ABSTRACT
Sialic acids, terminal structures of sialylated glycoconjugates, are widely distributed in animal tissues and are often involved in intercellular recognitions, including some bacteria and viruses. Mycoplasma mobile, a fish pathogenic bacterium, binds to sialyloligosaccharide (SO) through adhesin Gli349 and glides on host cell surfaces. The amino acid sequence of Gli349 shows no similarity to known SO-binding proteins. In the present study, we predicted the binding part of Gli349, produced it in Escherichia coli and proved its binding activity to SOs of fetuin using atomic force microscopy. Binding was detected with a frequency of 10.3% under retraction speed of 400 nm/s and was shown to be specific for SO, as binding events were competitively inhibited by the addition of free 3′-sialyllactose. The histogram of the unbinding forces showed 24 pN and additional peaks. These results suggested that the distal end of Gli349 constitutes a novel sialoadhesin domain and is directly involved in the gliding mechanism of M. mobile.
Keywords: atomic force microscope, atomic force microscopy, Mycoplasma, rupture force, sialic acid binding protein; adhesin
This study identified binding activity to sialyloligosaccharide in the foot domain of Gli349, a large 349 kDa molecular weight protein that works as a leg in Mycoplasma mobile gliding.
INTRODUCTION
Sialic acids are a family of negatively charged monosaccharides with a nine-carbon backbone that can be classified as N-acetylneuraminic acids (Neu5Ac), N-glycolylneuraminic or 2-keto-3-deoxynonic acids (Schauer 2000). Sialyloligosaccharides (SO) are oligosaccharides modified by sialic acid at a non-reducing end, which are found in the phyla Echinodermata and Vertebrata. SOs on the cell membrane are recognized by other cells in processes such as cell differentiation and immune response and also by pathogenic factors, such as the influenza virus, adenovirus, pneumococcus and Mycoplasma (Sobeslavsky, Prescott and Chanock 1968; Manchee and Taylor-Robinson 1969; Baseman, Banai and Kahane 1982; Roberts et al.1989; Nagai and Miyata 2006; Varki and Gagneux 2012). Mycoplasma, a parasitic bacterial group, is characterized by the lack of peptidoglycan and small cell and genome sizes (Razin, Yogev and Naot 1998; Razin and Hayflick 2010). A dozen species of Mycoplasma form a membrane protrusion at a cell pole, bind to SOs on solid surfaces through an SO-binding protein and glide in the direction of the protrusion (Miyata 2008; Nakane and Miyata 2012; Balish 2014a,b; Miyata and Hamaguchi 2016). A gliding mechanism has been proposed for Mycoplasma mobile, the fastest species, gliding at a speed up to 4.5 µm/s, as explained below (Fig. 1A and B). Gliding of M. mobile is caused by a repetitive cycle consisting of catching, pulling and releasing SOs, driven by a force that is generated by the intracellular hydrolysis of ATP and is transmitted to the legs on the cell surface (Uenoyama and Miyata 2005a; Miyata 2008, 2010; Kinosita et al.2014; Miyata and Hamaguchi 2016; Kinosita, Miyata and Nishizaka 2018). Gli349, the leg for gliding, is a membrane protein resembling the symbol for an eighth note in music and is composed of two 20-nm rods connected by a hinge, a 43-nm filamentous part and a 14-nm-long globular part designated as Foot, in this order from the N-terminus (Fig. 1A) (Adan-Kubo et al.2006). Although the amino acid sequence of Gli349 does not show any similarity to known SO-binding proteins, SO-binding activity was detected for Gli349 isolated from M. mobile (Lesoil et al.2010). The mycoplasma cell recognized Neu5Ac of SOs specifically, in a lock-and-key manner, with the highest affinity being toward the branched oligosaccharide with clustered sialic acids (Nagai and Miyata 2006; Kasai et al.2013; Kasai, Hamaguchi and Miyata 2015). These results suggest that Gli349 is a novel type of SO-binding protein (Miyata and Hamaguchi 2016). Another feature of this binding protein is the release of the ligand, because in this gliding mechanism, the legs are expected to release SOs after the stroke. Mycoplasmamobile is released from SOs more easily during forward movement than during backward movement, because of a catch bond-like behavior, the detachment probability of which depends on the tension exerted (Tanaka et al.2016). This unique binding is suggested to be assigned to the distal part of Gli349, Foot, because the legs of M. mobile cells seem to attach to the surface through the tip part under quick-freeze replica electron microscopy (Miyata and Petersen 2004), and a strain mutated on the Gli349 serine 2770 to leucine does not bind to solid surfaces (Miyata et al.2000; Uenoyama et al.2009; Mizutani et al.2018). Therefore, the structural and functional information about Foot would be a clue to clarify the unique mechanism of M. mobile gliding.
Figure 1.
