Abstract
The Streptococcus pyogenes CRISPR/Cas9 (SpCas9) nuclease has been widely applied in genetic engineering. Despite its importance in genome editing, aspects of the precise molecular mechanism of Cas9 activity remain ambiguous. In particular, because of the lack of a method with high spatio‐temporal resolution, transient interactions between Cas9 and DNA could not be reliably investigated. It therefore remains controversial how Cas9 searches for protospacer adjacent motif (PAM) sequences. We have developed single‐molecule Förster resonance energy transfer (smFRET) assays to monitor transient interactions of Cas9 and DNA in real time. Our study shows that Cas9 interacts with the PAM sequence weakly, yet probing neighboring sequences via facilitated diffusion. This dynamic mode of interactions leads to translocation of Cas9 to another PAM nearby and consequently an on‐target sequence. We propose a model in which lateral diffusion competes with three‐dimensional diffusion and thus is involved in PAM finding and consequently on‐target binding. Our results imply that the neighboring sequences can be very important when choosing a target in genetic engineering applications.
Keywords: CRISPR/Cas9, lateral diffusion, single‐molecule FRET, target search
Subject Categories: DNA Replication, Repair & Recombination; Methods & Resources
Introduction
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas systems are adaptive prokaryotic immune systems that provide bacteria and archaea with a defense mechanism against invading foreign genetic elements (Makarova et al, 2006, 2015; Barrangou et al, 2007; Wiedenheft et al, 2012; Marraffini, 2015; Mohanraju et al, 2016). Upon infection, fragments of the invader's DNA are incorporated into the CRISPR locus in the host genome (Bolotin et al, 2005; Mojica et al, 2005; van der Oost et al, 2014; Amitai & Sorek, 2016). Those fragments are then transcribed into short CRISPR RNAs (crRNAs), which assemble with CRISPR‐associated (Cas) proteins in order to recognize and destroy the invader when it returns (Brouns et al, 2008). The most famous of the discovered CRISPR systems is the type II system where the DNA of the invader is recognized and destroyed by the Cas9 protein, which assembles with two RNA molecules, namely crRNA and trans‐activating crRNA (tracrRNA) (Deltcheva et al, 2011; Gasiunas et al, 2012; Jinek et al, 2012; van der Oost et al, 2014; Makarova et al, 2015; Mohanraju et al, 2016). The most widely researched Cas9 ortholog, Streptococcus pyogenes Cas9, recognizes a 20‐nt target, which is flanked by a PAM (protospacer adjacent motif) sequence on the 3′ end of the target (Gasiunas et al, 2012; Jinek et al, 2012). The PAM sequence for SpCas9 is 5′‐NGG‐3′. CRISPR/Cas9 system has gained enormous attention due to its use in genome editing owing to its simplicity and programmability (Cong et al, 2013; Jiang et al, 2013; Hsu et al, 2014; Barrangou & Doudna, 2016). In order to edit a gene, Cas9 first has to find a small 23‐nt sequence in a genome containing kilo‐bases or mega‐bases of DNA, in a crowded cellular environment. This process has been demonstrated to be slow, with a single Cas9 protein requiring 6 h to locate a single target in a bacterial cell (Jones et al, 2017). This example shows that efficient targeting of specific genes requires an advanced knowledge of Cas9 target search mechanism.
A recent single‐molecule study using “DNA curtains” has shown that Cas9 uses only three‐dimensional diffusion to locate its target (Sternberg et al, 2014). Due to diffraction limit (~100 nm) of the DNA curtains technique, it remains unknown whether the model of exclusive three‐dimensional target search is valid for the length scale of nucleotides. Other single‐molecule studies have shown that RNA‐guided proteins such as Argonaute or CRISPR type I Cascade protein complex use facilitated one‐dimensional diffusion during their target search (Chandradoss et al, 2014; Dillard et al, 2018) Using single‐molecule FRET (Förster resonance energy transfer), we investigate the target search mechanism of SpCas9 and demonstrate that it uses 1D diffusion along the DNA strand during its target search.
Results
Single‐molecule observation of Cas9 PAM search
To visualize Cas9 target search process on a nanometer scale, we used single‐molecule Förster resonance energy transfer (smFRET) technique. Biotinylated Cas9 (Fig EV1A) was immobilized on a PEG‐coated quartz surface for long‐term observation (Fig 1A; Chandradoss et al, 2014). Biotinylation was shown not to affect Cas9 catalytic activity (Fig EV1B). The Cas9 protein was pre‐incubated with dye‐labeled crRNA and tracrRNA for 20 min before surface immobilization. Free‐floating molecules, such as any unattached RNA or Cas9 molecules, were washed away, and remaining immobilized Cas9:RNA complexes could be directly imaged using total internal reflection microscopy (Fig 1A). Synthetic DNA and crRNA substrates were labeled with Cy3 and Cy5 dyes respectively, such that the FRET efficiency between them would report on the position where Cas9 localizes on the DNA. Addition of Cy3‐labeled DNA substrates did not affect the immobilization of Cas9:RNA complexes (Fig 1A). When excited with a green laser, binding events appear as spots on the CCD image (Cy3 signal on the left side and Cy5 signal on the right side) (Fig 1B and C). In fluorescence time traces, binding events are characterized by increase in fluorescence intensity due to either direct excitation of donor or energy transfer from donor to acceptor (Fig 1B and C).
Figure EV1. Biotinylation and activity test of the SpCas9 protein.
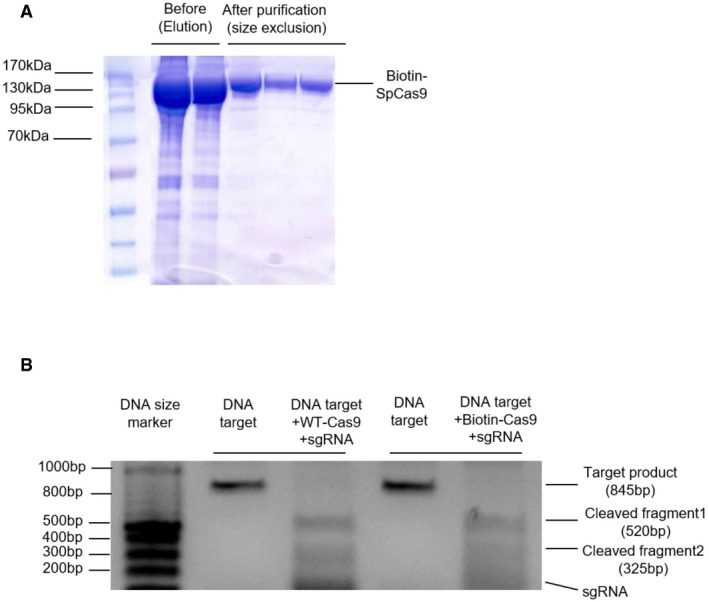
- SDS–PAGE data of the biotinylated SpCas9 protein.
- In vitro cleavage assay for activity validation of the biotinylated SpCas9. Target DNA strand was cleaved by equivalent molar ratio of the SpCas9 and biotinylated SpCas9. Cleaved fragments sizes are 520 bp and 325 bp, respectively. Primers used: CCR5 forward primer 5′‐GAGGTGAGAGGATTGCTTGAGCCC‐3′, CCR5 reverse primer 5′‐ATTGTCAGGAGGATGATGAAGAAG‐3′.
Figure 1. Single‐molecule assay for observation of Cas9‐DNA interactions.
