Supplemental Digital Content is available in the text.
Keywords: Microbiome, Polybrominated diphenyl ethers, Polychlorinated biphenyls
Background:
The gut microbiome is influenced by early-life exposures, but—despite potentially enormous implications for child health—is understudied in environmental epidemiology. This pilot study is one of the first to explore in utero exposures and long-term gut microbiome profiles. We examined the association between exposure to polybrominated diphenyl ethers (PBDEs) and polychlorinated biphenyls (PCBs) during pregnancy and the mid-childhood gut microbiome.
Methods:
We measured levels of PBDE-47, PBDE-99, PBDE-100, and PBDE-153 and PCB-138, PCB-153, and PCB-180 in maternal plasma during early pregnancy (n = 18) and at delivery (n = 25) in women of European descent who breastfed the child participant of the Gestation and Environment cohort in Sherbrooke, Québec (recruited 2007–2009). Bacteria in the mid-childhood (6–8 years) fecal microbiome were detected with 16S rRNA sequencing. To test for differences at the taxon level, we used the Microbiome Comprehensive Association Mapping algorithm.
Results:
Early pregnancy PCB-153, PCB-180, and the sum of PCBs (Σ3PCB) concentrations were associated with a higher relative abundance of Propionibacteriales and Propionibacteriaceae in mid-childhood. Higher PCB-180 and Σ3PCB were associated with higher relative abundance of Bacillales Family XI. Higher PBDE-99 exposure was associated with a decrease in uncultured bacteria within the Ruminococcaceae NK4A214 group and PBDE-47 was associated with differences in Ruminococcus 2. These taxon-level changes did not result in differences within- or between-subject diversity. Exposures at delivery were not associated with differences in taxa.
Conclusions:
Prenatal exposure to PCBs and PBDEs is associated with mid-childhood gut microbiome profiles. Larger studies are needed to confirm these results and explore health implications.
What this study adds
The gut microbiome is a complex “organ,” and disruption of its structure has been associated with several disease states. However, no epidemiological studies have been undertaken to explore the effects of environmental contaminants on the microbiome. Early studies have utilized in vitro or animal models to mimic the microbiome, but these models cannot adequately capture the complexities of the human gut. To our knowledge, this is the first article to explore the association between prenatal environmental chemicals, specifically polybrominated diphenyl ethers and polychlorinated biphenyls, and the childhood microbiome in an epidemiological study of healthy children.
Introduction
The human microbiome comprises millions of bacteria, viruses, and fungi found throughout the body with different compositions of species in different body sites. Of particular interest to epidemiological studies is the human gut bacterial microbiome because of its role as both a biomarker of environmental exposures and a mechanism by which they affect health and disease.1,2 Little research has been conducted to determine the effects of environmental chemicals on the balance and structure of the bacterial microbiome in human populations.3,4 In particular, the transfer of the microbiome from mother to infant is an area of active study owing to a limited understanding of how it occurs, and factors affecting that transfer and seeding of certain bacteria within the infant gut are vastly understudied.5 However, the establishment of the gut microbiome has potentially enormous health implications across the life-course.
Recently, the sterility of the intrauterine cavity during pregnancy has come into question. There have been several reports of bacterial detection in the uterine endometrium,6 placenta,7 and amniotic fluid.8 However, these results are controversial and others have not replicated these findings.9,10 Nonetheless, the maternal microbiome during pregnancy could have dramatic effects on what bacteria are able to colonize the fetal or neonatal gut. To date, studies have primarily examined the effects of diet and nutrient availability or disease status on the adult microbiome.5,11–14 However, factors such as environmental chemicals including endocrine disrupting chemicals and heavy metals have demonstrated pro and antimicrobial effects in animal and in vitro models.15 In contrast to antibiotics, which have broad spectrum effects on bacterial species, environmental toxicants seem to have more moderate effects, altering the balance of bacterial taxa present rather than eliminating bacteria indiscriminately. As such, the unique ways in which these chemicals affect established microbial community structures and the ability of different microbes to colonize naïve environments (e.g., the fetal gut) present a vital gap in the literature.