Schematics of M. mobile gliding and SO-binding proteins. (A) Mycoplasma mobile and Gli349. Left, whole-cell image of M. mobile. Gli349 molecules are aligned around the gliding machinery. Right, Gli349 molecule working as leg. Foot is a C-terminal region of Gli349 forming a hook-shaped fragment comprising 463 amino acid residues. (B) Mycoplasma mobile glides on glass surface coated by sialylated glycoconjugates. The pulling direction relative to SO changes before and after stroke, because continuous cell displacement caused by many legs pulls forward the leg after stroke. (C) Possible assignment of different unbinding forces to the pulling directions. The force exerted to the Foot is indicated by black arrows. (D) SO-binding proteins featured by repeat structures. Each binding protein molecule has a single binding domain at the N- or C-terminus. The extension produced by repeat sequences contributes to the flexible position toward the ligand, enabling effective binding.
MATERIALS AND METHODS
Protein production and purification
DNA encoding the C-terminal domain of Gli349 (Foot) was codon-optimized and synthesized to be expressed in Escherichia coli (GenScript, Piscataway, NJ, USA). The DNA fragment encoding Foot was inserted into pET15b (Novagen, Madison, WI, USA), using the NdeI and XhoI sites (supplementary Fig. S1 available online). The constructed plasmid was introduced into E. coli BL21(DE3), and the transformed cells were inoculated into 5 ml of LB medium (0.5% yeast extract, 1% Bacto tryptone, 1% NaCl) containing 50 µg/ml ampicillin. The overnight culture was inoculated into 1 l of LB medium containing ampicillin and grown at 37°C until the OD600 reached 0.4. The production of recombinant Foot (rFoot) was induced with isopropyl-β-d-1-thiogalactopyranoside at a final concentration of 1 mM, for 4 h at 37°C. Cells containing rFoot were collected by centrifugation at 18 800 × g for 5 min at 25°C. Harvested cells were washed twice using 10 mM Tris–HCl pH 8.0 and suspended in the same buffer containing 1 mM phenylmethylsulfonyl fluoride. The suspension was homogenized using a sonicator (Nippon Seiki, Tokyo, Japan) and centrifuged at 32 200 × g for 30 min at 4°C. The pellet was resuspended in an unfolding-binding buffer (UB-buffer; 10 mM Tris–HCl pH 8.0, 500 mM NaCl, 6 M urea) and sonicated. The suspension was ultracentrifuged at 287 000 × g for 30 min at 4°C, and the supernatant was loaded onto a HisTrap HP column (GE Healthcare, Little Chalfont, UK) equilibrated using the UB-buffer and connected to AKTA Pure (GE Healthcare). Bound proteins were washed with UB-buffer and eluted in a step-wise manner with UB-buffer containing 100, 200 or 300 mM imidazole. The eluted fractions were subjected to SDS–12.5% PAGE and stained with Coomassie Brilliant Blue (CBB). Fractions containing rFoot were collected and dialyzed against 10 mM Na-K phosphate buffer pH 6.3 containing 6 M urea. The dialyzed sample was loaded onto a HiTrap SP HP (GE Healthcare) equilibrated using the same buffer. The column was washed with the equilibration buffer, and the proteins were eluted with a linear gradient of 0–500 mM NaCl in 10 mM Na-K phosphate buffer containing 6 M urea. Fractions containing rFoot were collected, dialyzed against UB-buffer and loaded onto the HisTrap HP column again. The proteins bound to the column were eluted with a linear gradient of 0–300 mM imidazole in UB-buffer. rFoot fractions eluted using 50–100 mM imidazole were collected and used for refolding.
Gradient refolding
All steps were carried out at 4°C, unless stated otherwise. Purified rFoot was transferred into a dialysis tube and put into an empty XK 50/100 column (GE Healthcare) filled with 250 ml of 10 mM Tris–HCl pH 8.8 containing 4 M urea and 100 mM arginine (supplementary Fig. S2 available online). The refolding was performed with a 4–0 M urea linear gradient dialysis, controlled by a peristaltic pump, at a flow rate of 2.5 ml/min, for 540 min. Next, rFoot was dialyzed against 10 mM Tris–HCl pH 8.8, at a flow rate of 2.5 ml/min, for 360 min. The refolded Foot (reFoot) was centrifuged at 287 000 × g for 30 min at 4°C, and the supernatant was analyzed by gel filtration.
Biochemical analyses
For gel filtration analysis, reFoot was concentrated using an Amicon Ultra 30 K spin filter (Millipore, Bedford, MA, USA) and loaded at a flow rate of 1 ml/min onto a HiLoad 26/60 Superdex 200 pg column equilibrated using 10 mM Tris–HCl pH 8.0 containing 150 mM NaCl and 100 mM arginine and connected to AKTA purifier (GE Healthcare). CD spectroscopy was performed as previously described (Kenri et al.2019).