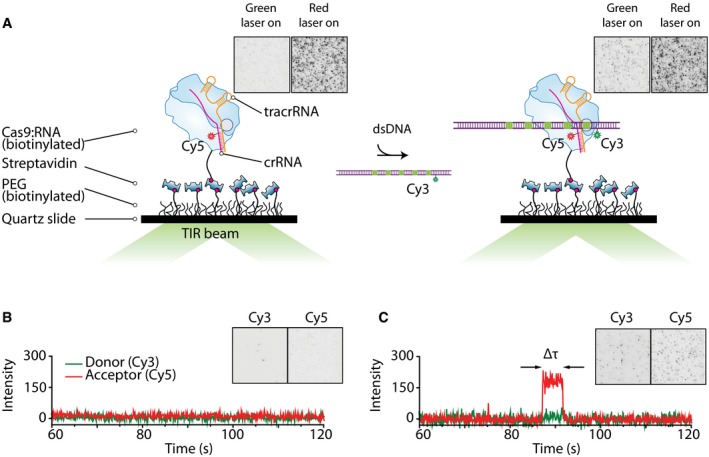
- Illustrated schematic representation of our single‐molecule assay. Cas9 assembled with tracrRNA and Cy5‐labeled crRNA is immobilized on the surface via biotin–streptavidin links (left). After addition of Cy3‐labeled DNA substrate, the interaction between Cas9 and DNA brings the dyes together resulting in FRET signal (right). EMCCD images show Cas9:RNA immobilized on the surface with green and red laser illumination (left). No fluorescence is seen when illuminated with green laser. Immobilized Cas9 can be seen when illuminated with red laser. After addition of Cy3‐labeled DNA, spots appear on the EMCCD in the green channel when illuminated with green laser (right). Size of EMCCD images is 25 × 25 μm.
- Example time trace when no binding event occurs (green laser excitation). EMCCD image shows signal from green and red channels before the addition of Cy3‐labeled DNA when illuminated with green laser. Size of EMCCD image is 25 × 50 μm.
- Example time trace showing a binding event (green laser excitation). EMCCD image shows signal from green and red channels after the addition of Cy3‐labeled DNA when illuminated with green laser. Size of EMCCD image is 25 × 50 μm.
Source data are available online for this figure.
The initial step in Cas9 target search is finding and recognizing a PAM sequence. It has been shown that, without encountering a cognate PAM, Cas9 cannot start R‐loop formation, despite the full complementarity between the guide RNA and the target (Szczelkun et al, 2014). Therefore, in order to elucidate the mechanism by which Cas9 finds and recognizes PAM alone, we first investigated how the protein interacts with DNA strands containing only PAM sequences when no target sequence is present. In particular, multiple binding sites in close proximity have been shown to cause a synergistic effect in another RNA‐guided target search system (Chandradoss et al, 2015). This effect emerges when the interaction between a searcher and a target is characterized by more than simple one‐step binding and dissociation (Grimson et al, 2007; Chandradoss et al, 2014). Such a synergistic effect, if observed, would be an indication that Cas9 uses an additional mechanism to 3D diffusion when searching for PAM sequences.
To investigate how Cas9 interacts with multiple PAM sites and whether such synergistic effect exists in the CRISPR/Cas9 system, we designed DNA constructs containing 0, 1, 2, 3, 4, and 5 equidistant PAM sites (Fig 2A). DNA was labeled with a Cy3 dye at position ‐8 with respect to the first guanine on the first PAM site on the 5′ end of the target DNA strand (Fig 2A). crRNA was labeled with a Cy5 dye outside the guide region such that Cas9:RNA complex binding to the first PAM site would yield a high‐FRET value (Fig 2A and B). Binding to other PAM sequences would increase distance between dyes and therefore was expected to yield lower and distinct FRET values for each site, thus allowing to distinguish which PAM site was bound (Fig 2B).
Figure 2. PAM search mechanism.
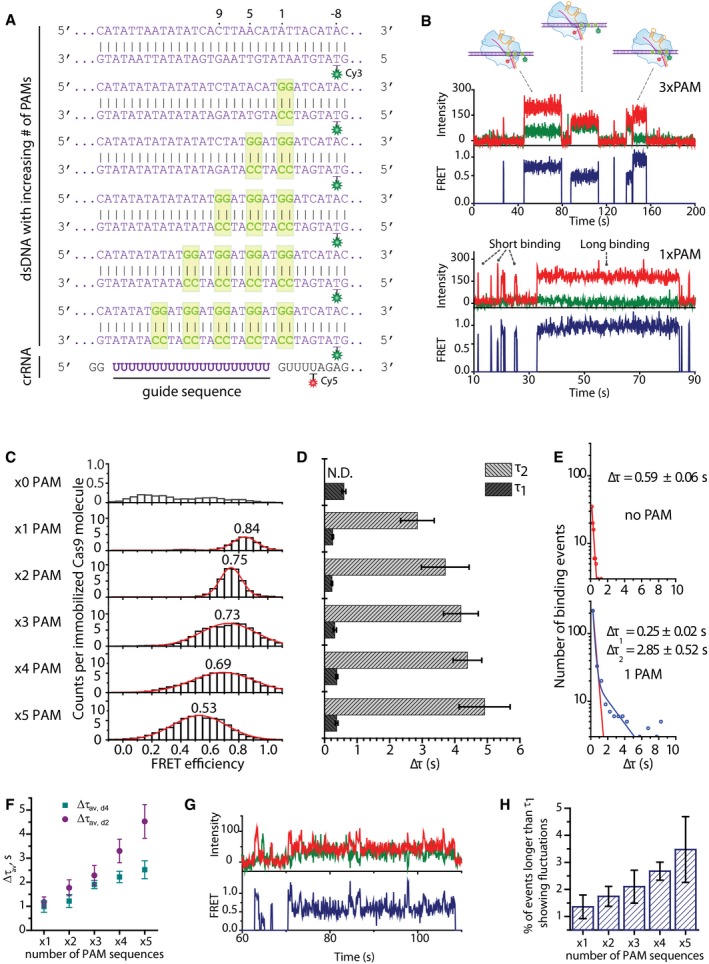
- DNA and crRNA sequences used for multiple PAM experiments with indicated labeling positions. Di‐nucleotide GG PAMs are shown in boxes.
- Example time traces when DNA containing 3 PAM sequences is added (top) and when DNA containing a single PAM is added showing both short and long binding events (bottom).
- FRET histograms of all binding events for each construct. Y‐axis represents total number of counts, divided by the number of immobilized molecules in the field of view.
- Bar plot of all dwelltime values for all constructs showing short binding events for all DNA and long events only for the constructs containing PAM sequences. The times shown are mean values of dwelltimes obtained during four different experiments on four different days. Error bars represent standard error of the mean.
- Example dwelltime histograms from negative control and a construct containing a single PAM. Negative control dwelltime distribution is characterized by a single‐exponential decay (top), and dwelltime distribution for PAM‐containing DNA is fitted by a double‐exponential decay (blue line) (bottom). Equation for the double‐exponential decay fit: y = A1e−t/τ1 + A2e−t/τ2; ΔA1 = 796.4 ΔA2 = 28.3. Red line shows what a single‐exponential decay fit for such distribution would be. Errors represent standard error of the mean. Dwelltime distributions for remaining DNA constructs are shown in Fig EV2C.
- Scatter plot showing the average dwelltime (Δτav) values for the cases of 2 (d2)‐ and 4 (d4)‐nucleotide separation between the PAM sites. The values are averages of four measurements over four different days for each case. Error bars represent the standard error of the mean. The average dwelltime was obtained using the equation Δτav = (A1Δτ1 2 + A2Δτ2 2)/(A1Δτ1 + A2Δτ2)
- Example trace showing direct FRET transitions for a 5 PAM‐containing DNA construct with separation between PAM sites being 4 nucleotides.
- Histogram showing the percentage of events that are longer than τ1 showing FRET fluctuations for multiple PAM constructs with PAM separation of 4 nucleotides. The values are averages of four measurements over four different days. The error bars represent standard error of the mean.
