D…. |
| XP_014028116.1 | PREDICTED: cyclin-G-associated kinase isoform X2 [Salmo salar] | GAK | Other | NAK | Ser1049 | ……S.D…. |
| XP_013989897.1 | PREDICTED: tyrosine-protein kinase HCK isoform X1 [Salmo salar] | HCK | TK | Src | Ser236 | ….R.S…… |
| XP_014022605.1 | PREDICTED: protein kinase C delta type-like [Salmo salar] | PKCd | AGC | PKC | Ser658 | …K..S…… |
| XP_014022605.1 | PREDICTED: protein kinase C delta type-like [Salmo salar] | PKCd | AGC | PKC | Ser297 | …K..S…… |
| XP_014022605.1 | PREDICTED: protein kinase C delta type-like [Salmo salar] | PKCd | AGC | PKC | Ser344 | …K..S…… |
| XP_013984775.1 | PREDICTED: ribosomal protein S6 kinase alpha-3-like [Salmo salar] | RSK2 | AGC | RSK | Ser363 | …K..S…… |
| XP_013984775.1 | PREDICTED: ribosomal protein S6 kinase alpha-3-like [Salmo salar] | RSK2 | AGC | RSK | Ser221 | …K..S…… |
Salmonicida infection in kidney of Atlantic Salmon by phosphoproteomics.
Network of phosphoproteins
Using the STRING database, interactions among 290 phosphoproteins from both the proteome and phosphoproteome were identified, including 111 increased and 2 decreased at the phosphorylation level, with filter criteria of log2 ratios > 2 representing increased proteins and log2 ratios < 2 representing decreased proteins (Supplemental Fig. 2). We had filtered different significantly up or down regulated phosphoproteins with specific motifs in previous analyses. Details of these proteins are summarized in Supplemental Table 2 (Supporting Information). Combined with previous findings, significantly up or down regulated phosphoproteins from both low-infected and high-infected groups, compared to controls, at 7 d were selected (Fig. 5A,B), including fibrinogen alpha chain (fga), prothrombin-like (f2), apolipoprotein A-I precursor (apoa1a), ribosomal protein S6 kinase alpha-3-like (rps6ka3a), hematopoietic cell-specific Lyn substrate 1 isoform X1 (hcls1), membrane-associated progesterone receptor component 1 (pgrmc1) and trifunctional enzyme subunit beta, mitochondrial (HADHB). Among the proteins identified, only fga clustered within the network acted as a central rod.
Figure 5.
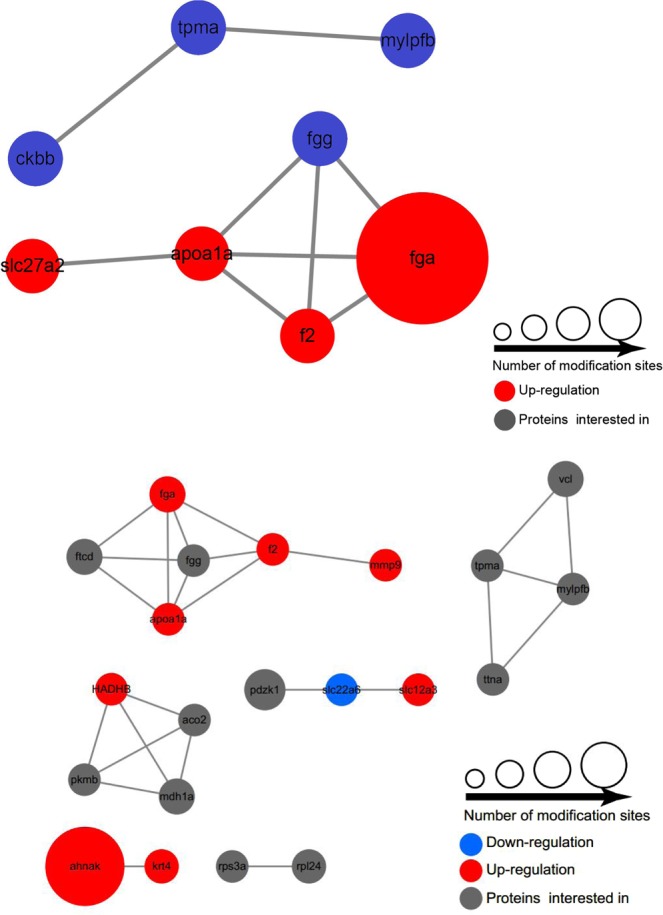
Interaction networks of A. salmonicida-responsive phosphoproteins. Functional and physical interactions among genes with A. salmonicida-responsive phosphosites were extracted from the STRING database (version 10.5). Upregulated proteins are represented by red nodes, downregulated proteins by blue nodes, and overlap between up- and downregulated sites by gray nodes. All genes are specified in Supplemental Table 1 (Supporting Information). (A) Proteins further selected in low-infected groups at 7 d. (B) Proteins further selected in high-infected groups at 7 d.
