Abstract
Organoids are self‐organizing 3D structures grown from stem cells that recapitulate essential aspects of organ structure and function. Here, we describe a method to establish long‐term‐expanding human airway organoids from broncho‐alveolar resections or lavage material. The pseudostratified airway organoids consist of basal cells, functional multi‐ciliated cells, mucus‐producing secretory cells, and CC10‐secreting club cells. Airway organoids derived from cystic fibrosis (CF) patients allow assessment of CFTR function in an organoid swelling assay. Organoids established from lung cancer resections and metastasis biopsies retain tumor histopathology as well as cancer gene mutations and are amenable to drug screening. Respiratory syncytial virus (RSV) infection recapitulates central disease features, dramatically increases organoid cell motility via the non‐structural viral NS2 protein, and preferentially recruits neutrophils upon co‐culturing. We conclude that human airway organoids represent versatile models for the in vitro study of hereditary, malignant, and infectious pulmonary disease.
Keywords: 3D culture, airway organoids, cystic fibrosis, lung cancer, respiratory syncytial virus
Subject Categories: Cancer, Methods & Resources, Molecular Biology of Disease
Introduction
To date, several approaches have been explored to generate mammalian airway organoids (Barkauskas et al, 2017). In 1993, Puchelle and colleagues described the first self‐organizing 3D structures of adult human airway epithelium in collagen (Benali et al, 1993). A first description of the generation of lung organoids from human iPS (induced pluripotent stem) cells was given by Rossant and colleagues and included the use of CFTR‐mutant iPS cells as a proof of concept for modeling CF (Wong et al, 2012). Snoeck and colleagues designed an improved four‐stage protocol (Huang et al, 2014) and later generated lung bud organoids from human pluripotent stem cells that recapitulate fetal lung development (Chen et al, 2017). Spence and colleagues (Dye et al, 2015) followed a modified trajectory to generate mature lung organoids, containing basal, ciliated, and club cells. These cultures were stable for up to several months and resembled proximal airways. Konishi et al (2016) improved on the iPS cell‐derived generation of multi‐ciliated airway cells in 3D, and McCauley et al (2017) generated CF patient iPS cell‐derived airway organoids for disease modeling. Hogan and colleagues reported the first adult stem cell‐based murine bronchiolar lung organoid culture protocol, involving Matrigel supplemented with EGF (Rock et al, 2009). Single basal cells isolated from the trachea grew into tracheospheres consisting of a pseudostratified epithelium with basal and ciliated luminal cells. These organoids could be passaged at least twice. No mature club, neuroendocrine, or mucus‐producing cells were observed (Rock et al, 2009). In a later study, this clonal 3D organoid assay was used to demonstrate that IL‐6 treatment resulted in the formation of ciliated cells at the expense of secretory and basal cells (Tadokoro et al, 2014). Tschumperlin and colleagues combined human adult primary bronchial epithelial cells, lung fibroblasts, and lung microvascular endothelial cells in 3D to generate airway organoids (Tan et al, 2017). Under these conditions, randomly seeded mixed cell populations underwent rapid condensation to self‐organize into discrete epithelial and endothelial structures that were stable up to 4 weeks of culture (Tan et al, 2017). Hild and Jaffe have described a protocol for the culture of bronchospheres from primary human airway basal cells. Mature bronchospheres are composed of functional multi‐ciliated cells, mucin‐producing secretory cells, and airway basal cells (Hild & Jaffe, 2016). Mou et al (2016) expanded basal cells of mouse and human airway epithelium in 2D that allowed subsequent differentiation under air–liquid interphase conditions. And finally, Nikolic et al (2017) designed conditions to expand human fetal lung epithelium as self‐renewing organoids.
Since none of these approaches allows long‐term expansion of pseudostratified airway epithelium from adult human individuals in vitro, we set out to establish such culture conditions and model a variety of pulmonary diseases.
Results
Generation and characterization of human airway organoids
We collected macroscopically inconspicuous lung tissue from non‐small‐cell lung cancer (NSCLC) patients undergoing medically indicated surgery and isolated epithelial cells through mechanical and enzymatic tissue disruption (see Materials and Methods). Following our experience with generating organoids from other adult human tissues (Sato et al, 2011; Karthaus et al, 2014; Boj et al, 2015; Huch et al, 2015; van de Wetering et al, 2015) and recent developments in the field (Mou et al, 2016; Tadokoro et al, 2016; Balasooriya et al, 2017), we embedded isolated cells in basement membrane extract (BME) and activated/blocked signaling pathways important for airway epithelium (Table EV1). Under optimized conditions, 3D organoids formed within several days (94% success rate, n = 18). The organoids were composed of a polarized, pseudostratified airway epithelium containing basal, secretory, and multi‐ciliated cells (Fig 1A and B, Appendix Fig S1A, Movie EV1) and were termed airway organoids (AOs). Cells that stained for basal cell marker keratin‐5 (KRT5), club cell marker secretoglobin family 1A member 1 (SCGB1A1), cilia marker acetylated α‐tubulin, or secretory cell marker mucin 5AC (MUC5AC) localized to their corresponding in vivo positions (Fig 1C, Appendix Fig S1B). Secretory cells as well as cilia were functional as evidenced by time‐lapse microscopy showing beating cilia and whirling mucus (Movies EV2 and EV3).
Figure 1. Characterization of airway organoids.
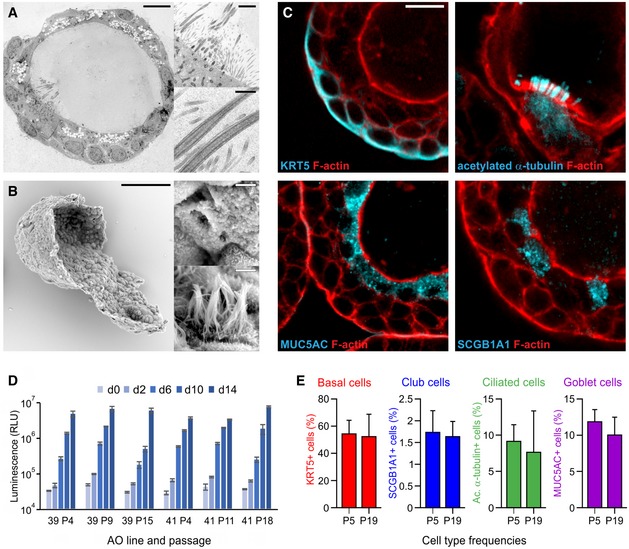
- Transmission electron micrograph of an AO cross section showing the polarized, pseudostratified epithelium containing basal, secretory, brush, and multi‐ciliated cells. Details display apical microvilli and cilia with their characteristic microtubule structure. Scale bars equal 10 μm, 2 μm, and 500 nm. See also Appendix Fig S1A and [Link], [Link], [Link].
- Scanning electron micrograph of a partially opened AO visualizing its 3D architecture, as well as basal and apical ultrastructure. Details display apical surfaces of secretory and multi‐ciliated cells. Scale bars equal 50 μm (overview) and 2 μm (details).
- Immunofluorescent sections of AOs showing markers for basal cells (KRT5), cilia (acetylated α‐tubulin), secretory cells (MUC5AC), and club cells (SCGB1A1). KRT5 is present exclusively in basally localized cells, while cilia, MUC5AC, and SCGB1A1 localize luminally. Counterstained is the actin cytoskeleton (red). Scale bar equals 10 μm. See Appendix Fig S1B for IHC images.
- Luminescent cell viability assay comparing proliferative capacity of two independently generated AO lines at early, mid‐, and late passage numbers. Per group, 3,000 cells were seeded and their expansion was measured at the indicated time points. Error bars represent standard deviations of technical triplicates.
- Quantification of cell types in AO lines at early and late passage (P5 vs. P19) as determined by immunofluorescence using the indicated markers. The number of basal cells, club cells, ciliated cells, and secretory cells does not differ significantly between early and late passage AOs. Data shown are representatives of at least three independent experiments. Error bars indicate s.e.m.
Airway organoids were passaged by mechanical disruption at 1:2 to 1:4 ratios every other week for > 1 year, proliferating at comparable rates regardless of passage number (Fig 1D) while retaining similar frequencies of basal, club, multi‐ciliated, and secretory cells (Fig 1E, Appendix Fig S1C). Comparative RNA sequencing of early and late passage AOs confirmed these findings with dozens of airway cell type‐specific genes retaining their respective expression patterns (Appendix Fig S1D and E, Table EV2). The airway epithelial composition of 10 independently established AO lines was validated by quantitative PCR (qPCR): While expressing the general lung marker NKX2‐1 and several airway‐specific markers, AOs expressed virtually no HOXA5 [a bona fide lung mesenchyme gene (Hrycaj et al, 2015)] or alveolar transcripts (Appendix Fig S2A). While AO transcriptomes were strongly enriched for bulk lung and small airway epithelial signature (Appendix Fig S2B) as shown by gene set enrichment analysis (GSEA), cell type‐specific signatures were limited to basal, club, and ciliated cells (Appendix Fig S2C, Table EV3). Accordingly, hallmark lung genes encoding for keratins, secretoglobins, dyneins, and others were consistently among the highest AO enriched genes (Appendix Fig S2D).
Elevated levels of WNT3A transcripts explained why AOs—in contrast to intestinal organoids (Sato et al, 2011)—did not require the addition of exogenous WNT3A to the culture media. Manipulation of WNT signaling resulted in dramatic changes in the expression of WNT target genes (Appendix Fig S2E). Withdrawal of the Wnt amplifier R‐spondin terminated AO expansion after 3–4 passages (Appendix Fig S2E), similar to withdrawal of fibroblast growth factors (Appendix Fig S2F).
Taken together, our culture conditions allow long‐term expansion of AOs while retaining major characteristics of the in vivo epithelium.
Airway organoids from patients with cystic fibrosis recapitulate central disease features and swell upon modulation of CFTR as well as activation of TMEM16A
Rectal organoids are being successfully used as functional model for cystic fibrosis (CF; Noordhoek et al, 2016), a multi‐organ disease with extensive phenotypic variability caused by mutations in the CF transmembrane conductance regulator gene (CFTR; Ratjen et al, 2015). Following opening of the CFTR channel by cAMP‐inducing agents (e.g., forskolin), anions and fluid are transported to the organoid lumen resulting in rapid organoid swelling (Dekkers et al, 2013), allowing personalized in vitro drug screenings (Dekkers et al, 2016). The current gold standard for modeling the primarily affected CF lung epithelium is air–liquid interface (ALI) culture of human bronchial epithelial cells, a system with limited cell expansion and lengthy differentiation protocols (Fulcher et al, 2005). While iPS cell‐derived AOs have been used for functional assessment of CFTR, their generation is also considerably long (McCauley et al, 2017). To assess adult stem cell AOs for CF disease modeling, we applied forskolin and observed a dose‐dependent swelling response that was largely, but not entirely, abrogated upon chemical inhibition of CFTR (Fig 2A, Appendix Fig S3A), indicating the presence of additional ion channels. Indeed, AOs—but not rectal organoids—swell upon addition of Eact (Fig 2B, Appendix Fig S3A), an activator of the chloride channel TMEM16A (Namkung et al, 2011; Sondo et al, 2014).
Figure 2. Airway organoids to study cystic fibrosis.
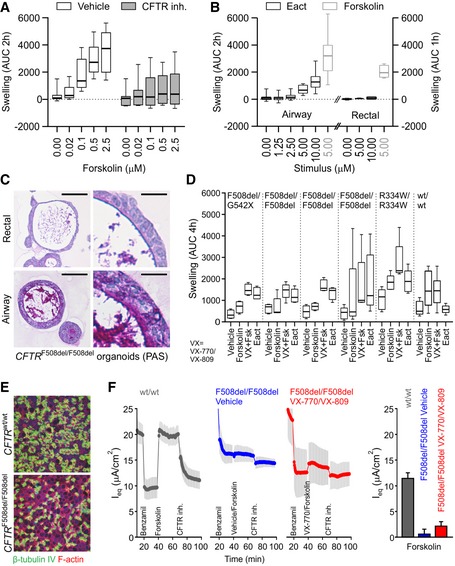
- Box‐and‐whisker plot showing concentration‐dependent forskolin‐induced swelling of AOs in the absence and presence of CFTR inhibitors CFTRinh‐172 and GlyH101. Upon CFTR inhibition, swelling is noticeably decreased but not absent. Shown are pooled data from three different AO lines used in each of three independent experiments. Whiskers indicate smallest and largest values, boxes indicate 25th to 75th percentile, and horizontal solid line indicates median. AUC, area under the curve.