Negative control construct containing no PAM sites showed a very low number of binding events with random FRET values (Figs 2C and EV2A). Binding to DNA containing a single PAM yielded a narrow FRET distribution centered at 0.84 (Fig 2C). Binding events showing different FRET states appeared when the number of PAM sites increased (Fig 2B). FRET histogram of all binding events for the construct containing 2 PAM sites shifted to a lower FRET value of 0.75 (Fig 2C). Lower FRET states appeared for DNA constructs containing 3 and 4 PAMs with FRET histograms broadening significantly and the peaks of the histograms shifting to 0.73 and 0.69, respectively (Fig 2C). Finally, a histogram of all binding events for the construct containing 5 PAM sites was broad and centered at 0.53—an average of all FRET values yielded by specific binding to any of the five PAM sites on the DNA strand (Fig 2C). Furthermore, multiple binding events showing FRET efficiencies corresponding to different PAMs could be observed in a single FRET trace showing that a single Cas9 can bind and dissociate from different PAM sites, as expected (Fig 2B).
Figure EV2. Evidence of synergistic effect and lateral diffusion between PAMs.
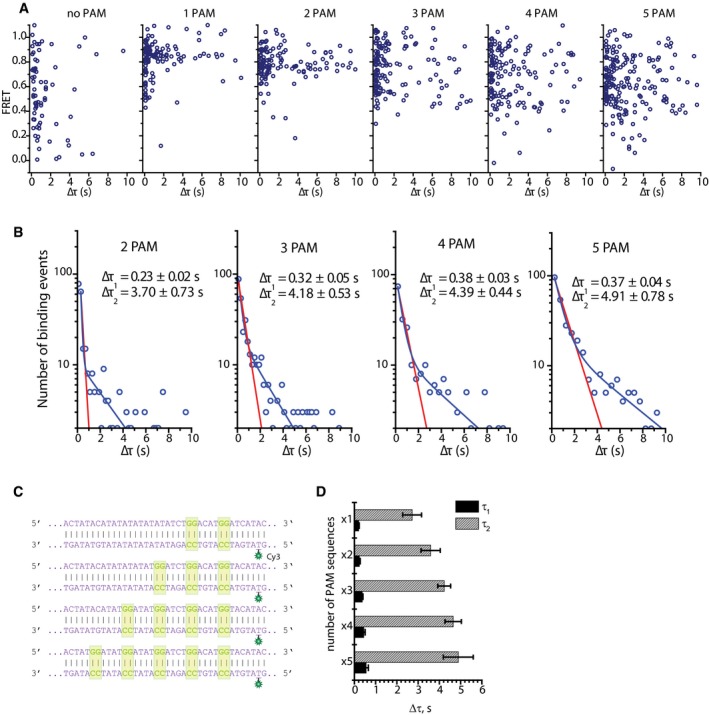
- A scatter plot showing FRET versus dwelltime for each construct containing 0, 1, 2, 3, 4, and 5 PAMs. No correlation between FRET values and dwelltimes is observed for either construct.
- Dwelltime distributions for constructs containing 2, 3, 4, and 5 neighboring PAMs. Blue line shows a double‐exponential decay fit, and red line shows what a single‐exponential decay fit would look like for these dwelltime distributions. Equation of the double‐exponential decay fit: y = A1e−t/τ1 + A2e−t/τ2.
- DNA sequences used in the multiple PAM experiments where the PAM separation is 4 nucleotides.
- Bar plot of all dwelltime values for all constructs (d4). The times shown are mean values of dwelltimes obtained during four different experiments on four different days. Error bars represent standard error of the mean.
In addition to broadening of the FRET histograms, the dwelltime was observed to increase with increasing number of PAM sequences (Fig 2D). Experiments on DNA constructs without any PAM or target sequences only showed short‐lived binding (Δτ1), with a dwelltime of 0.59 ± 0.06 s (Fig 2D and E). Short binding events were also observed with PAM‐containing DNA constructs ranging between 0.25±0.02 s and 0.37 ± 0.04 s—similar for all constructs, regardless of the number of PAM sequences (Figs 2D and EV2B). Interestingly, a second type of longer binding events was observed for the constructs containing PAM sites as opposed to the negative control (Figs 2B, D and E, and EV2B). It is further noted that the second dwelltime (Δτ2) increased with increasing number of adjacent PAM sites from 2.85 ± 0.52 s for the construct with a single PAM to 4.91 ± 0.78 s for the construct with 5 PAM sites (Fig 2D). In addition to Δτ2, the average dwelltime (Δτav) also was found to increase with increasing number of PAM sites from 1.18 ± 0.21 s for a single PAM to 4.53 ± 0.70 s for the construct containing 5 PAM sequences (Fig 2F). Further analysis showed no correlation between FRET values and dwelltimes, suggesting that it is not the position of a PAM site, but rather the number of PAM sites in close proximity that caused this increase (Fig EV2A).
The observation that even for a single PAM the binding time is characterized by a double‐exponential distribution suggests that Cas9 uses another mechanism in addition to 3D diffusion during its target search, as processes following exclusively 3D diffusion follow one‐step dissociation kinetics (Berg et al, 1981). Furthermore, the increase in τ2 implies that, due to the presence of multiple PAM sites in close proximity, Cas9 experiences a synergistic effect which causes it to stay bound to DNA for longer. We therefore hypothesize that upon encountering a PAM, Cas9 can follow two pathways. First, Cas9 can dissociate from DNA in a three‐dimensional fashion upon failing to form an RNA‐DNA R‐loop (corresponding to τ1). Second, Cas9 can locally diffuse in a one‐dimensional fashion probing adjacent PAM sites (corresponding to τ2).
To further investigate whether the observed increase in τ2 could indeed be the result of one‐dimensional diffusion between the PAM, DNA sequences where the PAM sites were placed further apart (4 nucleotides) were designed (Fig EV2C). In this case, the separation between the two furthermost PAM sites is 24 nucleotides. Dwelltime analysis revealed that the dwelltime distribution for each construct was again characterized by a double‐exponential decay. Further analysis has shown that values of τ1 were similar for all constructs, ranging between 0.20 ± 0.02 s and 0.55 ± 0.10 s (Fig EV2D). These values also are similar to those observed in the case of 2‐nucleotide separation between PAM sites (Fig 2D), further supporting the hypothesis that τ1 results from weak interactions with a single PAM site, followed by dissociation from the DNA strand. In addition, an almost identical increase in τ2 was observed as in the case of 2‐nucleotide separation (Fig EV2D). The values were found to increase from 2.72 ± 0.44 s for a single PAM to 4.89 ± 0.70 s for 5 PAMs (Fig EV2D). Average dwelltime analysis also revealed an increase in Δτav: from 1.22 ± 0.27 s for a single PAM to 2.52 ± 0.37 s for the construct containing 5 PAMs (Fig 2F). The increased separation between the PAMs results in a slower rise in Δτav, which further suggests that this increase is due to one‐dimensional diffusion, as increased distance between binding sites lowers the probability of finding a target by diffusing laterally along the DNA.
The increased separation between the PAM sites results in the greater separation of FRET efficiency values corresponding to binding to each individual PAM site. As a result, a type of binding event showing FRET fluctuations was observed in the case of 4‐nucleotide separation between the PAMs (Fig 2G). Furthermore, upon further analysis it was found that the percentage of binding events that show fluctuations and are longer than τ1 increases with increasing number of neighboring PAM sites from 1.36 ± 0.44% to 3.47 ± 1.21% (Fig 2H). These data directly show that Cas9 laterally diffuses between PAM sites, thus providing an explanation for the observed increase in τ2 and Δτav. The overall low occurrence of such events can be explained by the nature of these interactions. Cas9‐PAM interactions are intrinsically weak and three‐dimensional dissociation dominates, leading to a large population of short‐lived binding events characterized by a single FRET efficiency value.
