Abstract
Circular RNAs (circRNAs) are novel clusters of endogenous noncoding RNAs (ncRNAs) that are widely expressed in eukaryotic cells. In contrast to the generation of linear RNA transcripts, circRNAs undergo a “back-splicing” process to form a continuous, covalently closed, stable loop structure without 5ʹ or 3ʹ polarities and poly (A) tails during posttranscriptional modification. Due to the widespread availability of several technologies, especially high-throughput RNA sequencing, numerous circRNAs have been discovered not only in mammals but also in plants and insects. Notably, due to their abilities to serve as microRNA (miRNA) “sponges”, miRNA “reservoirs”, regulate gene expression and encode proteins, circRNAs participate in the development and progression of different immune responses and immune diseases by enriching various forms of epigenetic modification. CircRNAs have been demonstrated to be expressed in a tissue-specific and pathogenesis-related manner during the occurrence of multiple immune diseases. Additionally, because of their circular configurations, expression in blood and peripheral tissues and coexistence with exosomes, circRNAs show inherent conservation along with environmental resistance stability and may be regarded as potential biomarkers or therapeutic targets for some immune diseases. In this review, we summarize the characteristics, functions and mechanisms of circRNAs and their involvement in immune responses and diseases. Although our knowledge of circRNAs remains preliminary, this field is worthy of deeper exploration and greater research efforts.
Keywords: circRNAs, biogenesis, function, immune responses, immune diseases
Introduction
For decades, numerous experiments have focused on protein-coding genes and their associated transcripts in different species; however, these hotspots account for only approximately 2% of the entire human genome 1. In fact, the vast majority of the remaining sequences, up to 90%, cannot encode proteins, indicating that noncoding RNAs (ncRNAs) are dominant in the eukaryotic transcriptome 2. In accordance with the standard length of 200 nucleotides (nt), ncRNAs can be generally classified into long ncRNAs (lncRNAs), microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), small interfering RNAs (siRNAs) and circular RNAs (circRNAs) 3. Notably, the mechanisms underlying the activation or inhibition of ncRNAs in transcriptional and posttranscriptional regulation and the development of human diseases are gradually being discovered.
As members of the ncRNA family, circRNAs, which were once believed to be redundant byproducts of gene splicing, are not fully understood because circRNAs have only been recognized in the past 3-5 years due to the extensive utilization of high-throughput RNA sequencing and microarray analyses 4. By undergoing a unique “back-splicing” process, circRNAs acquire a covalently closed loop without 5ʹ or 3ʹ polarities and a poly (A) tail configuration 5; these features represent the basic requirements for RNase resistance and inherent conservation 6. CircRNAs are predominantly localized in the cytoplasm 7 and are originated mainly from exons and partially from introns 8 and exon-introns 9. In addition to the canonical property of miRNA “sponges”, circRNAs also possess a series of biological functions, including miRNA “reservoirs”, gene expression regulation and protein coding, that allow circRNAs to be involved in the extra- or intracellular modulation and occurrence of different diseases 10. Similarly, many studies have suggested that circRNAs are more inclined to display tissue-dependent expression and are frequently expressed in several diseases 11, highlighting the essential correlation between circRNAs and certain diseases and the necessity to clarify the functions and mechanisms of circRNAs in these diseases.
The immune system is responsible for the surveillance and defense against the invasiveness of different exogenous pathogens and the maintenance of internal homeostasis by maintaining proper immune tolerance and regulation. In fact, immune surveillance and defense are the main immune responses. Based on the types of exogenous antigens, immune responses can exert antiviral, antibacterial and antitumor functions depending on the diverse immunocytes that synthetically construct the immune defense. Elements, such as the inappropriate exposure to self-antigens, dysregulation of immune responses and stimulation of cross antigens, may launch autoimmunity and promote the generation of certain immune diseases. Although most recent studies have concentrated on proteins, several studies have reported that ncRNAs, particularly lncRNAs and circRNAs, can also be novel candidates involved in the regulation of immune responses and immune diseases 12.
In this review, we summarize the identification, biogenesis and functions of circRNAs. We also elucidate the correlation and regulatory mechanisms of circRNAs in various immune responses and diseases. Finally, we discuss some of the limitations of current explorations and highlight the potential clinical value of circRNAs as novel diagnostic biomarkers and potential therapeutic strategies.
Identification, biogenesis and functions of circRNAs
Identification: circRNAs are novel gene-splicing products with high stability and inherent conservation
In 1979, Hsu and Coca-Prados published a report presenting the first conclusive evidence of the existence of circRNAs in the cytoplasm of eukaryotic cells by electron microscopy 13, opening a new era of the understanding of the abundance of RNA transcripts. However, unfortunately, studies investigating circRNAs stagnated for the following 20-30 years due to insufficient attention after the initial interest. CircRNAs were even considered byproducts of gene splicing for a long time. Due to the advent of transcriptome sequencing technology and improved computational approaches 14, circRNAs were sequentially discovered in a series of organisms, including humans 15, Drosophila 16, plants 17, Cryptococcus 18, Zebrafish 19 and protists 20. In addition to their broad range of expression, circRNAs appear to be more stable than linear RNAs in vivo because of their covalently closed loop structures; this stability results in resistance against RNase and RNA exonucleases 6. Moreover, circRNAs display high levels of conservation regardless of the evolutionary distance between species. For example, approximately 20% of murine circRNAs are orthologous to those in humans 21. Based on the above characteristics, an increasing number of studies have demonstrated that circRNAs are involved in multiple physical processes, such as synaptic development and neuronal differentiation 11. Taken together, these data suggest that circRNAs are novel, rather than simply nonfunctional, products of gene splicing and that they are highly stable and intrinsically conserved.
Biogenesis: circRNAs are formed through a special splicing pattern and are normally divided into three categories
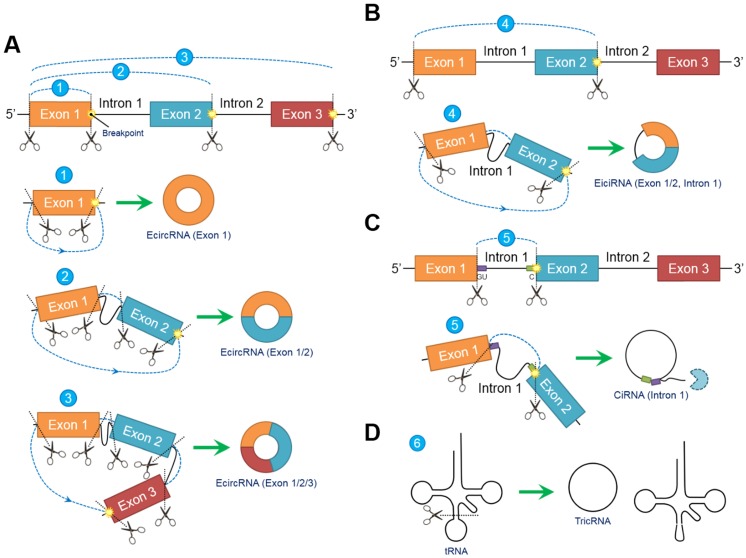
In contrast to conventional linear splicing forming a linear and exoteric 5ʹ to 3ʹ structure of joint exons, circRNAs contain a loop configuration that is covalently closed and terminally deficient 22. Both “back-splicing” and “exon-skipping” have been confirmed in vitro and in vivo, respectively, and are canonical splicing patterns of circRNAs 14, 23. Nevertheless, accumulating evidence suggests that “back-splicing” seems to be more important than “exon-skipping” because this pattern is more frequently observed 15. Briefly, following the appearance of a breakpoint, the ligation of the downstream 5ʹ splicing donor site to the upstream 3ʹ splicing acceptor site in a reverse confirmation is induced, followed by the alternative trimming of intron sequences and the final generation of circRNAs 24. Different breakpoint locations determine the diversity of the circRNA components, indicating that circRNAs can be circularized by exons, introns, intergenic regions, nontranscriptional areas or random combinations through “back-splicing” 25. In fact, circRNAs can be generally classified into the following three categories according to their inner elements: exonic circRNAs (ecircRNAs) 26, exon-intron circRNAs (eiciRNAs) 9 and circular intronic RNAs (ciRNAs) 27. EcircRNAs, which exclusively comprise one or more exons, account for most (over 80%) 6 identified circRNAs and selectively arise from “lariat-driven circularization” 28 or “intron-pairing-driven circularization” 23 (Figure 1A). EiciRNAs, which are circularized via “exon-skipping” events, harbor flanking intron sequences at the offside of core exons that should generally be spliced; therefore, EiciRNAs are referred to as eiciRNAs or retained-intron circRNAs 9 (Figure 1B). CiRNAs are also produced by a lariat-derived mechanism relying on a consensus GU-rich domain near the 5ʹ splice site of pre-mRNA and a C-rich domain near the breakpoint 8. Intriguingly, the GU-rich domain can protect the C-rich domain from branching or degrading, thus generating stable ciRNAs or so-called intron-derived circRNAs 29 (Figure 1C). Additionally, pre-tRNAs can be cleaved by the tRNA splicing endonuclease complex, and the spliced exons and introns are ligated to form tRNA intronic circular RNAs (tricRNAs) 30 (Figure 1D).
Figure 1.
Biogenesis models of circular RNAs (circRNAs). Due to the emergence of differentially located breakpoints, primary RNA transcripts undergo “back-splicing” to produce the 5ʹ splicing donor site and 3ʹ splicing acceptor site. Subsequently, the 5ʹ splicing donor site is combined with the 3ʹ splicing acceptor site in reverse order to form a covalently closed loop without 5ʹ or 3ʹ polarities and poly (A) tails. (A) Exonic circRNAs (ecircRNAs) are composed exclusively of exons without flanking introns. The number of exons ranges from one to two or more depending on the breakpoints. Over 80% of circRNAs arise from ecircRNAs. (B) Exons (at least one) accompanied by flanking introns that have not been degraded during “back-splicing” compose exon-intron circRNAs (eiciRNAs). (C) Intron-derived circRNAs (ciRNAs) are produced through a lariat-derived mechanism depending on a consensus GU-rich domain near the 5ʹ splicing site and a C-rich domain near the breakpoint. The remaining noncircularized introns are sequestered. (D) tRNA intronic circRNAs (tricRNAs) originate from the exons and introns of pre-tRNAs cleaved by the tRNA splicing endonuclease complex. Abbreviations: ciRNA: circular intronic RNA; ecircRNA: exonic circRNA; eiciRNA: exon-intron circRNA; tricRNA: tRNA intronic circular RNA.
Although the basic mechanisms of circRNA biogenesis have already been verified, some factors, including the sequence properties, protein regulators and transcriptional phases, may also influence the efficiency or products of “back-splicing”. 1) Flanking intronic elements are much longer, have more complementary matches or repeats and have been reported to promote circRNA circularization 16. Nevertheless, in some cases, short intron sequences (30- to 40-nt inverted repeats), especially Alu repetitive elements, can enhance the accessibility of two separated pre-mRNA splice sites in a base-pairing manner, ultimately facilitating circRNA formation 31. 2) Circularized exons are much longer than noncircularized exons created by linear splicing 6, and the exon length is positively related to the efficiency of circRNA biogenesis 31. For example, single-exon circRNAs contain exons that are on average 3-fold longer than those of linear RNAs 32. 3) Hyperedited RNAs possess unique and sufficient sequences for circularization 33. 4) Different splicing-associated proteins are critical regulators of cyclization. The RNA binding proteins (RBPs) muscleblind (MBL) and Quaking (QKI) bind and enhance the proximity of splicing sites, thus driving circularization 34, 35. However, adenosine deaminase acting on RNA 1 (ADAR1) is negatively correlated with circRNA production due to its high affinity to double-stranded RNAs (dsRNAs) and melting of the stem structure 33. Additionally, heterogeneous nuclear ribonucleoprotein L (HNRNPL), FUS RNA binding protein (FUS) and DExH-box helicase 9 (DHX9) can regulate circRNA formation 36-38. 5) There is no absolute distinction between circRNA generation and cotranscription or posttranscription. Recent studies have reported that the cotranscriptional circularization of circRNAs relies on repetitive intron sequences longer than 300-nt, but sequences with intron repeats shorter than 40-nt are likely to induce posttranscriptional “back-splicing” 39, indicating that circRNAs can arise from cotranscription or posttranscription.