AFM tip modification
Soft ‘Biolever’ cantilevers (Olympus, Tokyo, Japan) with a nominal spring constant of 6 pN/nm were used. The spring constant of each cantilever was determined using the thermal oscillation method (Hutter and Bechhoefer 1993). Atomic force microscope (AFM) tips pre-cleaned by a UV Ozone Cleaner (Bioforce Nanosciences, Ames, IA, USA) were incubated with 0.5 mg/ml of thiol-carboxylic PEG (SH-PEG-COOH, MW = 10 000; Nanocs, New York, NY, USA) in water for 2 h at 25°C to form a self-assembled monolayer, followed by rinsing in ethanol and water, and drying with nitrogen gas. The tips were activated using water-soluble carbodiimide (WSC) and N-hydroxysuccinimide (NHS), by using the Amine Coupling Kit (Dojindo, Kumamoto, Japan). NHS-activated tips were incubated with 0.05 mg/ml calf fetuin, as a sialylated glycoprotein for 30 min, and washed with phosphate-buffered saline (PBS). The coupling reaction was quenched by the addition of a blocking buffer for 30 min and washed with PBS. The fetuin-coated AFM tips were stored in 10 mM HEPES pH 7.5 at 4°C and used for force detection.
Immobilization of reFoot on gold surface
The freshly stripped gold surface (Stamou et al.1997) was modified with SH-PEG-COOH, as described above. reFoot at 0.2 mg/ml was dialyzed against 10 mM HEPES pH 8.0 twice, and the protein solution was dropped onto the gold surface activated by using the Amine Coupling Kit. After immobilization, PBS was replaced by 10 mM HEPES pH 7.5 before force measurement.
Force measurement by AFM
Force measurements were performed using MFP1D AFM (Asylum Research, Santa Barbara, CA, USA). The AFM tip coated with fetuin was repeatedly approached and retracted from the gold plate coated with reFoot at a velocity of 400 nm/s, a dwell time of 0.5 s, a distance of 400 nm and a pushing force of 400 pN. After every 100 force curve measurements, the relative position of the cantilever to the gold substrate was shifted for fresh surface area. In control experiments, 3′-sialyllactose (Neu5Ac α2–3 Gal β1–4 Glc) (3′-SL) at 30 or 300 µM was added to the reaction fluid as a competitive inhibitor. Events before the 80-nm retraction of the cantilever from the gold surface were not applied to the subsequent analyses to exclude non-specific binding and the effect of metal contact.
Sequence analyses
The secondary structure was predicted using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/). Domains and tertiary structure were predicted by Ginzu and Robetta, respectively (http://robetta.bakerlab.org/). Sequence similarity was searched by PSI-BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) against NCBI non-redundant protein sequences. Intrinsically disordered and low complexity regions were predicted by DISOPRED (http://bioinf.cs.ucl.ac.uk/psipred/?disopred=1) and SEG (http://mendel.imp.ac.at/METHODS/seg.server.html), respectively. Predicted tertiary structures were visualized using UCSF Chimera 1.13.1 (https://www.cgl.ucsf.edu/chimera/).
RESULTS
Isolation and refolding of rFoot
The 3183 amino acid sequence of Gli349 is divided into three parts, in the following order from the N-terminus: a transmembrane segment, a middle region including 18-repeated sequences and a remaining C-terminal 463-amino acid-long region (Metsugi et al.2005). The C-terminal region including serine 2770, essential for SO binding, has been suggested to correspond to Foot, the globular moiety observed by electron microscopy. To examine the putative binding activity to SOs, M. mobile Foot, composed of 463 amino acids, was tagged at the N-terminus with a histidine cluster, produced and purified. As rFoot was produced as inclusion bodies, rFoot was solubilized by the addition of urea and isolated by using His-tag affinity chromatography. rFoot was then subjected to dialysis, cation exchange chromatography, another dialysis and another His-tag affinity chromatography (Fig. 2A). Purified rFoot was refolded by dialysis against a gradient of urea, from 4 to 0 M, in the presence of 100 mM arginine: a dialysis tube containing 40 ml of 0.2 mg/ml rFoot was set inside a column filled with a 4 M urea solution containing 100 mM arginine, and the 250 ml of outer solution was replaced, at a flow at 2.5 ml/min using a peristaltic pump, by 0 M urea after 540 min (supplementary Fig. S2 available online). The urea gradient was formed in the column, enabling the slow removal of urea in a well-controlled condition. Refolded rFoot, designated ‘reFoot’ was found entirely in the supernatant after ultracentrifugation, showing that the in-column gradient refolding is a useful method. reFoot was concentrated to 1.3 mg/ml and subjected to gel filtration in the presence of arginine (Fig. 2B). reFoot was not found in the void; it was found in the fractions corresponding to two symmetrical peaks at the positions of 3.8 and 4.9 nm Stokes radii and in a preceding broad peak, suggesting that reFoot folded uniformly and polymerized. The molecular mass of reFoot calculated from the amino acid sequence was 53.6 kDa, and a globular protein with 53.6 kDa was expected to elute at the position of 3.1 nm Stokes radius in this gel filtration, suggesting that the structure of reFoot is not completely spherical. The sharp peaks may correspond to the monomer and dimer because the cube ratio of the Stokes radii was approximately 2.1. The monomer peak is unlikely to contain dimer and trimer reFoot as shown by the result of Gaussian fitting curves. Far-UV CD spectrum for the reFoot monomer was obtained, and the secondary structure content was estimated (Fig. 2C). The estimated values were consistent with the ones predicted from the amino acid sequence.