Discussion
In the current investigation, quantitative proteomics was applied to analyze the phosphoproteome in Atlantic salmon infected with A. salmonicida. To our knowledge, this is the first study to characterize the phosphorylation events in infected Atlantic salmon. Based on proteomic data obtained from our previous experiments6 in combination with phosphoproteomic data, we identified four proteins (epoxide hydrolase 1, neuroblast differentiation-associated protein AHNAK-like, matrix metalloproteinase-9 precursor, stanniocalcin and lymphocyte-specific protein 1-like) that were significantly altered following infection.
The soluble epoxide hydrolase (sEH), a ubiquitous enzyme in vertebrates that transforms epoxides to their corresponding diols implicated in the inflammatory cascade, plays broad biological roles by virtue of its activity against lipid epoxide substrates and localization in not only liver but also kidneys and vascular tissue18–21. sEH is a bifunctional enzyme with C-terminal hydrolase activity and N-terminal phosphatase activity22. Interestingly, Skugor et al. (2009) observed elevated expression of biotransformation genes, such as ephx1, in Atlantic salmon with high resistance against A. salmonicida infection, highlighting a possible role in tissue protection and toxic metabolite clearance23.
Matrix metalloproteinase-9 (MMP-9), one of the members of the MMP family, functions in normal and neoplastic invasive processes, cleaves numerous structural components and participates in inflammatory pathways24,25. MMP-9 cleaves a diverse range of substrates, such as extracellular matrix (ECM) proteins, ligands and receptors, and is mainly produced by inflammatory cells, including macrophages, leukocytes and monocytes. The protein plays important roles in embryogenesis, innate immune defense and apoptosis26,27. In earlier studies, expression of MMP-9 could be induced by bacterial pathogens in fish, suggesting its involvement in fish immune responses28–30. It is believed that these two proteins contribute to defense against A. salmonicida infection in Atlantic salmon, but further research is required to validate this hypothesis. Few studies have characterized the roles of neuroblast differentiation-associated protein, AHNAK-like and stanniocalcin and lymphocyte-specific protein 1-like during bacterial or viral infection in fish species such as Atlantic salmon. Using earlier results combined with integrated whole proteome and phosphoproteome data, we focused on the potential functions of these four proteins in subsequent experiments.
Motif-X analysis led to the identification of prospective novel kinases, including inactive serine/threonine-protein kinase VRK3-like, cyclin-G-associated kinase isoform X2, tyrosine-protein kinase HCK isoform X1, protein kinase C delta type-like and ribosomal protein S6 kinase alpha-3-like, which are well-known key modulators and initiators of the global phosphorylation response to A. salmonicida infection in Atlantic salmon.
Two proteins containing the common SxDx motif were identified. Vaccinia-related kinase 3 (VRK3), a member of the novel VRK family is located in the nucleus31,32. VRK3 displays alterations in the critical motifs essential for kinase activity, and includes two active vertebrate paralogs (VRK1 and VRK2), one or two orthologs in all metazoans, and an ortholog in most poxviruses, including the founding member, vaccinia virus B1R31,33–37. The VRK3 structure may be maintained to provide a binding surface for other proteins, a known function of many pseudokinases38. VRK3 also shows putative kinase activity in a cellular context39. The protein is highly expressed during development and promotes cell cycle progression by phosphorylating the nuclear envelope protein barrier to autointegration factor (BAF)40. Moreover, VRK3 suppresses extracellular signal regulated kinase (ERK) activity through direct binding to the MAP kinase phosphatase (MKP), vaccinia H1-related (VHR)41. In our experiments, phosphorylation of VRK3 at Ser118 and Ser151 was increased significantly in A. salmonicida-infected fish at 7 d. However, the underlying mechanisms and specific functions of these phosphorylation events are yet to be defined.