Functions
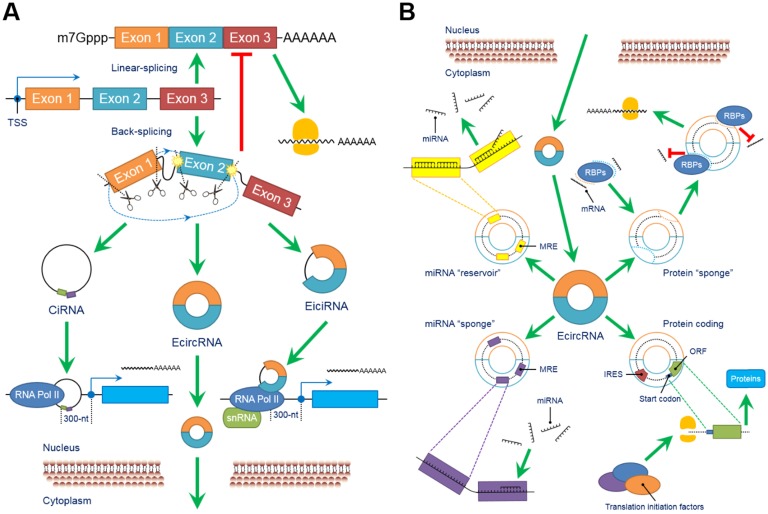
The biological functions of circRNAs, such as serving as miRNA “sponges” or miRNA “reservoirs”, regulating gene expression and encoding proteins, have been successively clarified in recent years (Figure 2).
Figure 2.
CircRNAs perform a series of biological functions. (A) Since primary RNA transcripts are generated by transcription, these immature products undergo “back-splicing”, which competitively suppresses linear splicing, producing circRNAs, including eiciRNAs, ciRNAs and ecircRNAs, at the cost of losing their linear equivalents. EiciRNAs and ciRNAs can conversely activate their parental genes by forming transcription complexes and binding the 300-nt region upstream of the transcription start site (TSS). Both eiciRNA and ciRNA transcription complexes require RNA polymerase II, but U1 snRNAs are also needed to complete the eiciRNA transcription complex. Notably, eiciRNAs and ciRNAs are primarily located in the nucleus, whereas ecircRNAs dominantly translocate to the cytoplasm to perform other tasks. (B) EcircRNAs contain a certain number of miRNA response elements (MREs) that create fundamental structures to interact with target miRNAs as specific miRNA “sponges”, which significantly interrupt the expression and behaviors of downstream miRNAs. In contrast, by using MREs, ecircRNAs can also store miRNAs to stabilize their expression and release under certain conditions, serving as miRNA “reservoirs”. Additionally, ecircRNAs harbor some internal ribosome entry sites (IRESs) to launch the cap-independent translation, open reading frames (ORFs) and start codons to initiate the “protein coding” in polymerases. Various translation initiation factors, such as N6-methyladenosine (m6A), eukaryotic initiation factor 3 (eIF3), eIF4G2, YTH N6-methyladenosine RNA binding protein 3 (YTHDF3) and methyltransferase like 14 (METTL14), are involved in this process. Moreover, many ecircRNAs can combine with RNA binding proteins (RBPs) via complimentary sequences, blocking the binding between RBPs and mRNAs/miRNAs as protein “sponges” and cooperatively regulating the procedures of translation. In conclusion, circRNAs broadly participate as critical links in intracellular events and form an intricate regulatory network. Abbreviations: ciRNA: circular intronic RNA; ecircRNA: exonic circRNA; eiciRNA: exon-intron circRNA; IRES: internal ribosome entry site; miRNA: microRNA; MRE: miRNA response element; ORF: open reading frame; RBP: RNA binding protein; snRNA: small nuclear RNA.
1. CircRNAs suppress miRNA expression by functioning as specific miRNA “sponges”
MiRNAs are large clusters of small ncRNAs that negatively regulate the level of target mRNAs by directly binding their 3ʹ-untranslated regions (3ʹ-UTRs) and interfering with the translation process 40. The complementary sequences of miRNAs in the 3ʹ-UTRs of target mRNAs are commonly referred to as miRNA response elements (MREs) 41. Similarly, circRNAs, which are mainly located in the cytoplasm, contain different MREs that can interact with target miRNAs, serving as competing endogenous RNAs (ceRNAs) 42. In addition, without the 5ʹ or 3ʹ terminal ends, circRNAs are free from RNase digestion, facilitating their functions as miRNA “sponges” 43.
Despite abundant unknowns regarding the functions and mechanisms of most discovered circRNAs, the miRNA “sponge” property has already been mentioned in several studies. CDR1as (also known as ciRS-7) has been previously identified to sequester and impair the functions of miR-7 by possessing more than 70 conserved MREs and collaborating with the argonaute (AGO) protein to influence miR-7 actions in the central nervous system (CNS) 44. Sex determining region Y (Sry)-derived circRNA harboring 16 putative miR-138 interacting sites significantly diminishes the presence of miR-138, followed by an increase in the level of miR-138 downstream genes 45. The ubiquitination effects of itchy E3 ubiquitin protein ligase (ITCH) are amplified following the overexpression of circ-ITCH, which sustainably functions as a “sponge” of miR-7, miR-17 and miR-214 46. Circ-TCF25, i.e., the “sponge” of miR-103 and miR-107, is associated with some malignant phenotypes of bladder cancer cells and promotes the expression of cyclin-dependent kinase 6 (CDK6) 47. A considerable number of identified circRNAs have been confirmed or predicted to be “sponges” of specific miRNAs 48-50. Furthermore, numerous targets of these miRNAs also participate in the modulation of the immune system, highlighting the potential roles of circRNAs in immune responses and immune diseases.
However, studies and bioinformatic analyses indicate that certain circRNAs with various MREs are actually in the minority. Many circRNAs only possess a few or even no miRNA target sites 21, 35. In addition, accumulating evidence has clarified that the lack of MRE polymorphisms does not prevent the sequestering of target miRNAs 10. Moreover, the amount of specific circRNAs has been restricted, but the expression of their target miRNAs is not altered or even decreased in the presence of some potential negative feedback loops, significantly limiting circRNA functioning as miRNA “sponges” 51. Therefore, the hypothesis that circRNAs induce miRNA suppression by behaving as specific miRNA “sponges” remains controversial.
2. CircRNAs act as designated miRNA “reservoirs” to stabilize miRNA functions
In addition to serving as specific inhibitors of target miRNAs by functioning as miRNA “sponges”, circRNAs harbor the opposite ability to stabilize or activate the functions of miRNAs, which is so-called miRNA “reservoirs”. The canonical pattern of miRNA “reservoirs” was discovered and identified in the regulatory network of ciRS-7, miR-7 and miR-671. Intriguingly, the miR-671-dependent Ago2-involved cleavage results in the turnover of ciRS-7, leading to the disconnection between ciRS-7 and miR-7 and finally the release of miR-7. In this scenario, miR-671 could play as an activator that liberates miR-7, but ciRS-7 accumulates and stores miR-7 as a putative miR-7 “reservoir”, resulting in an immediate elevation in miR-7 activity and a substantial repression of miR-7 targets 52. Similarly, a recent study reported that circ-HIAT1, originating from its parental gene Hippocampus Abundant Transcript 1 (HIAT1), protects three of its targets, i.e., miR-195-5p, miR-29a-3p and miR-29c-3p, from the inhibition of the androgen receptor (AR) via a binding interaction, thereby functioning as an miRNA “reservoir” to stabilize the expression of target miRNAs and suppress the downstream CDC42 level 53. Although current explorations of miRNA “reservoirs” are limited and more studies are still needed to comprehensively understand the mechanisms of circRNA-miRNA networks 54, this property, which differs from the miRNA “sponge” property, enriches the diversity of the regulation of the interaction between circRNAs and miRNAs and proposes potentially novel strategies for disease therapy.
3. CircRNAs are involved in the modulation of gene expression
Universally, due to their intracellular location and special structure, circRNAs can regulate the expression of their parental or downstream genes by cooperating with RNA polymerases, competing with linear splicing and sequestering proteins as “sponges”, ultimately producing alterations in gene expression during the transcription, posttranscription and translation phases.
3.1. CircRNAs combine with RNA polymerases to organize the transcription complex
The intracellular distribution of ciRNAs and eiciRNAs is primarily enriched in the nucleus, suggesting that both ciRNAs and eiciRNAs are likely to function at the transcriptional level 45. The mRNA levels of sirtuin 7 (SIRT7), minichromosome maintenance complex component 5 (MCM5) and ankyrin repeat domain 52 (ANKRD52) are notably decreased following the depletion of their corresponding ciRNAs, i.e., ci-sirt7, ci-mcm5 and ci-ankrd52, respectively 8. In fact, the abovementioned ciRNAs play critical roles in transcription by collaborating with RNA polymerase II and initiating the transcription of their parental genes 55. Similarly, eukaryotic translation initiation factor 3 subunit J (EIF3J) and poly (A) binding protein interacting protein 2 (PAIP2) are positively correlated with their corresponding eiciRNAs, i.e., eici-eif3j and eici-paip2, respectively, because the transcriptional processes of these two parental genes is triggered by an interaction among their eiciRNAs, the U1 small nuclear RNA (snRNA) (U1A and U1C) and RNA polymerase II to establish an activating complex and their binding the promoter region 300-nt upstream of the transcription start site (TSS) 56.
3.2. “Back-splicing” competitively inhibits the classical procedure of linear splicing
In contrast to their activator roles of elevating gene expression, circRNAs switch their characteristics to inhibit the parental genes via a unique “back-splicing” pattern. First, after “back-splicing” successfully generates circRNAs, the remaining primary RNA transcripts are forced to undergo alternative splicing (AS) 57. Then, “back-splicing” also serves as an endogenous competitor of canonical linear splicing because of the common need for the spliceosome machinery 58. Furthermore, several circRNAs, such as circ-FMN, circ-DMD and circ-HIPK2/3, are able to disguise translation start codons, preventing linear coding transcripts from being translated 14, 59, 60. Thus, protein-coding mRNAs are confronted with a “survival crisis” caused by circRNAs derived from the same pre-mRNAs and appear to be suppressed. Therefore, circRNA synthesis incurs the cost of losing the linear equivalents of the circRNAs 61.
3.3. CircRNAs are special forms of protein “sponges”
Similar to the miRNA “sponge” property, circRNAs can utilize specific regions to interact with some proteins, acting as protein “sponges”. RNA binding proteins (RBPs) widely affect the cyclic parts of RNA transcripts, such as mRNAs and miRNAs, from creation to degradation. Particularly, RBPs harbor complementary sequences to bind their RNA targets, allowing circRNAs to combine with the structures. Thus, similar to the relationship among poly (A) binding protein nuclear 1 (PABPN1), circ-PABPN1 and the RBP Human antigen R (HuR), the translation of target mRNAs is partially or completely blocked 62. In addition, recent studies have demonstrated that circ-Amotl1 can determine the subcellular translocation of several RBPs, such as PDK1, MYC, and STAT3 63, and that circ-Foxo3 functions as a scaffold to assemble the functional proteins P21 and CDK1, forming a large complex that regulates translation 64, significantly influencing target gene expression and enriching the methods by which circRNAs regulate RBPs.
4. CircRNAs have the ability to encode proteins
In accordance with conventional opinions, the 5ʹ-cap and 3ʹ-poly (A) tail are important elements that initiate and drive translation. Nevertheless, circRNAs form a stable loop without 5ʹ and 3ʹ ends, which is likely the key to their noncoding property. Intriguingly, internal ribosome entry sites (IRESs) have been discovered in some circRNAs 65. IRESs are symbols of cap-independent translation 66, and in vitro assays have revealed that circRNAs may acquire translation abilities once the IRESs are inserted into their loops 67. In addition, several circRNAs arising from exons or exon-introns may possess open reading frames (ORFs), thereby conferring translation potential. Circ-ZNF609 can be translated into a protein in a splicing-dependent and cap-independent manner mainly depending on its two start codons in the 735-nt ORF, which are closely correlated with polysomes 68. Furthermore, certain translation initiation factors, such as N6-methyladenosine (m6A), eukaryotic initiation factor 3 (eIF3), eIF4G2, YTH N6-methyladenosine RNA binding protein 3 (YTHDF3) and methyltransferase-like 14 (METTL14) 69, are able to trigger circRNA translation. Although several circRNAs have successively been proven to encode proteins 70-72, several studies question this result. Ribosome footprint detection shows no evidence of the presence of translatable circRNAs in osteosarcoma 21. Thus, whether and how circRNAs can encode proteins require further and deeper exploration.