Figure 2.

Isolation of reFoot. (A) The fractions collected after each step were subjected to SDS-12.5% PAGE and visualized by CBB staining. Lane 1: E. coli whole cell lysate; Lane 2: soluble fraction; Lanes 3 and 4: supernatant and pellet after urea treatment; Lanes 5–9: fractions after nickel affinity, cation exchange, and nickel affinity chromatographies, after refolding, and gel filtration chromatography, respectively. The position of rFoot band is indicated by a black triangle. (B) ReFoot was loaded onto a Superdex 200 26/60 column chromatography. Stokes radii of thyroglobulin (8.5 nm, 669 kDa), ferritin (6.1 nm, 440 kDa), aldolase (4.8 nm, 158 kDa), and ovalbumin (3.1 nm, 44 kDa) are shown by red circles, respectively from left to right. Fitted peaks are shown in blue lines. (C) Far-UV CD spectra of reFoot. Secondary structure contents were predicted and estimated by PSIPRED and BESTSEL, respectively.
Binding activity of reFoot to SOs
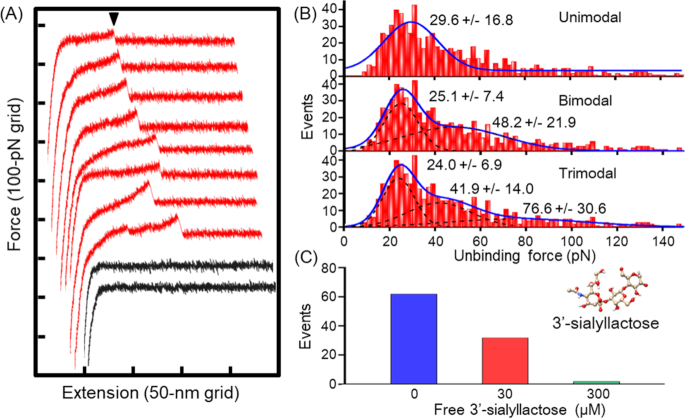
Next, we examined the SO-binding activity of the reFoot monomer by AFM, which can detect interactions larger than 5 pN (Florin, Moy and Gaub 1994; Sekiguchi et al.2003). We used bovine fetuin as the binding target, because gliding of M. mobile has been analyzed in detail on fetuin-coated glass (Kasai, Hamaguchi and Miyata 2015). Bovine fetuin is a serum-derived glycoprotein of 48.4 kDa, with three N- and O-type sugar chains, each of which is attached by an α2,3- and α2,6-linked Neu5Ac at the non-reducing terminal (Green et al.1988). reFoot and fetuin were immobilized on an SH-PEG-COOH-coated gold base and an AFM tip, respectively, by chemical crosslinking with WSC and NHS, and mounted on the AFM setup (Fig. 3A). Each surface was coated with numerous molecules. We succeeded in detecting binding activity in a process where the fetuin-coated cantilever was moved toward the reFoot-coated gold substrate at 400 nm/s from the position at 400 nm, and then retracted at the same speed after a dwell of 0.5 s on the substrate. This speed was used in the previous study (Lesoil et al.2010) and in the range of smooth gliding of intact cells (Miyata, Ryu and Berg 2002; Uenoyama et al.2009; Kinosita et al.2014). A typical trace is shown in Fig. 3B, where the interaction was detected when the cantilever bent downward and the deflection increased as the AFM tip was separated from the gold surface. When the tip rose up, a dissociation event occurred, probably because the force acting on the cantilever exceeded the rupture force of the interaction. The unbinding event was then detected as a peak after the detachment of the cantilever from the solid surface. In the absence of interaction, the deflection signal after the retraction of the cantilever from the gold base was zero. SH-PEG-COOH, with a molecular weight of 10 000, is predicted to extend to a maximum of 82 nm, based on its chemical structure, when the force of several hundreds of pN is applied (Rief et al.1997). To eliminate non-specific interactions occurring on the gold substrate surface, only the unbinding events occurring at more than half of the sum of both spacer lengths, i.e. more than 80 nm, were defined as specific binding events, as the PEG linker should be partially extended because of its elasticity with several tens of pN. In the 6127 approaches that were made, 632 peaks were detected as specific binding events, i.e. the frequency of binding events was 10.3% (Fig. 4A). The distribution of the rupture forces calculated from the deflections of the peak and initial positions were fitted by the sum of the Gaussian distributions, and the three Gaussian distributions were well-matched with the histogram of force distribution (Fig. 4B). The fitting peaks for the trimodal model were 24.0 ± 6.9, 41.9 ± 14.0 and 76.6 ± 30.6 pN, in the order of event frequency. The smallest peak should correspond to the binding of a single unit, because of the largest frequency of unbinding events. Next, we examined whether binding of reFoot is specific for SOs on fetuin by analyzing the inhibitory effect of 3′-SL, which is the direct binding target of the M. mobile cell (Nagai and Miyata 2006; Kasai et al.2013). Addition of 30 and 300 µM 3′-SL decreased the frequency of binding events from 60 to 32 and 2, respectively, in 400 retractions (Fig. 4C), and only non-specific binding events with more than 200 pN were detected when 300 µM of 3′-SL were added. Based on these results, we concluded that Foot is the SO-binding domain in Gli349.