- Box‐and‐whisker plot showing concentration‐dependent Eact‐induced swelling of AOs, but not rectal organoids (black outlines). Forskolin causes swelling in both organoid types (gray outlines). Shown are pooled data from three different AO and two different rectal organoid lines used in three to four independent experiments. Whiskers indicate smallest and largest values, boxes indicate 25th to 75th percentile, and horizontal solid line indicates median. Swelling was linear for 2 h for AOs, but only 1 h for rectal organoids. See Appendix Fig S3A for respective time course plots.
- Representative histological sections of periodic acid–Schiff (PAS)‐stained organoids from a CF patient with CFTR F508del/F508del mutation. Note the thick layer of PAS‐positive polysaccharides apically lining the airway epithelium. Rectal organoids were generated from rectal biopsies; AOs were generated from broncho‐alveolar lavages (BALs). Scale bars equal 50 μm. See Appendix Fig S3B for PAS‐stained wild‐type and CFTR R334W/R334W organoid sections.
- Box‐and‐whisker plot showing swelling assays of several CF patient AO lines carrying the indicated CFTR mutations (G542X is a premature stop associated with severe disease and no functional CFTR protein; F508del is the most common CFTR mutation in subjects with CF and severely reduces apical trafficking and function, leading to severe disease (high sweat chloride, high pancreas insufficiency, high pseudomonas infection rate); R334W is a milder CFTR mutation associated as indicated by lower pseudomonas infection rates and pancreas sufficiency with reduced ion channel conductivity, normal apical expression, and some residual function). Forskolin‐induced swelling rarely exceeds vehicle controls in CF AOs, but increases in the presence of the CFTR modulating drugs VX‐770 and VX‐809. Eact‐induced swelling exceeds forskolin‐induced swelling to a similar extent as pre‐treatment with VX‐770 and VX‐809 in four out of five CF AO lines. In contrast, BAL‐derived WT AOs swell following forskolin stimulation independent of the presence of VX‐770 and VX‐809. Of note, Eact‐induced swelling is not observed in this AO line. Shown are pooled data of four to five independent experiments (CF AOs) or two independent experiments (WT AOs). Whiskers indicate smallest and largest values, boxes indicate 25th to 75th percentile, and horizontal solid line indicates median. See Appendix Fig S3C for selected time course plots.
- Whole‐mounted immunofluorescence analysis of BAL AO‐derived ALI cultures. After 3 weeks of differentiation, ALI cultures show ciliated airway epithelium (green, β‐tubulin). Counterstained are the actin cytoskeleton (red) and nuclei (blue).
- Transepithelial electrical measurements of BAL AO‐derived ALI cultures. Shown are equivalent current (I eq) traces of a CFTR wt/wt (gray) and CFTR F508del/F508del donor, treated with vehicle (blue) or VX‐809 and VX‐770 (red). Inhibition of the epithelial sodium channel (ENaC) with benzamil was observed in all traces (declining I eq). Forskolin increased the current in CFTR wt/wt and CFTR F508del/F50del donor treated with VX‐809 and VX‐770, but not in the vehicle treated. A decline was observed with CFTRinh‐172 in all examined conditions. The bar graphs quantify the peak forskolin activated‐ and CFTRinh‐172 inhibited‐currents. Shown are pooled data of four independent measurements for each donor. Results are shown as mean ± s.d.
We next established AO lines from fresh or cryopreserved broncho‐alveolar lavage fluids including five CF patients (62% success rate, n = 8). CF AOs presented a much thicker layer of apical mucus compared to patient‐matched rectal organoids and wild‐type AOs (Fig 2C, Appendix Fig S3B), recapitulating aspects of the in vivo CF phenotype. Forskolin‐induced swelling was reduced in CF compared to wild‐type AOs, correlated with severity of tested CFTR genotypes (Sosnay et al, 2013), and could be augmented with the CFTR modulators VX‐770 and VX‐809 (Fig 2D, Appendix Fig S3C) in agreement with clinical data (Kuk & Taylor‐Cousar, 2015). Eact stimulation generally caused a swelling response like stimulation with forskolin ± VX‐770 and VX‐809 (Fig 2D, Appendix Fig S3C). It remains to be determined if AO swelling is mediated by recently described rare ionocytes present in vivo as well as traditional ALI cultures (Montoro et al, 2018; Plasschaert et al, 2018).
Both CF and wild‐type AOs could be used to generate ALI cultures on transwell membranes. The AO‐derived ALIs displayed accessible multi‐ciliated airway epithelia (Fig 2E) and allowed studying CFTR function in Ussing chambers (Fig 2F). Taken together, our culture method allows expansion of limited CF patient material to be subjected to traditional ALI cultures and, by recapitulating central disease features, provides an alternative option for in vitro modeling of CF, a classic example of a monogenic disease.
Lung cancer organoids recapitulate tumor histopathology, mutation status, and allow xenotransplantations and in vitro drug screening
We next tested if AOs can be used to model lung cancer, a global respiratory disease burden (Chen et al, 2014; Ferkol & Schraufnagel, 2014). We have previously observed that remnants of normal epithelium in carcinoma samples will rapidly overgrow tumor tissue (Karthaus et al, 2014; van de Wetering et al, 2015). While we successfully generated organoid lines in the majority of cases (88% success rate, n = 16), we could not selectively expand lung tumoroids by removing a single medium component [such as Wnt3A in colon tumoroids (van de Wetering et al, 2015)], due to the diversity of mutated signaling pathways in lung cancer (Chen et al, 2014). We reasoned that Nutlin‐3a (Vassilev et al, 2004) could drive TP53 wild‐type AOs into senescence or apoptosis and would allow outgrowth of tumoroids with mutant p53, present in a large proportion of NSCLCs (Deben et al, 2016).
Using Nutlin‐3a selection, we then generated pure lung tumoroid lines from different NSCLC subtypes recapitulating fundamental histological characteristics of the respective primary tumors (Fig 3A). In addition, we established pure tumoroid lines from needle biopsies of metastatic NSCLCs, which circumvented the need for Nutlin‐3a selection (Appendix Fig S4A) due to the absence of normal lung tissue (28% success rate, n = 18). Upon orthotopic transplantation into immunocompromised mice, tumor AOs gave rise to lung cancer in 30% of the cases (n = 12) after 2 months (Fig 3B). Whole‐genome sequencing (WGS) of primary tumor tissues and corresponding tumor AO lines identified matching missense and oncogenic mutations in cancer‐associated genes including ALK, EMR1, MDN1, and others (Fig 3C, Appendix Table S1). Hotspot DNA sequencing of selected cancer genes in additional tumor AO lines revealed loss‐ (e.g., STK11, TP53) and gain‐of‐function mutations (e.g., KRAS, ERBB2; Fig 3C, Appendix Table S2).
Figure 3. Modeling lung cancer using airway organoids.
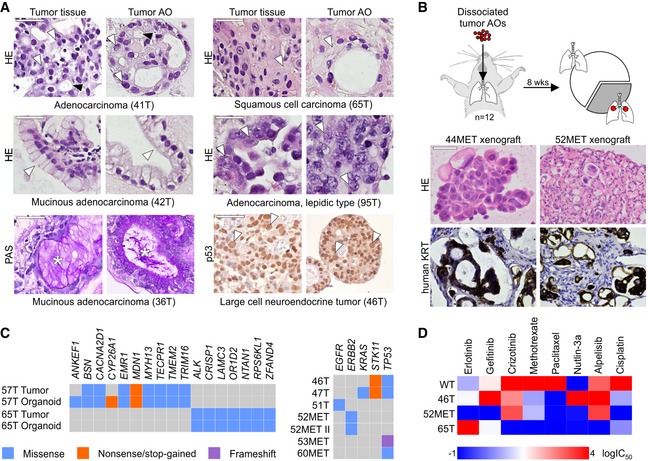
- Tumor AOs derived from different resected primary lung cancer types recapitulate their respective histopathological features. Examples include adenocarcinoma (41T, white arrowheads: multiple nucleoli, black arrowheads: tubule formation), mucinous adenocarcinoma (42T/36T, arrowheads: mucinous glands, stars: PAS‐positive mucus), adenocarcinoma lepidic type (previously known as broncho‐alveolar carcinoma, 95T, arrowheads: distinct large nuclei), squamous cell carcinoma (65T, arrowheads: multi‐layered, keratinization), and large cell neuroendocrine tumor (46T, arrowheads: large nuclei, p53 immunolabeling). Scale bars equal 50 μm.
- Tumor AOs derived from metastatic adenocarcinomas give rise to lung cancer in vivo following intratracheal instillation into immunocompromised mice. 30% of injected mice (n = 12) developed visible tumors 8 weeks after instillation. Tumors show features of adenocarcinomas (HE stainings) and are integrated within murine stroma (human keratin immunolabeling). Scale bars equal 50 μm.
- WGS reveals largely conserved mutation status of lung cancer genes between matched tumor–organoid pairs (left plot, Appendix Table S1). The right plot shows mutation status of selected lung cancer genes in several additional tumor AO lines derived from primary as well as metastatic lung cancer. Of note, ERBB2 mutation frequencies in 52MET and 52MET II tumor AOs were almost identical (67 and 70%) despite originating from independent biopsies taken 3 months apart. See Appendix Table S2.
- Heat map of IC50 values of selected anti‐cancer drugs indicating differential responses of tumor AOs. For example, 46T (mutant TP53) is resistant to Nutlin‐3a treatment, 65T (mutant ALK1) is resistant to treatment with crizotinib, and 52MET (mutant ERBB2) is sensitive to treatment with erlotinib and gefitinib. See Appendix Fig S4F for individual dose–response curves.
We next showed that tumor AOs can be used for drug screening (Weeber et al, 2017). As shown in Fig 3D, individual tumor AOs varied greatly in their respective responses in line with their mutational profile. For example, and as expected, TP53 mutant line 46T was resistant to treatment with the p53‐stabilizing drug Nutlin‐3a, the ERBB2 mutant line 52MET was sensitive to EGFR/ERBB2 inhibitors, and the ALK1 mutant line 65T was sensitive to ALK/ROS inhibition (see also Appendix Fig S4B). In conclusion, we provide basic protocols for the selective outgrowth of p53 mutant tumor AOs from primary and metastatic NSCLCs that recapitulate tumor histology as well as mutation status and can be used for in vitro drug screens.