Functions and mechanisms of circRNAs in immune responses
Immune responses are protective reactions of the immune system against exogenous invasiveness to maintain internal homeostasis. Depending on different immunocytes, the immune system can induce appropriate responses to resist viruses, bacteria and tumors. Notably, circRNAs have recently been considered participants in the regulation of immunocytes and various immune responses (Table 1).
Table 1.
CircRNAs are closely associated with different types of immune responses.
| Immune Responses | Subjects | CircRNAs | Species | Tissues | Sponge Targets |
Potential Functions / Biogenesis | References |
|---|---|---|---|---|---|---|---|
| Immunocytes | Macrophages | Hsa_circ_003780 | Murine | Bone Marrow | - | Contributes to the process of macrophages' polarization |
Zhang et al (2017) 74 |
| Hsa_circ_010056 | - | ||||||
| Hsa_circ_010231 | - | ||||||
| Hsa_circ_003424 | - | ||||||
| Hsa_circ_013630 | - | ||||||
| Hsa_circ_001489 | - | ||||||
| Hsa_circ_018127 | - | ||||||
| Circ-RasGEF1B | Murine | Cell Line | - | Elevates expression of ICAM-1; Maintains appropriate activation of macrophages when exposed to LPS |
Ng et al (2016) 75 Ng et al (2017) 76 |
||
| Circ-ANRIL | Homo Sapiens | Endarterium | - | Combines with PES1 to impede the generation of pre-rRNA and ribosome; Inhibits biogenesis of macrophages by TP53 |
Holdt et al (2016) 77 | ||
| Circ-ZC3H4 | Homo Sapiens | Alveolar | - | Participates in the SiO2-induced stimulation of pulmonary macrophages |
Yang et al (2018) 78 | ||
| Murine | Cell Line | ||||||
| Neutrophils | Circ-TLR6 | Homo Sapiens | Blood | - | Influences functions of parental genes | Maass et al (2017) 79 | |
| Circ-MYO1F | - | ||||||
| CD8+T cells | Hsa_circ_100783 | Homo Sapiens | PBMC | - | Induces loss of CD28 during aging of CD8+T cells |
Wang et al (2015) 80 | |
| Anti-Virus | Virus | Circ-RNP | Homo Sapiens | Cell Line | - | Can be formed via nuclear export of NF90/NF110 during virus infection |
Li et al (2017) 86 |
| Ebola Virus | Circ-chr19 | Homo Sapiens | Cell Line | miR-30b | Serves as miR-30b “sponge” to enhance expression of CLDN18 |
Wang et al (2017) 88 | |
| Anti-Bacteria | TB | Hsa_circ_001937 | Homo Sapiens | PBMC | - | Modulates the NF-κB signaling; Represents grades and stages of TB |
Qian et al (2018) 81 Huang et al (2018) 83 |
| Anti-Tumor | CRC | Hsa_circ_0020397 | Homo Sapiens | Cell Line | miR-138 | Serves as miR-138 “sponge” to promote expression of TERT and PD-L1 |
Zhang et al (2017) 90 |
| Circ-FAT1 | Homo Sapiens | Cell Line | - | Can be transferred to exosomes; Serves as a promising cancer biomarker |
Dou et al (2016) 95 |
Abbreviations: CLDN18: claudin 18; CRC: colorectal cancer; ICAM-1: intercellular adhesion molecule 1; LPS: lipopolysaccharide; PBMC: peripheral blood mononuclear cell; PES1: pescadillo ribosomal biogenesis factor 1; TB: tuberculosis; TERT: telomerase reverse transcriptase.
CircRNAs and immunocytes
Immunocytes are the core components of immune responses that implement immune surveillance, defense and regulation. Current studies investigating circRNAs in immunocytes predominantly focus on macrophages, which play critical and indispensable roles in immune responses.
According to their phenotypes and functions in response to different immune microenvironments, macrophages can be authoritatively classified into the following two groups: canonically activated macrophages (M1) and alternatively activated macrophages (M2) 73. Zhang et al reported that 189 circRNAs were differentially expressed between M1 and M2 macrophages, strongly indicating that circRNAs are indeed acquired during macrophage polarization 74. Additionally, circRNAs may influence the antigen presentation process of macrophages. For instance, circ-RasGEF1B positively regulates the expression of intercellular adhesion molecule 1 (ICAM-1), which is an adhesion protein that anchors on the surface of the cell membranes of macrophages and promotes cell-cell interactions during antigen presentation by modulating the stability of mature ICAM-1 mRNA 75. Moreover, circ-RasGEF1B is necessary for the proper activation of macrophages exposed to lipopolysaccharide (LPS) 76. Holdt et al confirmed that circ-ANRIL could associate with pescadillo ribosomal biogenesis factor 1 (PES1) to impede exonuclease-mediated pre-rRNA generation and ribosome formation, thus elevating apoptosis and suppressing proliferation in macrophages by inducing the expression of Tp53 77. Additionally, circ-ZC3H4 participates in the SiO2-induced stimulation of pulmonary macrophages 78. Therefore, circRNAs broadly affect macrophages from development to activity.
In addition to macrophages, circRNAs can also contribute to the biogenesis and behaviors of other immunocytes. Maass et al reported that toll-like receptor 6 (TLR6) and myosin IF (MYO1F), which are two key genes allowing neutrophils to complete innate immune responses, possess a circRNA that probably influences the functions of its parental gene 79. Wang et al exhibited that hsa_circ_100783 induced the loss of CD28 in CD8+T cells during their aging by affecting phosphoprotein-related signaling transduction 80. In addition, some studies have reported that circRNAs are closely associated with the diverse signaling transduction pathways in peripheral blood mononuclear cells (PBMCs) 81-83. However, explorations of circRNAs in immunocytes remain preliminary, and there are still many unknowns.
CircRNAs and antiviral immune responses
Innate immunity is the body's first line of defense against viral infection. RNA viruses initiate various signaling pathways to fulfill infection by viral nucleic acids. The 5ʹ-triphosphate (5ʹ-ppp) and double-stranded segments of specific viral RNAs can be recognized and bound by several dsRNA-binding proteins, including retinoic acid-inducible gene-I (RIG-I), laboratory of genetics and physiology 2 (LGP2), melanoma differentiation-associated gene 5 (MDA5) and toll-like receptor 3 (TLR3) 84, leading to the interruption of viral gene activation. In contrast, the antiviral immune responses triggered through exogenous circRNAs are independent of the 5ʹ-ppp and double-stranded structure. Recent studies have shown that endogenous circRNAs can be distinguished from exogenous circRNAs through biosynthesis. Self-circRNAs are frequently programmed by introns and are combined with RBPs that mark their origins. Conversely, few proteins, such as HDV and Viroids, bind exogenous circRNAs. Hence, nonself circRNAs are differentiated, and antiviral immune responses can be triggered by the RIG-I-mediated pathway, which provides an essential antiviral signal 85. However, the mechanisms by which RIG-I recognizes exogenous circRNAs require further investigation. Intriguingly, antiviral immune responses affect circRNAs. Li et al verified that immune responses and virus infection influence the biogenesis of host circRNAs. NF90/NF110 promotes circ-RNP formation in the nucleus, while virus infections inhibit circ-RNP biogenesis by inducing the nuclear export of NF90/NF110. Notably, a competitive binding relationship exists between host circRNAs and viral mRNAs 86, indicating that circRNAs are likely to resist viral infection. Additionally, circRNAs play a putative role in controlling multiple signaling pathways, such as the Toll-like receptor, Tp53 and Wnt pathways, in the cellular response to SV40 (a DNA virus) infection by miRNA “sponges” 87. Wang et al verified that circ-chr19 enhances claudin-18 (CLDN18) expression by targeting miR-30b-3p, thus functioning as a ceRNA in Ebola virus infection 88. Considering that CLDN18 influences cell permeability, which is the basis of Ebola pathogenesis, circ-chr19 may serve as a potential biotarget for Ebola virus treatment. Furthermore, alterations in circRNAs have been confirmed to participate in immune resistance to Avian Leukosis virus infection in chicken 89, expanding our knowledge of how circRNAs modulate antiviral immune responses.
CircRNAs and antibacterial immune responses
In addition to antiviral immune responses, circRNAs are associated with antibacterial immune responses. Mycobacterium tuberculosis is known as the root cause of tuberculosis (TB), and many circRNAs are dysregulated in patients with TB 82. CircRNAs in PBMCs from TB patients are significantly upregulated in the cytokine-cytokine receptor and chemokine-mediated signaling pathways 81. Notably, hsa_circ_001937 is markedly increased in PBMCs from TB specimens compared with that in specimens from patients who are healthy or have other respiratory diseases, such as chronic obstructive pulmonary disease (COPD), lung carcinogenesis and pneumonia. In addition, the potential miRNA targets of hsa_circ_001937 participate in antibacterial immune responses by modulating the NF-κB signaling pathway. Particularly, the level of hsa_circ_001937 can represent the grades and stages of TB in patients 83. Thus, hsa_circ_001937 possesses some features of the ideal biomarker for TB diagnosis.
CircRNAs and antitumor immune responses
The close relationship between circRNAs and miRNAs lays the foundation for circRNAs to participate in the regulation of antitumor immune responses. As previously reported, hsa_circ_0020397 induces the expression of telomerase reverse transcriptase (TERT) and PD-L1 by binding and inhibiting the activity of miR-138 in colorectal cancer (CRC) cells 90. In fact, the interaction between PD-1 and PD-L1 on specific tumor cell surfaces exhausts immunocytes and promotes immune escape. Circ-Amotl1 can indirectly restrain the presence of miR-17-5p by upregulating Dnmt3a, which may induce a highly DNA methylation enriched condition in the promoter region of miR-17, leading to the elevated expression and subcellular translocation of STAT3, launching tumor-mediated immune inhibition 63, 91. These studies suggest that circRNAs can modulate the expression and functions of immune-related miRNAs at the transcriptional or posttranscriptional level in immune responses against tumors. Recently, studies have increasingly clarified the functions and mechanisms of miRNAs in antitumor immune responses. Further explorations focusing on circRNA-miRNA interactions are likely to provide new insight into tumor immunotherapy.
In addition to the circRNA-miRNA network, circRNAs are also able to influence antitumor immune responses in other manners. First, by affecting the stability of some proteins, circRNAs indirectly participate in antitumor immune responses; for example, the complex of circ-Foxo3 and MDM2 induces the degradation of Tp53, which is obligatory for immune responses during tumorigenesis 7, 92. Second, circRNAs may function as potential tumor antigens that can be transported via exosomes and extracellular vesicles to regulate cell-to-cell communication between immunocytes and tumor cells 93, 94. For instance, Dou et al reported that KRAS mutant CRC cells can transfer several circRNAs into exosomes 95, and Lasda et al observed that circRNAs may be coprecipitated with extracellular vesicles 96. Third, via complementary sequences, circRNAs can bind some tumor-specific miRNAs and mRNAs to form a novel type of tumor antigen that coexists with these RNAs in exosomes to enhance their stability and release once arriving at the destined immunocytes 97. Fourth, some exogenous circRNAs may activate immunocytes, promoting their functions in the fight against tumors. A recent study has shown that purified circRNAs can stimulate the expression of RIG-1 to trigger innate immune responses in the tumor microenvironment 92, 98. Taken together, this evidence indicates that circRNAs are widely involved and play various roles in the modulation of antitumor immune responses.
Collectively, circRNAs participate in all key processes of host immunity by monitoring the balance of inner homeostasis and exerting efficient protection against microbial infection and malignant tumors. Based on these data, circRNAs can be considered biomarkers or therapy targets. However, the underlying mechanisms of circRNAs in this field remain largely unknown and should be further explored and elucidated.