One‐dimensional diffusion used for PAM and target search
The observation that Cas9 stays bound to a DNA strand for longer when multiple neighboring PAM sequences are present and that the incidence of events showing FRET fluctuations increases with increasing number of PAM sequences suggested that this effect could be caused by lateral diffusion between the PAM sites (Berg et al, 1981). To investigate the possibility that Cas9 is able to scan PAM sequences in a 1D fashion and to explore whether such probing could lead to binding to a neighboring target site, we designed DNA constructs containing a partial target of 9 nucleotides and an increasing number of PAM sites adjacent to it: 1xPAM, 3xPAM, 5xPAM, 7xPAM, and 9xPAM (Fig 3A). DNA was labeled at position +13 on the target strand and crRNA at position +10 relative to the first complementary nucleotide, such that high FRET efficiency would only be observed upon productive binding to the partial target site (Fig 3A). Partial complementarity was chosen in order to allow for observation of multiple binding events (Szczelkun et al, 2014; Singh et al, 2016).
Figure 3. Multiple PAM influence on binding to a target site.
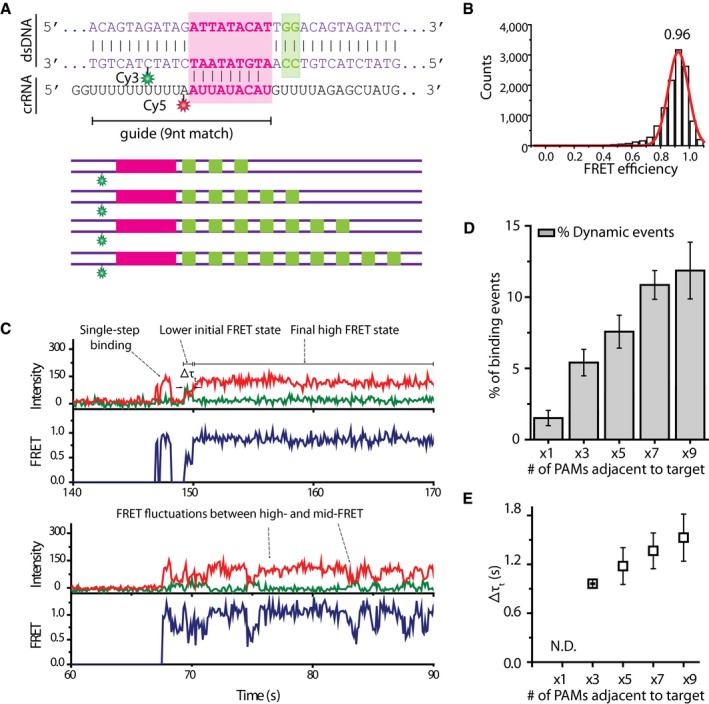
- DNA and crRNA sequence used for experiments of a single‐target and multiple PAMs. DNA sequence is shown for the single‐target single‐PAM construct, and schematic representation is shown for multiple‐PAM‐containing constructs.
- FRET histogram of all binding events from the construct containing a single PAM next to the partial target. Y‐axis represents total number of counts.
- Example FRET traces from single‐target multiple‐PAM experiments. Top trace shows an example of a single‐step binding event and a binding event that starts at a lower FRET state before transitioning to productive binding high‐FRET state. Bottom trace shows an example of FRET fluctuations between the high‐FRET state and lower FRET states.
- A histogram showing the percentage of dynamic events. The percentage values were obtained from five experiments on five different days. Error bars represent standard error of the mean. The percentage of events showing dynamic binding behavior was calculated by dividing the number of binding events that showed such behavior by the total number of binding events.
- A scatter plot showing the time Cas9 spends in a lower FRET state before transitioning to a high‐FRET state. The dwelltimes increase with the number of PAMs adjacent to the target. Such dwelltime could not be determined for a construct containing a single PAM. The values were obtained from five experiments on five different days. Error bars represent standard error of the mean.
Binding to DNA containing a single PAM next to the partial target resulted in single‐step events showing a stable expected high FRET efficiency of 0.96 (Fig 3B and C). However, increasing number of adjacent PAM sites next to the target displayed an increasing percentage of binding events that either start at a lower FRET state before transitioning to the productive binding FRET state or show fluctuations between a clearly defined high FRET state and various lower FRET states (Fig 3C and D). In particular, the percentage of events that show either fluctuations or two‐step binding (“dynamic events”) shows a sixfold increase from 2.03 ± 1.61% for the 1xPAM construct to 13.9 ± 2.51% for the 9xPAM construct (Fig 3D). Furthermore, the time Cas9 spends in an initial low FRET state before transitioning to high FRET state (Δτt), which indicates on‐target binding, was found to increase with increasing number of PAM sequences adjacent to the target (Fig 3E). Together with the observation of the increasing dwelltime when the number of neighboring PAM sites increases in Fig 2D, these results further suggest that Cas9 does not exclusively use 3D diffusion alone during target search, but also can find a target site by laterally probing neighboring PAM sites. Therefore, upon failing to form a stable R‐loop Cas9 does not necessarily dissociate from the DNA strand but can go back to scanning the PAM sequences in a one‐dimensional fashion.
Mechanism of lateral diffusion
The observations of increasing τ2 with increasing number of neighboring PAM sequences together with the observed transitions from a lower FRET state to a high FRET state when a partial target was present suggest that lateral diffusion may indeed be involved in Cas9 target search. In order to investigate this mechanism more systematically, we designed tandem‐target DNA constructs, where identical partial targets were placed at different distances: 6, 9, 12, and 23 bp (Fig 4A). In this assay, binding to one target (H) would yield a high FRET value and binding to the second target (L) would correspond to a lower value (Fig 4A). We used a partial match (3 nt) between crRNA and DNA target to investigate how the presence of a second target site influences Cas9 binding dynamics and whether it can laterally diffuse between the two target sites. Control experiments with a single target at each distance showed no fluctuations in FRET efficiency (Fig EV3A).
Figure 4. Mechanism of lateral diffusion.

- DNA sequences used in tandem‐target experiments. Complementary sequences to crRNA are marked in pink boxes, and PAMs are marked in green boxes. Distances between the protospacers are indicated above the sequences. An illustration of Cas9 binding to each target site resulting in either high FRET (target H) or low FRET (target L) is shown on the right.
- Example fluorescence traces showing Cas9 transitioning between H and L targets with increasing target distances (right). FRET histograms constructed from events that show fluctuations showing two FRET peaks corresponding to binding to either target. High FRET peak stays the same in each histogram, but lower FRET peak moves to lower FRET values as the distance between targets increases.
- A scatter plot showing the probability of Cas9 transitioning between targets before dissociation. The values are averages from five measurements, and error bars represent standard error of the mean. The transition probability (p = T/(T+D)) was calculated by summing up all the number of FRET transitions (T) and dividing T by the sum of T and the total number of dissociation events (D).
- A histogram showing all dwelltimes from single‐target controls and tandem‐target experiments. τ1 is similar for single‐target control and tandem‐target experiments. τ2 only appears in tandem‐target experiments and is over 20‐fold higher than τ1 or τst. The values represent the mean dwelltimes that were obtained by fitting dwelltime distributions by either single or double‐exponential decay function from five measurements. Error bars represent standard error of the mean.
Figure EV3. Single‐target control experiments.