Figure 3.
Experimental scheme of the binding assay by AFM. (A) Surface modification of AFM tip and gold base. Both AFM tip and gold base were coated by carboxylated self-assembled monolayer with 82-nm linkers. reFoot and fetuin were immobilized by amine coupling on the gold base and cantilever, respectively. Unbinding events were scanned repeatedly. (B) Typical force-extension curves with and without interaction. After approach, the AFM tip was retracted for the detection of interaction between reFoot and fetuin. The rupture force at the dissociation was estimated from the center of the dissociation peak and the center of the peak of the initial position. Traces are shown in arbitrary positions.
Figure 4.
Unbinding force between reFoot and fetuin. (A) Typical force-extension curves. The final rupture peak corresponding to an unbinding event between reFoot and fetuin is marked by a black triangle. Traces with and without interaction are shown in red and black, at arbitrary positions, respectively. (B) Distribution of the unbinding force that occurred after 80-nm extension from the zero position. The frequency of unbinding events was 10.3% (632 events in 6127 scans). The whole fittings (blue) for uni-, bi- and trimodal are shown as the sum of the Gaussian distributions (black dots). Bandwidth of each bin is 2 pN. (C) Frequencies of unbinding events in presence of 0 (n = 400), 30 (n = 400) and 300 µM (n = 400) 3′-SL.
Analyses of amino acid sequence of Foot
We succeeded in domain parsing of the Foot by a domain detection tool, Ginzu, based on the alignment of amino acid sequences, low complexity and intrinsically disordered regions (Fig. 5) (Dunker et al.2001). Then, a possible structure was provided by using a full-chain-protein-structure prediction server, named Robetta, that can predict the tertiary structures of each domain by comparative and ab initio modeling. In the process, the amino acid sequence was divided into three parts, 2721–2997, 2998–3057 and 3058–3183, with similarities to a hypothetical leucine-rich repeats (LRR) protein from Eubacterium ventriosum, F-actin cross-linking gelation factor from the soil-living amoeba Dictyostelium discoideum and a hypothetical protein from the parasitic nematode Anisakis simplex, respectively (Fig. 5A) by Ginzu, and then their tertiary structures were predicted by Robetta (Fig. 5B). The N-terminal region of the Foot, 2721–2997, contained LRRs consisting of 20–30 amino acid residues (Fig. 5A and C). The predicted structure of this region was consistent with the previous prediction by Phyre2, based on only homology/analogy modeling (Miyata and Hamaguchi 2016). The connections between the predicted structures of each domain remain unknown. Generally, LRRs form a horseshoe fold that is often found in protein–protein interactions; it is present, for instance, in the ribonuclease inhibitor, tropomodulin, toll-like receptor and internalin from Listeria (Kobe and Deisenhofer 1994; Enkhbayar et al.2004). Although repetitive β-strands are connected through α-helices in many LRRs, the β-strands of Foot were predicted to be connected by long loops in Foot. The set of gliding genes can be found in the genome of Mycoplasma pulmonis, a mouse pathogen related to M. mobile (Uenoyama and Miyata 2005b). As this species forms a protrusion at a cell pole and glides with comparable speed to M. mobile, it is expected to glide by a common mechanism with M. mobile, although the details have not been well characterized (Bredt and Radestock 1977). MYPU2110 from M. pulmonis is the only protein among all the proteins reported so far showing sequence similarity to Gli349, which features a high-similarity region, 623–2818, with an e-value of 4e-121 and 25% identity. However, Foot is not well conserved: although the LRR region found in MYPU2110 showed similarity to that of Gli349 with an e-value of 1e-13 and 43% identity, most of the remaining parts were not similar (Fig. 5A). The N-terminal part of Foot comprising amino acid residues 2721 to 2818 may be involved in the binding activity through the horseshoe-fold characteristic for LRRs, which is consistent with the position of essential serine residue.
Figure 5.
Structural characteristics of Gli349 Foot and comparison with MYPU2110. Serine 2770 and corresponding residues essential for the binding activity are indicated by triangles. (A) Schematic presentation of amino acid sequences. Left: Gli349 and Foot. Right: MYPU2110 from M. pulmonis and region corresponding to the Foot. Predicted transmembrane segment, α-helix and β-strand are shown in black, red and blue bars, respectively. Hatched pink regions show similarity between Gli349 and MYPU2110, with an e-value of 4e-121 for the whole part. Leucine rich repeats are shown by open boxes marked as LRRs. Sequence fragmentation based on predicted domains are shown short bars and amino acid numbers. Templates used to predict the tertiary structures of each domain were 4h09 (a hypothetical LRR protein from E. ventriosum), 1ksr (F-actin cross-linking gelation factor from D. discoideum) and 2mar (hypothetical protein from A. simplex) for Foot, 4fdw (leucine rich hypothetical protein from Bacteroides ovatus), 5ods (phosphoprotein from Mus musculus) and 5ts2 (phosphopantetheine adenylyltransferase from Pseudomonas aeruginosa) for MYPU2110, respectively. (B) Predicted tertiary structures of domain containing LRRs region. The direction of structures are adjusted according to the axis of LRR domain, using MatchMaker command of Chimera. (C) Amino acid sequences of homologous regions from Foot and MYPU2110. Upper: M. mobile. Lower: M. pulmonis. The amino acid numbers in each open reading frame are shown in the left. The region 2722–2818 in the M. mobile sequence is similar to the 2691–2781 region in MYPU2110. Identical amino acids are highlighted by asterisks. The LRR regions are colored red. Intrinsically disordered and low complexity regions are indicated by light blue and yellow hatching, respectively.