Although many environmental chemicals have the potential to impact the balance of bacteria present in the human gut, polybrominated diphenyl ethers (PBDEs) and polychlorinated biphenyls (PCBs) are of particular interest because they are found nearly ubiquitously in human populations, have known health effects whose mechanisms are not fully described, and have been associated with alterations in microbial community structure in environmental samples and/or in animal models. PBDEs are persistent flame retardant chemicals that were applied to many consumer products and are found nearly universally in U.S. populations.16 These chemicals have been associated with adverse health outcomes including impaired thyroid function with resulting IQ deficits,17–19 ADHD,20,21 and obesity.22,23 PBDE degradation has been shown to increase the abundance of Clostridia and Bacilli genera in environmental and in vitro samples.24,25 Although the detected genera are present in human stools, these findings are yet to be translated to rodent or human studies. Similarly, PCBs were used throughout the 1970s in coolant fluids and as common plasticizers, and were banned due to their toxicity and mutagenicity, but most pregnant women and children continue to be exposed today owing to its environmental persistence.16 Adverse outcomes associated with PCB exposure include decreased in utero growth, immunosuppression, decreased thyroid function, and carcinogenesis.26–32 Oral PCB exposure has been shown to change the structure of the gut microbiome in mice and zebrafish—affecting a broad range of genera.33,34 However, whether these effects are also seen in humans and the clinical significance of such changes remains unknown.
The purpose of the present pilot study was to explore the long-term effects of in utero exposure to PBDEs and PCBs on the mid-childhood microbiome. We first determined whether in utero exposure to these compounds was associated with the number of taxa within a child’s stool sample, known as alpha diversity.35 We then examined whether exposure to PBDEs and PCBs was associated with differences in the phylogenetic distances between subjects, known as beta diversity.35 Finally, we explored which gut bacterial taxa experienced changes in abundance with these exposures.
Methods
Study participants
To examine the relationship between PBDEs/PCBs and the mid-childhood microbiome, we utilized the Gestation and Environment (GESTE) longitudinal birth cohort based in Sherbrooke, Quebec, Canada. Between 2007 and 2009, mothers were recruited at less than 20 weeks pregnancy and followed until delivery. At 6–8 years of age, children were recalled and a pilot group provided stool samples for microbiome analysis. Maternal stool samples were not collected. The population is ethnically and socioeconomically homogenous, with most subjects identifying as French-Canadian and middle-class. The pilot participants included a convenience sample of children who were willing and able to provide a stool sample. To improve precision of effect estimates for this pilot study, all analyses were restricted to children whose mothers were identified as white French-Canadian, who ever breastfed the child subject, and who did not smoke or drink during pregnancy. The pregnancy pilot (n = 18) comprised mother–child pairs where the mother’s PBDE levels were measured in early pregnancy and the child participated in the stool collection pilot. The delivery pilot (n = 24) comprised mother–child pairs where the mother’s PBDE levels were measured in at delivery and the child participated in the stool collection pilot. Nine subjects were included in both pilot populations. Written informed consent was obtained from parents. All study protocols were approved by the Institutional Review Boards of the University of Sherbrooke, Harvard T.H. Chan School of Public Health, and Columbia University.