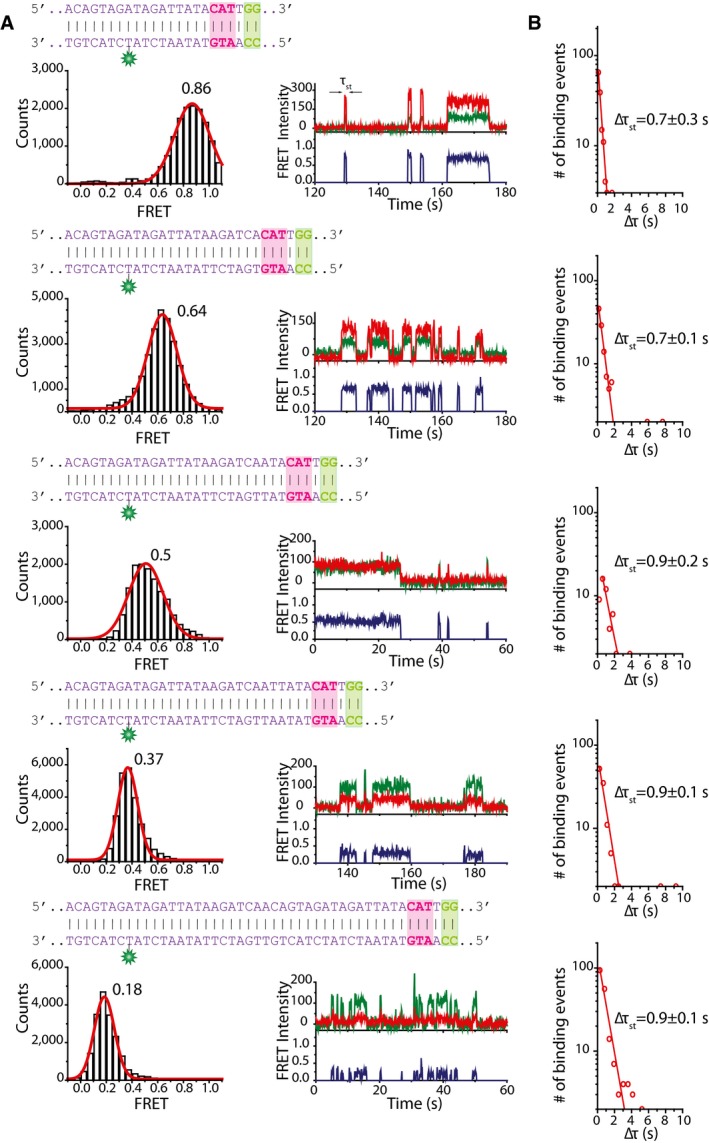
- DNA sequences used in single‐target control experiments, complementary sequence is marked in pink box, and PAMs are marked in green box. Corresponding FRET histograms and example FRET traces are shown below each DNA sequence.
- Dwelltime distributions for each DNA construct fitted with a single‐exponential decay function.
In the tandem‐target assay, Cas9 was directly observed to switch between two FRET states in a single binding event for each target separation (Fig 4B). The observed two FRET peaks in histograms from transition events agree with the values from single‐target controls (Figs 4B and EV3A), confirming that the fluctuations are arising due to Cas9 shuttling between two target sites. The probability of translocating to a neighboring target before dissociation that arose due to 1‐D diffusion was highest at ~0.35 when the distance between protospacers was 6 bp (Fig 4C). At 9‐bp and 12‐bp separation, the probability dropped to ~0.19 and ~0.13, respectively. At 23‐bp distance, the probability dropped to ~0.07—a fivefold decrease compared to 6‐bp separation.
The measured dwelltimes for a single target at each distance from the dye (Figs 4D and EV3B) followed a single‐exponential decay with dwelltimes (τst) lower than 1 s which is in agreement with literature (Singh et al, 2016). In contrast, the dwelltime distribution for tandem‐target constructs was characterized by a double‐exponential decay (Figs 4D and EV4A). The dwelltimes for target binding were obtained by measuring the binding times of events that have a FRET value, corresponding to on‐target binding, which allowed any non‐specific or PAM‐only interactions to be excluded from analysis. Short binding events with a similar dwelltime as single‐target controls were observed with all constructs regardless of the distance between protospacers (τ1). However, a second type of events observed had strikingly long dwelltimes (τ2) of 20–30 s (Fig 4D). The presence of such binding events with more than a 20‐fold increase in dwelltime compared to a single target suggests that Cas9 is experiencing a strong synergistic effect when two targets are neighboring each other.
Figure EV4. Tandem‐target dwelltime and single‐target binding rate.
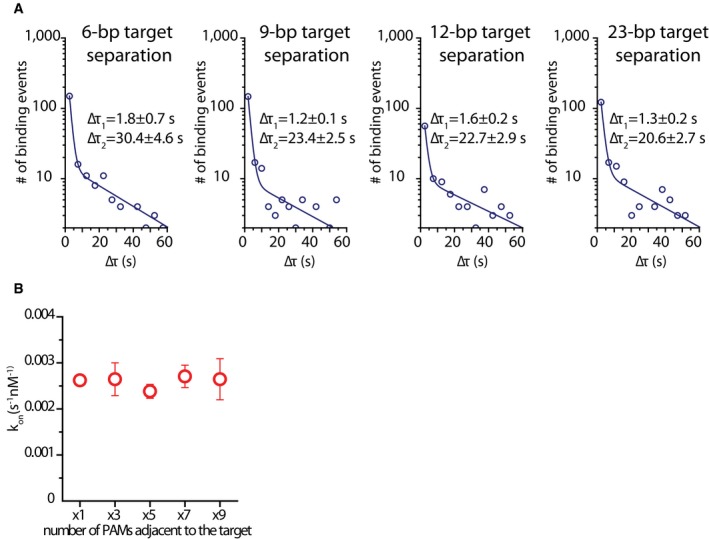
- Dwelltime distributions for tandem‐target construct for each target separation, fitted with a double‐exponential decay function. Equation of the fit: y = A1e−t/τ1 + A2e−t/τ2. A values for each tandem‐target construct: d6 ‐ A1 = 640.3, A2 = 8.0; d9 ‐ A1 = 654.7, A2 = 8.8; d12 ‐ A1 = 144.6, A2 = 6.4; d23 – A1 = 635.3 A2 = 5.6. Average dwelltime for each construct: d6 ‐ τav = 12.3 s; d9 ‐ τav = 6.9 s; d12 ‐ τav = 10.7 s; d23 ‐ τav = 7.6 s. Average dwelltimes were calculated using equation
- Scatter plot showing the total binding rate (kon) for each single‐target, multiple PAM construct. Binding rate stays constant with increasing number of adjacent PAMs. The values are averages of four measurements over 3 days. Error bars represent standard error of the mean. kon was calculated using the equation kon = 1/(CxΔτon) where C is the molarity of DNA target in the channel.
PAM multiplicity delays on‐target binding
While the results indicate that lateral diffusion is involved in PAM and consequently target search, it remains unclear whether the presence of multiple PAM sites next to a target would promote on‐target binding or act as a decoy binding site, thus delaying target recognition. To investigate the effects PAM multiplicity has on the on‐target binding, we designed 3 tandem‐target constructs. A single PAM was always adjacent to the first target, while the second target had 1, 3, and 5 neighboring PAMs (Fig 5A). As in the previous tandem‐target experiments (target separation 12 bp), binding to the first target resulted in a high FRET state (~0.86) and binding to the second target resulted in a lower FRET state (~0.5) (Fig 4B). Complementarity between crRNA and DNA was chosen to be 9 nucleotides for greater binding stability. Analysis of individual binding events revealed that for the symmetric case where both target sites are flanked by a single PAM, the binding events that begin at either target are equally distributed: 52.9 ± 3.4% begin at the first target site and 47.1 ± 3.4% begin at the second target site (Fig 5B). When the number of PAM sites adjacent to the second target site was increased to 3 and consequently 5 PAMs, the distribution of events changed as in both cases more than 60% of events now started at the first target site having a single PAM (Fig 5B). These results suggest that having multiple PAM sites to the target can deter the protein from binding the target site.