DISCUSSION
Unbinding force of the reFoot molecule
In the present study, reFoot showed binding activity toward SOs on bovine fetuin, with the rupture forces distributing around three peaks, around 24.0, 41.9 and 76.6 pN (Fig. 4B). The binding events were detected with a frequency of 10.3% in retractions, suggesting that they occurred as single-unit events, as generally the binding caused by multiple sites shows a frequency higher than 30% (Parreira et al.2014). Moreover, peaks smaller than 24.0 pN did not appear, showing that, at least a rupture force of approximately 24.0 pN seems to arise from the binding of one reFoot molecule to one SO molecule (Fig. 4C). The binding events observed herein should be related to the binding to solid surfaces during cell gliding, because binding was inhibited by 300 µM free 3′-SL for reFoot, as previously observed for M. mobile cells (Fig. 4C) (Kasai et al.2013). The larger rupture force peaks, around 41.9 and 76.6 pN, are unlikely to be the result of a simultaneous dissociation of the smallest one, because the traces of unbinding events did not show any step-wise processes (Fig. 4A). Therefore, we suggest that the reFoot molecule has three different unbinding forces.
Directed binding of reFoot
Mycoplasma mobile cells can detach more easily forward than backward along the gliding direction, which may derive from the characteristics of Foot and enable directed gliding (Fig. 1B) (Tanaka et al.2016). In a previous analysis of Gli349 isolated from Mycoplasma cells (Lesoil et al.2010), using the same rate of probe retraction as the present study, only a single unbinding force was detected, around 67.7 pN, which is larger than 24.0 pN, the major force of reFoot. Possibly the largest force, 76.6 pN, detected as a Gaussian peak in the present study, may correspond to this force. The two smaller forces, 24.0 and 41.9 pN, were possibly caused by the direct fixation of reFoot onto the surface. The unbinding force should depend on the pulling direction of SO relative to reFoot, because the unbinding force of a starved cell depends on the pulling direction (Tanaka et al.2016). In the case of Gli349 analyses, the molecule is likely to be fixed onto the surface through its flexible part, resulting in the unbinding of Foot in the direction of the largest force (Lesoil et al.2010). Recently, the stall force of M. mobile cells was reported to be 113 pN, which is only 1.5-fold of the possible largest unbinding force of reFoot observed herein (Mizutani et al.2018). Considering that the gliding machinery has 450 legs, the unbinding force of reFoot appears much higher than the expected value. This apparent inconsistency can be explained if we consider that the stall force of a cell reflects the force generating a step rather than the integrated unbinding force of the feet (Mizutani et al.2018). The binding of Foot depends on the lock-and-key recognition manner, which has been proven previously using various SOs (Kasai et al.2013; Kasai, Hamaguchi and Miyata 2015). In general, the lock-and-key recognition manner of SO-binding proteins can be classified into two types, zipper and all-or-none type, of which influenza virus hemagglutinin and selectin are examples, respectively (Griffin et al.1975; Griffin, Griffin and Silverstein 1976; Sieben et al.2012). The zipper type shows small dissociation steps rather than a single and larger jump, which is observed in the all-or-none type (Griffin et al.1975; Griffin, Griffin and Silverstein 1976). Binding of Foot appears to occur in the all-or-none manner, because the dissociation occurred in a single step in the AFM analyses (Fig. 4A). The structure of predicted Foot is featured by a rather rigid part including LRRs and an essential serine residue (Fig. 5C). This rigidity may cause the directed binding by acting like a lever arm. The previous study suggested the involvement of catch bonds in the binding of cells to SOs, where the bond is stabilized by the application of a pulling force (Tanaka et al.2016). This catch bond may be caused by the distortion of the rather rigid structure.