Cyclin G-associated kinase (GAK) is a protein containing highly conserved serine/threonine kinases with multiple functional domains42. GAK was initially reported to be associated with cyclin G and shown to play an important role in uncoating clathrin-coated vesicles (CCVs) in non-neuronal cells43. Another study demonstrated a role of GAK in clathrin-mediated endocytosis/vesicle trafficking in the cytoplasm and nucleus44. In addition, GAK has been implicated as an androgen receptor-interacting transcriptional coactivator45. GAK interacts specifically with interleukin 12 receptor β2 (IL-12Rβ2)46 that plays a central role in the initiation and control of cell-mediated immune responses to suppress IL-12-induced production of IFN-γ. Moreover, GAK is one of the two active kinases present in clathrin-coated vesicles and its Ser/Thr kinase activity is directed towards the l2 components of CCVs47. Based on integrated data from the current study and earlier experiments, we propose that GAK containing a phosphorylation site at Ser1049 has the potential to modulate immune responses in Atlantic salmon. The mechanisms linking GAK activity with immune pathways for development of a defense system against A. salmonicida in Atlantic salmon remain to be resolved.
Another protein, hematopoietic cell kinase (hck) containing the RxSx motif, is a member of the src family of tyrosine kinases and primarily expressed in myeloid cells. Expression of the gene is principally limited to cells of monocyte/macrophage and granulocyte lineages48–51. Levels of the gene increase with differentiation along either the macrophage or granulocyte pathways52–54 and are further augmented by activation stimuli, including LPS, in mature monocytes and macrophages51,53,55. A number of studies support a role of hck in the terminal differentiation of macrophages and/or signal transduction pathways of mature monocytes, macrophages lymphocytes, mast cells, and basophils56–59. We observed increased phosphorylation of this protein only at the Ser236 site with bacterial treatment.
The two other selected proteins had a common KxxS motif. The first was protein kinase C delta (PKCδ), which is involved in cytoskeletal regulation and apoptosis as a member of the PKC family60,61. Proteolytic activation of PKCδ is considered responsible for apoptotic execution62,63. Previous studies have focused on the mechanisms underlying PKCδ involvement in regulating cellular apoptosis and the cytoskeleton60,63,64. Notably, we observed significantly increased phosphorylation of this protein at the Ser297, Ser344 and Ser658 sites.
The second protein, ribosomal S6 protein kinase (RSK), phosphorylates serum response factors in vitro at an in vivo serum-stimulated phosphorylation site65. The protein is also regulated by extracellular signal regulated kinase (ERK) to inhibit the proapoptic Bcl2-family member, BCL2, an antagonist of cell death and caspase66. In this study, phosphorylation of RSK at Ser221 and Ser363 was increased significantly in infected fish at 7 d.
In conclusion, the five prospective novel kinases identified in our analyses (VRK3, GAK, HCK, PKCδ and RSK6) potentially contribute to resistance against bacterial propagation in fish. These proteins have common functions in regulation of apoptosis and the cytoskeleton in inflammatory and cellular pathways. However, further research is required to clarify whether these proteins participate in the defense mechanism against microbial pathogens and the underlying mechanisms.
Our findings, in conjunction with previous data, show that fga is the most central protein in the pathogenic network and may serve a potential biomarker of A. salmonicida infection in Atlantic salmon. In keeping with this proposal, earlier proteomic profiling studies support the utility of fga as a diagnostic marker of lung squamous cell carcinoma67. We therefore hypothesize the protein with well-established key functions than other proteins obtained from STRING-based functional network analysis in fish against bacteria.
In summary, we have presented an integrated analysis of the whole proteome and phosphoproteome showing that specific proteins are overexpressed and collectively drive the immune response against high-dose infection of A. salmonicida in kidneys of Atlantic salmon at 7 d. These data provide a framework for further investigation of Atlantic salmon resistance to A. salmonicida, and selection of indicators to improve resistance.