RSV infection causes dramatic epithelial remodeling in airway organoids
Respiratory infections pose a major global disease burdens (Ferkol & Schraufnagel, 2014). RSV infections alone cause millions of annual hospital admissions and hundreds of thousands of deaths among young children (Nair et al, 2010) due to bronchiolitis, edema, and abnormalities of the airway epithelium (necrosis, sloughing; Borchers et al, 2013). iPS‐derived human airway epithelium can be infected by RSV virus (Chen et al, 2017), as can human airway epithelium grown under ALI conditions (Persson et al, 2014). We tested if AOs can serve as in vitro model for RSV infection. RSV replicated readily in multiple AO lines (Fig 4A, Movie EV4) but failed to do so after pre‐incubation with commercial palivizumab (an antibody against the RSV F‐glycoprotein that prevents RSV–cell fusion) indicating specific virus–host interaction (Fig 4B; Huang et al, 2010). Morphological analysis of RSV‐infected AOs revealed massive epithelial abnormalities (Fig 4C) that recapitulated in vivo phenomena including cytoskeletal rearrangements, apical extrusion of infected cells, and syncytia formation (Fig 4C and D). We used time‐lapse microscopy to visualize the underlying dynamics and surprisingly found RSV‐infected AOs to rotate and move through BME, often fusing with neighboring AOs (Fig 4E, Movie EV4). Organoid motility was caused by increased motilities of all cells within RSV‐infected AOs (Fig 4F, Appendix Fig S5A–C, Movies EV5 and EV6). Mathematical modeling and simulation suggested organoid rotation to be the macroscopic manifestation of coordinated cell motilities (Appendix Fig S5D–G, Movie EV7). RNA sequencing of RSV‐ vs. mock‐infected AOs showed strong enrichment for genes involved in interferon α/β signaling (particularly cytokines), which correlated with enrichment of migratory as well as of antiviral response genes (Fig 4G and H, Table EV4). Enzyme‐linked immunosorbent assays confirmed the secretion of significant quantities of cytokines such as IP‐10 and RANTES from RSV‐infected AOs (Fig 4I). To test whether secreted cytokines attract neutrophils [known to accumulate at RSV infection sites in vivo (Geerdink et al, 2015)], we co‐cultured RSV‐infected AOs with freshly isolated primary human neutrophils. Of note, Yonker et al (2017) have described co‐culture of neutrophils with RSV‐infected human airway epithelium grown in 2D ALI culture. Time‐lapse microscopy indeed showed preferential neutrophil recruitment to RSV‐infected AOs compared to mock controls (Fig 4J, Movies EV8 and EV9) allowing experimental dissection of immune cell interaction with RSV‐infected airway epithelium ex vivo. Mechanistically, the non‐structural protein NS2 has been shown to induce the shedding of dead cells in vivo, leading to airway obstructions as well as to be required for efficient propagation of the virus in vivo (Teng & Collins, 1999; Liesman et al, 2014). RSVΔNS2 indeed replicated less efficiently than RSVwt in AOs (Fig 4K, Movie EV10), while inducible overexpression of NS2 alone in non‐infected AOs induced motility and fusion of RSV‐infected AOs, underlining a previously unappreciated role of the protein in the modification of the behavior of the infected cell (Fig 4L, Appendix Fig S6, Movie EV11). Taken together, RSV‐infected AOs recapitulate central disease features and reveal their underlying highly dynamic epithelial remodeling.
Figure 4. Modeling RSV infection with airway organoids.
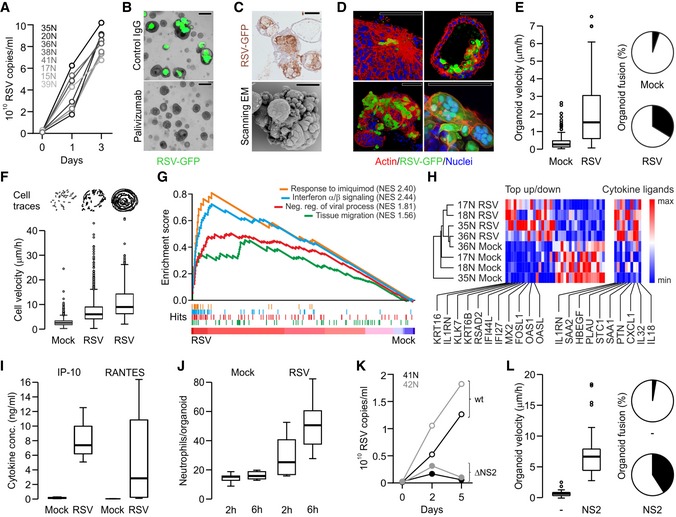
- Quantitative PCR showing RSV replication kinetics in the indicated AO lines.
- Representative phase‐contrast/GFP overlays of RSV‐infected AOs (5 days post‐infection). GFP signal is absent following pre‐incubation with palivizumab (fusion blocking antibody) but not control IgG. Scale bars equal 100 μm.
- Immunohistochemical RSV staining (top) and scanning electron micrograph (bottom) of RSV‐infected AOs (5 days post‐infection) show organoid fusion and blebbing. Scale bars equal 50 μm.
- 3D reconstructions of immunolabeled AOs 3 days after RSV infection showing (clockwise) cytoskeletal rearrangements, apical extrusion of infected cells, syncytia formation, and organoid fusion. Scale bars equal 50 μm.
- RSV infection causes increased organoid velocity (Tukey boxplot) and organoid fusion (pie chart). Shown are data from three independent experiments. Horizontal solid line indicates median, box indicate 25th to 75th percentile, whiskers indicate 25th percentile minus 1.5 times inter‐quartile distance (IQR) or 75th percentile plus 1.5 times IQR, and circles indicate individual values greater than 1.5 times IQR. See also Movie EV4.
- Cells within RSV‐infected organoids are more motile than cells in control organoids irrespective of induced organoid rotation. Cell traces above individual Tukey boxplots are from representative organoids. Shown are data from two to three independent experiments. Horizontal solid line indicates median, box indicates 25th to 75th percentile, whiskers indicate 25th percentile minus 1.5 times inter‐quartile distance (IQR) or 75th percentile plus 1.5 times IQR, and circles indicate individual values greater than 1.5 times IQR. See also Appendix Fig S5A and B, and Movies EV5 and EV6.
- GSEA plots showing strong enrichment of indicated gene signatures in transcriptomes of four independently RSV‐ vs. mock‐infected AO lines. NES, normalized enrichment score. See Table EV4 for signatures and leading‐edge genes.
- Hierarchical clustering of the indicated AO lines displaying the 36 most differentially expressed genes of RSV‐ vs. mock‐infected AO transcripts as well as selected cytokines (see Table EV4). Gradients depict the relative maximum and minimum values per transcript. Names and ranks of selected genes involved in migration (KRT16, KRT6B), interferon signaling (IL1RN, IFI44L), and viral response (MX2, OAS1) are indicated.
- ELISA‐based quantification of cytokines secreted by mock‐ vs. RSV‐infected AOs. Shown are data from two independent experiments. Whiskers indicate smallest and largest values, boxes indicate 25th to 75th percentile, and horizontal solid line indicates median.
- Box‐and‐whisker plot showing increased numbers of primary human neutrophils populating RSV‐ compared to mock‐infected AOs. Shown are data from two independent experiments. Whiskers indicate smallest and largest values, boxes indicate 25th to 75th percentile, and horizontal solid line indicates median. See also Movies EV8 and EV9.
- Quantitative PCR showing replication kinetics of wild‐type (wt) and mutant RSV lacking NS2 (ΔNS2) in the indicated AO lines. See also Movie EV10.
- Inducible overexpression of NS2 causes increased organoid velocity (box‐and‐whisker plot) and fusion (pie chart). Shown are data from three independent experiments. Horizontal solid line indicates median, box indicates 25th to 75th percentile, whiskers indicate 25th percentile minus 1.5 times inter‐quartile distance (IQR) or 75th percentile plus 1.5 times IQR, and circles indicate individual values greater than 1.5 times IQR. See Appendix Fig S6, Movie EV11.
Discussion
Here, we describe a versatile approach to establishing adult human airway epithelial organoids, containing all major cellular elements. Major differences to other lung organoid models are the relative ease with which AOs can be grown from small amounts of routinely obtained patient materials (lavages, biopsies, resections) and their long‐term expansion from healthy individuals, as well as from patients with a hereditary (CF) or malignant lung disease (NSCLC). We show that organoids derived from individual CF and lung cancer patients are amenable to medium‐throughput drug screening. Such organoid drug screening is already proving its value in personalized medicine. In the Netherlands, a personalized swelling test on rectal organoids formally allows prescription of the CF drug Orkambi® to patients with rare variants of CF (Technology, 2017). Excitingly, Vlachogiannis et al (2018) recently demonstrated convincingly that tumoroids are robust predictors of drug response in patients with gastrointestinal malignancies. The current protocol provides similar opportunities for personalized approaches in CF (bypassing the need for rectal biopsies) and lung cancer. Indeed, our AO protocol has been successfully used to test individualized cancer immunotherapy by co‐culture of peripheral blood lymphocytes and tumor AOs (Dijkstra et al, 2018). Finally, we show that AOs readily allow modeling of viral infections such as RSV and present in vitro evidence for the direct effects of the viral protein NS2 on cell mobility and fusion and we demonstrate the possibility to study neutrophil–epithelium interaction in an organoid model. Validating the robustness of AOs as model for human pulmonary infections are recent studies with enterovirus (van der Sanden et al, 2018), influenza virus (Hui et al, 2018; Zhou et al, 2018), and cryptosporidium (Heo et al, 2018). Taken together, we anticipate that human AOs will continue to find broad applications in the study of adult human airway epithelium in health and disease.
Materials and Methods
Reagents and Tools table
| Reagent/resource | Reference or source | Identifier or catalog number |
|---|---|---|
| Experimental models | ||
| NSG (Mus musculus) | Netherlands Cancer Institute | NOD.Cg‐PrkdcscidIl2rgtm1Wjl/SzJ |
| Recombinant DNA | ||
| pCSCMV:tdTomato | Addgene | 30530 |
| pLVX‐TetOne™‐Puro | Clontech | 631849 |
| Antibodies | ||
| APC anti‐human EPCAM | Biolegend | 369810 |
| PE anti‐human NGFR | Biolegend | 345106 |
| 488 anti‐human CD24 | Biolegend | 311108 |
| Rabbit anti‐keratin 14 | Biolegend | 905301 |
| Goat anti‐SCGB1A1 | Santa Cruz | sc‐9773 |
| Mouse anti‐acetylated α‐tubulin | Santa Cruz | sc‐23950 |
| Mouse anti‐mucin 5AC (IF) | Santa Cruz | sc‐21701 |
| Mouse anti‐Ki67 | Monosan | MONX10283 |
| Rabbit anti‐keratin 5 | Covance | PRB 160P‐100 |
| Mouse anti‐mucin 5AC (IHC) | Novocastra Leica | NCL‐HGM‐45M1 |
| Mouse anti‐p53 | Santa Cruz | sc‐126 |
| Mouse anti‐cytokeratin | Becton Dickinson | 345779 |
| Rabbit anti‐GFP | Thermo Fisher | A11122 |
| Mouse anti‐RSV | Abcam | ab35958 |
| Mouse anti‐β‐tubulin IV | Biogenex | MU178‐UC |
| Rabbit anti‐GAPDH | Abcam | ab‐9485 |
| Oligonucleotides and other sequence‐based reagents | ||
| qPCR primers | This study | Appendix Table S3 |
| Chemicals, enzymes, and other reagents | ||
| Advanced DMEM/F12 | Thermo Fisher | 12634‐028 |
| Collagenase | Sigma | C9407 |
| Fetal bovine serum (FBS) | Thermo Fisher | 10270106 |
| Red blood cell lysis buffer | Roche | 11814389001 |
| Sputolysin | Boehringer Ingelheim | SPUT0001 |
| Gentamicin/amphotericin solution | Thermo Fisher | R‐015‐10 |
| BME | Trevigen | 3533‐010‐02 |
| TrypLE Express | Invitrogen | 12605036 |
| Nutlin‐3a | Cayman Chemicals | 10004372 |
| Organoid culture components | Table EV1 | Table EV1 |
| Glutaraldehyde | Sigma | G5882 |
| Hexamethyldisilazane (HMDS) | Sigma | 440191 |
| Triton X‐100 | Sigma | T8532 |
| Bovine serum albumin (BSA) | Thermo Fisher | AM2616 |
| DAPI stain | Thermo Fisher | D1306 |
| Phalloidin‐atto 647 stain | Sigma | 65906 |
| Vectashield | Vectorlabs | H1400 |
| Wnt inhibitor IWP‐2 | Stemgent | 130‐095‐584 |
| Chir99021 | Stemgent | 130‐095‐555 |
| VX‐809 | Selleck Chemicals | s1565 |
| VX‐770 | Selleck Chemicals | s1144 |
| Calcein | Thermo Fisher | C3100MP |
| Forskolin | Tocris | 1099 |
| DMSO | Sigma | D8418 |
| Semi‐permeable transwell membranes | Corning | 3378 |