Gli349, an SO-binding protein in the gliding machinery of M. mobile
The Gli349 sequence is composed of a transmembrane segment, an ∼100-amino acid region of 18 repeats including a flexible part and an SO-binding domain, in this order from the N-terminus. This constitution is common to other SO-binding proteins such as selectin, with the sushi domain (SCR), and Siglec, with the C2-set Ig domain (Fig. 1D), and has been suggested to be advantageous in the efficient catch of various sugar chains in fluids and also for leukocyte rolling on vessel surfaces (Preston et al.2016). Gli349 of M. mobile might have originated as a static binding protein, with similar features to other SO-binding proteins, and then evolved as the leg used for gliding. These observations support our suggestion that an accidental combination between a static binding protein and an F-type ATPase became the origin of M. mobile gliding (Miyata and Hamaguchi 2016). In the M. mobile gliding mechanism, the force generated in the internal structure, based on ATP energy, is transmitted across the membrane along several proteins and reaches Gli349, resulting in the repeated pulling of SOs (Fig. 1B) (Nakane and Miyata 2007; Miyata and Hamaguchi 2016; Kinosita et al.2018; Mizutani et al.2018). The two N-terminal rigid rods, characteristics of Gli349, might be a major modification from the ancestral binding protein to efficiently transmit that force to Foot (Adan-Kubo et al.2006).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Masaki Mizutani, at Osaka City University, for data analysis, and to Ritsuko Fujii and Nami Yamano, at Osaka City University, for technical help with CD spectroscopy.
FUNDING
This work was supported by a Grant-in-Aid for Scientific Research on the Innovative Area ‘Harmonized Supramolecular Motility Machinery and Its Diversity’ (MEXT KAKENHI Grant Number 24117002) and by Grant-in-Aids for Scientific Research (B) and (A) (MEXT KAKENHI Grant Numbers 24390107 and 17H01544) to MM.
Conflicts of interest. None declared.
REFERENCES
- Adan-Kubo J, Uenoyama A, Arata T et al. Morphology of isolated Gli349, a leg protein responsible for Mycoplasma mobile gliding via glass binding, revealed by rotary shadowing electron microscopy. J Bacteriol. 2006;188:2821–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balish MF. Giant steps toward understanding a mycoplasma gliding motor. Trends Microbiol. 2014a;22:429–31. [DOI] [PubMed] [Google Scholar]
- Balish MF. Mycoplasma pneumoniae, an underutilized model for bacterial cell biology. J Bacteriol. 2014b;196:3675–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseman JB, Banai M, Kahane I. Sialic acid residues mediate Mycoplasma pneumoniae attachment to human and sheep erythrocytes. Infect Immun. 1982;38:389–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt W, Radestock U. Gliding motility of Mycoplasma pulmonis. J Bacteriol. 1977;130:937–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. [DOI] [PubMed] [Google Scholar]
- Enkhbayar P, Kamiya M, Osaki M et al. Structural principles of leucine-rich repeat (LRR) proteins. Proteins. 2004;54:394–403. [DOI] [PubMed] [Google Scholar]
- Florin E, Moy V, Gaub H. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264:415–7. [DOI] [PubMed] [Google Scholar]
- Green ED, Adelt G, Baenziger JU et al. The asparagine-linked oligosaccharides on bovine fetuin. Structural analysis of N-glycanase-released oligosaccharides by 500-megahertz 1H NMR spectroscopy. J Biol Chem. 1988;263:18253–68. [PubMed] [Google Scholar]
- Griffin FM, Griffin JA, Leider JE et al. Studies on the mechanism of phagocytosis. I. Requirements for circumferential attachment of particle-bound ligands to specific receptors on the macrophage plasma membrane. J Exp Med. 1975;142:1263–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin FM, Griffin JA, Silverstein SC. Studies on the mechanism of phagocytosis. II. The interaction of macrophages with anti-immunoglobulin IgG-coated bone marrow-derived lymphocytes. J Exp Med. 1976;144:788–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter JL, Bechhoefer J. Calibration of atomic-force microscope tips. Rev Sci Instrum. 1993;64:1868–73. [Google Scholar]
- Kasai T, Hamaguchi T, Miyata M. Gliding motility of Mycoplasma mobile on uniform oligosaccharides. J Bacteriol. 2015;197:2952–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai T, Nakane D, Ishida H et al. Role of binding in Mycoplasma mobile and Mycoplasma pneumoniae gliding analyzed through inhibition by synthesized sialylated compounds. J Bacteriol. 2013;195:429–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenri T, Kawakita Y, Kudo H et al. Production and characterization of recombinant P1 adhesin essential for adhesion, gliding, and antigenic variation in the human pathogenic bacterium, Mycoplasma pneumoniae. Biochem Biophys Res Commun, 2019;508:1050-5, 10.1016/j.bbrc.2018.11.132. [DOI] [PubMed] [Google Scholar]
- Kinosita Y, Miyata M, Nishizaka T. Linear motor driven-rotary motion of a membrane-permeabilized ghost in Mycoplasma mobile. Sci Rep. 2018;8:11513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinosita Y, Nakane D, Sugawa M et al. Unitary step of gliding machinery in Mycoplasma mobile. Proc Natl Acad Sci USA. 2014;111:8601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–21. [DOI] [PubMed] [Google Scholar]