Figure 5. PAM multiplicity delays on‐target binding.
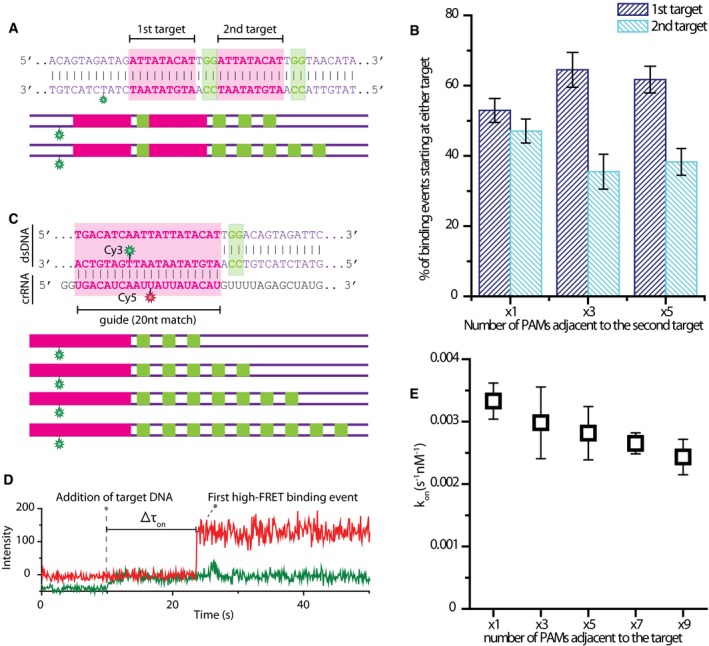
- DNA and crRNA sequence used for experiments with tandem‐target multiple PAM constructs. DNA sequence is shown for the symmetric construct with each target having only one adjacent PAM, and schematic representation is shown for multiple‐PAM constructs adjacent to the second target‐containing constructs.
- Histogram showing the percentage of binding events starting at either target. The values are averages of five measurements over three different days. Error bars represent standard error of the mean.
- DNA and crRNA sequence used for experiments with a full target and multiple PAMs. DNA sequence is shown for the single‐target single‐PAM construct, and schematic representation is shown for multiple‐PAM‐containing constructs.
- An example trace showing the addition of Cy3‐labeled DNA target and the first binding event. Δτon is indicated.
- Scatter plot showing the on‐target binding rate (kon‐target) for each construct. Binding rate decreases moderately with increasing number of adjacent PAMs. The values are averages of four measurements over 3 days. Error bars represent standard error of the mean. kon‐target was calculated using the equation kon‐target = 1/(CxΔτon) where C is the molarity of DNA target in the channel.
To further investigate this effect, we designed constructs with full complementarity (20 nt) between DNA and crRNA and an increasing number of neighboring PAM sites: x1, x3, x5, x7, and x9 (Fig 5C). Flow experiments were performed, and for each construct, the binding rate to the target (kon‐target) was obtained by measuring the time between the addition of DNA to the flow chamber and the first high FRET binding event (Fig 5D). The binding rate values were found to decrease moderately with increasing number of PAM sites (Fig 5E). Once a high FRET state was achieved indicating on‐target binding, no further FRET fluctuations were observed. This indicates that although multiple PAM sites cause the binding rate to the target to go down, they cannot compete with target binding once the target has been recognized. When all events, such as zero‐ or low‐FRET peaks were included in the analysis, the binding (kon‐total) rate was found to remain constant for all constructs (Fig EV4B). Together with the results showing that a target with a single neighboring PAM is preferred in tandem‐target experiments, these data hint that while PAM multiplicity does not affect overall binding behavior, it delays on‐target binding with PAM clusters acting as decoy binding sites for Cas9.
Discussion
As a means of prokaryotic defense against invading foreign genetic elements, Cas9 has to be able to find its target in a crowded cellular environment, among kilo‐bases of DNA. The target search becomes even more complicated when Cas9 is applied in eukaryotic cells as a genome engineering tool (Cong et al, 2013; Hsu et al, 2014). In such situations where a protein needs to sample a myriad of sequences before finding a cognate target, facilitated diffusion has been shown to speed up target search as opposed to three‐dimensional diffusion alone (Riggs et al, 1970; Berg et al, 1981; Halford & Marko, 2004; Gorman et al, 2012; Hammar et al, 2012; Leith et al, 2012; Ragunathan et al, 2012). We propose that, once Cas9 finds a PAM sequence by 3D collisions, it is able to diffuse laterally on a DNA strand. By competing with the dissociation process, this lateral diffusion mode intervenes in PAM finding and consequently target recognition. We determined that lateral diffusion of Cas9 primarily occurs in a local manner of ~20 basepairs, when searching for both, the PAM and partial complementarity between DNA and crRNA. This explains the disagreement with previous studies which suggested lateral diffusion does not occur in Cas9 target search, since such distances could not be investigated due to the diffraction limit of other microscopy techniques (Sternberg et al, 2014).
We speculate that a limiting factor for Cas9 diffusion may not only be distance, but also the need to open the DNA duplex which is energetically unfavorable if a protein without a helicase domain were to laterally diffuse long distances. Structural data showed that Cas9 interacts with PAM sites directly, without opening up the double‐stranded structure or the involvement of DNA‐RNA interactions (Anders et al, 2014). Thereby, the lateral diffusion for PAM search would be more effective than for PAM and partial complementarity. This speculation is supported by our observation that multiple neighboring PAM sites provide a binding site for Cas9 and allow Cas9 to interrogate an adjacent target site. This observation is in contrast to rapid dissociation from a PAM when an adjacent target is not present (Singh et al, 2016). In addition, we show evidence of Cas9 laterally diffusing between individual PAM sites, further supporting the hypothesis that lateral diffusion is used for PAM search. Our data also provide explanation as to why PAM‐rich DNA stretches can be efficiently bound in vivo even if no target is present nearby (O'Geen et al, 2015). Our work is also in agreement with DNA curtains studies, which show that Cas9 localizes on PAM‐rich regions on λ‐DNA(Sternberg et al, 2014).
Based on our findings, we propose a model in which PAM sequences drive lateral diffusion as the protein directly interacts with them, as shown by structural studies (Fig 6; Anders et al, 2014). If upon binding to a PAM site a matching target is not found, Cas9 can dissociate or diffuse locally on the DNA strand until another PAM site is found. If a matching DNA sequence flanks the PAM, Cas9 checks for complementarity and if it is not sufficient for stable binding, it can dissociate or again diffuse laterally until another PAM is found, as shown by our tandem‐target assays. Such a process repeats until a target with a sufficiently high degree of complementarity (> 12nt) is found and Cas9 cannot further dissociate. Therefore, we expand the knowledge of Cas9 target search mechanism by showing that it is a combination of three‐dimensional and one‐dimensional diffusion along the DNA strand and that the PAM sequences are not only important as the first step of target recognition but also drive target search.
Figure 6. Model for Cas9 target search.
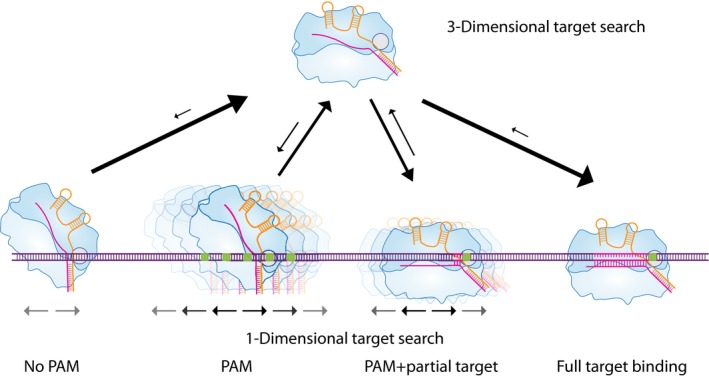
Upon binding to DNA without PAM, Cas9 rapidly dissociates. Upon binding to a PAM, Cas9 can dissociate or diffuse locally until another PAM site is found. If a matching DNA sequence flanks the PAM, Cas9 checks for complementarity, and if it is not sufficient for stable binding, it can dissociate or diffuse laterally until another PAM is found. Such a process repeats until a target with a high enough degree of complementarity (9–12 nt) is found and Cas9 cannot further dissociate.