Supplementary information
Acknowledgements
This research was supported by the National Key R&D Program of China (Grant No. 2017YFD0701700), National Natural Science Foundation of China (31671376), Chinese State Key Projects for Basic Research (“973 Program”, 2015CB910700 and 2014CBA02001), Innovation project (16CXZ027), the Beijing Nova Program (Z161100004916148), and the State Key Laboratory of Proteomics (SKLP-O201704, SKLP-O201507). Thanks for the help from Dr. Xiaoni Li.
Author Contributions
Experiment and article was finished by Dr. Peng-fei Liu and instructed by Professor Ying Liu. We are indebted to Dr. Yishuai Du, M.S. Lingjie Meng and Dr. Xian Li for their kind comments about our experiments, and also thank the support from associate professor Dong Yang in Beijing Proteome Research Center for their excellent assistance with the data analysis of the phosphoproteomic. All authors had reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dong Yang, Email: yangdongbprc@163.com.
Ying Liu, Email: yingliu@dlou.edu.cn.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-38890-3.
References
- 1.Dallaire-Dufresne S, Tanaka KH, Trudel MV, Lafaille A, Charette SJ. Viru-lence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet. Microbiol. 2014;169:1–7. doi: 10.1016/j.vetmic.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 2.Ogut H, Reno PW. Evaluation of an experimental Aeromonas salmonicida epidemic in chinook salmon, Oncorhynchus tshawytscha (Walbaum) J. Fish. Dis. 2005;28:263–269. doi: 10.1111/j.1365-2761.2005.00626.x. [DOI] [PubMed] [Google Scholar]
- 3.Orozova P, Barker M, Austin DA, Austin B. Identification and pathogenicity to rainbow trout, Oncorhynchus mykiss (Walbaum), of some aeromonads. J. Fish. Dis. 2009;32:865–871. doi: 10.1111/j.1365-2761.2009.01065.x. [DOI] [PubMed] [Google Scholar]
- 4.Burr SE, Pugovkin D, Wahli T, Segner H, Frey J. Attenuated virulence of an Aeromonas salmonicida subsp. salmonicida type III secretion mutant in a rainbow trout model. Microbiology. 2005;151:2111–2118. doi: 10.1099/mic.0.27926-0. [DOI] [PubMed] [Google Scholar]
- 5.Dos Santos NM, et al. The gill is a major organ for antibody secreting cell production following direct immersion of sea bass (Dicentrarchus labrax, L.) in a photobacterium damselae ssp piscicida bacterin: an ontogenetic study. Fish Shellfish Immunol. 2001;11:65–74. doi: 10.1006/fsim.2000.0295. [DOI] [PubMed] [Google Scholar]
- 6.Liu P-fei, et al. Proteomic analysis in kidneys of Atlantic salmon infected with Aeromonas salmonicida by iTRAQ. Dev. Comp.Immunol. 2017;58:292–301. doi: 10.1016/j.dci.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Huttlin EL, et al. A tissue specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendriks IA, Vertegaal ACO. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016;17:581–595. doi: 10.1038/nrm.2016.81. [DOI] [PubMed] [Google Scholar]
- 9.Delom F, Chevet E. Phosphoprotein analysis: from proteins to proteomes. Proteome Sci. 2006;4:15. doi: 10.1186/1477-5956-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engholm-Keller K, Larsen MR. Technologies and challenges in large-scale phosphoproteomics. Proteomics. 2013;13:910–931. doi: 10.1002/pmic.201200484. [DOI] [PubMed] [Google Scholar]
- 11.Zhou H, et al. Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography.[J] Nature Protocols. 2013;8(3):461–480. doi: 10.1038/nprot.2013.010. [DOI] [PubMed] [Google Scholar]
- 12.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szklarczyk D, et al. STRINGv10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 15.Xue Y, et al. GPS 2.1: enhanced prediction of kinase-spec ific phosphorylation sites with an algorithm of motif length selection. Protein Eng. Des. Sel. 2011;24:255–260. doi: 10.1093/protein/gzq094. [DOI] [PubMed] [Google Scholar]
- 16.Dos Santos NM, et al. The gill is a major organ for antibody secreting cell production following direct immersion of sea bass (Dicentrarchus labrax, L.) in a photobacterium damselae ssp piscicida bacterin: an ontogenetic study. Fish Shellfish Immunol. 2001;11:65–74. doi: 10.1006/fsim.2000.0295. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson HW, Claxton MJ, Moccia RD, Wilkie EJ. The quantitative clearance of bacteria of from the bloodstream of rainbow trout (salmo gaindneri) Veterinary Pathology. 1982;19:687–699. doi: 10.1177/030098588201900614. [DOI] [PubMed] [Google Scholar]
- 18.Gill, S. S., Hammock, B. D., Yamamoto, I. & Casida, J. E. In Insect Juvenile Hormones: Chemistry and Action, eds Menn, J. J. & Beroza, M. (Academic, New York), 177–189 (1972).