| Bovine collagen type I | Purecol | 5005 |
| Benzamil | Sigma | B2417 |
| Direct PCR (tail) | Viagen | 102‐T |
| Complete protease inhibitors | Roche | 11836170001 |
| PVDF membranes | Millipore | IPVH00010 |
| Paclitaxel | Sigma | T7402 |
| Methotrexate | Sigma | A6770 |
| Crizotinib | Sigma | PZ0240 |
| Cisplatin | Sigma | C2210000 |
| Erlotinib | Santa Cruz | S1023 |
| Alpelisib | LC Laboratories | A‐4477 |
| Gefitinib | Selleck Chemicals | S1025 |
| Cell recovery solution | Corning | 734‐0107 |
| RPMI 1640 | Thermo Fisher | 72400‐054 |
| Penicillin–streptomycin | Thermo Fisher | 15140122 |
| Hoechst33342 | Thermo Fisher | H3570 |
| Ficoll‐Paque PLUS | GE Healthcare | 17‐1440‐02 |
| Puromycin | Invivogen | ant‐pr‐1 |
| Doxycycline | Sigma | 324385 |
| Software | ||
| LAS AF software | https://www.leica-microsystems.com/products/microscope-software/details/product/leica-las-x-ls/ | |
| Hokawo 2.1 imaging software | https://camera.hamamatsu.com/eu/en/download_center/index.html | |
| ImageJ | https://imagej.nih.gov/ij/index.html | |
| Corel Draw X7 | https://www.coreldraw.com/en/pages/coreldraw-x7/ | |
| In‐house RNA analysis pipeline v2.1.0 | https://github.com/CuppenResearch/RNASeq | |
| Adobe Creative Cloud | https://www.adobe.com/gr_en/creativecloud/desktop-app.html | |
| Gene Set Enrichment Analysis (GSEA) | http://software.broadinstitute.org/gsea/index.jsp | |
| Morpheus | https://software.broadinstitute.org/morpheus | |
| Volocity 6.1.1 | https://download.cnet.com/Volocity/3000-2053_4-51574.html | |
| Zen Blue | https://www.zeiss.com/microscopy/int/products/microscope-software/zen-lite.html | |
| GraphPad Prism 5 and 6 | https://www.graphpad.com/ | |
| GATK HaplotypeCaller V3.5 | https://software.broadinstitute.org/gatk/download/archive | |
| GATK‐Queue V3.5 | https://software.broadinstitute.org/gatk/download/archive | |
| GATK VariantFiltration V3.5 | https://github.com/UMCUGenetics/IAP/blob/develop/settings/hartwig_kg.ini | |
| xPONENT version 4.2 | https://www.luminexcorp.com/download/manual-ivd-xponent-4-2-for-magpix-software-user/ | |
| Bio‐Plex Manager, version 6.1.1 | http://www.bio-rad.com/en-uk/product/bio-plex-manager-software-standard-edition?ID=5846e84e-03a7-4599-a8ae-7ba5dd2c7684 | |
| Other | ||
| FACSAriaII | BD Biosciences | |
| FACSJazz | BD Biosciences | |
| EVOS FL inverted microscope | Thermo Fisher | |
| Microplate luminometer | Berthold Technologies | |
| EM AFS2 | Leica | |
| UC6 ultramicrotome | Leica | |
| T12 Spirit electron microscope | Tecnai | |
| SZX9 light microscope | Olympus | |
| Q150R sputter coater | Quorum Technologies | |
| Phenom PRO tabletop scanning electron microscope | Phenom‐World | |
| AF7000 microscope | Leica | |
| DM4000 microscope | Leica | |
| Eclipse E600 microscope | Leica | |
| C9300‐221 high‐speed CCD camera | Hamamatsu Photonics | |
| SP8X confocal microscope | Leica | |
| LSM710 confocal microscope | Zeiss | |
| LSM800 confocal microscope | Zeiss | |
| SP5 confocal microscope | Leica | |
| Ultraview VoX spinning disk | Perkin Elmer | |
| CellTiter‐Glo 3D Cell Viability Assay | Promega | G9683 |
| RNeasy mini kit | Qiagen | 74104 |
| Bioanalyzer2100 RNA Nano 6000 chips | Agilent | 5067‐1511 |
| Truseq Stranded Total RNA kit set A | Illumina | RS‐122‐2201 |
| Truseq Stranded Total RNA kit set B | Illumina | RS‐122‐2202 |
| Bioanalyzer2100 DNA High Sensitivity chips | Agilent | 5067‐4626 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher | Q32854 |
| Illumina Nextseq | Illumina | |
| GoScript Reverse Transcriptase kit | Promega | A5003 |
| QIAquick PCR purification kit | Qiagen | 28104 |
| pGEM‐T Easy vector syst. I | Promega | A3600 |
| Nanodrop Lite | Thermo Fisher | |
| TruSeq Nano kit | Illumina | 20015965 |
| Illumina HiSeqX | Illumina | |
| DNeasy blood & tissue kit | Qiagen | 69506 |
| D300e Digital Dispenser | Tecan | |
| Micromanipulator | Narishige | M‐152 |
| Microinjector | Narishige | IM‐5B |
| Stereomicroscope | Leica | MZ75 |
| FlexMAP3D | Bio‐Rad Laboratories | |
Methods and Protocols
Procurement of human material and informed consent
The collection of patient data and tissue for the generation and distribution of airway organoids has been performed according to the guidelines of the European Network of Research Ethics Committees (EUREC) following European, national, and local law (Lanzerath, 2011). In the Netherlands, the responsible accredited ethical committees reviewed and approved the studies in accordance with the “Wet medisch‐wetenschappelijk onderzoek met mensen” (medical research involving human subjects act; Borst‐Eilers & Sorgdrager, 1998). The medical ethical committee UMC Utrecht (METC UMCU) approved protocols 07‐125/C (isolation and research use of neutrophils from healthy donors), TCBio 15‐159 (isolation and research use of broncho‐alveolar lavage fluid of CF patients), and TCBio 14‐008 (generation of organoids from rectal biopsies of CF patients). The “Verenigde Commissies Mensgebonden Onderzoek” of the St. Antonius Hospital Nieuwegein approved protocol Z‐12.55 (collection of blood, generation of normal and tumor organoids from resected surplus lung tissue of NSCLC patients). The Medical Ethics Committees of the Netherlands Cancer Institute Amsterdam approved PTC14.0929/M14HUP (collection of blood, generation of normal and tumor organoids from resected surplus lung tissue of NSCLC patients), and PTC14.0928/M14HUM (generation of tumor organoids from biopsies of metastatic NSCLC). All patients participating in this study signed informed consent forms approved by the responsible authority. In all cases, patients can withdraw their consent at any time, leading to the prompt disposal of their tissue and any derived material. AOs established under protocols Z‐12.55, PTC14.0929/M14HUP, and PTC14.0928/M14HUM were biobanked through Hubrecht Organoid Technology (HUB, www.hub4organoids.nl). Future distribution of organoids to any third (academic or commercial) party will have to be authorized by the METC UMCU/TCBio at request of the HUB in order to ensure compliance with the Dutch medical research involving human subjects’ act.
Tissue processing
Processing of solid lung tissue:
Solid lung tissue was minced, washed with 10 ml AdDF+++ (Advanced DMEM/F12 containing 1× Glutamax, 10 mM HEPES, and antibiotics), and digested in 10 ml AO medium (Table EV1) containing 1–2 mg ml−1 collagenase (Sigma‐C9407) on an orbital shaker at 37°C for 1–2 h.
The digested tissue suspension was sequentially sheared using 10‐ and 5‐ml plastic and flamed glass Pasteur pipettes. After every shearing step, the suspension was strained over a 100‐μm filter with retained tissue pieces entering a subsequent shearing step with ˜10 ml AdDF+++.
2% FCS was added to the strained suspension before centrifugation at 400 rcf. The pellet was resuspended in 10 ml AdDF+++ and centrifuged again at 400 rcf.
In case of a visible red pellet, erythrocytes were lysed in 2 ml red blood cell lysis buffer (Roche‐11814389001) for 5 min at room temperature before the addition of 10 ml AdDF+++ and centrifugation at 400 rcf.
Processing of broncho‐alveolar lavage fluid:
Broncho‐alveolar lavage fluid was collected and incubated with 10 ml airway organoid medium containing 0.5 mg ml−1 collagenase (Sigma‐C9407), 0.5% (w·v−1) Sputolysin (Boehringer Ingelheim‐SPUT0001), 250 ng ml−1 amphotericin B, and 10 μg ml−1 gentamicin on an orbital shaker at 37°C for 10–30 min.
The suspension was mildly sheared using a 1‐ml pipette tip and strained over a 100‐μm filter.
2% FCS was added to the strained suspension before centrifugation at 400 rcf. The pellet was resuspended in 10 ml AdDF+++ and centrifuged again at 400 rcf.
Organoid culture
Lung cell pellets were resuspended in 10 mg ml−1 cold Cultrex growth factor reduced BME type 2 (Trevigen‐3533‐010‐02), and 40 μl drops of BME‐cell suspension were allowed to solidify on pre‐warmed 24‐well suspension culture plates (Greiner‐M9312) at 37°C for 10–20 min.
Upon completed gelation, 400 μl of AO medium was added to each well and plates transferred to humidified 37°C/5% CO2 incubators at ambient O2.
Medium was changed every 4 days and organoids were passaged every 2 weeks: Cystic organoids were resuspended in 2 ml cold AdDF+++ and mechanically sheared through flamed glass Pasteur pipettes. Dense (organoids were dissociated by resuspension in 2 ml TrypLE Express (Invitrogen‐12605036), incubation for 1–5 min at room temperature, and mechanical shearing through flamed glass Pasteur pipettes.
Following the addition of 10 ml AdDF+++ and centrifugation at 300 or 400 rcf respectively, organoid fragments were resuspended in cold BME and reseeded as above at ratios (1:1–1:6) allowing the formation of new organoids. Single‐cell suspensions were initially seeded at high density and reseeded at a lower density after ˜1 week. Success rate was determined by dividing the number of successfully established, expanded, and cryopreserved AO lines by the number of attempts.
NSCLC organoids could be distinguished from normal regular cystic organoids by morphology (size, irregular shape, thick organoid walls, dense) as well as histology.
Separation from normal AOs was achieved by manual separation and in case of TP53 mutations by the addition of 5 μM Nutlin‐3a (Cayman Chemicals‐10004372) to the culture medium. For the R‐spondin withdrawal assay, established organoid lines were trypsinized to single cells and grown in AO medium ± R‐spondin until organoids were depleted. Intestinal organoids were cultured as previously described (Sato et al, 2011).
Unless specified, airway organoids were analyzed after at least 7 days post‐splitting at the indicated passage.
Luminescent viability assay
Single‐cell suspensions from AOs were generated as described and counted. 3 × 103 cells per replicate were plated in BME and incubated in AO medium as described. At the indicated time points, the medium was removed, and organoids were lysed in CellTiter‐Glo 3D (Promega) according to the manufacturer's instructions. Luminescence was measured using a microplate luminometer (Berthold Technologies).
Electron microscopy
Transmission EM:
Organoids were placed in BME on 3 mm diameter and 200 μm depth standard flat carriers for high‐pressure freezing and immediately cryoimmobilized using a Leica EM high‐pressure freezer (equivalent to the HPM10) and stored in liquid nitrogen until further use.
They were freeze‐substituted in anhydrous acetone containing 2% osmium tetroxide and 0.1% uranyl acetate at −90°C for 72 h and warmed to room temperature at 5°C per hour (EM AFS‐2, Leica). The samples were kept for 2 h at 4°C and 2 h at room temperature.
After several acetone rinses (4 × 15 min), samples were infiltrated with Epon resin for 2 days (acetone: resin 3:1—3 h; 2:2—3 h; 1:3—overnight; pure resin—6 h + overnight + 6 h + overnight + 3 h). Resin was polymerized at 60°C for 48 h.
Ultrathin sections from the resin blocks were obtained using an UC6 ultramicrotome (Leica) and mounted on Formvar‐coated copper grids. Grids were stained with 2% uranyl acetate in water and lead citrate.
Sections were observed in a Tecnai T12 Spirit electron microscope equipped with an Eagle 4kx4k camera (FEI Company), and large EM overviews were collected using the principles and software described earlier (Faas et al, 2012).
Scanning EM:
Organoids were removed from BME, washed with excess AdDF+++, fixed for 15 min with 1% (v/v) glutaraldehyde (Sigma) in phosphate‐buffered saline (PBS) at room temperature, and transferred onto 12‐mm poly‐L‐lysine coated coverslips (Corning).