Involvement and biological effects of circRNAs in immune diseases
Due to the rapid advances in high-throughput sequencing and microarray technologies, many circRNAs have been successively identified. In addition, circRNAs have been reported to participate in the pathogenesis and diagnosis of different immune diseases, such as primary immunodeficiency diseases (PIDs), autoimmune diseases (AIDs) and other immune diseases. Performing bioinformatics analyses using databases, such as circRNA and the disease-related database circRNA Disease (http://cgga.org.cn:9091/circRNADisease/) 99, can also provide clues explaining and predicting the potential correlation between circRNAs and certain immune diseases. However, the detailed functions of the specific circRNAs in these diseases have not been elucidated to date (Table 2).
Table 2.
CircRNAs are widely involved in different immune diseases.
| Types of Diseases | Diseases | CircRNAs | Species | Tissues | Sponge Targets | Expression Pattern |
Potential Functions / Applications | References |
|---|---|---|---|---|---|---|---|---|
| PIDs | SCID | Circ-CDC42BPA | Homo Sapiens | Bone Marrow |
- | Upregulated | Disrupts transduction of B cell signaling to induce occurrence of SCID |
Maass et al (2017) 79 Brigida et al (2014) 103 |
| Circ-TNFRSF11A | Homo Sapiens | Blood | - | Upregulated | Participates in the SCID mediated alteration of different signaling pathways | Maass et al (2017) 79 Cassani et al (2008) 105 |
||
| WAS | Circ-ROBO1 | Homo Sapiens | Cell Line | - | Upregulated | Activates the pathogenesis of WAS | Maass et al (2017) 79 Sheldon et al (2009) 102 |
|
| Circ-CDC42BPA | Homo Sapiens | Blood | - | Upregulated | Disrupts transduction of B cell signaling to induce occurrence of WAS |
Maass et al (2017) 79 | ||
| AIDs | SLE | CDR1as / ciRS-7 | Homo Sapiens | Blood | miR-7 | Downregulated | Serves as the miR-7 “sponge” to increase expression of PTEN and restrain hyper-responsiveness of B cells |
Wu et al (2014) 109 Zhao et al (2016) 110 |
| Hsa_circ_102584 | Homo Sapiens | Plasma | - | Upregulated | Can be developed as novel non-invasive biomarkers for SLE |
Li et al (2018) 111 | ||
| Hsa_circ_400011 | Homo Sapiens | Plasma | - | Upregulated | ||||
| Hsa_circ_101471 | Homo Sapiens | Plasma | - | Upregulated | ||||
| Hsa_circ_100226 | Homo Sapiens | Plasma | - | Downregulated | ||||
| RA | Hsa_circ_104871 | Homo Sapiens | PBMC | - | Upregulated | Serves as potential biomarkers for diagnosis other than representing severity or pathological process of RA |
Ouyang et al (2017) 112 | |
| Hsa_circ_003524 | Homo Sapiens | PBMC | - | Upregulated | ||||
| Hsa_circ_101873 | Homo Sapiens | PBMC | - | Upregulated | ||||
| Hsa_circ_103047 | Homo Sapiens | PBMC | - | Upregulated | ||||
| Hsa_circ_0057980 | Homo Sapiens | Plasma | miR-181d | Downregulated | Serves as the miR-181d “sponge” to suppress the development of RA |
Wang et al (2015) 113 Zheng et al (2017) 116 |
||
| Hsa_circ_0088088 | Homo Sapiens | Synovium | miR-16 | Downregulated | Serves as the miR-16 “sponge” to suppress the development of RA |
Murata et al (2015) 114 Zheng et al (2017) 116 |
||
| Hsa_circ_0001045 | Homo Sapiens | Synovium | miR-30a | Upregulated | Serves as the miR-30a “sponge” to promote the biogenesis of RA |
Xu et al (2013) 115 Zheng et al (2017) 116 |
||
| MS | GSDMB ecircRNA | Homo Sapiens | PBMC | miR-1275 miR-149 |
Upregulated | Serves as the miR-1275 and miR-149 “sponges” to induce MS |
Paraboschi et al (2014) 118 | |
| Hsa_circ_0005402 | Homo Sapiens | PBMC | - | Downregulated | Both are derived from the ANXA2; Can be developed as MS biomarkers; Negatively regulate the biogenesis of MS |
Iparraguirre et al (2017) 121 | ||
| Hsa_circ_0035560 | Homo Sapiens | PBMC | - | Downregulated | ||||
| PBC | Hsa_circ_402458 | Homo Sapiens | Cell Line | miR-522 miR-943 |
Upregulated | Can be suitable for PBC diagnosis; Serves as the miR-522 and miR-943 “sponges” to resist chronic inflammation and aberrant TGF-β signaling of PBC |
Zheng et al (2017) 123 Kang et al (2017) 124 Danza et al (2017) 125 |
|
| Immuno-compromised Diseases | OA | Circ-Atp9b | Murine | Articular Chondrocytes | miR-138 | Upregulated | Serves as the miR-138 “sponge” to promote extracellular matrix degradation | Zhou et al (2018) 129 |
| Hsa_circ_0005105 | Homo Sapiens | Articular Cartilage | miR-26a | Upregulated | Serves as the miR-26a “sponge” to promote extracellular matrix degradation | Wu et al (2017) 130 | ||
| Hsa_circ_0045714 | Homo Sapiens | Articular Cartilage | miR-193b | Downregulated | Elevates the proliferation of cartilage cells; Serve as miR-193b “sponge” to increase the level of type II collagen and aggrecan |
Li et al (2017) 132 | ||
| IVDD | Circ-VMA21 | Homo Sapiens; Murine |
Nucleus Pulposus | miR-200c | Downregulated | Alleviates apoptosis of nuclei pulposus cells and catabolism of extracellular matrix through a miR-200c-XIAP pathway | Cheng et al (2018) 133 | |
| Silicosis | Circ-ZC3H4 | Homo Sapiens | Brain | miR-212 | Upregulated | Serves as the miR-212 “sponge” to promote proliferation and migration of alveolar macrophages as well as fibroblasts | Yang et al (2018) 78 | |
| Hepatic Fibrosis | Hsa_circ_0071410 | Homo Sapiens | Hepatic Stellate Cells | miR-9 | Upregulated | Suppresses the miR-9 to activate HSCs | Chen et al (2017) 135 | |
| Myocardial Fibrosis | Hsa_circ_010567 | Murine | Cardiac Fibroblasts | miR-141 | Upregulated | Suppresses the miR-141 by initiating TGF-β1 signaling |
Zhou et al (2017) 48 | |
| Neuro- inflammation |
Circ-HIPK2 | Murine | Astrocytes | miR-124-2HG | Upregulated | Serves as the miR-124-2HG “sponge” to activate astrocytes |
Huang et al (2017) 138 | |
| MMD | Hsa_circ_062557 | Homo Sapiens | Blood | - | Upregulated | Can be developed as potential biomarkers for MMD |
Zhao et al (2017) 139 | |
| Hsa_circ_067130 | Homo Sapiens | Blood | - | Upregulated | ||||
| Hsa_circ_067209 | Homo Sapiens | Blood | - | Upregulated | ||||
| Hsa_circ_100914 | Homo Sapiens | Blood | - | Downregulated | ||||
| Hsa_circ_089761 | Homo Sapiens | Blood | - | Downregulated | ||||
| Hsa_circ_089763 | Homo Sapiens | Blood | - | Downregulated |
Abbreviations: AIDs: autoimmune diseases; ANXA2: annexin A2; HSCs: hepatic stellate cells; IVDD: intervertebral disc degeneration; MMD: moyamoya; MS: multiple sclerosis; OA: osteoarthritis; PBC: primary biliary cirrhosis; PBMC: peripheral blood mononuclear cell; PIDs: primary immunodeficiency diseases; PTEN: phosphatase and tensin homolog; RA: rheumatoid arthritis; SCID: severe combined immunodeficiency disease; SLE: systemic lupus erythematosus; WAS: Wiskott-Aldrich syndrome; XIAP: X-linked inhibitor of apoptosis.
CircRNAs and primary immunodeficiency diseases
Primary immunodeficiency diseases (PIDs) are agnogenic diseases that partially or completely impair the differentiation and functions of immunocytes, tissues and organs, leading to disorders or deficiencies in the entire immune system. Severe combined immunodeficiency disease (SCID) and Wiskott-Aldrich syndrome (WAS) are two representative PIDs that involve mutated adenosine deaminase (ADA) and WAS genes, respectively 100, 101. Maass et al compared SCID and WAS patient specimens with their corresponding controls and identified that the levels of circRNAs derived from roundabout guidance receptor 1 (ROBO1) and CDC42 binding protein kinase alpha (CDC42BPA) are significantly increased in both patient groups 79. The ROBO family members can activate the pathogenesis of WAS 102, and CDC42BPA disrupts the transduction of B-cell signaling, thus inducing WAS and SCID 103, 104. In addition, TNF receptor superfamily member 11a (TNFRSF11A)-produced circRNA is notably upregulated in SCID patients 79. According to the experimental data reported by Cassani et al, the receptors of the TNF superfamily are closely correlated with the SCID-mediated alteration in different signaling pathways 105. These results provide a novel approach to studying the pathogenesis and therapeutics of PIDs.
CircRNAs and autoimmune diseases
Autoimmune diseases (AIDs) mainly manifest as local or systemic symptoms of inflammation caused by aberrant immune responses to self-antigens. Here, we list some typical AIDs, such as systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), multiple sclerosis (MS) and primary biliary cirrhosis (PBC), to further elucidate the regulatory roles of circRNAs associated with AIDs.
1. CircRNAs and systemic lupus erythematosus
Systemic lupus erythematosus (SLE), which predominantly affects young females, is implicated in connective tissues in multiple systems and organs 106. SLE is characterized by the overproduction of autoantibodies against specific self-antigens, excessive depositions of immune complexes, complement exhaustion and frequent inflammatory reoccurrence 107 and induces damage ranging from local minimal lesions to serious injuries in major organs in the urinary, circulatory, respiratory and central nervous systems 108. Based on clinical experience and laboratory criteria, the diagnosis and therapy of SLE patients remain limited. Due to their ability to serve as miRNA “sponges”, circRNAs probably possess novel strategies that could be used to combat this dilemma. MiR-7 restricts the functions of PTEN and contributes to B-cell hyperresponsiveness in SLE 109, whereas the overexpression of CDR1as leads to a reduction in miR-7 and promotion of its target mRNAs 110. In addition, through high-throughput RNA-sequencing, 207 circRNAs, including 113 upregulated and 94 downregulated circRNAs, were discovered in SLE patients compared with normal plasma specimens. Additionally, most miRNAs interacting with the altered circRNAs were determined to regulate SLE in this study. Furthermore, four markedly dysregulated circRNAs in SLE patients, i.e., hsa_circ_102584, hsa_circ_400011, hsa_circ_101471 and hsa_circ_100226, are predicted to act as novel noninvasive biomarkers of SLE 111. Further investigations are required to identify the functions and mechanisms of these circRNAs and their target miRNAs in the biogenesis of SLE.
2. CircRNAs and rheumatoid arthritis
Rheumatoid arthritis (RA) is a chronic autoimmune syndrome that progressively impairs periphery joints and leads to irreversible disability or even death. PBMCs are closely related to the development of RA. Ouyang et al discovered 9 elevated and 3 repressed circRNAs in RA-associated PBMCs compared with healthy controls and found that hsa_circ_104871, hsa_circ_003524, hsa_circ_101873 and hsa_circ_103047 in PBMCs have diagnostic value in RA. However, the alterations in the levels of these PBMC-related circRNAs are not parallel to the changes observed in the erythrocyte sedimentation rate (ESR), C-reactive protein (CRP) levels, health assessment questionnaire (HAQ) scores and disease activity score (DAS28), suggesting that these circRNAs cannot represent the severity or pathological processes of RA 112. Similarly, Zheng et al found 584 differentially expressed circRNAs, including 255 overexpressed and 329 knocked-down circRNAs, between RA patients and healthy controls. In particular, these authors also proposed that different identified circRNAs may act as potential suppressors or inducers during the occurrence of RA. The markedly enhanced expression of the RA promoter miR-181d 113 is partially due to the repression of hsa_circ_0057980, which is a miR-181d “sponge”. The altered expression of hsa_circ_0088088 is related to a high level of miR-16, which is an activator of ESR and CRP 114. In contrast, hsa_circ_0001045 was increased in samples of RA synovialis, which is directly associated with and seriously inhibited miR-30a, thereby reducing cell apoptosis and contributing to RA progression 115. Intriguingly, the corresponding parental genes of the significantly changed circRNAs in the RA patients, such as polynucleotide kinase 3'-phosphatase (PNKP), ArfGAP with GTPase domain, hydroxysteroid dehydrogenase like 2 (HSDL2), ankyrin repeat and PH domain 1 (AGAP1) and protein kinase C beta (PRKCB), also participate in the regulation of RA 116.