- 19.Imig JD, Zhao X, Capdevila JH, Morisseau C, Hammock BD. Soluble epoxide hydrolase inhibition lowers arterial blood pressure in angiotensin II hypertension. Hypertension. 2002;39:690–694. doi: 10.1161/hy0202.103788. [DOI] [PubMed] [Google Scholar]
- 20.Fang X, et al. Pathways of epoxyeicosatrienoic acid metabolism in endothelial cells. J. Biol. Chem. 2001;276:14867–14874. doi: 10.1074/jbc.M011761200. [DOI] [PubMed] [Google Scholar]
- 21.Moghaddam MF, et al. Bioactivation of leukotoxins to their toxic diols by epoxide hydrolase. Nat. Med. 1997;3:562–566. doi: 10.1038/nm0597-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman JW, Morisseau C, Harris TR, Hammock BD. The soluble epoxide hydrolase encoded by EPXH2 is a bifunctional enzyme with novel lipid phosphate phosphatase activity. Proc Natl Acad Sci USA. 2003;100:1558–1563. doi: 10.1073/pnas.0437724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.kugor S, Jorgensen SM, Gjerde B, Krasnov A. Hepatic gene expression profiling reveals protective responses in Atlantic salmon vaccinated against furunculosis. BMC Genomics. 2009;10:503. doi: 10.1186/1471-2164-10-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandooren J, Van den Steen PE, Opdenakker G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): the next decade. Crit. Rev. Biochem. Mol. Biol. 2013;48(3):222–272. doi: 10.3109/10409238.2013.770819. [DOI] [PubMed] [Google Scholar]
- 26.Yabluchanskiy A, Ma Y, Iyer RP, Hall ME, Lindsey ML. Matrix metalloproteinase-9: many shades of function in cardiovascular disease. Physiol. (Bethesda) 2013;28(6):391–403. doi: 10.1152/physiol.00029.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu XP, Chen ZH, Wang Y, Yamada Y, Steffensen B. Functional basis for the overlap in ligand interactions and substrate specificities of matrix metalloproteinases-9 and-2. Biochem. J. 2005;392(1):127–134. doi: 10.1042/BJ20050650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu XY, et al. Characterization of MMP-9 gene from grass carp (Ctenopharyngodon idella): an Aeromonas hydrophila-inducible factor in grass carp immune system. Fish Shell fish Immunol. 2013;35(3):801–807. doi: 10.1016/j.fsi.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 29.Matsuyama T, et al. Microarray analyses of gene expression in Japanese flounder Paralichthys olivaceus leucocytes during monogenean parasite Neo-heterobothrium hirame infection. Dis. Aquat. Organ. 2007;75(1):79–83. doi: 10.3354/dao075079. [DOI] [PubMed] [Google Scholar]
- 30.Chadzinska M, Baginski P, Kolaczkowska E, Savelkoul HF, Kemenade BM. Expression profiles of matrix metalloproteinase 9 in teleost fish provide evidence for its active role in initiation and resolution of inflammation. Immunology. 2008;125(4):601–610. doi: 10.1111/j.1365-2567.2008.02874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nichols RJ, Traktman P. Characterization of three paralogous members of the mammalian vaccinia related kinase family. J. Biol. Chem. 2004;279:7934–7946. doi: 10.1074/jbc.M310813200. [DOI] [PubMed] [Google Scholar]
- 32.Vega FM, Gonzalo P, Gaspar ML, Lazo PA. Expression of the VRK (vaccinia-related kinase) gene family of p53 regulators in murine hematopoietic development. FEBS Lett. 2003;544:176–180. doi: 10.1016/s0014-5793(03)00501-5. [DOI] [PubMed] [Google Scholar]