Samples were subsequently serially dehydrated by consecutive 10‐min incubations in 2 ml of 10% (v/v), 25% (v/v) and 50% (v/v) ethanol–PBS, 75% (v/v) and 90% (v/v) ethanol–H2O, and 100% ethanol (2×), followed by 50% (v/v) ethanol–hexamethyldisilazane (HMDS) and 100% HMDS (Sigma).
Coverslips were removed from the 100% HMDS and air‐dried overnight at room temperature.
Organoids were manipulated with 0.5‐mm tungsten needles using an Olympus SZX9 light microscope and mounted onto 12‐mm specimen stubs (Agar Scientific).
Following gold‐coating to 1 nm using a Q150R sputter coater (Quorum Technologies) at 20 mA, samples were examined with a Phenom PRO tabletop scanning electron microscope (Phenom‐World).
Time‐lapse microscopy
Bright‐field AO time‐lapse movies were recorded at 37°C and 5% CO2 on an AF7000 microscope equipped with a DFC420C camera using LAS AF software (all Leica). Bright‐field cilia movement was recorded using the same setup equipped with a Hamamatsu C9300‐221 high‐speed CCD camera (Hamamatsu Photonics) at 150 frames per second using Hokawo 2.1 imaging software (Hamamatsu Photonics). Confocal imaging was performed using the following microscopes at 37°C and 5% CO2: SP8X (Leica), LSM710, LSM800 (both Zeiss), and Ultraview VoX spinning disk (Perkin Elmer).
Fixed immunofluorescence microscopy and immunohistochemistry
Organoids were removed from BME, washed with excess AdDF+++, fixed for 15 min in 4% paraformaldehyde, permeabilized for 20 min in 0.2% Triton X‐100 (Sigma), and blocked for 45 min in 1% BSA. Organoids were incubated with primary antibodies overnight at 4°C (anti‐keratin 14, Biolegend 905301; anti‐SCGB1A1, Santa Cruz sc‐9773; anti‐acetylated α‐tubulin, Santa Cruz sc‐23950; anti‐mucin 5AC, Santa Cruz sc‐21701), washed three times with PBS, incubated with secondary antibodies (Invitrogen) overnight at 4°C, washed two times with PBS, incubated with indicated additional stains (DAPI, Life Technologies D1306, phalloidin‐atto 647, Sigma 65906), washed two times with PBS, and mounted in VECTASHIELD hard‐set antifade mounting medium (Vectorlabs). Samples were imaged on SP5 and SP8X confocal microscopes using LAS X software (all Leica) and processed using ImageJ. For histological analysis, tissue and organoids were fixed in 4% paraformaldehyde followed by dehydration, paraffin embedding, sectioning, and standard HE and PAS stainings. Immunohistochemistry was performed using antibody against Ki67 (Monosan, MONX10283), keratin 5 (Covance, PRB 160P‐100), SCGB1A1 (Santa Cruz, sc‐9773), acetylated α‐tubulin (Santa Cruz, sc‐23950), Mucin 5AC (Novocastra Leica, NCL‐HGM‐45M1), P53 (Santa Cruz, sc‐126), human keratin (Becton Dickinson, 345779), GFP (Life Technologies, A11122), and RSV (Abcam, ab35958). Images were acquired on Leica DM4000 and Leica Eclipse E600 microscopes and processed using Corel Draw X7 and Adobe Creative Cloud software packages. Whole‐mount staining of air–liquid interface cultured‐cultured AOs was performed as previously described (Amatngalim et al, 2015) using anti‐β‐tubulin IV (Biogenex‐MU178‐UC).
RNA sequencing
Airway organoids derived from three independent human donors were collected 6 days after splitting (at passage 16, 18, and 19, respectively). Small intestinal organoids were also derived from three different human donors and collected 2 days after splitting (passage 7). Total RNA was isolated from the collected organoids using RNeasy kit (QIAGEN) according to manufacturer's protocol including DNaseI treatment. Quality and quantity of isolated RNA were checked and measured with Bioanalyzer2100 RNA Nano 6000 chips (Agilent, Cat. 5067‐1511). Library preparation was started with 500 ng of total RNA using the Truseq Stranded Total RNA kit with Ribo‐Zero Human/Mouse/Rat set A and B by Illumina (Cat. RS‐122‐2201 and RS‐122‐2202). After preparation, libraries were checked with Bioanalyzer2100 DNA High Sensitivity chips (Cat. 5067‐4626) as well as Qubit (Qubit® dsDNA HS Assay Kit, Cat. Q32854); all samples had a RIN value of 10. Libraries were equimolarly pooled to 2 nM and sequenced on the Illumina Nextseq, 1 × 75 bp high output (loaded 1.0–1.4 pM of library pools). Samples were sequenced to an average depth of 9.4 million mapped reads (SD 2.6 million). After sequencing, quality control, mapping, and counting analyses were performed using our RNA analysis pipeline v2.3.0 (https://github.com/UMCUGenetics?RNASeq/tree/2.3.0), which based on best practices guidelines (https://software.broadinstitute.org/gatk/guide/article?id=3891). In short, sequence reads were checked for quality by FastQC (v0.11.4) after which reads were aligned to GRCh37 using STAR (v2.4.2a) and add read groups using Picard perform quality control on generated BAM files using Picard (v1.141). Samples passing QC were then processed to count reads in features using HTSeq‐count (v0.6.1). Read counting for genes (ENSEMBL definitions GRCh37, release 74) resulted in a per sample count matrix with Ensembl gene identifiers. DeSeq (v1.18.0) was used for read normalization and differential expression analysis. Gene set enrichment analysis (GSEA) was performed using signature gene lists for airway basal cells (Dvorak et al, 2011), club cells (Treutlein et al, 2014), cilia (Dvorak et al, 2011), small airway epithelial cells (Hackett et al, 2012), and lung (Lindskog et al, 2014) against normalized RNA‐seq reads of three AO and three SIO lines using GSEA software v3.0 beta2 (Mootha et al, 2003; Subramanian et al, 2005). Normalized RNA‐seq reads were averaged per organoid type and sorted by the log2‐transformed ratio of AO over SIO. The 500 highest and 250 lowest transcripts were plotted using Morpheus (https://software.broadinstitute.org/morpheus) and CorelDraw X7.
RNA preparation and qRT–PCR
Total RNA was isolated from three independent lung organoid stains after 6, 12, 24, and 48 h upon either Wnt activation or Wnt inhibition by using RNeasy kit (QIAGEN). To inhibit Wnt signaling, 6 days after splitting the culture media of lung organoids was changed to media lacking R‐spondin and included IWP2 (3 μM, Stemgent). To activate Wnt signaling, Chir (3 μM) was added to the culture media 6 days after splitting. cDNA was synthesized from 1 μg of total RNA using GoScript (Promega). Quantitative PCR was performed in triplicate using the indicated primers, SYBR green, and Bio‐Rad systems. Gene expression was quantified using the ΔΔC t method and normalized by HPRT using the following primers:
HPRT (5′‐AAGAGCTATTGTAATGACCAGT‐3′ and 5′‐CAAAGTCTGCATTGTTTTGC‐3′), NKX2‐1 (5′‐ACCAAGCGCATCCAATCTCA‐3′ and 5′‐CAGAGCCATGTCAGCACAGA‐3′), HOXA5 (5′‐CGAGCCACAAATCAAGCACA‐3′ and 5′‐GAATTGCTCGCTCACGGAAC‐3′), CDH1 (5′‐TTACTGCCCCCAGAGGATGA‐3′ and 5′‐TGCAACGTCGTTACGAGTCA‐3′), CLDN1 (5′‐CTGTCATTGGGGGTGCGATA‐3′ and 5′‐CTGGCATTGACTGGGGTCAT‐3′), KRT5 (5′‐GCATCACCGTTCCTGGGTAA‐3′ and 5′‐GACACACTTGACTGGCGAGA‐3′), SCGB1A1 (5′‐TCCTCCACCATGAAACTCGC‐3′ and 5′‐AGGAGGGTTTCGATGACACG‐3′), DNAH5 (5′‐AGAGGCCATTCGCAAACGTA‐3′ and 5′‐CCCGGAAAATGGGCAAACTG‐3′), NPHP1 (5′‐CAGAGCCACATGGCAACCTA‐3′ and 5′‐ACCCAGCCACAGCTTAACTC‐3′), AGR2 (5′‐TCAGAAGCTTGGACCGCATC‐3′ and 5′‐AGTGTAGGAGAGGGCCACAA‐3′), UCHL1 (5′‐GACGAATGCCTTTTCCGGTG‐3′ and 5′‐AGAAGCGGACTTCTCCTTGC‐3′), ID2 (5′‐GCAGCACGTCATCGACTACA‐3′ and 5′‐TTCAGAAGCCTGCAAGGACA‐3′), ABCA3 (5′‐CACCAGGGGCTCTCTAGACT‐3′ and 5′‐ACAGCCATCGTCTTGCTGAA‐3′), SFTPA1 (5′‐CAGACGGGACCCCTGTAAAC‐3′ and 5′‐CCTGTCATTCCACTGCCCAT‐3′), SFTPC (5′‐ATGGATGTGGGCAGCAAAGA‐3′ and 5′‐CAGCAGGGAATGCCAAATCG‐3′), LGR5 (5′‐CACCGCTCTCAGTCACTGGATAAGC‐3′ and 5′‐AAACGCTTATCCAGTGACTGAGAGC‐3′), LGR6 (5′‐CCAAGGACAGTTTCCCAAAA‐3′ and 5′‐GACTCCTCATCATCAAGGTGAA‐3′), AXIN2 (5′‐AGCTTACATGAGTAATGGGG‐3′ and 5′‐AATTCCATCTACACTGCTGTC‐3′), TROY (5′‐TGATGAAAGTAGGCAGGGCTGTGT‐3′ and 5′‐TCTCCAGCCAGTGTTTGTCCTTGA‐3′).
Functional organoid swelling assay
Organoid swelling assays were performed as previously described (Dekkers et al, 2013). Intestinal organoids were cultured for 7 days, collected, and disrupted. Smallest fragments were selected and seeded for swelling assays.
AOs were sheared, passed through a 70‐μm strainer, and cultured for 4 days. Organoids were harvested and seeded for swelling assays in 96‐well plates (Greiner). We seeded 50–100 organoids in 5 μl drops (50% BME) per well overlaid with 100 μl culture medium after gel solidification. Cultures were incubated for 24 h with or without CFTR‐targeting drugs VX‐809 and VX‐770 (3 and 1 μM, respectively, Selleck Chemicals).
The next day, cultures were pre‐incubated for 3 h with CFTR inhibitors (150 μM CFTRinh‐172 and 150 μM GlyH101, Cystic Fibrosis Foundation Therapeutics) as indicated.
Calcein green (3 μM, Invitrogen) was added 1 h prior to stimulation with forskolin (Selleck Chemicals), Eact (Calbiochem), or DMSO (Sigma) at the indicated concentrations. Organoids were imaged per well (every 10 min, for 60–120 min in total or every 30 min, for 270 min in total) using confocal live cell microscopy (Zeiss LSM710 and LSM800) at 37°C and 5% CO2. Data were analyzed using Volocity 6.1.1 (Perkin Elmer), Zen Blue (Zeiss), and Prism 5 and 6 (GraphPad).
Air–liquid interface cultures and CFTR function measurement
Air–liquid interface cultures were established from BAL fluid‐derived AOs from a CFTR wt/wt and CFTR F508del/F508del donor.
AOs were dissociated into single cells using trypsin–EDTA (0.25%; Gibco‐25200).
250,000 cells were seeded on semi‐permeable transwell membranes (Corning‐3378) coated with bovine collagen type I (30 μg ml−1; Purecol; Advanced BioMatrix‐#5005). Single cells were seeded in AO growth medium supplemented with 25 ng ml−1 recombinant human epidermal growth factor (Peprotech‐AF‐100‐15) and cultured in submerged conditions.
After 4 days, confluent monolayers were cultured in air‐exposed conditions using differentiation medium adapted from Neuberger et al (2011). Medium was changed every 4 days.