3. CircRNAs and multiple sclerosis
Multiple sclerosis (MS) is a known CNS disease that frequently relapses with inflammatory demyelination in white matter and infiltration by immunocytes, such as macrophages and B and T lymphocytes 117. As previously reported, aberrant AS isoforms, particularly the AS of gasdermin B (GSDMB), are correlated with the pathogenesis of MS 118. Exons 4 and 5 of GSDMB are subjected to “back-splicing” to form a head-to-tail splicing junction and generate a circular product named GSDMB ecircRNA, which is a newly described RNA species in MS. The potential targets of this GSDMB ecircRNA, i.e., miR-1275 and miR-149, have been found to be significantly deregulated in MS 119. Furthermore, increasing the level of GSDMB ecircRNA resulted in a 2.8-fold elevation in PBMCs from MS patients compared with the controls 120. Additionally, RNA profiling has been used to identify 406 differentially expressed circRNAs in peripheral blood leucocytes from patients with MS and demonstrates that hsa_circ_0005402, which is associated with hsa_circ_0035560, both of which arise from annexin A2 (ANXA2), is a negative regulator of MS 121, further supporting that ectopic RNA metabolism is an intrinsic characteristic feature of MS.
4. CircRNAs and primary biliary cirrhosis
Primary biliary cirrhosis (PBC), which is also called primary biliary cholangitis, primarily destroys the small intrahepatic bile ducts, leading to hepatic fibrosis or cirrhosis 122. In general, patients with PBC are always in a severe disease state at diagnosed due to their typically asymptomatic conditions during the early stages. Hence, the identification of sensitive biomarkers for the early diagnosis of PBC is urgently needed. Zheng et al performed a circRNA microarray analysis of plasma from patients with PBC and healthy individuals and identified 18 upregulated and 4 downregulated circRNAs that were possibly associated with PBC. Subsequently, among the 18 elevated circRNAs, hsa_circ_402458 was demonstrated to be a biomarker of PBC 123. Two miRNAs, i.e., miR-522 and miR-943, are putative biotargets of hsa_circ_402458. Studies have validated that miR-522 124 plays a critical role in chronic inflammatory disorder and that miR-943 125 affects TGF-β signaling to participate in PBC development. Hsa_circ_402458 may competitively suppress the activity of miR-522 and miR-943 and is involved in the regulation of PBC. In fact, due to its properties of high stability and inherent conservation 126, hsa_circ_402458 is suitable for early PBC diagnosis by using patient exosomes and blood plasma.
CircRNAs and other immune diseases
The loss of inner homeostasis and accumulation of disorders in immune responses can cause different immune diseases. Recent studies have elucidated that circRNAs are critical regulatory factors of the development and progression of such diseases, including osteoarthritis (OA), fibrosis and CNS lesions.
1. CircRNAs and osteoarthritis
Osteoarthritis (OA) is a chronic joint-retrogressive lesion with articular cartilage degradation and inflammation. Studies have reported that circRNAs mediate the pathogenesis of OA in an IL-1β-induced manner by sponging certain miRNAs 127, 128. Circ-Atp9b and hsa_circ_0005105 suppress the expression of type II collagen, enhance the functions of matrix metallopeptidase 13 (MMP-13) and promote the generation of IL-6 and IL-8 by targeting miR-138-5p and miR-26a, respectively, to activate the biogenesis of OA 129, 130. Additionally, circRNAs participate in the signaling pathway of TNF-α, which is a vital inflammatory factor in OA, to induce damage to cartilage cells 131. Intriguingly, circRNAs not only play a pathogenic role but also protect our bodies from OA lesions. Li et al verified that hsa_circ_0045714 elevates proliferation in cartilage cells and increases the expression of type II collagen and aggrecan by sponging miR-193b, which strengthened the functions of insulin like growth factor 1 receptor (IGF1R), a biotarget of miR-193b and a negative regulator of OA 132. Similarly, circ-VMA21 can alleviate inflammatory cytokine-induced apoptosis in nuclei pulposus cells and catabolism of the extracellular matrix (ECM) through the miR-200c-XIAP pathway during intervertebral disc degeneration (IVDD) 133. In summary, these data indicate that circRNAs are important regulators of and potential therapeutic targets for OA.
2. CircRNAs and fibrosis
Fibrosis is the comprehensive result of chronic inflammation caused by constant stimulation by infection, allergic response and tissue injury 134. Studies have revealed that circRNAs induce fibrosis progression in diverse organs. The long-term inhalation of large amounts of silica is a key inducer of lung fibrosis. SiO2-induced pulmonary fibrosis is accompanied by a high expression level of circ-ZC3H4. Circ-ZC3H4 can increase the presence of zinc finger CCCH-type containing 4 (ZC3H4) by restricting miR-212 as an endogenous competitor through its binding sites, thereby elevating proliferation and migration in alveolar macrophages and fibroblasts after exposure to SiO2 78. Liver fibrosis is inseparable from the activation of hepatic stellate cells (HSCs). HSCs are the dominant source of the ECM and can transform into myofibroblastic-like cells, highlighting their critical pro-fibrosis functions. The prolonged stimulation of pro-inflammatory factors dramatically initiates the activation of HSCs. Studies have clarified that the inhibition of hsa_circ_0071410 increases the expression of miR-9-5p, leading to the attenuation of HSC activation in several immune-associated pathways 135. Hsa_circ_010567 can also promote myocardial fibrosis by inhibiting the expression level of miR-141 through the initiation of TGF-β1 signaling 48 and expanding the effects of circRNAs on fibrosis and the cardiovascular system. Further exploration is required to investigate the roles played by circRNAs in organ fibrosis.
3. CircRNAs and central nervous system lesions
CircRNAs are closely correlated with normal and aberrant conditions in the CNS 136. Similarly, astrocytes generally maintain homeostasis in the CNS but become anomalously activated in response to multiple lesions, such as those associated with stroke, trauma, neuro-inflammation and neuro-degenerative diseases 137. Recently, Huang et al found that circ-HIPK2 could increase the sigma nonopioid intracellular receptor 1 (SIGMAR1) expression level by sponging endogenous miR-124-2HG and cooperating with autophagy and endoplasmic reticulum (ER) stress, thereby significantly stimulating astrocytes. The inhibition of circ-HIPK2 can restrict activated astrocytes induced by methamphetamine or LPS in vivo 138, providing novel therapeutic strategies for neuro-inflammation lesions. Both vasculopathy and cerebral ischemia-reperfusion injury (CIRI) account for a large proportion of CNS lesions. Studies have revealed that aberrantly expressed circRNAs appear to be closely correlated with overactivated or non-activated immune reactions in the CNS, leading to the occurrence of moyamoya disease (MMD) 139 or oxygen-glucose deprivation (OGD)/reoxygenation 140. These data highlight the regulatory functions of circRNAs and provide a new insight into CNS lesions.
Diagnosis and therapeutic properties of circRNAs in clinical application
CircRNAs serve as ideal biomarkers for the diagnosis of immune diseases
Currently, immune diseases are attracting increasing attention due to their high morbidity, indefinite pathogenesis and poor prognosis. In particular, numerous patients with immune diseases are typically asymptomatic during the early stage and become seriously ill at diagnosis, missing the ideal therapeutic time window. Hence, early diagnosis is highly significant and essential for immune disease therapy. Due to the constant emergence of novel diagnosis methods and tremendous efforts by researchers in this field, numerous circRNAs have successively been found to play potential roles in the etiology and pathology of certain immune diseases. Thus, these circRNAs may possibly be biomarkers of these diseases. Notably, circRNAs contain some remarkable properties that could be helpful while diagnosing patients. 1) CircRNAs have a highly stable inner structure. Due to their continuous, covalently closed loop structure without 5ʹ or 3ʹ polarities and poly (A) tails, circRNAs are able to fight against the degradation effects of RNase and RNA exonucleases 6. Thus, circRNAs appear to be more stable than linear RNAs, and this feature seems to be a key element of a biomarker. 2) circRNAs exhibit a high level of inherent conservation. CircRNAs are highly inherently conserved and infrequently mutated in different species regardless of the evolutionary distance 21, which is beneficial for detection. 3) CircRNAs exhibit broad-spectrum expression. CircRNAs can be discovered not only in a series of organisms but also in different organs, tissues, and even blood and saliva 141, which is suitable for acquisition and extraction. 4) CircRNAs exhibit longer half-lives. The half-lives of circRNAs are approximately 2.5-fold longer than those of their corresponding linear counterparts on average, making it possible for continuous inspection 142. 5) CircRNAs exist in exosomes and extracellular vesicles. CircRNAs can be found in exosomes 95 and are coprecipitated with extracellular vesicles 96, indicating that they are likely to be used in the diagnosis of secretion-related diseases. 6) CircRNAs have a highly specific expression pattern. Tissue-specificity and stage-association represent the main expression pattern of circRNAs 21; thus, circRNAs are suitable for differential diagnosis and prediction of prognosis. 7) The abundance of circRNAs is consistent. The correlation between circRNAs and their canonical linear counterparts is close because both are present at comparable levels with relative consistency. Taken together, all these characteristics suggest that circRNAs have the potential to be sensitive and precise biomarkers for the diagnosis of immune diseases.
CircRNAs exhibit therapeutic advantages for immune diseases
In addition to representing promising biomarkers for diagnosis, circRNAs also display some advantages for therapy in immune diseases. 1) CircRNAs serve as therapeutic targets. As widely expressed molecules in multiple immune processes in the human body, natural circRNAs are of significance in maintaining the inner homeostasis of the immune system. Therefore, aberrant circRNAs are likely to be regarded as therapeutic targets. Directly controlling the level of certain circRNAs in cells, local tissues or organs, such as via gene-editing therapy, is a therapeutic option. Actually, this strategy may yield less side effects than exogenously synthesized compounds, such as interfering RNAs (RNAis) and chemical drugs 10. 2) circRNAs could be implemented as therapy methods. Developing and optimizing antisense approaches could be applied to alter RNA splicing patterns to create more circRNAs or circRNA analogues that better exert miRNA “sponge” or miRNA “reservoir” properties. Potential artificial sponges and reservoirs could be designed according to the complementary sequences by which circRNAs interact with miRNAs to ultimately regulate their functions, rending these circRNAs promising drugs targeting miRNAs. 3) CircRNAs could have reduced “off-target” effects. Because of their stable structure and conservation, circRNAs can reduce off-target effects, which is superior to miRNAs and siRNAs since both of these oligonucleotides have frequent off-target effects due to their short length. In addition, circRNAs are likely to decrease the off-target effects of their target miRNAs 52. In conclusion, although limited studies have investigated therapeutic strategies using circRNAs, their potential and prospects are immeasurable in this field.
Limitations and prospects
The immune system comprises diverse components and monitors, defends against and eliminates pathogenic invaders to maintain the inner homeostasis of the human body. Aberrant immune responses, particularly the misrecognition of self-antigens, seriously destroy the balance within the immune system and create advantageous conditions for the occurrence of immune diseases. Although multiple traditional biomarkers have been identified and applied for decades, the clinical diagnosis and control of immune diseases remain unsatisfactory. The identification of sensitive and effective biomarkers has become increasingly important.