In order to increase efficiency in genome editing, an in‐depth knowledge of how Cas9 finds its target is crucial. Therefore, our results will be important when choosing target sites during genome editing. Neighboring PAM sites close to a desired target will cause the protein to stay bound to the PAM cluster for longer, thereby delaying on‐target binding in a case where Cas9 expression levels are high and therefore the likelihood of finding a target by 3‐D collisions is high, thus greatly decreasing total search time (Jones et al, 2017). If Cas9 expression levels are low, it is likely that by keeping Cas9 bound to a neighboring region for longer, PAM‐rich sites could increase the chance of a Cas9 molecule finding the target faster via 1D diffusion. If no target is present next to a PAM‐rich DNA site, such PAM clusters could be used as decoy binding sites by phages in order to prevent Cas9 binding to a cognate target during Cas9 DNA interference. In addition to importance in genetic engineering, our results suggest that the strong interaction and lateral diffusion between PAM sites could be important in bacterial defense against phages. Cas9 has been shown to be important in recognizing the PAM during the CRISPR adaptation step, together with Cas1‐Cas2‐Csn2 complex (Heler et al, 2015). Therefore, PAM density in the invader's genome could potentially play a role in selecting which targets will be integrated in the CRISPR locus. Further in vivo studies will provide an answer to whether PAM clusters are beneficial for the invader, by acting as decoys and delaying target recognition, or for the host, by increasing the efficiency of functional spacer selection during CRISPR adaptation.
Materials and Methods
Recombinant SpCas9 purification
The pET plasmid encoding (6x)His‐tagged Cas9 was transformed into BL21 (DE3), Rosetta. Transformed bacterial cells were moved to a 400 ml of fresh LB medium containing 50 μg/ml kanamycin. The culture was incubated with shaking (200 rpm) at 18°C for 24 h. Optical density was monitored, and Cas9 protein expression was induced (A550 = 0.6) by using 0.5 mM IPTG at 18°C for 24 h. After the cells were harvested by centrifugation (5,000× g) for 10 min (at 4°C), bacterial cells were resuspended with lysis buffer [20 mM Tris–HCl (pH 8.0), 400 mM NaCl, 10 mM b‐mercaptoethanol, 1% Triton X‐100, 50 mg aprotinin, 50 mg antipain, 50 mg bestatin, 1 mM PMSF (phenylmethylsulfonyl fluoride)] (Sigma‐Aldrich) and sonicated on ice. The lysate was centrifuged at 5000 × g for 10 min(4°C), and supernatant solution was mixed with 2 ml of Ni‐NTA slurry (Qiagen) at 4°C for 1 and half hour. The lysate/Ni‐NTA mixture was loaded onto a column (Bio‐Rad) with capped bottom outlet. Loaded sample was washed multiple times with pre‐made wash buffer [20 mM Tris–HCl (pH 8.0), 400 mM NaCl, 10 mM b‐mercaptoethanol], and (6×)His‐tagged SpCas9 was eluted with elution buffer [20 mM Tris–HCl (pH 8.0), 400 mM NaCl, 10 mM b‐mercaptoethanol, 200 mM Imidazole] (Fig EV1A). Finally, buffer containing eluted SpCas9 protein was changed to storage buffer [10 mM HEPES‐KOH (pH 7.5), 250 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, 7 mM b‐mercaptoethanol and 20% glycerol] by using centrifugal filter (Amicon Ultra 100K). The purified SpCas9 protein was frozen with liquid nitrogen and stored at −80°C.
Biotinylation of the recombinant SpCas9
The process of linking biotin to the recombinant protein was carried out in vitro and proceeded during the process of protein purification. After loading the SpCas9 over‐expressed bacterial lysate and Ni‐NTA mixture onto a column (Bio‐Rad), mixed sample was washed multiple times with wash buffer [20 mM Tris–HCl (pH 8.0), 400 mM NaCl]. Then we added 10‐fold molar excess of maleimide–biotin (Sigma‐Aldrich) to SpCas9 solution and incubated for overnight at 4°C (mix gently with rotator). To get rid of unbound maleimide–biotin chemicals, mixed sample was washed sufficiently with wash buffer [20 mM Tris–HCl (pH 8.0), 400 mM NaCl]. Finally, biotinylated SpCas9 protein was eluted with elution buffer [20 mM Tris–HCl (pH 8.0), 400 mM NaCl, 200 mM Imidazole], and then, the protein concentration was measured by spectrophotometer (NanoDrop 2000; Thermo Fisher Scientific). Eluted SpCas9 protein was further purified with size exclusion chromatography. The biotinylation degree of the wild‐type SpCas9 protein was calculated with commercial kit (Pierce), and it reached about 100% for two cysteine sites (Cys80/Cys574). Biotinylated SpCas9 protein was stored in storage buffer [10 mM HEPES‐KOH (pH 7.5), 250 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, 7 mM b‐mercaptoethanol and 20% glycerol], and purified protein was frozen in liquid nitrogen and stored at −80°C.
Preparation of the single‐guide RNA
We used in vitro RNA transcription (DNA template, T7 RNA polymerase (NEB) 5 μl, 10× buffer 10 μl, rNTP mix (2.5 mM each), MgCl2 10 mM, DTT 1 mM, H2O up to 100 μl, RNase inhibitor (NEB) 0.5 μl, total 100 μl reaction) to generate single‐guide RNAs. DNA template contains X20 target protospacer sequence, which is complementary to the RNA strand. After RNA transcription, DNA template was removed by DNase (NEB) treatment. Then pure single‐guide RNA was purified, and concentration was measured by spectrophotometer (NanoDrop 2000; Thermo Fisher Scientific).
In vitro DNA cleavage assay with wild‐type and biotinylated SpCas9
In vitro cleavage experiments were performed with sgRNA and SpCas9 proteins purified at high purity (Fig EV2). DNA containing the target site was prepared by PCR, and higher molar concentration of the SpCas9 protein and biotinylated SpCas9 was treated at the same molarity. The sgRNA was added at a molar ratio three times greater than the protein (final molar ratio, DNA: protein: sgRNA = 1:3:9) with a complementary sequence to the target site. Target DNA, SpCas9 protein, and sgRNA were mixed and incubated at 37°C for 1 h. The cleaved DNA product was separated on the 1.5% agarose gel, and cleavage ratio was calculated by ImageJ software.
Labeling of nucleic acids
Nucleic acids were labeled using NHS‐ester chemistry. DNA and RNA strands with a C6 amine modification on a thymine or uracil base were ordered synthetic from companies Ella Biotech and IBA Lifesciences, respectively. 1 mM DNA or RNA samples was mixed with ~20 mM dye (GE healthcare) and labeling buffer (Sodium bicarbonate, 8.4 mg/ml) in a volume ratio 1:1:5 and incubated for 6 h at room temperature with gentle mixing in the dark. Full labeling procedure can be found in Joo and Ha (2012).