PBDE and PCB measurement
Blood was collected from mothers during the early pregnancy and in the hospital at delivery. Plasma levels of PBDEs (PBDE-47, PBDE-99, PBDE-100, PBDE-153) and PCBs (PCB-153, PCB-180, PCB-138) were measured as previously described.36 Briefly, we employed a gas chromatography-tandem mass spectrometer (GC-MS/MS) method with a limit of detection defined as three times the level of noise (0.1 pg/µL for PBDEs, 0.02 pg/µL for PCBs). Total plasma lipid content was measured using the sulfophosphovanillin colorimetric method.37 Exposures were normalized by concurrent plasma lipid content, and z-transformed for comparability and to create a sum of exposures for each group of chemicals (i.e., Σ4PBDE, Σ3PCB). To assess the effect of outlying concentrations, we conducted an analysis of the association between the exposures of interest and alpha diversity where exposures were log-transformed before z-standardizing. The PBDE congeners were chosen because they are nearly ubiquitously detected in the U.S. population and also they are abundant components of the commercial penta-PBDE mixtures (DE-71 and Bromkal 70-5DE).16,38 Similarly, the PCB congeners selected have the highest of the PCB congeners measured in a U.S. national representative sample,16 and contribute most to the total PCB burden in Canadian blood bank samples.39
DNA extraction and microbiome composition analysis
Stool was collected at home by children with parental assistance. After the child defecated into a compostable receptacle nested in their toilet, parents, wearing gloves, transferred the stool from the receptacle to a sterile glass vial with a wooden tongue depressor. Stools were then stored at −20°C in home freezers until study staff picked up the stool (within 48 hours), at which point the stools were frozen at −80°C for long-term storage. This follows the gold standard protocol, but even significant deviations have been shown to produce consistent sequencing results such that interindividual variability is significantly greater than the intraindividual variability arising from different storage methods.40 Total DNA was extracted from children’s stools using the QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA) following the manufacturer’s protocol with the addition of a bead beating step after the addition of lysis buffer.41 Sequencing of the 16S rRNA V1-V2 hypervariable region (primer sequences: 8F: AGAGTTTGATCCTGGCTCAG, 338R: GCTGCCTCCCGTAGGAGT) was performed by the UNC Microbiome Core (Durham, NC).42 Paired-end sequences were cleaned using the Divisive Amplicon Denoising Algorithm (DADA2) and paired to Operational Taxonomic Units (OTUs) using the SILVA v128 classifier in the Quantitative Insights Into Microbial Ecology (QIIME) pipeline.43–45 Singleton sequences and chimeras were removed.
Statistical analyses
Bacterial alpha diversity was calculated using the Shannon Index and Faith’s Phylogenetic Diversity (PD) in QIIME. The Shannon Index accounts for species richness and evenness, while the Faith’s PD accounts for the phylogenetic distance between taxa within each sample. Both metrics are unitless. Each exposure and the sum of each class of exposures was linearly regressed against each alpha diversity metric adjusting for delivery mode and socioeconomic status. Because the microbiome stabilizes by 2 years of age, the exact age at time of collection is not expected to affect the microbiome in mid-childhood. Therefore, age was not included as a covariate.
Associations between age and alpha diversity metrics can be found in Supplementary Table 1, http://links.lww.com/EE/A33. To determine whether exposure to PBDEs and PCBs had a statistically significant impact on beta diversity, we used the optimal microbiome-based association test (OMiAT) package to estimate a P-value for each exposure, adjusting for delivery mode and socioeconomic status.46 Briefly, OMiAT conducts microbiome regression-based kernel association tests (MiRKATs) for Bray-Curtis dissimilarities, and unweighted, weighted, and four generalized UniFrac distances, and an optimal MiRKAT test as well as adaptive Sum of Powered Score (aSPU) tests with five parameters for γ. In addition to reporting P-values for these tests, OMiAT selects the minimum P-value among these tests as the test statistic to estimate a final outcome P-value. To provide a comprehensive view of these results, we report P-values for tests using unweighted and weighted UniFrac distances, the optimal MiRKAT, and OMiAT.
To determine whether individual OTUs were different at 6–8 years of age depending on exposure level, we utilized the Microbiome Comprehensive Association Mapping (MiCAM) function with 30,000 permutations to obtain stable P-values for the associations adjusted for delivery mode and socioeconomic status.46 This method utilizes kernel machine methods, and therefore does not estimate an effect size for each OTU. However, the directionality for lowest-level taxa is estimated using linear regression methods. Because this function treats OTUs as the exposures and the chemical exposures as the outcome, the directionality of the association and P-values obtained cannot be interpreted as causal. OTUs were considered significantly associated if they met a false discovery rate of 0.05 using the Benjamini-Hochberg correction. All statistical analyses were conducted in R version 3.4.2 (R Core Team, Vienna, Austria).