Due to considerable advances in RNA sequencing analyses, many circRNAs have been sequentially discovered, thereby enriching our knowledge regarding transcriptional and posttranscriptional regulation and developing a novel area of epigenetic mechanisms. CircRNAs have recently been confirmed to extensively participate in the regulation of different immune responses and immune diseases that are inseparable from their biological functions. Their role as miRNA “sponges” has been the most studied function to date. For instance, hsa_circ_0020397 targets miR-138 to control antitumor immune responses; CDR1as, hsa_circ_0001045 and GSDMB ecircRNA are capable of influencing several autoimmune diseases by sponging miR-7, miR-30a, miR-1275 and miR-149, respectively. In secondary immunodeficiency diseases, hsa_circ_0045714 and hsa_circ_0005105 are involved in disease regulation by targeting the corresponding miRNAs. In addition to acting as miRNA “sponges”, circRNAs can also combine with proteins to function as protein “sponges” in immune regulation, such as circ-ANRIL and PES1 complex-induced aberrant immune responses; CircRNAs derived from ROBO1 or CDC42 can bind CDC42BPA to promote congenital immune deficiency diseases. Notably, numerous circRNAs have been found to participate in the biogenesis of immune diseases, and some circRNAs have even been confirmed as potential biomarkers or therapeutic targets; however, studies investigating circRNA functions in immune diseases are still in their infancy. Except for their role as miRNA “sponges”, the other functions of circRNAs in immune disease regulation, such as miRNA “reservoirs” and protein coding, are not fully understood or even reported, but these functions provide novel insight for further explorations of circRNAs in immune diseases along the directions of their functions.
Recently, multiple studies have reported that following the forced overexpression or downregulation of specific circRNAs, immune responses and immune diseases can be differentially influenced, and several procedures may even be partially enhanced or reversed, highlighting the potential of circRNAs in the modulation of immune responses and immune diseases, which is highly significant for clinical application (Table 3).
Table 3.
Effects of overexpression or knockdown of circRNAs in immune responses and immune diseases.
| CircRNAs | Treatments | Immune Responses/ Diseases | Effects | References |
|---|---|---|---|---|
| Circ-RasGEF1B | Knockdown | Responses to LPS | Reduces the expression of ICAM-1 | Ng et al (2017) 75 |
| Circ-ANRIL | Overexpression | Macrophage Biogenesis | Inhibits the biogenesis of macrophages | Holdt et al (2016) 77 |
| Hsa_circ_0020397 | Overexpression | Colorectal Cancer | Promotes the expression of TERT and PD-L1 | Zhang et al (2017) 90 |
| Circ-HIPK2 | Knockdown | Neuro-inflammation | Inhibits astrocyte activation | Huang et al (2017) 138 |
| Hsa_circ_010567 | Knockdown | Myocardial Fibrosis | Increases miR-141 and inhibits the TGF-β1 signaling |
Zhou et al (2017) 48 |
| Hsa_circ_0071410 | Knockdown | Hepatic Fibrosis | Increases miR-9-5p and attenuates HSC activation |
Chen et al (2017) 135 |
| Circ-VMA21 | Overexpression | Intervertebral Disc Degeneration | Increases the expression of XIAP | Cheng et al (2018) 133 |
| Hsa_circ_0045714 | Knockdown | Osteoarthritis | Decreases the expression of XIAP | Li et al (2017) 132 |
| Overexpression | Promotes IGF1R expression and increases chondrocyte proliferation |
|||
| Hsa_circ_0005105 | Overexpression | Osteoarthritis | Promotes ECM degradation by regulating the miR-26a/NAMPT |
Wu et al (2017) 130 |
| Circ-Atp9b | Knockdown | Osteoarthritis | Increases the synthesis of type II collagen and reduces the level of MMP-13 in chondrocytes | Zhou et al (2018) 129 |
| Circ-Foxo3 | Knockdown | Mesenchymal Stem Cells | Promotes cell proliferation | Du et al (2016) 7 Huang et al (2014) 92 |
However, several limitations persist in this field. First, while the levels of certain circRNAs may be known, the roles, functions and underlying mechanisms of these circRNAs are unclear. Second, while several parental genes from which these circRNAs are encoded or spliced have been confirmed, the potential association or functional relationship between them is not often correlated as expected. Third, only a small subset of altered circRNAs in sequencing profiles meets the standards of a biomarker; furthermore, these circRNAs, i.e., so-called biomarkers, have rarely been tested in vitro and in vivo or in clinical trials to confirm their possible application in diagnosis, prognosis and therapy. Fourth, bioinformatics predictions specifically determining the targets of circRNAs remain nascent. Fifth, there is no formal rule for naming circRNAs 42. Finally, in addition to the classic splicing events of circRNAs, i.e., “back-splicing” and “exon-skipping”, other patterns of splicing or posttranscriptional modifications may exist. Therefore, further investigation is still required.
Studies investigating circRNAs are also hampered by several challenges that should be conquered. 1) To discover more novel circRNAs involved in immune responses and diseases, high-throughput sequencing is regarded as an unbiased approach that highly relies on the quality and library preparations of RNA samples 143. However, most available RNA-seq databases are pre-treated by a poly (A) purification step that may wipe eliminate circRNAs. Additionally, this technology is frequently limited to a small sample size and a shortage of tissue types, leading to the loss of a thorough evaluation by an experienced pathologist 144. 2) For the identification and verification of differentially expressed circRNAs in RNA-seq, most studies prefer to use qRT-PCR instead of Northern blotting as qRT-PCR is relatively convenient and efficient. However, in a larger cohort, the divergent qRT-PCR primers flanking the back-splicing junction are occupied and cannot detect the selected circRNAs. Moreover, reverse transcription can cause template switching 15, and a single, inappropriate normalized gene may give rise to a false positive or negative result. In addition, circRNAs can only be distinguished from the exon repetition through pretreatment with RNase or poly (A) enrichment in qRT-PCR 145. 3) Many bioinformatic algorithms have been successively used to further investigate circRNAs, and some algorithms require gene annotation lists while others perform de novo investigations. This divergence may decrease the presence of false positives while preventing the detection of several specific circRNAs without annotation because a standard naming system for circRNAs is not available 42. By bioinformatic prediction, hypothetical interactions between miRNAs and circRNAs can be tested in accordance with stoichiometric relationships, but this method is challenging due to the simultaneous analysis of a large number of cells with differential expression of miRNAs and circRNAs. Notably, competitive binding reportedly exists between the canonical 6-nt and 8-nt binding sites, which is rarely evaluated 146. In fact, some classical assays determining direct binding, such as luciferase reporter assays, are rarely applied in circRNAs studies. 4) Both genomic arrangement and mutations play vital roles in the regulation of the functions of certain genes. If these negative alterations occur in genes responsible for splicing, such as splicing factor 3b subunit 1 (SF3B1) 147 and QKI 35, the generation of circRNAs may be affected. Meanwhile, if such changes exist in the elements of regulatory regions of circRNAs, in theory, the biogenesis and function of downstream circRNAs are likely to be influenced 144. Intriguingly, the amplification of genomic DNA is positively correlated with the elevated expression of circ-PVT1 148, indicating that genomic amplification or deletion can also drive the activation of circRNAs. Despite the nascent state of circRNA studies, these challenges still provide multiple novel and open questions that deserve more and deeper explorations in the future.
Although some limitations and challenges exist that are likely to persist, innovations in molecular biological techniques are constantly emerging, which may promote the integrated development of circRNAs studies.
Recently, several databases were built based on the results of RNA-seq, such as circBase 149, circNet 150 and deepBase v2.0 151, which help fill in the gaps in our knowledge of the annotations of circRNAs. More importantly, by using these databases, it could be beneficial to create a reasonable and effective naming system to unify the IDs of discovered and undiscovered circRNAs. Meanwhile, databases, such as starBase v2.0 152 and CircInteractome 65, show verified or potential networks between miRNAs and circRNAs and between proteins and circRNAs, providing directions for further investigations of the functions of circRNAs, especially the behaviors of miRNA “reservoirs” and protein coding, which are newly discovered and not well understood. In addition, Circ2Traits 153 and circRNA Disease 99 provide data obtained directly from patients with different diseases, which are more clinically relevant. These clues can be used to evaluate the possibility of certain circRNAs serving as biomarkers and predictors of prognosis in some diseases. Notably, similar to the challenges faced in the former technologies, a bioinformatic approach may inevitably cause some errors. These databases should be updated in a timely manner and utilized comprehensively to maximize the potentials of these databases, particularly in increasing the veracity of hypotheses for further investigation.
In addition to the advances in bioinformatic approaches, detection methods are also further developed in circRNA studies. Compared with qRT-PCR, Northern blotting is more accurate and intuitive and does not cause template switching; thus, this approach should be the gold standard. In situ hybridization (ISH) and fluorescence in situ hybridization (FISH) solve the issues of both the expression and distribution of circRNAs by directly providing visualized spatial information. Single-cell separation and sequencing can effectively eliminate issues with the detection of differentially expressed circRNAs in a group of cells or tissues. Clustered regularly interspaced short palindromic repeats/Cas9 (CRISPR-cas9) can be used to assess the correlation between circRNAs and their parent genes regardless of the off-target effects of foreign synthesis oligonucleotides. Intriguingly, the novel technology nCounter platform from NanoString Technologies (Seattle, USA) 154 can simplify the protocol of RNA qualification by removing RNA amplification and reverse transcription, which seems to be useful in circRNA detection.
Notably, several controversial phenomena have been observed during recent explorations of circRNAs. Furthermore, a few conclusions may even challenge the status of traditional opinions; for example, the lack or loss of MREs does not influence the suppressive effects of circRNAs on target miRNAs 10, questioning the structural basis of miRNA “sponges”. No translatable circRNAs are found in osteosarcoma through ribosome footprint detection 21, querying the accuracy of the protein coding ability. We suppose that in addition to their role as miRNA “sponges”, circRNAs can inhibit their target miRNAs, which are mainly predicted by bioinformatic algorithms in other manners, such as by forming a transcription complex to repress the generation of pri-miRNA and coding relevant proteins to restrain the level of target miRNAs. Additionally, the translation of circRNAs is likely to be limited by space and time. Whether translatable circRNAs exist in a specific microenvironment, such as a tumor, whether regulators of translation, such as translation initiation factors and RBPs, are expressed, and whether the timing of their presence can affect the process of coding proteins are unknown. In addition, some unknown positive or negative feedback loops may participate in the control of circRNA functions. Meanwhile, we speculate that circRNAs may perform some novel functions that have not been discovered, such as serving as precursors of miRNAs and piRNAs, influencing epigenetic modifications and forming dimers with RNA transcripts to create endogenous siRNAs by Dicer-induced digestion.
Although our current knowledge of circRNAs is preliminary, it is reasonable to speculate about the promising perspective of deeper explorations of these ncRNAs regarding gene regulation and immune disease occurrence. The observed differential expression levels of circRNAs between normal tissues and diseased tissue indicate that circRNAs probably play important physiological and pathological roles. CircRNAs are highly conserved, quite stable and very appropriate as biomarkers for some immune diseases. Moreover, circRNAs can be exploited as therapeutic targets because the overexpression of specific circRNAs can suppress the pro-immune disease miRNA level via their miRNA “sponge” property. Optimizing antisense approaches to alter the splicing pattern in order to produce more beneficial circRNAs that block or postpone the development and progression of immune diseases might be considered a feasible strategy 155. It is expected that the development of new technologies will contribute to the discovery of more diagnosis- and prognosis-related circRNAs and that therapeutics depending on circRNAs will be formulated, thereby providing new perspectives and directions for the identification and treatment of immune diseases.
Acknowledgments
Our work was supported by grants 81773003 and 81472633 from the National Natural Science Foundation of China.