- 33.Boyle KA, Traktman P. Members of a novel family of mammalian protein kinases complement the DNA-negative phenotype of a vaccinia virus ts mutant defective in the B1 kinase. J. Virol. 2004;78:1992–2005. doi: 10.1128/JVI.78.4.1992-2005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 35.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 36.Nezu J, Oku A, Jones MH, Shimane M. Identification of two novel human putative serine/threonine kinases, VRK1 and VRK2, with struc-tural similarity to vaccinia virus B1R kinase. Genomics. 1997;45:327–331. doi: 10.1006/geno.1997.4938. [DOI] [PubMed] [Google Scholar]
- 37.Vega FM, Gonzalo P, Gaspar ML, Lazo PA. Expression of the VRK (vaccinia-related kinase) gene family of p53 regulators in murine hemato-poietic development. FEBS Lett. 2003;544:176–180. doi: 10.1016/s0014-5793(03)00501-5. [DOI] [PubMed] [Google Scholar]
- 38.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Kang TH, Kim KT. Negative regulation of ERK activity by VRK3-mediated activation of VHR phosphatase. Nat. Cell Biol. 2006;8:863–869. doi: 10.1038/ncb1447. [DOI] [PubMed] [Google Scholar]
- 40.Park CH, et al. Presumed pseudokinase VRK3 functions as a BAF kinase. Biochim. Biophys. Acta. 2015;1853:1738–1748. doi: 10.1016/j.bbamcr.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Yuvaniyama J, Denu JM, Dixon JE, Saper MA. Crystal structure of the dual specificity protein phosphatase VHR. Science. 1996;272:1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]
- 42.Kanaoka Y, et al. GAK: a cyclin G associated kinase contains a tensin/auxilin-like domain. FEBS Lett. 1997;402:73–80. doi: 10.1016/s0014-5793(96)01484-6. [DOI] [PubMed] [Google Scholar]
- 43.Greener T, et al. Role of cyclin G-associated kinase in uncoating clathrin-coated vesicles from non-neuronal cells. J. Biol. Chem. 2000;275:1365–1370. doi: 10.1074/jbc.275.2.1365. [DOI] [PubMed] [Google Scholar]
- 44.Sato J, et al. GAK, a regulator of clathrin-mediated membrane trafficking, localizes not only in the cytoplasm but also in the nucleus. Genes Cells. 2009;14:627–641. doi: 10.1111/j.1365-2443.2009.01296.x. [DOI] [PubMed] [Google Scholar]
- 45.Ray MR, et al. Cyclin G-associated kinase: a novel androgen receptor-interacting transcriptional coactivator that is overexpressed in hormone refractory prostate cancer. Int. J. Cancer. 2006;118:1108–1119. doi: 10.1002/ijc.21469. [DOI] [PubMed] [Google Scholar]
- 46.Lin Y, et al. Cyclin G associated kinase interacts with interleukin 12 receptor beta2 and suppresses interleukin 12 induced IFN-gamma production. FEBS Lett. 2007;581:5151–5157. doi: 10.1016/j.febslet.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 47.Korolchuk VI, Banting G. CK2 and GAK/auxilin2 are major protein kinases in clathrin-coated vesicles. Traffic. 2002;3:428–439. doi: 10.1034/j.1600-0854.2002.30606.x. [DOI] [PubMed] [Google Scholar]
- 48.Ziegler SE, Marth JD, Lewis DB, Perlmutter KM. Novel protein-tyrosine kinase gene (hck) preferentially expressed in cells of hematopoietic origin. Mol. Cell. Biol. 1987;7:2276. doi: 10.1128/mcb.7.6.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quintrell N, et al. Identification of a human gene HCI0 that encodes a protein-tyrosine kinase and is expressed in hematopoietic cells. Mol. Cell. Biol. 1987;7:2267. doi: 10.1128/mcb.7.6.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holtzman DA, Cook WD, Dunn AK. Isolation and sequence of a eDNA corresponding to a src-related gene expressed in murine hematopoietic cells. Proc. Natl. Acad. Sci. USA. 1987;84:8325. doi: 10.1073/pnas.84.23.8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziegler SF, Wilson CB, Perlmutter KM. Aug-mented expression of a myeloid-specific protein tyrosine kinase gene (hck) after macrophage activation. J. Extx Med. 1988;168:1801. doi: 10.1084/jem.168.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quintrell N, et al. Identification of a human gene (HCI 0 that encodes a protein-tyrosine kinase and is expressed in hematopoietic cells. Mol. Cell. Biol. 1987;7:2267. doi: 10.1128/mcb.7.6.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yi T, Willman CL. Cloning of the murine c-fgr proto-oncogene cDNA and induction of c-fgr expression by proliferation and activation factors in normal bone marrow-derived monocytic cells. Oncogene. 1989;4:1081. [PubMed] [Google Scholar]