Results
Study participant characteristics
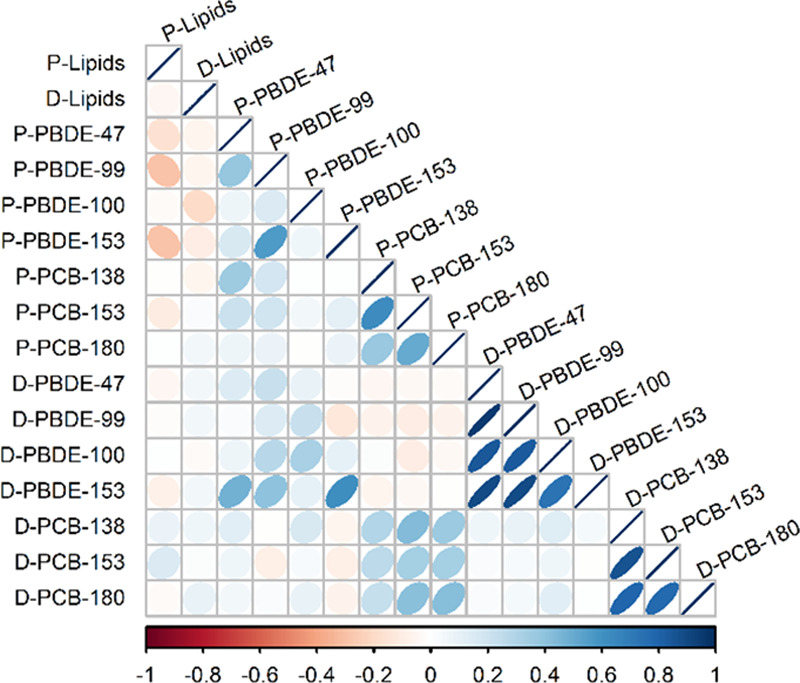
Characteristics of the overall GESTE population were similar to the pilot population characteristics except that the average concentration of PCB-153 was lower in the pregnancy pilot population (P = 0.02) and the average concentration of PBDE-153 was marginally lower in the delivery pilot population (P = 0.05) (Table 1). The exposures were correlated within class of chemical at each time point, with the highest correlations occurring among PBDEs at delivery. PBDEs were positively correlated with each other between the two time points (correlation coefficients ranging from 0.14 to 0.61, P-values ranging from 0.15 to <0.01) in the full GESTE population, as were concentrations of PCBs (correlation coefficients ranging from 0.29 to 0.42, P-values all <0.01). Normalized exposure concentrations were not correlated with serum lipid content (Figure 1). Concentrations for the exposures varied by an order of magnitude depending on the congener. Thus we created z-scores for each exposure to normalize the effect estimates for within- and between-subject diversity. After transformation, the rank-ordering of subjects was different for each exposure, and there was no clear pattern in bacterial profiles at the phylum level with higher exposures (Figure 2).
Table 1.
Characteristics of the GESTE microbiome pilot populations compared to the overall population (expressed as mean ± SD unless otherwise indicated).
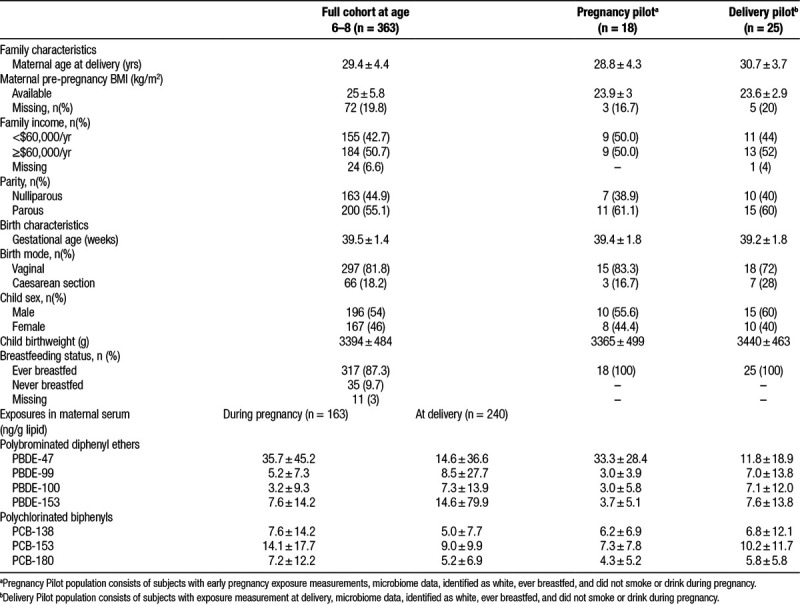
Figure 1.