Abbreviations
- 3ʹ-UTRs
3ʹ-untranslated regions
- AIDs
autoimmune diseases
- Amotl1
angiomotin like 1
- AS
alternative splicing
- ATP9B
ATPase phospholipid transporting 9B
- CDR1as
CDR1 antisense RNA
- ceRNAs
competing endogenous RNAs
- circRNAs
circular RNAs
- CIRI
cerebral ischemia-reperfusion injury
- ciRNAs
circular intronic RNAs
- CNS
central nervous system
- CRP
C-reactive protein
- DMD
dystrophin
- ecircRNAs
exonic circRNAs
- ECM
extracellular matrix
- eiciRNAs
exon-intron circRNAs
- ESR
erythrocyte sedimentation rate
- FMN
formin
- Foxo3
forkhead box O3
- HIPK 2
homeodomain interacting protein kinase 2
- HSCs
hepatic stellate cells
- IRES
internal ribosome entry sites
- IVDD
intervertebral disc degeneration
- LPS
lipopolysaccharide
- miRNAs
microRNAs
- MMD
moyamoya disease
- MREs
miRNA response elements
- MS
multiple sclerosis
- MYC
MYC proto-oncogene, bHLH transcription factor
- ncRNAs
noncoding RNAs
- nt
nucleotides
- OA
osteoarthritis
- OGD
oxygen-glucose deprivation
- ORFs
open reading frames
- PBC
primary biliary cirrhosis
- PBMCs
peripheral blood mononuclear cells
- PD-1
programmed cell death 1
- PDK1
pyruvate dehydrogenase kinase 1
- PIDs
primary immunodeficiency diseases
- RA
rheumatoid arthritis
- RasGEF1B
RasGEF domain family member 1B
- RBPs
RNA binding proteins
- RNP
RNA binding region (RNP1, RRM) containing 3
- SCID
severe combined immunodeficiency disease
- SLE
systemic lupus erythematosus
- snRNA
small nuclear RNA
- STAT3
signal transducer and activator of transcription 3
- TB
tuberculosis
- TCF25
transcription factor 25
- TGF-β
transforming growth factor-beta
- tricRNAs
tRNA intronic circular RNAs
- TSS
transcription start site
- VMA21
vacuolar ATPase assembly factor
- WAS
Wiskott-Aldrich syndrome
- XIAP
X-linked inhibitor of apoptosis
- ZNF609
zinc finger protein 609.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J. et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–40. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 3.St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–51. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franz A, Rabien A, Stephan C, Ralla B, Fuchs S, Jung K. et al. Circular RNAs: a new class of biomarkers as a rising interest in laboratory medicine. Clin Chem Lab Med. 2018;56:1992–2003. doi: 10.1515/cclm-2018-0231. [DOI] [PubMed] [Google Scholar]
- 5.Dragomir M, Calin GA. Circular RNAs in Cancer - Lessons Learned From microRNAs. Front Oncol. 2018;8:179. doi: 10.3389/fonc.2018.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du WW, Yang WN, Liu E, Yang ZG, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH. et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 10.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S. et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58:870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 12.Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18:962–72. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–40. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 14.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–61. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PloS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H. et al. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–87. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huo L, Zhang P, Li CX, Rahim K, Hao XR, Xiang BY, Genome-Wide Identification of circRNAs in Pathogenic Basidiomycetous Yeast Cryptococcus neoformans Suggests Conserved circRNA Host Genes over Kingdoms. Genes; 2018. p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y, Guo X, Wang W. Identification and characterization of circular RNAs in zebrafish. FEBS Lett. 2017;591:213–20. doi: 10.1002/1873-3468.12500. [DOI] [PubMed] [Google Scholar]
- 20.Broadbent KM, Broadbent JC, Ribacke U, Wirth D, Rinn JL, Sabeti PC. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genomics. 2015;16:454. doi: 10.1186/s12864-015-1603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta Gene Regul Mech. 2016;1859:1245–51. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–47. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu SB, Zhong Y, Shang RZ, Zhang X, Song WJ, Kjems J. et al. The emerging landscape of circular RNA in life processes. Rna Biol. 2017;14:992–9. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563–79. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L. Splicing noncoding RNAs from the inside out. Wiley Interdiscip Rev RNA. 2015;6:651–60. doi: 10.1002/wrna.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matera AG, Wang ZF. A day in the life of the spliceosome. Nat Rev Mol Cell Bio. 2014;15:108–21. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang XO, Wang HB, Zhang Y, Lu XH, Chen LL, Yang L. Complementary Sequence-Mediated Exon Circularization. Cell. 2014;159:134–47. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Noto JJ, Schmidt CA, Matera AG. Engineering and expressing circular RNAs via tRNA splicing. Rna Biol. 2017;14:978–84. doi: 10.1080/15476286.2017.1317911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang DM, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–47. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH. et al. Exon Circularization Requires Canonical Splice Signals. Cell Rep. 2015;10:103–11. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR. et al. Analysis of Intron Sequences Reveals Hallmarks of Circular RNA Biogenesis in Animals. Cell Rep. 2015;10:170–7. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 34.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M. et al. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 35.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA. et al. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell. 2015;160:1125–34. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Fei T, Chen YW, Xiao TF, Li W, Cato L, Zhang P. et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. P Natl Acad Sci USA. 2017;114:E5207–E15. doi: 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D. et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aktas T, Avsar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G. et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–9. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 39.Kramer MC, Liang DM, Tatomer DC, Gold B, March ZM, Cherry S. et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–82. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chi Y, Zhou D. MicroRNAs in colorectal carcinoma-from pathogenesis to therapy. J Exp Clin Cancer Res. 2016;35:43. doi: 10.1186/s13046-016-0320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA Translation and Stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, Zhao Y. Circular RNAs: Characteristics, function, and role in human cancer. Histol Histopathol. 2018;33:887–93. doi: 10.14670/HH-11-969. [DOI] [PubMed] [Google Scholar]
- 43.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 44.Zheng XB, Zhang M, Xu MQ. Detection and characterization of ciRS-7: a potential promoter of the development of cancer. Neoplasma. 2017;64:321–8. doi: 10.4149/neo_2017_301. [DOI] [PubMed] [Google Scholar]
- 45.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 46.Li F, Zhang LY, Li W, Deng JQ, Zheng J, An MX. et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–13. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun. 2017;487:769–75. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 49.Zhu X, Wang X, Wei S, Chen Y, Fan X, Han S. et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284:2170–82. doi: 10.1111/febs.14132. [DOI] [PubMed] [Google Scholar]
- 50.Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W. et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7:5428. doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–22. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in Cancer. Cancer Res. 2013;73:5609–12. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 53.Wang KF, Sun Y, Tao W, Fei X, Chang CS. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12. doi: 10.1016/j.canlet.2016.12.036. [DOI] [PubMed] [Google Scholar]
- 54.Xu ZJ, Yan YL, Zeng SS, Dai S, Chen X, Wei J. et al. Circular RNAs: clinical relevance in cancer. Oncotarget. 2018;9:1444–60. doi: 10.18632/oncotarget.22846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Bio. 2016;17:205–11. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 56.Eidem TM, Kugel JF, Goodrich JA. Noncoding RNAs: Regulators of the Mammalian Transcription Machinery. J Mol Biol. 2016;428:2652–9. doi: 10.1016/j.jmb.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–42. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chao CW, Chan DC, Kuo A, Leder P. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med. 1998;4:614–28. [PMC free article] [PubMed] [Google Scholar]
- 60.Gualandi F, Trabanelli C, Rimessi P, Calzolari E, Toffolatti L, Patarnello T. et al. Multiple exon skipping and RNA circularisation contribute to the severe phenotypic expression of exon 5 dystrophin deletion. J Med Genet. 2003;40:e100. doi: 10.1136/jmg.40.8.e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelly S, Greenman C, Cook PR, Papantonis A. Exon Skipping Is Correlated with Exon Circularization. J Mol Biol. 2015;427:2414–7. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 62.Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. Rna Biol. 2017;14:361–9. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Du WW, Zhang C, Yang WN, Yong TQ, Awan FM, Yang BB. Identifying and Characterizing circRNA-Protein Interaction. Theranostics. 2017;7:4183–91. doi: 10.7150/thno.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hansen TB, Veno MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44:e58. doi: 10.1093/nar/gkv1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dudekulay DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. Rna Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Komar AA, Hatzoglou M. Cellular IRES-mediated translation The war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–40. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–9. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y. et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–41. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H. et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L. et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21. doi: 10.1016/j.molcel.2017.02.021. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst; 2018. p. 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–61. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y, Li X, Zhang M, Lv K. Microarray analysis of circular RNA expression patterns in polarized macrophages. Int J Mol Med. 2017;39:373–9. doi: 10.3892/ijmm.2017.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY, Ea CK. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. Rna Biol. 2016;13:861–71. doi: 10.1080/15476286.2016.1207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ng WL, Marinov GK, Chin YM, Lim YY, Ea CK. Transcriptomic analysis of the role of RasGEF1B circular RNA in the TLR4/LPS pathway. Sci Rep. 2017;7:12227. doi: 10.1038/s41598-017-12550-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W. et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang X, Wang J, Zhou Z, Jiang R, Huang J, Chen L. et al. Silica-induced initiation of circular ZC3H4 RNA/ZC3H4 pathway promotes the pulmonary macrophage activation. FASEB J. 2018;32:3264–77. doi: 10.1096/fj.201701118R. [DOI] [PubMed] [Google Scholar]
- 79.Maass PG, Glazar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L. et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl) 2017;95:1179–89. doi: 10.1007/s00109-017-1582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang YH, Yu XH, Luo SS, Han H. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing. 2015;12:17. doi: 10.1186/s12979-015-0042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qian ZQ, Liu H, Li MS, Shi JC, Li N, Zhang Y. et al. Potential Diagnostic Power of Blood Circular RNA Expression in Active Pulmonary Tuberculosis. Ebiomedicine. 2018;27:18–26. doi: 10.1016/j.ebiom.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang X, Zhu M, Yang R, Zhao W, Hu X, Gan J. Identification and comparison of novel circular RNAs with associated co-expression and competing endogenous RNA networks in pulmonary tuberculosis. Oncotarget. 2017;8:113571–82. doi: 10.18632/oncotarget.22710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang ZK, Yao FY, Xu JQ, Deng Z, Su RG, Peng YP. et al. Microarray Expression Profile of Circular RNAs in Peripheral Blood Mononuclear Cells from Active Tuberculosis Patients. Cell Physiol Biochem. 2018;45:1230–40. doi: 10.1159/000487454. [DOI] [PubMed] [Google Scholar]
- 84.Sun L, Liu S, Chen ZJ. SnapShot: pathways of antiviral innate immunity. Cell. 2010;140:436–e2. doi: 10.1016/j.cell.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang M, Yu F, Wu W, Zhang Y, Chang WG, Ponnusamy M. et al. Circular RNAs: A novel type of non-coding RNA and their potential implications in antiviral immunity. Int J Biol Sci. 2017;13:1497–506. doi: 10.7150/ijbs.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF. et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell. 2017;67:214–27. doi: 10.1016/j.molcel.2017.05.023. e7. [DOI] [PubMed] [Google Scholar]