- 54.Lichtenberg U, Quintrell N, Bishop JM. Human protein-tyrosine kinase gene lICK: expression and structural analysis of the promoter region. Oncogene. 1992;7:849. [PubMed] [Google Scholar]
- 55.Boulet I, et al. Lipopolysaccharide- and interferon-gamma-induced expression of hck and lyn tyrosine kinases in murine bone marrow-derived macrophages. Oncogene. 1992;7:703. [PubMed] [Google Scholar]
- 56.English BK, Ihle JN, Myracle A, Yi T. Hck tyrosine kinase activity modulates tumor necrosis factor production by murinemacrophages. J. Exp. Med. 1993;178:1017–1022. doi: 10.1084/jem.178.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghazizadeh S, Bolen JB, Fleit HB. Physical and functional association of Src-related protein tyrosine kinases with Fc gamma RII in monocytic THP-1 cells. J. Biol. Chem. 1994;269:8878–8884. [PubMed] [Google Scholar]
- 58.Lowell CA, Soriano P, Varmus HE. Functional overlap in the src gene family: inactivation of hck and fgr impairs natural immunity. Genes Dev. 1994;8:387–398. doi: 10.1101/gad.8.4.387. [DOI] [PubMed] [Google Scholar]
- 59.Wang AV, Scholl PR, Geha RS. Physical and functional association of the high affinity immunoglobulin G receptor (Fc gamma RI) with the kinases Hck and Lyn. J. Exp. Med. 1994;180:1165–1170. doi: 10.1084/jem.180.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liedtke CM, Hubbard M, Wang X. Stability of actin cytoskeleton and PKCδ binding to actin regulate NKCC1 function in airway epithelial cells. Am J Physiol Cell Physiol. 2003;284:C487–496. doi: 10.1152/ajpcell.00357.2002. [DOI] [PubMed] [Google Scholar]
- 61.You B. J. et al. Rottlerin inhibits Lonicera japonica-induced photokilling in human lung cancer cells through cytoskeleton-related signaling cascade. Evid Based Complement Alternat Med2011, 193842 Epub, Feb8 (2011). [DOI] [PMC free article] [PubMed]
- 62.Reyland ME, Anderson SM, Matassa AA, Barzen KA, Quissell DO. Protein kinase C δ is essential for etoposide-induced apoptosis in salivary gland acinar cells. J Biol Chem. 1999;274:19115–19123. doi: 10.1074/jbc.274.27.19115. [DOI] [PubMed] [Google Scholar]
- 63.Mitsutake N, et al. PKCδ mediates ionizing radiation-induced activation of c-Jun NH(2)-terminal kinase through MKK7 in human thyroid cells. Oncogene. 2001;20:989–996. doi: 10.1038/sj.onc.1204179. [DOI] [PubMed] [Google Scholar]
- 64.Xie MJ, et al. Evidence that apoptotic signalling in hypertrophic cardiomyocytes is determined by mitochondrial pathways involving protein kinase Cδ. Clin Exp Pharmacol Physiol. 2010;37:1120–1128. doi: 10.1111/j.1440-1681.2010.05447.x. [DOI] [PubMed] [Google Scholar]
- 65.Rivera, V. M. et al. Mol. Cell. Biol. 13, 6260–6273 (1993). [DOI] [PMC free article] [PubMed]
- 66.Hindley A, Kolch W. Extracellular signal regulated kinase (ERK)/mitogen activated protein kinase (MAPK)-independent functions of Raf kinases. J Cell Sci. 2002;115:1575–1581. doi: 10.1242/jcs.115.8.1575. [DOI] [PubMed] [Google Scholar]
- 67.Li XJ, He DL, Fu JK. Proteomic profiling of the serum from stage i lung squamous cell carcinoma patients [J] Research Journal of Biotechnology. 2014;9(2):71–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.