Correlation matrix of lipids and lipid-adjusted polybrominated diphenyl ethers (PBDE) -47, -99, -100, -153 and polychlorinated biphenyls (PCB) -138, -153, -180 during pregnancy (P-) and at delivery (D-) in the GESTE population. Stronger correlations are indicated by darker color and tighter ellipse. Positive correlations are blue and negative correlations are red.
Figure 2.
Relative abundance of phyla in each subject during the early pregnancy and at delivery ordered by lipid-adjusted exposure concentration.
Alpha diversity
The mean Shannon Index and Faith’s PD in the pregnancy pilot population were 6.9 ± 0.7 and 25.6 ± 4.9, respectively. Similarly, in the delivery pilot population the averages were 7 ± 0.6 and 29.2 ± 5.8 for the Shannon Index and Faith’s PD, respectively. Exposures in early pregnancy and at delivery were not significantly associated with a difference in alpha diversity measured by the Shannon Index (Figure 3A). Effect estimates for early pregnancy exposure ranged from −0.24 to 0.35 per standard deviation increase in exposure across the different PBDE congeners and from −0.26 to −0.08 per standard deviation increase in exposure for the different PCB congeners for an association with the Shannon Index. For exposures at delivery, these estimates ranged from −0.035 to 0.04 per standard deviation increase in exposure across PBDE congeners and from 0.02 to 0.12 per standard deviation increase in exposure for PCB congeners for an association with the Shannon Index. Similarly, exposures in early pregnancy and at delivery were not associated with a significant difference in alpha diversity measured by Faith’s PD (Figure 3B). Effect estimates for an association between early pregnancy exposure and Faith’s PD ranged from −2.01 to −0.24 and from −0.23 to 1.71 per standard deviation increase in exposure for PBDEs and PCBs, respectively, for an association with Faith’s PD. The estimates for an association between exposures at delivery and Faith’s PD ranged from −2.12 to −0.28 and from 0.14 to 0.44 per standard deviation increase in exposure for PBDE and PCB congeners, respectively. None of the estimates reached statistical significance. Associations between delivery mode/socioeconomic status and alpha diversity metrics are presented in Supplementary Table 2, http://links.lww.com/EE/A33. Results remained null when exposures were log-transformed before z-standardization (Supplementary Table 3, http://links.lww.com/EE/A33).