After 3 weeks of differentiation, CFTR‐dependent chloride conductance was determined using the open‐circuit TECC‐24 assay (EP design). CFTR F508del/F508del cultures were treated prior to CFTR function measurements with VX‐809 (3 μM) or vehicle for 24 h. Measurements were conducted by sequentially adding benzamil (Sigma‐B2417; 5 μM), forskolin (5 μM) with VX‐770 (1 μM) or vehicle, and CFTRinh‐172 (25 μM). The equivalent current (I eq) was calculated from the measured transepithelial voltage (VT) and transepithelial resistance (RT) according to Ohm's law (I eq = VT·RT−1).
In vivo orthotopic transplantations
Experiments on NSG mice were carried out at the Netherlands Cancer Institute according to local and international regulations and ethical guidelines and were approved by the local animal experimental committee at the Netherlands Cancer Institute (DEC‐NKI OZP: 13.022, WP 5418).
Human tumor AO lines carrying lentiviral luciferase were expanded in their corresponding selection media.
Following trypsinization, 50–100,000 single cells were resuspended in 20 μl BME and injected intratracheally into anesthetized (100 mg kg−1 ketamine, 10 mg kg−1 sedazine) NOD SCID gamma (NSG; NOD.Cg‐PrkdcscidIl2rgtm1Wjl/SzJ) mice (≥ 6 injections per condition) placed on a hanging device.
Mouse experiments spanned over an 8‐week period after which lungs were resected for examination.
Randomly distributed males and females (aged 8–10 weeks at the start of the experiment; weights, ∼30 g for males and ∼25 g for females) were used, and all animals were included in the analysis. Ear clipping was used for animal recognition. Animals were caged together and treated in the same way.
Whole‐genome sequencing and read alignment
Genomic DNA was isolated using an automated setup (QiaSymphony). Sequencing libraries were constructed using the TruSeq Nano kit according to manufacturer's instructions (Illumina) with 50–200 ng input DNA. Sequencing was performed by paired‐end sequencing (2 × 150 bp) on Illumina HiSeqX to a minimal depth of 30× base coverage. Sequence reads were mapped against human reference genome GRCh37 using Burrows–Wheeler Aligner V0.7.17 mapping tool (Li & Durbin, 2009). The read‐quality scores of the mapped sequence reads were recalibrated using GATK BaseRecalibrator V3.5 (DePristo et al, 2011). Alignments from different libraries of the same sample were combined into a single BAM file. The BAM files of 57 AO‐tumor and 65AO‐tumor samples were downsampled to a read depth of 33.82 and 29.88, respectively, resulting in an average read depth of 33.81 ± 2.67 for all the samples. The WGS data ID is EGAS00001002899.
Characterization of the COSMIC IDs
Raw variants were called by using the GATK HaplotypeCaller V3.5 and GATK‐Queue V3.5 with default settings and additional option “EMIT_ALL_CONFIDENT_SITES”. The quality of variant and reference positions was evaluated by using GATK VariantFiltration V3.5 with options as described in https://github.com/UMCUGenetics/IAP/blob/develop/settings/hartwig_kg.ini. The obtained variants were annotated using four germline databases: PONvs2 (1.762 individuals—WGS; https://github.com/hartwigmedical), GoNL V5 (750 individuals—WGS; Boomsma et al, 2014), GNOMAD (15.496 individuals—WGS; Lek et al, 2016), and dbSNP 137.b37 (Sherry et al, 2001). Any variant present in any of these databases was filtered out. To compare mutational load and COSMIC IDs between matching organoids and tissue, only the filtered variants present in the intersected callable loci regions of both samples were considered. COSMIC release version 76 was used to annotate the filtered variants.
Hotspot sequencing
AO gDNA was isolated using the DNeasy blood & tissue kit (Qiagen). A minimum of 100 ng gDNA was submitted for sequencing cancer hotspots of Ion AmpliSeq™ Cancer Hotspot Panel v2Plus at the NGS Core of the UMC Utrecht. The following genes were (partially) covered: ABL1, AKT1, ALK, APC, ARAF, ATM, BRAF, CALR, CDH1, CDKN2A, CRAF (RAF1), CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, IDH2, JAK2, JAK3, KDR, KIT, KRAS, MDM2, MET, MLH1, MPL, MYD88, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, TP53, and VHL.
Drug screening
Organoids were mechanically dissociated by pipetting before being resuspended in 5% BME/AO media (15–20,000 organoids ml−1) and dispensed into 384‐well microplates (Greiner).
Each compound was dispensed at 1–100 μM using a TECAN D300e Digital Dispenser, and after 5 days, cell viability was assayed as described above.
Paclitaxel (T7402), methotrexate (A6770), crizotinib (PZ0240), and cisplatin (C2210000) were obtained from Sigma; Nutlin‐3a (10004372) was obtained from Cayman chemicals; erlotinib (S1023) from Santa Cruz; alpelisib (A‐4477) from LC Laboratories; and gefitinib (S1025) from Selleck Chemicals. Averages of IC50s from at least three independent experiments were calculated and visualized using GraphPad Prism, version 6.0, Morpheus (https://software.broadinstitute.org/morpheus) and CorelDraw X7.
RSV infection
rgRSV224 (GFP version), rrRSV (RFP version), rgRSV P‐eGFP‐M (RSVwt), and rgRSV∆NS2 13A ∆NS2 6120 P‐eGFP‐M (RSVΔNS2) were produced as previously described (Hallak et al, 2000; Guerrero‐Plata et al, 2006; Liesman et al, 2014).
AOs were washed in cold AdDF+++ and sheared with a flamed Pasteur pipette. Per infection, 2 μl virus (˜5 × 107 pfu ml−1) was placed in a U‐bottom suspension 96‐well and 50 μl of sheared organoids (˜500,000 cells) in AdDF+++ was added. Empty wells were filled with PBS to prevent dehydration.
Plates were incubated for 5 h at 37°C and 5% CO2.
Afterward, contents of the wells were taken up in AdDF+++ and washed three times with excess AdDF+++.
Organoids were seeded as described before.
Alternatively, individual AOs were microinjected with ˜250 nl virus in AdDF+++ (˜5 × 107 pfu ml−1) using a micromanipulator and microinjector (Narishige, M‐152 and IM‐5B) under a stereomicroscope (Leica, MZ75).
Single‐cell tracking
Cells were manually tracked by following the center of mass of their nuclei using custom‐written image analysis software. Random cells were selected on the initial time frame so that they uniformly covered the organoid surface. In infected organoids, particularly when rotating rapidly, a small fraction of cells could not be identified between some consecutive time frames due to their fast movement, limiting our ability to track the fastest‐moving cells. In Fig 4F and Appendix Fig S5B, three outliers were beyond the limits of the plot, reaching a maximum value of 117 μm h−1. Due to the difficulty of tracking fast‐moving cells, outliers above 50 μm h−1 are likely underrepresented. For each cell at each time point, RFP intensity was calculated by summing the pixel intensities within a disk of a radius of 5 pixels, corresponding to 3.22 μm, on the XY plane around the center of mass of the nucleus. To distinguish between RFP− and RFP+ cells, an intensity threshold was manually chosen so that all nuclei in non‐infected organoids were categorized as RFP−.
Mathematical modeling of collective cell motility
To understand how local interactions between individual cells could give rise to collective rotational movement on the level of the organoid, we built a mathematical model of cells migrating within the constraints of the tissue they are embedded in. In our model, cells move on the surface of a sphere to mimic the organoid geometry. Each cell has a polarity vector that represents the direction of migratory force production. However, cells are constrained by adhesive connections to neighboring cells. In our model, these connections are modeled by harmonic springs. In general, due to these adhesive constraints the total force acting on each cell has a different orientation than the intrinsic polarity . We assume that cells change their internal polarity over time to align with the direction of . Crucially, this interaction causes neighboring cells to align their movement. To reproduce the random motility observed in non‐infected cells, we assume the polarity direction also fluctuates in a stochastic manner. This effect perturbs synchronization of movement between neighboring cells. Cells are initially distributed uniformly over the surface of sphere, with random orientation for their polarity vectors. The force on cell i is given by the sum of the propulsive force in the direction of the polarity and the spring forces due to the neighboring cells j:
| (1) |
The spring force between a pair of cells at positions and is given by:
| (2) |
where k is the spring constant. Here, σ corresponds approximately to the radius of the cell and, hence, the spring force is attractive when it is extended beyond its natural length 2σ and repulsive when compressed. The model is then described fully by two sets of ordinary differential equations, for the cell positions and the cell polarities :
| (3) |
| (4) |
Here, cell movement is assumed to be overdamped, with μ the mobility. is the projection of the total force in the local plane of cell i, i.e., , with the unit vector orthogonal to the surface of the sphere at . The polarity vector has length and rotates with fixed angular velocity J in the direction that aligns it with . This direction is given by δ i = cos θ i/|cos θ i|, where θ i is the angle between and and . In addition, we incorporate intrinsic stochastic fluctuations to the direction of cell polarity by the noise term ξ(t), with noise strength η. In the absence of cell–cell communication, i.e., for J = 0, the polarity vector will undergo a random walk that causes the polarity to deviate from its original polarity over time. Specifically, the correlation in polarity direction decreases in time as , where τ = 2/η 2 is the polarity persistence time.
Simulation of the mathematical model
To generate random initial configurations of N particles distributed approximately equidistantly on a sphere with radius , where φ is the cellular packing fraction, particles were first placed at random positions on the sphere and then propagated using the spring force in equation (2) between nearest neighbors until global mechanical equilibrium was attained.
At the start of the simulation, we assigned each particle i a polarity vector with a random orientation in the tangent plane of the sphere at position and determined the identity of the neighboring particles connected by a spring to particle i using Delauney triangulation. Equations (3) and (4) were integrated using the Euler–Maruyama method, with time step μk·Δt = 5 × 10−3.
To ensure that the position and polarity vector remain well defined throughout the simulation, after each time step we projected onto the tangent plane of the sphere at the new position and normalized the position and polarity so that and . Simulations were performed with N = 100, φ = 1 and f 0/σ k = 1, while we varied the parameters J/μk and η/μk. While we use dimensionless parameters here, in the main text we use, without any loss of generality, parameters values for μ, k, σ = 1.
Multiplex Immunoassays
AOs were infected with/without RSV as described.
3 days after infection equal numbers of AOs were released from Matrigel using Cell Recovery Solution (Corning), washed, and replated overnight in 30 μl AO medium as hanging drops.
The next morning supernatants were collected and selected cytokines measured using an in‐house developed and validated multiplex immunoassay (Laboratory of Translational Immunology, UMC Utrecht) based on Luminex technology (xMAP, Luminex, Austin, TX, USA). The assay was performed as described previously (de Jager et al, 2003).
Acquisition was performed with the Bio‐Rad FlexMAP3D (Bio‐Rad laboratories, Hercules, CA, USA) in combination with xPONENT software version 4.2 (Luminex). Data were analyzed by 5‐parametric curve fitting using Bio‐Plex Manager software, version 6.1.1 (Bio‐Rad).
Neutrophil isolation and co‐culture with AOs
Human neutrophils were isolated from sodium–heparin anticoagulated venous blood of healthy donors by density gradient centrifugation with and Ficoll (Amersham Biosciences).
Erythrocytes were lysed in ammonium chloride buffer (155 mM NH4Cl; 10 mM KHCO3; 0.1 mM EDTA in double‐distilled H2O; pH = 7.2), and neutrophils were resuspended in RPMI 1640 (Life Technologies, Paisley, UK) supplemented with 10% (v/v) HI FBS (Biowest, Nuaillé, France) and 50 U/ml penicillin–streptomycin (Thermo Fisher Scientific, Waltham, MA, USA).
Purity of isolated neutrophils was analyzed using the CELL‐DYN Emerald (Abbott Diagnostics, Illinois, USA).
100,000 freshly isolated neutrophils were co‐seeded in BME with AOs that had been infected with/without RSV 3 days earlier.
Immediately after BME solidification, drops were overlaid with AO medium and live imaged as described.
For quantification, neutrophils were labeled with 1 μM Hoechst33342 prior to imaging with 3D confocal microscopy and the number of neutrophils attached to AOs with/without RSV counted 2 and 6 h post‐cell organoid mixing.