- 87.Shi J, Hu N, Li J, Zeng Z, Mo L, Sun J. et al. Unique expression signatures of circular RNAs in response to DNA tumor virus SV40 infection. Oncotarget. 2017;8:98609–22. doi: 10.18632/oncotarget.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang ZY, Guo ZD, Li JM, Zhao ZZ, Fu YY, Zhang CM. et al. Genome-Wide Search for Competing Endogenous RNAs Responsible for the Effects Induced by Ebola Virus Replication and Transcription Using a trVLP System. Front Cell Infect Microbiol. 2017;7:479. doi: 10.3389/fcimb.2017.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang XH, Yan YM, Lei XY, Li AJ, Zhang HM, Dai ZK. et al. Circular RNA alterations are involved in resistance to avian leukosis virus subgroup-J-induced tumor formation in chickens. Oncotarget. 2017;8:34961–70. doi: 10.18632/oncotarget.16442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41:1056–64. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 91.Yang ZG, Awan FM, Du WW, Zeng Y, Lyu JJ, Wu D. et al. The Circular RNA Interacts with STAT3, Increasing Its Nuclear Translocation and Wound Repair by Modulating Dnmt3a and miR-17 Function. Mol Ther. 2017;25:2062–74. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang Y, Yu P, Li W, Ren G, Roberts AI, Cao W. et al. p53 regulates mesenchymal stem cell-mediated tumor suppression in a tumor microenvironment through immune modulation. Oncogene. 2014;33:3830–8. doi: 10.1038/onc.2013.355. [DOI] [PubMed] [Google Scholar]
- 93.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM. et al. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell. 2016;166:1055–6. doi: 10.1016/j.cell.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 94.Li P, Liu C, Yu Z, Wu M. New Insights into Regulatory T Cells: Exosome- and Non-Coding RNA-Mediated Regulation of Homeostasis and Resident Treg Cells. Front Immunol. 2016;7:574. doi: 10.3389/fimmu.2016.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM. et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lasda E, Parker R. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS One. 2016;11:e0148407. doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu Z, Li P, Fan L, Wu M. The Potential Role of circRNA in Tumor Immunity Regulation and Immunotherapy. Front Immunol. 2018;9:9. doi: 10.3389/fimmu.2018.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cadena C, Hur S. Antiviral Immunity and Circular RNA: No End in Sight. Mol Cell. 2017;67:163–4. doi: 10.1016/j.molcel.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao Z, Wang K, Wu F, Wang W, Zhang K, Hu H. et al. circRNA disease: a manually curated database of experimentally supported circRNA-disease associations. Cell Death Dis. 2018;9:475. doi: 10.1038/s41419-018-0503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jin Y, Mazza C, Christie JR, Giliani S, Fiorini M, Mella P. et al. Mutations of the Wiskott-Aldrich Syndrome Protein (WASP): hotspots, effect on transcription, and translation and phenotype/genotype correlation. Blood. 2004;104:4010–9. doi: 10.1182/blood-2003-05-1592. [DOI] [PubMed] [Google Scholar]
- 101.Sauer AV, Di Lorenzo B, Carriglio N, Aiuti A. Progress in gene therapy for primary immunodeficiencies using lentiviral vectors. Curr Opin Allergy Cl. 2014;14:527–34. doi: 10.1097/ACI.0000000000000114. [DOI] [PubMed] [Google Scholar]
- 102.Sheldon H, Andre M, Legg JA, Heal P, Herbert JM, Sainson R. et al. Active involvement of Robo1 and Robo4 in filopodia formation and endothelial cell motility mediated via WASP and other actin nucleation-promoting factors. FASEB J. 2009;23:513–22. doi: 10.1096/fj.07-098269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brigida I, Sauer AV, Ferrua F, Giannelli S, Scaramuzza S, Pistoia V. et al. B-cell development and functions and therapeutic options in adenosine deaminase-deficient patients. J Allergy Clin Immunol. 2014;133:799–806. doi: 10.1016/j.jaci.2013.12.1043. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pala F, Morbach H, Castiello MC, Schickel JN, Scaramuzza S, Chamberlain N. et al. Lentiviral-mediated gene therapy restores B cell tolerance in Wiskott-Aldrich syndrome patients. J Clin Invest. 2015;125:3941–51. doi: 10.1172/JCI82249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cassani B, Mirolo M, Cattaneo F, Benninghoff U, Hershfield M, Carlucci F. et al. Altered intracellular and extracellular signaling leads to impaired T-cell functions in ADA-SCID patients. Blood. 2008;111:4209–19. doi: 10.1182/blood-2007-05-092429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fairhurst AM, Wandstrat AE, Wakeland EK. Systemic lupus erythematosus: multiple immunological phenotypes in a complex genetic disease. Adv Immunol. 2006;92:1–69. doi: 10.1016/S0065-2776(06)92001-X. [DOI] [PubMed] [Google Scholar]
- 107.D'Cruz DP, Khamashta MA, Hughes GRV. Systemic lupus erythematosus. Lancet. 2007;369:587–96. doi: 10.1016/S0140-6736(07)60279-7. [DOI] [PubMed] [Google Scholar]
- 108.Smith PP, Gordon C. Systemic lupus erythematosus: clinical presentations. Autoimmun Rev. 2010;10:43–5. doi: 10.1016/j.autrev.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 109.Wu XN, Ye YX, Niu JW, Li Y, Li X, You X. et al. Defective PTEN Regulation and Function Contributes to B Cell HyperResponsiveness in Systemic Lupus Erythematosus. Arthritis Rheumatol. 2014;66:S858–S. doi: 10.1126/scitranslmed.3009131. [DOI] [PubMed] [Google Scholar]
- 110.Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer's Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7) Genes; 2016. p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li H, Li K, Lai W, Li X, Wang H, Yang J. et al. Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin Chim Acta. 2018;480:17–25. doi: 10.1016/j.cca.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 112.Ouyang QQ, Wu J, Jiang ZL, Zhao JJ, Wang R, Lou AJ. et al. Microarray Expression Profile of Circular RNAs in Peripheral Blood Mononuclear Cells from Rheumatoid Arthritis Patients. Cell Physiol Biochem. 2017;42:651–9. doi: 10.1159/000477883. [DOI] [PubMed] [Google Scholar]
- 113.Wang WH, Zhang YY, Zhu B, Duan TH, Xu QG, Wang R. et al. Plasma microRNA expression profiles in Chinese patients with rheumatoid arthritis. Oncotarget. 2015;6:42557–68. doi: 10.18632/oncotarget.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Murata K, Yoshitomi H, Tanida S, Ishikawa M, Nishitani K, Ito H. et al. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12:R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu K, Xu P, Yao JF, Zhang YG, Hou WK, Lu SM. Reduced apoptosis correlates with enhanced autophagy in synovial tissues of rheumatoid arthritis. Inflamm Res. 2013;62:229–37. doi: 10.1007/s00011-012-0572-1. [DOI] [PubMed] [Google Scholar]
- 116.Zheng FP, Yu XQ, Huang JH, Dai Y. Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol Med Rep. 2017;16:8029–36. doi: 10.3892/mmr.2017.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ponath G, Park C, Pitt D. The Role of Astrocytes in Multiple Sclerosis. Front Immunol. 2018;9:217. doi: 10.3389/fimmu.2018.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Paraboschi EM, Rimoldi V, Solda G, Tabaglio T, Dall'Osso C, Saba E. et al. Functional variations modulating PRKCA expression and alternative splicing predispose to multiple sclerosis. Hum Mol Genet. 2014;23:6746–61. doi: 10.1093/hmg/ddu392. [DOI] [PubMed] [Google Scholar]
- 119.Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M. et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One. 2009;4:e7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cardamone G, Paraboschi EM, Rimoldi V, Duga S, Solda G, Asselta R. The Characterization of GSDMB Splicing and Backsplicing Profiles Identifies Novel Isoforms and a Circular RNA That Are Dysregulated in Multiple Sclerosis. Int J Mol Sci; 2017. p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Iparraguirre L, Munoz-Culla M, Prada-Luengo I, Castillo-Trivino T, Olascoaga J, Otaegui D. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis. Hum Mol Genet. 2017;26:3564–72. doi: 10.1093/hmg/ddx243. [DOI] [PubMed] [Google Scholar]
- 122.Carey EJ, Ali AH, Lindor KD. Primary biliary cirrhosis. Lancet. 2015;386:1565–75. doi: 10.1016/S0140-6736(15)00154-3. [DOI] [PubMed] [Google Scholar]
- 123.Zheng J, Li Z, Wang T, Zhao Y, Wang Y. Microarray Expression Profile of Circular RNAs in Plasma from Primary Biliary Cholangitis Patients. Cell Physiol Biochem. 2017;44:1271–81. doi: 10.1159/000485487. [DOI] [PubMed] [Google Scholar]
- 124.Kang GJ, Lee HJ, Byun HJ, Kim EJ, Kim HJ, Park MK. et al. Novel involvement of miR-522-3p in high-mobility group box 1-induced prostaglandin reductase 1 expression and reduction of phagocytosis. Biochim Biophys Acta. 2017;1864:625–33. doi: 10.1016/j.bbamcr.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 125.Danza K, De Summa S, Pinto R, Pilato B, Palumbo O, Carella M. et al. TGFbeta and miRNA regulation in familial and sporadic breast cancer. Oncotarget. 2017;8:50715–23. doi: 10.18632/oncotarget.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P. et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhou Z, Du D, Chen A, Zhu L. Circular RNA expression profile of articular chondrocytes in an IL-1beta-induced mouse model of osteoarthritis. Gene. 2018;644:20–6. doi: 10.1016/j.gene.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 128.Liu Q, Zhang X, Hu XQ, Dai LH, Fu X, Zhang JY, Circular RNA Related to the Chondrocyte ECM Regulates MMP13 Expression by Functioning as a MiR-136 'Sponge' in Human Cartilage Degradation. Sci Rep; 2016. p. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou ZB, Du D, Huang GX, Chen AM, Zhu L. Circular RNA Atp9b, a competing endogenous RNA, regulates the progression of osteoarthritis by targeting miR-138-5p. Gene. 2018;646:203–9. doi: 10.1016/j.gene.2017.12.064. [DOI] [PubMed] [Google Scholar]
- 130.Wu Y, Zhang Y, Zhang Y, Wang JJ. CircRNA hsa_circ_0005105 upregulates NAMPT expression and promotes chondrocyte extracellular matrix degradation by sponging miR-26a. Cell Biol Int. 2017;41:1283–9. doi: 10.1002/cbin.10761. [DOI] [PubMed] [Google Scholar]
- 131.Liu Q, Zhang X, Hu X, Yuan L, Cheng J, Jiang Y. et al. Emerging Roles of circRNA Related to the Mechanical Stress in Human Cartilage Degradation of Osteoarthritis. Mol Ther Nucleic Acids. 2017;7:223–30. doi: 10.1016/j.omtn.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li BF, Zhang Y, Xiao J, Wang F, Li M, Guo XZ. et al. Hsa_circ_0045714 regulates chondrocyte proliferation, apoptosis and extracellular matrix synthesis by promoting the expression of miR-193b target gene IGF1R. Hum Cell. 2017;30:311–8. doi: 10.1007/s13577-017-0177-7. [DOI] [PubMed] [Google Scholar]
- 133.Cheng X, Zhang L, Zhang K, Zhang G, Hu Y, Sun X. et al. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis. 2018;77:770–9. doi: 10.1136/annrheumdis-2017-212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen Y, Yuan B, Wu Z, Dong Y, Zhang L, Zeng Z. Microarray profiling of circular RNAs and the potential regulatory role of hsa_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene. 2017;629:35–42. doi: 10.1016/j.gene.2017.07.078. [DOI] [PubMed] [Google Scholar]
- 136.Chen W, Schuman E. Circular RNAs in Brain and Other Tissues: A Functional Enigma. Trends Neurosci. 2016;39:597–604. doi: 10.1016/j.tins.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 137.Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huang RR, Zhang Y, Han B, Bai Y, Zhou RB, Gan GM. et al. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy. 2017;13:1722–41. doi: 10.1080/15548627.2017.1356975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhao M, Gao FL, Zhang D, Wang S, Zhang Y, Wang R. et al. Altered expression of circular RNAs in Moyamoya disease. J Neurol Sci. 2017;381:25–31. doi: 10.1016/j.jns.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 140.Lin SP, Ye S, Long YM, Fan YX, Mao HF, Chen MT. et al. Circular RNA expression alterations are involved in OGD/R-induced neuron injury. Biochem Biophys Res Commun. 2016;471:52–6. doi: 10.1016/j.bbrc.2016.01.183. [DOI] [PubMed] [Google Scholar]
- 141.Bonizzato A, Gaffo E, Te Kronnie G, Bortoluzzi S. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J. 2016;6:e483. doi: 10.1038/bcj.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–83. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Panda AC, De S, Grammatikakis I, Munk R, Yang X, Piao Y. et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017;45:e116. doi: 10.1093/nar/gkx297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–65. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rigatti R, Jia JH, Samani NJ, Eperon IC. Exon repetition: a major pathway for processing mRNA of some genes is allele-specific. Nucleic Acids Res. 2004;32:441–6. doi: 10.1093/nar/gkh197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Denzler R, McGeary SE, Title AC, Agarwal V, Bartel DP, Stoffel M. Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Mol Cell. 2016;64:565–79. doi: 10.1016/j.molcel.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chabot B, Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol. 2016;212:13–27. doi: 10.1083/jcb.201510032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen J, Li Y, Zheng QP, Bao CY, He J, Chen B. et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–19. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 149.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–70. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM. et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209–D15. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zheng LL, Li JH, Wu J, Sun WJ, Liu S, Wang ZL. et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2016;44:D196–D202. doi: 10.1093/nar/gkv1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL. et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 155.Lee T, Awano H, Yagi M, Matsumoto M, Watanabe N, Goda R, 2'-O-Methyl RNA/Ethylene-Bridged Nucleic Acid Chimera Antisense Oligonucleotides to Induce Dystrophin Exon 45 Skipping. Genes; 2017. p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]