Figure 3.
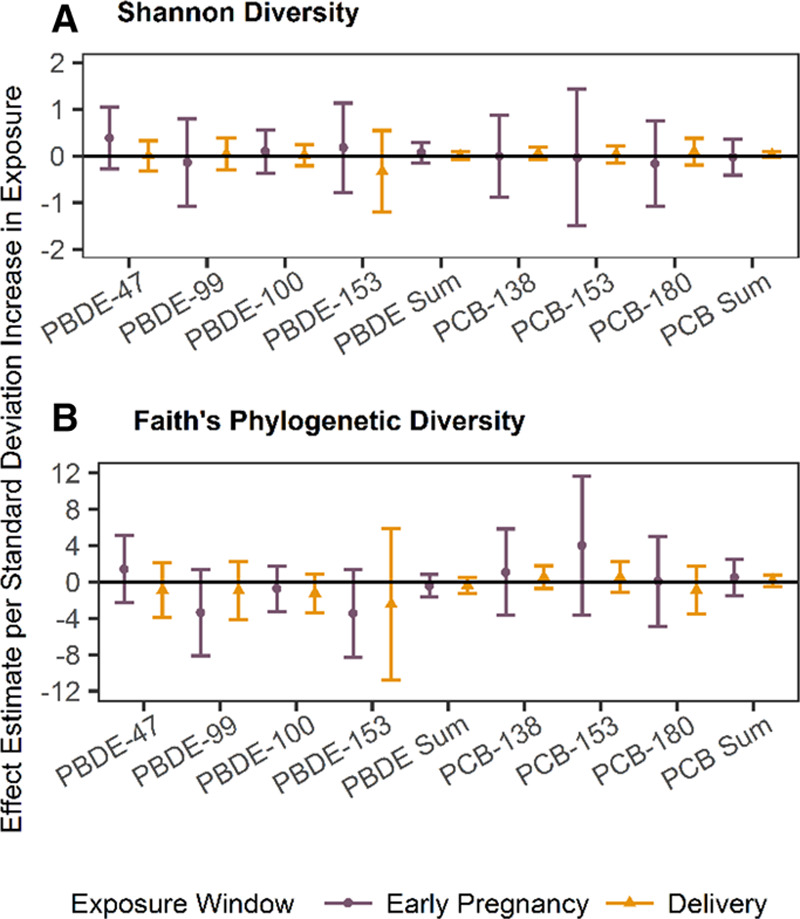
Associations of exposure to polybrominated diphenyl ethers (PBDE) -47, -99, -100, -153 and polychlorinated biphenyls (PCB) -138, -153, -180, and the sum of each group of compounds with age 6– year gut microbial alpha diversity for early pregnancy and perinatal exposure measured by the Shannon Index (A) and Faith’s Phylogenetic Diversity (PD) Index (B). All analyses are restricted to women who did not smoke or drink during pregnancy, identified as white, and breastfed, and are adjusted for C-section and socioeconomic status. Point estimates are the result of linear regression; whiskers represent 95% confidence intervals.
Beta diversity
Neither early pregnancy nor delivery exposure was statistically, significantly associated with differences in beta diversity in mid-childhood (Figure 4). However, exposure to BDE-99, PCB-180, and Σ3PCB showed a trend of association (P < 0.2) when considering unweighted UniFrac distances, indicating possible differences in rare taxa. In general, any exposure during early pregnancy was more significantly associated with differences in beta diversity than the same exposure at delivery, regardless of the dissimilarity metric used (Figure 4).
Figure 4.
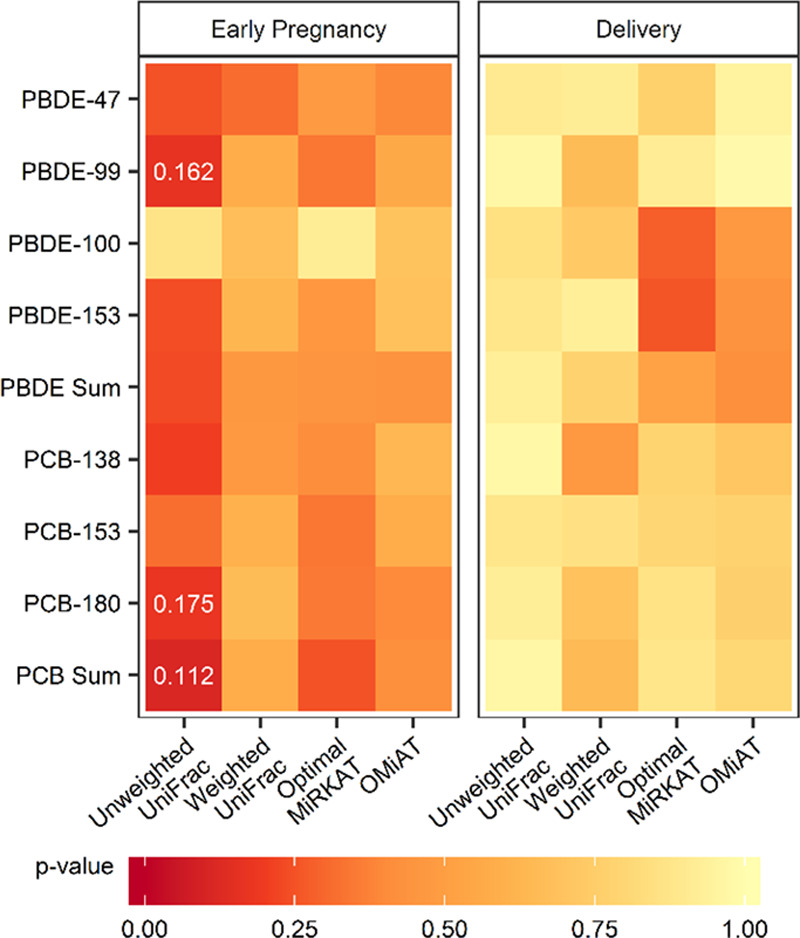
The significance of the association of exposure to polybrominated diphenyl ethers (PBDE) -47, -99, -100, -153 and polychlorinated biphenyls (PCB) -138, -153, -180, and the sum of each group of compounds with age 6–8 year gut microbial beta diversity measured using unweighted and weighted UniFrac values as well as the optimal MiRKAT and OMiAT algorithms. All analyses are restricted to women who did not smoke or drink during pregnancy, identify as white, and breastfed, and are adjusted for mode of delivery and socioeconomic status. P-values <0.2 are displayed.