Single‐molecule two‐color FRET
Single‐molecule fluorescence measurements were performed with a prism‐type total internal reflection fluorescence microscope. 0.1 mg/ml streptavidin was added to a polyethylene glycol‐coated quartz surface and incubated for 2 min before being washed with T50 (10 mM Tris–HCl (pH 8.0), 50 mM NaCl). Biotinylated Cas9 was pre‐incubated with Cy5‐labeled crRNA and tracrRNA (ratio 1:2:4) at 37 degrees for 20 min in NEB buffer 3 (100 mM NaCl, 50 mM Tris–HCl, 10 mM MgCl2, 1 mM DTT) and then added to the chamber containing streptavidin. After 2‐min incubation, unbound Cas9 and RNA molecules were washed away with an imaging buffer (50 mM HEPES‐NaOH [pH7.5], 10 mM NaCl, 2 mM MgCl2, 1% glucose (Dextrose monohydrate), 1 mM Trolox (2.5 mg/10 ml), 1 mg/ml glucose oxidase [Sigma], 170 μg/ml catalase [Merck]). 8 nM Cy3‐labeled DNA substrate in imaging buffer was added to the channel. A reference video of immobilized Cy5‐labeled Cas9:RNA complexes was made. Following the reference video, Cy3‐labeled DNA molecules were excited using a 532 nm diode laser. Fluorescence signals of Cy3 and Cy5 were collected through a 60× water immersion objective (UplanSApo, Olympus) with an inverted microscope (IX73, Olympus). The 532 nm laser scattering was blocked out by a 532‐nm long‐pass filter (LPD01‐532RU‐25, Semrock). The Cy3 and Cy5 signals were separated with a dichroic mirror (635 dcxr, Chroma) and imaged using an EMCCD camera (iXon Ultra, DU‐897U‐CS0‐#BV, Andor Technology). RNA and DNA sequences used can be found in Appendix Tables S1 and S2.
Data acquisition and analysis
Using a custom‐made program written in Visual C++ (Microsoft), a series of CCD images of time resolution 0.1 s was recorded. The time traces were extracted from the CCD image series using IDL (ITT Visual Information Solution) employing an algorithm that looked for fluorescence spots with a defined Gaussian profile and with signals above the average of the background signals. Colocalization between Cy3 and Cy5 signals was carried out with a custom‐made mapping algorithm written in IDL. The extracted time traces were processed using MATLAB (MathWorks) and Origin (OriginLab).
Author contributions
VG and SHL performed single‐molecule experiments; VG and SHL performed data analysis; SHL and TB performed protein purification; VG, SHL, CJ and J‐SK wrote and discussed the manuscript.
Conflict of interest
J.‐S.K. is a co‐founder of and holds stock in ToolGen, Inc. The remaining authors declare no competing interests.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 1
Acknowledgements
We would like to acknowledge Luuk Loeff and Sungchul Kim for their assistance in setting up the experiments. We are grateful to Chun Heung Wong, Iasonas Katechis, and Sabina Colombo for critical reading of the manuscript. C.J. was funded Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) (Netherlands Organisation for Scientific Research) VIDI grant (864.11.005) and the Frontiers of Nanoscience program (NWO). J.‐S.K. was supported by grants from the Institute for Basic Science (IBS‐R021‐D1).
The EMBO Journal (2019) 38: e99466
See also: A Santiago‐Frangos et al (February 2019)
Contributor Information
Jin‐Soo Kim, Email: jskim01@snu.ac.kr.
Chirlmin Joo, Email: c.joo@tudelft.nl.
References
- Amitai G, Sorek R (2016) CRISPR‐Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol 14: 67–76 [DOI] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M (2014) Structural basis of PAM‐dependent target DNA recognition by the Cas9 endonuclease. Nature 513: 569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Barrangou R, Doudna JA (2016) Applications of CRISPR technologies in research and beyond. Nat Biotechnol 34: 933–941 [DOI] [PubMed] [Google Scholar]
- Berg OG, Winter RB, von Hippel PH (1981) Diffusion‐driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry 20: 6929–6948 [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151: 2551–2561 [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandradoss SD, Haagsma AC, Lee YK, Hwang JH, Nam JM, Joo C (2014) Surface passivation for single‐molecule protein studies. J Vis Exp e50549, 10.3791/50549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandradoss SD, Schirle NT, Szczepaniak M, MacRae IJ, Joo C (2015) A dynamic search process underlies MicroRNA targeting. Cell 162: 96–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E (2011) CRISPR RNA maturation by trans‐encoded small RNA and host factor RNase III. Nature 471: 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard KE, Brown MW, Johnson NV, Xiao Y, Dolan AE, Hernandez ET, Dahlhauser S, Kim Y, Myler LR, Anslyn E, Ke A, Finkelstein I (2018) Assembly and translocation of a CRISPR‐Cas primed acquisition complex. Cell 175: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9‐crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109: E2579–E2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J, Wang F, Redding S, Plys AJ, Fazio T, Wind S, Alani EE, Greene EC (2012) Single‐molecule imaging reveals target‐search mechanisms during DNA mismatch repair. Proc Natl Acad Sci USA 109: E3074–E3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett‐Engele P, Lim LP, Bartel DP (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford SE, Marko JF (2004) How do site‐specific DNA‐binding proteins find their targets? Nucleic Acids Res 32: 3040–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J (2012) The lac repressor displays facilitated diffusion in living cells. Science 336: 1595–1598 [DOI] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA (2015) Cas9 specifies functional viral targets during CRISPR‐Cas adaptation. Nature 519: 199–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR‐Cas9 for genome engineering. Cell 157: 1262–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA‐guided editing of bacterial genomes using CRISPR‐Cas systems. Nat Biotechnol 31: 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Leroy P, Unoson C, Fange D, Curic V, Lawson MJ, Elf J (2017) Kinetics of dCas9 target search in Escherichia coli . Science 357: 1420–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, Ha T (2012) Labeling DNA (or RNA) for single‐molecule FRET. Cold Spring Harb Protoc 2012: 1005–1008 [DOI] [PubMed] [Google Scholar]
- Leith JS, Tafvizi A, Huang F, Uspal WE, Doyle PS, Fersht AR, Mirny LA, van Oijen AM (2012) Sequence‐dependent sliding kinetics of p53. Proc Natl Acad Sci USA 109: 16552–16557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV (2006) A putative RNA‐interference‐based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R et al (2015) An updated evolutionary classification of CRISPR‐Cas systems. Nat Rev Microbiol 13: 722–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA (2015) CRISPR‐Cas immunity in prokaryotes. Nature 526: 55–61 [DOI] [PubMed] [Google Scholar]
- Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J (2016) Diverse evolutionary roots and mechanistic variations of the CRISPR‐Cas systems. Science 353: aad5147 [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez‐Villasenor C, Garcia‐Martinez J, Soria E (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60: 174–182 [DOI] [PubMed] [Google Scholar]
- O'Geen H, Henry IM, Bhakta MS, Meckler JF, Segal DJ (2015) A genome‐wide analysis of Cas9 binding specificity using ChIP‐seq and targeted sequence capture. Nucleic Acids Res 43: 3389–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B (2014) Unravelling the structural and mechanistic basis of CRISPR‐Cas systems. Nat Rev Microbiol 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragunathan K, Liu C, Ha T (2012) RecA filament sliding on DNA facilitates homology search. Elife 1: e00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD, Bourgeois S, Cohn M (1970) The lac repressor‐operator interaction. 3. Kinetic studies. J Mol Biol 53: 401–417 [DOI] [PubMed] [Google Scholar]
- Singh D, Sternberg SH, Fei J, Doudna JA, Ha T (2016) Real‐time observation of DNA recognition and rejection by the RNA‐guided endonuclease Cas9. Nat Commun 7: 12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA (2014) DNA interrogation by the CRISPR RNA‐guided endonuclease Cas9. Nature 507: 62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R (2014) Direct observation of R‐loop formation by single RNA‐guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA 111: 9798–9803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA (2012) RNA‐guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 1