Generation of inducible dTomato and RSV‐NS2/dTomato overexpressing AO lines
The following sequence comprising humanized NS1, GS linker, FLAG tag, HA tag, P2A sequence, humanized NS2, GS linker, V5 tag, 6xHis tag, and T2A sequence flanked by EcoRI restriction sites was synthesized by Integrated DNA Technologies (Leuven, Belgium) and cloned into IDT Vector Amp Blunt: gaattcgccaccATGGGCAGCAACTCCCTGAGCATGATCAAAGTGCGGCTGCAGAACCTGTTCGACAACGACGAAGTCGCACTGCTCAAGATCACATGCTACACCGACAAGCTCATCCACCTGACCAACGCCCTGGCAAAGGCAGTGATCCACACTATCAAACTGAACGGTATCGTGTTCGTGCACGTCATCACCAGCAGCGACATCTGCCCTAACAACAACATCGTCGTCAAGTCCAACTTCACAACAATGCCCGTGCTGCAGAACGGCGGCTACATCTGGGAGATGATGGAGCTCACACACTGCTCCCAGCCCAACGGACTGATCGACGACAACTGCGAGATCAAGTTCTCCAAGAAGCTGAGCGACTCCACCATGACCAACTACATGAACCAGCTCTCCGAGCTGCTGGGATTCGACCTCAACCCTggcggcggcggctcgggaggaggaggaagcgactacaaggacgacgatgacaagTACCCCTACGACGTGCCCGACTACGCCggaagcggaGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGACCTATGGACACCACCCACAACGACACCACTCCACAGCGGCTGATGATCACCGACATGCGGCCACTGTCCCTGGAGACCACCATCACCTCCCTGACCCGCGACATCATCACCCACCGGTTCATCTACCTGATCAACCACGAGTGCATCGTGCGGAAGCTGGACGAGCGGCAGGCCACCTTCACCTTCCTGGTGAACTACGAGATGAAGCTGCTGCACAAAGTCGGCAGCACCAAGTACAAGAAGTACACTGAGTACAACACCAAATACGGCACCTTCCCAATGCCCATCTTCATCAACCACGACGGCTTCCTGGAGTGCATCGGCATCAAGCCCACAAAGCACACTCCCATCATCTACAAATACGACCTCAACCCTggtggtggtggttcaggaggaggatcgGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGCATCATCACCATCACCACggaagcggaGAGGGCAGAGGAAGTCTGCTAACATGCGGTGACGTCGAGGAGAATCCTGGACCTgaattc.
Humanized RSV‐NS sequences were from Lo et al (2005), 2A sequences from Kim et al (2011). EcoRI‐tdTomato‐NotI was amplified from pCSCMV:tdTomato using primers aagaattcatggtgagcaagggcgagg and ctgcggccgcttacttgtacagctcgtccat and cloned into pLVX‐TetOne™‐Puro (Clontech) yielding an inducible dTomato lentiviral overexpression plasmid.
EcoRI‐hNS2‐GS‐V5‐His‐T2A‐EcoRI was amplified from the IDT Vector using primers taccctcgtaaagaattcgccaccATGGACACCACCCACAACG and cttgctcaccatgaattcAGGTCCAGGATTCT and cloned into pLVX‐TetOne‐Puro‐tdTomato yielding an inducible hNS2/dTomato lentiviral overexpression plasmid.
Virus was generated and organoids infected as previously described (Koo et al, 2011). Infected organoids were selected with 1 μg ml−1 puromycin (Sigma). Expression was induced with 5 μM doxycycline (Sigma).
Statistical analysis
Statistical analysis was performed with the GraphPad Prism 7 software. Two‐tailed Student's t‐test analysis was used to assess the statistical significance of differences between two experimental groups.
Author contributions
NS designed, performed, and analyzed experiments and wrote the manuscript. NS and DDZO designed, performed, and analyzed swelling experiments. GDA performed and analyzed ALI experiments. AP performed and analyzed tumor AO experiments. NP, MV, and JJ performed and analyzed xenotransplantations. JFD and ACR helped with immunofluorescent analysis. NS, AP, IH, AL, JL, EC, and AO performed and/or analyzed RNA sequencing experiments. AH and EC analyzed WGS experiments. LB performed and analyzed viability assays. NS, DK, FW, MFMO, KKD, EFS, EEV, CHMM, CKE, SFB, RGV, and JMB organized lung tissue collection. GH‐P, EPO, SJT, and JSZ tracked and modeled single‐cell migration in RSV‐infected organoids. NS, LT, and SD performed RSV infection experiments. MCV and FEC quantified RSV replication. LM and LJB analyzed and supervised the RSV exp. JK and HB performed histology. MFMO, GJO, and DJW classified lung tumors and evaluated organoid histology. KK and SD produced virus stocks. NI and PJP performed transmission electron microscopy. MCV quantified RSV replication. NS, AP, LB, ER, and SJ generated organoid lines. ML isolated neutrophils. HC supervised the study.
Conflict of interest
N.S., S.F.B., H.C., J.M.B., and C.K.E. are inventors on patents/patent applications related to organoid technology.
Supporting information
Appendix
Table EV1
Table EV2
Table EV3
Table EV4
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Movie EV8
Movie EV9
Movie EV10
Movie EV11
Review Process File
Acknowledgements
We would like to acknowledge Carmen Lopez‐Iglesias for supporting high‐pressure freezing and freeze substitution. We thank Raimond Ravelli for providing transmission EM overview software and Antoni P.A. Hendrickx for help with scanning EM. We acknowledge Anko de Graaff and the Hubrecht Imaging Center for supporting light microscopy. We thank Inez Bronsveld for obtaining intestinal biopsies. We thank people from the Preclinical Intervention Unit of the Mouse Clinic for Cancer and Ageing (MCCA) at the NKI for performing the intervention studies. We acknowledge UBEC and USEQ. We thank the WKZ Pediatric Pulmonology Department for obtaining lavage fluid. We are grateful to Stieneke van den Brink and Nilofar Ehsani for preparing media. Genetically modified RSV variants were generously provided by Peter Collins, Mark E. Peeples, and Ulla Buchholz. We thank the Multiplex Core facility of the LTI UMC Utrecht for developing, validating, and performing immunoassays. Work at the Hubrecht Institute was supported by MKMD and VENI grants from the Dutch Organization of Scientific Research NWO (ZonMw 114021012 and 916.15.182), a Stand Up to Cancer International Translational Cancer Research Grant (a program of the Entertainment Industry Foundation administered by the AACR) and an Alpe d'Huze/KWF program grant (2014‐7006). This work is part of the Oncode Institute which is partly financed by the Dutch Cancer Society and was funded by the gravitation program CancerGenomiCs.nl from the Netherlands Organisation for Scientific Research (NWO). The spinning disk microscope is funded by NWO equipment grant 834.11.002. Work in the laboratory of S.J.T. is part of the research program of the Foundation for Fundamental Research on Matter (FOM), as part of NWO. The work of G.H. and J.S.Z. was supported by a NWO VIDI grant (680‐47‐529) and a HFSP Career Development Award (CDA00076/2014‐C). Work at the St. Antonius Hospital was supported by the Prof. Dr. Jaap Swierenga Foundation (development of human lung organoids). J.M.B., D.Z., and C.K.E. were supported by a grant from The Netherlands Organisation for Health Research and Development (ZonMW 40‐00812‐98‐14103) and the Dutch Cystic Fibrosis Society (HIT‐CF program).
The EMBO Journal (2019) 38: e100300
See also: M Paschini & CF Kim (February 2019)
Data availability
Whole‐genome sequencing data of matching tumor tissue and tumor organoid and RNA sequencing have been deposited in the EGA repository: https://ega-archive.org/studies/EGAS00001002899.
References
- Amatngalim GD, van Wijck Y, de Mooij‐Eijk Y, Verhoosel RM, Harder J, Lekkerkerker AN, Janssen RA, Hiemstra PS (2015) Basal cells contribute to innate immunity of the airway epithelium through production of the antimicrobial protein RNase 7. J Immunol 194: 3340–3350 [DOI] [PubMed] [Google Scholar]
- Balasooriya GI, Goschorska M, Piddini E, Rawlins EL (2017) FGFR2 is required for airway basal cell self‐renewal and terminal differentiation. Development 144: 1600–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Chung MI, Fioret B, Gao X, Katsura H, Hogan BL (2017) Lung organoids: current uses and future promise. Development 144: 986–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benali R, Tournier JM, Chevillard M, Zahm JM, Klossek JM, Hinnrasky J, Gaillard D, Maquart FX, Puchelle E (1993) Tubule formation by human surface respiratory epithelial cells cultured in a three‐dimensional collagen lattice. Am J Physiol 264: L183–L192 [DOI] [PubMed] [Google Scholar]
- Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz‐Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T, Yu KH, van Boxtel R, Huch M, Rivera KD, Wilson JP, Feigin ME, Ohlund D, Handly‐Santana A et al (2015) Organoid models of human and mouse ductal pancreatic cancer. Cell 160: 324–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma DI, Wijmenga C, Slagboom EP, Swertz MA, Karssen LC, Abdellaoui A, Ye K, Guryev V, Vermaat M, van Dijk F, Francioli LC, Hottenga JJ, Laros JF, Li Q, Li Y, Cao H, Chen R, Du Y, Li N, Cao S et al (2014) The Genome of the Netherlands: design, and project goals. Eur J Hum Genet 22: 221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchers AT, Chang C, Gershwin ME, Gershwin LJ (2013) Respiratory syncytial virus–a comprehensive review. Clin Rev Allergy Immunol 45: 331–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst‐Eilers E, Sorgdrager W (1998) Wet medisch‐wetenschappelijk onderzoek met mensen [PubMed]
- Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK (2014) Non‐small‐cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 14: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YW, Huang SX, de Carvalho A, Ho SH, Islam MN, Volpi S, Notarangelo LD, Ciancanelli M, Casanova JL, Bhattacharya J, Liang AF, Palermo LM, Porotto M, Moscona A, Snoeck HW (2017) A three‐dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol 19: 542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deben C, Deschoolmeester V, Lardon F, Rolfo C, Pauwels P (2016) TP53 and MDM2 genetic alterations in non‐small cell lung cancer: evaluating their prognostic and predictive value. Crit Rev Oncol Hematol 99: 63–73 [DOI] [PubMed] [Google Scholar]
- Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter‐de Groot KM, Brandsma AM, de Jong NW, Bijvelds MJ, Scholte BJ, Nieuwenhuis EE, van den Brink S, Clevers H, van der Ent CK, Middendorp S, Beekman JM (2013) A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med 19: 939–945 [DOI] [PubMed] [Google Scholar]
- Dekkers JF, Berkers G, Kruisselbrink E, Vonk A, de Jonge HR, Janssens HM, Bronsveld I, van de Graaf EA, Nieuwenhuis EE, Houwen RH, Vleggaar FP, Escher JC, de Rijke YB, Majoor CJ, Heijerman HG, de Winter‐de Groot KM, Clevers H, van der Ent CK, Beekman JM (2016) Characterizing responses to CFTR‐modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci Transl Med 8: 344ra84 [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ (2011) A framework for variation discovery and genotyping using next‐generation DNA sequencing data. Nat Genet 43: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi L, Slagter M, van der Velden DL, Kaing S, Kelderman S, van Rooij N, van Leerdam ME, Depla A, Smit E, Hartemink K, Groot R, Wolkers M, Sachs N, Snaebjornsson P, Monkhorst K et al (2018) Facilitating individualized T cell therapy by co‐culture of peripheral blood lymphocytes and tumor organoids. Cell 174: 1586–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak A, Tilley AE, Shaykhiev R, Wang R, Crystal RG (2011) Do airway epithelium air‐liquid cultures represent the in vivo airway epithelium transcriptome? Am J Respir Cell Mol Biol 44: 465–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BR, Hill DR, Ferguson MA, Tsai YH, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, White ES, Deutsch GH, Spence JR (2015) In vitro generation of human pluripotent stem cell derived lung organoids. Elife 4: e05098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faas FG, Avramut MC, van den Berg BM, Mommaas AM, Koster AJ, Ravelli RB (2012) Virtual nanoscopy: generation of ultra‐large high resolution electron microscopy maps. J Cell Biol 198: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferkol T, Schraufnagel D (2014) The global burden of respiratory disease. Ann Am Thorac Soc 11: 404–406 [DOI] [PubMed] [Google Scholar]
- Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH (2005) Well‐differentiated human airway epithelial cell cultures. Methods Mol Med 107: 183–206 [DOI] [PubMed] [Google Scholar]