Community structure
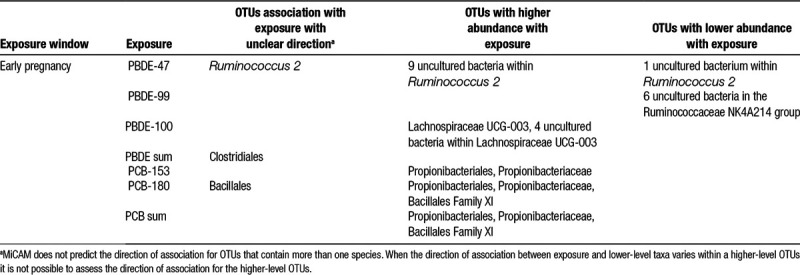
Exposure to PBDEs and PCBs in early pregnancy was significantly associated with differences in bacterial structure (Table 2). Higher exposure to PCB-153, PCB-180, and Σ3PCB was associated with higher relative abundance of Propionibacteriales and the family Propionibacteriaceae. PCB-180 and Σ3PCB were also associated with higher relative abundance of Bacillales Family XI, and PCB-180 was additionally associated with changes in the greater Bacillales order. However, the direction of the association was not consistent among lower level taxa and cannot be estimated for higher-level taxa when nested taxa have mixed effects. Higher PBDE-47 concentrations were associated with higher relative abundances of nine uncultured Ruminococcus 2 species and a lower relative abundance of one uncultured species in the same genus. These differences were consistent with statistically significant differences at the genus level. Higher PBDE-99 exposure in early pregnancy was associated with decreased relative abundance of six uncultured bacteria in the Ruminococcaceae NK4A214 group. Higher PBDE-100 exposure was associated with higher relative abundance of four uncultured bacteria within Lachnospiraceae UCG-003, as well as increased abundance of the genus as a whole. Finally, Σ4PBDE exposure was associated with changes in the Clostridiales order, but the direction of effect varied by lower level taxa. Exposure at delivery was not associated with changes in the community structure.
Table 2.
MiCAM results for operational taxonomic units (OTUs).
Discussion
In this pilot study, we first explored the potential that PBDEs and PCBs may have associations on inter- and intra-sample diversity. Although some chemicals may act as antimicrobial agents and decrease within-subject diversity, we found this was not the case with PBDE and PCB exposure in either early pregnancy or the perinatal period. This was in conjunction with our hypothesis that PBDEs and PCBs likely act on the microbiome by promoting the growth of certain bacterial species over others rather than universally increasing or decreasing diversity. Similarly, the OTU-specific differences with PBDE and PCB exposure during gestation did not result in significant alterations in between-subject diversity using several distance metrics. However, the OTU-specific associations in this pilot sample indicate that in utero exposure to environmental chemicals may result in changes in the composition of the childhood microbiome. Depending on the functional abilities that are gained or lost as a result of these changes, the microbiome may mediate the adverse effects of in utero exposure to environmental chemicals. The results of our pilot study also suggest the relative importance of early pregnancy exposures compared with exposures at delivery. These findings may indicate that the examined exposures affect bacterial taxa by limiting or enhancing their ability to colonize a naïve environment, but future in vitro studies are needed to confirm this hypothesis.