- Lesoil C, Nonaka T, Sekiguchi H et al. Molecular shape and binding force of Mycoplasma mobile’s leg protein Gli349 revealed by an AFM study. Biochem Biophys Res Commun. 2010;391:1312–7. [DOI] [PubMed] [Google Scholar]
- Manchee RJ, Taylor-Robinson D. Studies on the nature of receptors involved in attachment of tissue culture cells to mycoplasmas. Br J Exp Pathol. 1969;50:66–75. [PMC free article] [PubMed] [Google Scholar]
- Metsugi S, Uenoyama A, Adan-Kubo J et al. Sequence analysis of the gliding protein Gli349 in Mycoplasma mobile. Biophysics (Oxf). 2005;1:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata M. Centipede and inchworm models to explain Mycoplasma gliding. Trends Microbiol. 2008;16:6–12. [DOI] [PubMed] [Google Scholar]
- Miyata M. Unique centipede mechanism of Mycoplasma gliding. Annu Rev Microbiol. 2010;64:519–37. [DOI] [PubMed] [Google Scholar]
- Miyata M, Hamaguchi T. Prospects for the gliding mechanism of Mycoplasma mobile. Curr Opin Microbiol. 2016;29:15–21. [DOI] [PubMed] [Google Scholar]
- Miyata M, Petersen JD. Spike structure at the interface between gliding Mycoplasma mobile cells and glass surfaces visualized by rapid-freeze-and-fracture electron microscopy. J Bacteriol. 2004;186:4382–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata M, Ryu WS, Berg HC. Force and velocity of Mycoplasma mobile gliding. J Bacteriol. 2002;184:1827–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata M, Yamamoto H, Shimizu T et al. Gliding mutants of Mycoplasma mobile: Relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology. 2000;146:1311–20. [DOI] [PubMed] [Google Scholar]
- Mizutani M, Tulum I, Kinosita Y et al. Detailed analyses of stall force generation in Mycoplasma mobile gliding. Biophys J. 2018;114:1411–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai R, Miyata M. Gliding motility of Mycoplasma mobile can occur by repeated binding to N-acetylneuraminyllactose (sialyllactose) fixed on solid surfaces. J Bacteriol. 2006;188:6469–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane D, Miyata M. Cytoskeletal jellyfish; structure of Mycoplasma mobile. Proc Natl Acad Sci USA. 2007;104:19518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane D, Miyata M.. Mycoplasma mobile cells elongated by detergent and their pivoting movements in gliding. J Bacteriol. 2012;194:122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parreira P, Shi Q, Magalhaes A et al. Atomic force microscopy measurements reveal multiple bonds between Helicobacter pylori blood group antigen binding adhesin and Lewis b ligand. J R Soc Interface. 2014;11:2014.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston RC, Jakob RP, Binder FPC et al. E-selectin ligand complexes adopt an extended high-affinity conformation. J Mol Cell Biol. 2016;8:67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin S, Hayflick L. Highlights of mycoplasma research–an historical perspective. Biologicals. 2010;38:183–90. [DOI] [PubMed] [Google Scholar]
- Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M, Oesterhelt F, Heymann B et al. Single molecule force spectroscopy on polysaccharides by atomic force microscopy. Science. 1997;275:1295–7. [DOI] [PubMed] [Google Scholar]
- Roberts DD, Olson LD, Barile MF et al. Sialic acid-dependent adhesion of Mycoplasma pneumoniae to purified glycoproteins. J Biol Chem. 1989;264:9289–93. [PubMed] [Google Scholar]
- Schauer R. Achievements and challenges of sialic acid research. Glycoconj J. 2000;17:485–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi H, Arakawa H, Taguchi H et al. Specific interaction between GroEL and denatured protein measured by compression-free force spectroscopy. Biophys J. 2003;85:484–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieben C, Kappel C, Zhu R et al. Influenza virus binds its host cell using multiple dynamic interactions. Proc Natl Acad Sci USA. 2012;109:13626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobeslavsky O, Prescott B, Chanock RM. Adsorption of Mycoplasma pneumoniae to neuraminic acid receptors of various cells and possible role in virulence. J Bacteriol. 1968;96:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamou D, Gourdon D, Liley M et al. Uniformly flat gold surfaces: Imaging the domain structure of organic monolayers using scanning force microscopy. Langmuir. 1997;13:2425–8. [Google Scholar]
- Tanaka A, Nakane D, Mizutani M et al. Directed binding of gliding bacterium, Mycoplasma mobile, shown by detachment force and bond lifetime. MBio. 2016;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uenoyama A, Miyata M. Gliding ghosts of Mycoplasma mobile. Proc Natl Acad Sci USA. 2005a;102:12754–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uenoyama A, Miyata M. Identification of a 123-kilodalton protein (Gli123) involved in machinery for gliding motility of Mycoplasma mobile. J Bacteriol. 2005b;187:5578–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uenoyama A, Seto S, Nakane D et al. Regions on Gli349 and Gli521 protein molecules directly involved in movements of Mycoplasma mobile gliding machinery, suggested by use of inhibitory antibodies and mutants. J Bacteriol. 2009;191:1982–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Gagneux P. Multifarious roles of sialic acids in immunity. Ann N Y Acad Sci. 2012;1253:16–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.