- Geerdink RJ, Pillay J, Meyaard L, Bont L (2015) Neutrophils in respiratory syncytial virus infection: a target for asthma prevention. J Allergy Clin Immunol 136: 838–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero‐Plata A, Casola A, Suarez G, Yu X, Spetch L, Peeples ME, Garofalo RP (2006) Differential response of dendritic cells to human metapneumovirus and respiratory syncytial virus. Am J Respir Cell Mol Biol 34: 320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett NR, Butler MW, Shaykhiev R, Salit J, Omberg L, Rodriguez‐Flores JL, Mezey JG, Strulovici‐Barel Y, Wang G, Didon L, Crystal RG (2012) RNA‐Seq quantification of the human small airway epithelium transcriptome. BMC Genom 13: 82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallak LK, Collins PL, Knudson W, Peeples ME (2000) Iduronic acid‐containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 271: 264–275 [DOI] [PubMed] [Google Scholar]
- Heo I, Dutta D, Schaefer DA, Iakobachvili N, Artegiani B, Sachs N, Boonekamp KE, Bowden G, Hendrickx APA, Willems RJL, Peters PJ, Riggs MW, O'Connor R, Clevers H (2018) Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol 3: 814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hild M, Jaffe AB (2016) Production of 3‐D airway organoids from primary human airway basal cells and their use in high‐throughput screening. Curr Protoc Stem Cell Biol 37: IE 9 1–IE 9 15 [DOI] [PubMed] [Google Scholar]
- Hrycaj SM, Dye BR, Baker NC, Larsen BM, Burke AC, Spence JR, Wellik DM (2015) Hox5 genes regulate the Wnt2/2b‐Bmp4‐signaling axis during lung development. Cell Rep 12: 903–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Incognito L, Cheng X, Ulbrandt ND, Wu H (2010) Respiratory syncytial virus‐neutralizing monoclonal antibodies motavizumab and palivizumab inhibit fusion. J Virol 84: 8132–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SX, Islam MN, O'Neill J, Hu Z, Yang YG, Chen YW, Mumau M, Green MD, Vunjak‐Novakovic G, Bhattacharya J, Snoeck HW (2014) Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat Biotechnol 32: 84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubrecht Organoid Technology (2017) HUB and Dutch health insurance companies to validate use of organoids for Cystic Fibrosis. In Vries R (ed) www.hub4organoids.eu
- Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt J, van de Wetering M, Sasaki N, Boers SJ, Kemperman H, de Jonge J, Ijzermans JN, Nieuwenhuis EE, Hoekstra R, Strom S, Vries RR et al (2015) Long‐term culture of genome‐stable bipotent stem cells from adult human liver. Cell 160: 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui KPY, Ching RHH, Chan SKH, Nicholls JM, Sachs N, Clevers H, Peiris JSM, Chan MCW (2018) Tropism, replication competence, and innate immune responses of influenza virus: an analysis of human airway organoids and ex‐vivo bronchus cultures. Lancet Respir Med 6: 846–854 [DOI] [PubMed] [Google Scholar]
- de Jager W, te Velthuis H, Prakken BJ, Kuis W, Rijkers GT (2003) Simultaneous detection of 15 human cytokines in a single sample of stimulated peripheral blood mononuclear cells. Clin Diagn Lab Immunol 10: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, Vries RG, Cuppen E, Chen Y, Sawyers CL, Clevers HC (2014) Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY (2011) High cleavage efficiency of a 2A peptide derived from porcine teschovirus‐1 in human cell lines, zebrafish and mice. PLoS One 6: e18556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi S, Gotoh S, Tateishi K, Yamamoto Y, Korogi Y, Nagasaki T, Matsumoto H, Muro S, Hirai T, Ito I, Tsukita S, Mishima M (2016) Directed induction of functional multi‐ciliated cells in proximal airway epithelial spheroids from human pluripotent stem cells. Stem Cell Reports 6: 18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BK, Stange DE, Sato T, Karthaus W, Farin HF, Huch M, van Es JH, Clevers H (2011) Controlled gene expression in primary Lgr5 organoid cultures. Nat Methods 9: 81–83 [DOI] [PubMed] [Google Scholar]
- Kuk K, Taylor‐Cousar JL (2015) Lumacaftor and ivacaftor in the management of patients with cystic fibrosis: current evidence and future prospects. Ther Adv Respir Dis 9: 313–326 [DOI] [PubMed] [Google Scholar]
- Lanzerath D (2011) EUREC ‐ Information ‐ Germany
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell‐Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce‐Hoffman E, Berghout J, Cooper DN et al (2016) Analysis of protein‐coding genetic variation in 60,706 humans. Nature 536: 285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesman RM, Buchholz UJ, Luongo CL, Yang L, Proia AD, DeVincenzo JP, Collins PL, Pickles RJ (2014) RSV‐encoded NS2 promotes epithelial cell shedding and distal airway obstruction. J Clin Invest 124: 2219–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindskog C, Fagerberg L, Hallstrom B, Edlund K, Hellwig B, Rahnenfuhrer J, Kampf C, Uhlen M, Ponten F, Micke P (2014) The lung‐specific proteome defined by integration of transcriptomics and antibody‐based profiling. FASEB J 28: 5184–5196 [DOI] [PubMed] [Google Scholar]
- Lo MS, Brazas RM, Holtzman MJ (2005) Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J Virol 79: 9315–9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley KB, Hawkins F, Serra M, Thomas DC, Jacob A, Kotton DN (2017) Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of Wnt signaling. Cell Stem Cell 20: 844–857 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, Rogel N, Burgin G, Tsankov AM, Waghray A, Slyper M, Waldman J, Nguyen L, Dionne D, Rozenblatt‐Rosen O, Tata PR et al (2018) A revised airway epithelial hierarchy includes CFTR‐expressing ionocytes. Nature 560: 319–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D et al (2003) PGC‐1alpha‐responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273 [DOI] [PubMed] [Google Scholar]
- Mou H, Vinarsky V, Tata PR, Brazauskas K, Choi SH, Crooke AK, Zhang B, Solomon GM, Turner B, Bihler H, Harrington J, Lapey A, Channick C, Keyes C, Freund A, Artandi S, Mense M, Rowe S, Engelhardt JF, Hsu YC et al (2016) Dual SMAD signaling inhibition enables long‐term expansion of diverse epithelial basal cells. Cell Stem Cell 19: 217–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW et al (2010) Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta‐analysis. Lancet 375: 1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkung W, Yao Z, Finkbeiner WE, Verkman AS (2011) Small‐molecule activators of TMEM16A, a calcium‐activated chloride channel, stimulate epithelial chloride secretion and intestinal contraction. FASEB J 25: 4048–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger T, Burton B, Clark H, Van Goor F (2011) Use of primary cultures of human bronchial epithelial cells isolated from cystic fibrosis patients for the pre‐clinical testing of CFTR modulators. Methods Mol Biol 741: 39–54 [DOI] [PubMed] [Google Scholar]
- Nikolic MZ, Caritg O, Jeng Q, Johnson JA, Sun D, Howell KJ, Brady JL, Laresgoiti U, Allen G, Butler R, Zilbauer M, Giangreco A, Rawlins EL (2017) Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long‐term self‐renewing organoids. Elife 6: e26575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordhoek J, Gulmans V, van der Ent K, Beekman JM (2016) Intestinal organoids and personalized medicine in cystic fibrosis: a successful patient‐oriented research collaboration. Curr Opin Pulm Med 22: 610–616 [DOI] [PubMed] [Google Scholar]
- Persson BD, Jaffe AB, Fearns R, Danahay H (2014) Respiratory syncytial virus can infect basal cells and alter human airway epithelial differentiation. PLoS One 9: e102368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert LW, Zilionis R, Choo‐Wing R, Savova V, Knehr J, Roma G, Klein AM, Jaffe AB (2018) A single‐cell atlas of the airway epithelium reveals the CFTR‐rich pulmonary ionocyte. Nature 560: 377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratjen F, Bell SC, Rowe SM, Goss CH, Quittner AL, Bush A (2015) Cystic fibrosis. Nat Rev Dis Primers 1: 15010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL (2009) Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA 106: 12771–12775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sanden SMG, Sachs N, Koekkoek SM, Koen G, Pajkrt D, Clevers H, Wolthers KC (2018) Enterovirus 71 infection of human airway organoids reveals VP1‐145 as a viral infectivity determinant. Emerg Microbes Infect 7: 84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H (2011) Long‐term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141: 1762–1772 [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29: 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondo E, Caci E, Galietta LJ (2014) The TMEM16A chloride channel as an alternative therapeutic target in cystic fibrosis. Int J Biochem Cell Biol 52: 73–76 [DOI] [PubMed] [Google Scholar]
- Sosnay PR, Siklosi KR, Van Goor F, Kaniecki K, Yu H, Sharma N, Ramalho AS, Amaral MD, Dorfman R, Zielenski J, Masica DL, Karchin R, Millen L, Thomas PJ, Patrinos GP, Corey M, Lewis MH, Rommens JM, Castellani C, Penland CM et al (2013) Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet 45: 1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T, Wang Y, Barak LS, Bai Y, Randell SH, Hogan BL (2014) IL‐6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc Natl Acad Sci USA 111: E3641–E3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T, Gao X, Hong CC, Hotten D, Hogan BL (2016) BMP signaling and cellular dynamics during regeneration of airway epithelium from basal progenitors. Development 143: 764–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Choi KM, Sicard D, Tschumperlin DJ (2017) Human airway organoid engineering as a step toward lung regeneration and disease modeling. Biomaterials 113: 118–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng MN, Collins PL (1999) Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein. J Virol 73: 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR (2014) Reconstructing lineage hierarchies of the distal lung epithelium using single‐cell RNA‐seq. Nature 509: 371–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA (2004) In vivo activation of the p53 pathway by small‐molecule antagonists of MDM2. Science 303: 844–848 [DOI] [PubMed] [Google Scholar]
- Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez‐Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R, Rata M, Koh DM, Tunariu N, Collins D, Hulkki‐Wilson S, Ragulan C, Spiteri I, Moorcraft SY, Chau I, Rao S et al (2018) Patient‐derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359: 920–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber F, Ooft SN, Dijkstra KK, Voest EE (2017) Tumor organoids as a pre‐clinical cancer model for drug discovery. Cell Chem Biol 24: 1092–1100 [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor‐Weiner A, Kester L, McLaren‐Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VS, Seepo S, Sekhar Pedamallu C, Cibulskis K et al (2015) Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161: 933–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AP, Bear CE, Chin S, Pasceri P, Thompson TO, Huan LJ, Ratjen F, Ellis J, Rossant J (2012) Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat Biotechnol 30: 876–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonker LM, Mou H, Chu KK, Pazos MA, Leung H, Cui D, Ryu J, Hibbler RM, Eaton AD, Ford TN, Falck JR, Kinane TB, Tearney GJ, Rajagopal J, Hurley BP (2017) Development of a primary human co‐culture model of inflamed airway mucosa. Sci Rep 7: 8182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li C, Sachs N, Chiu MC, Wong BH, Chu H, Poon VK, Wang D, Zhao X, Wen L, Song W, Yuan S, Wong KK, Chan JF, To KK, Chen H, Clevers H, Yuen KY (2018) Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc Natl Acad Sci USA 115: 6822–6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Table EV1
Table EV2
Table EV3
Table EV4
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Movie EV8
Movie EV9
Movie EV10
Movie EV11
Review Process File
Data Availability Statement
Whole‐genome sequencing data of matching tumor tissue and tumor organoid and RNA sequencing have been deposited in the EGA repository: https://ega-archive.org/studies/EGAS00001002899.