In addition to examining associations with subject diversity, we assessed associations between exposures and individual taxa. In this pilot study, we found significant associations between maternal PCB exposure in early pregnancy and higher relative abundance of the families Propionibacteriaceae and Bacillales in the gut microbiome of offspring eight years later. These novel results demonstrate the importance of environmental chemicals in shaping the long-term gut microbiome potentially by favoring or hindering the growth of certain bacteria after the initial seeding. There are two mechanisms by which in utero exposure to PBDEs and PCBs may affect the microbiome of a child years later. Specifically, the exposures may modify the maternal microbiome, which is then passed to the infant and establishes a community trajectory that is detectable in mid-childhood. Alternatively, PBDEs and PCBs, which are both known to cross the placenta,47,48 could establish a naïve environment that is more hospitable to certain bacteria, promoting their growth at the expense of others. Future in vitro research should explore both of these possibilities. In addition to the better understanding the mechanism of action of these chemicals on the microbiome, it is important to place our findings in the context of the health effects associated with these bacterial families.
Exposure to higher concentrations of PCB-153, PCB-180, and Σ3PCB were associated with higher relative abundance of the family Propionibacteriaceae as well as its order Propionibacteriales. Propionibacteria are abundant skin microbes,49 but are also found in the feces of animals and humans.50 In fact, several Propionibacterium species found in dairy products have been identified as potential probiotics to counteract intestinal inflammation.51,52 Relatedly, subjects who have taken ibuprofen in the last 30 days have higher levels of Propionibacteriaceae compared to subjects who have taken no prescription or nonprescription drugs over that period, indicating a potential antiinflammatory effect.53 If our observed association is causal, it is possible that Propionibacteriales and Propionibacteriaceae increase in response to the proinflammatory environment caused by higher PCB exposure in utero.54 However, without further data, including inflammatory status of the mother during pregnancy, this hypothesis cannot be confirmed.
In addition to Propionibacteriales, higher PCB-180 and Σ3PCB exposure was associated with higher abundances of Bacillales, specifically an unnamed family (Family XI) that contains the genus Gemella. While the family is understudied, Gemella was overrepresented in colorectal tumor tissue compared with adjacent tissue in a Spanish population.55 Additionally, G. sanguinis was found to be more abundant in children who went on to develop type I diabetes.56 However, Gemella species were found to be more abundant in healthy individuals than in subjects with irritable bowel disease and negatively correlated with inflammatory markers in saliva.57 Owing to the lack of literature on Gemella and the larger Bacillales Family XI in the human intestine, we cannot draw conclusions about the functional consequences of its potential increased abundance with higher PCB exposure.
Higher early pregnancy exposure to PBDE-47 was associated with changes in the genus Ruminococcus 2, with nine species increasing and one decreasing significantly. The genus comprises species that were previously classified under different genera, but clustered based on sequencing. In addition to many uncultured and thus unnamed species, Ruminococcus 2 includes a species previously classified as Ruminococcus bromii. R. bromii is increased with resistant starch and high-flavonoid fruit and vegetable intake,58 and decreased with an animal-based diet.59 Despite its association with a traditionally healthy diet, R. bromii is increased in patients with Parkinson’s disease, indicating an important link to the nervous system.60 Ruminococcus 2 also includes uncultured bacteria previously classified as Papillibacter. Although Papillibacter spp. are found to be more abundant in healthy colonic specimens compared to patients with inflammatory bowel disease,61 some species have also been linked to Parkinson’s.60 While these associations are interesting, more research is needed to uncover the function of the uncultured bacteria within Ruminococcus 2, and whether this genus is in fact associated with PBDE-47 exposure.
Like PBDE-47, PBDE-99 and PBDE-100 were associated with changes in genera that conglomerate species previously classified in other genera or families. Six uncultured bacteria within the Ruminococcaceae NK4A214 group had lower relative abundance with higher concentrations of PBDE-99 in early pregnancy. This group was found to decrease in the rumen of cows with increased thiamine supplementation.62 Ruminococcaceae NK4A214 includes species that were previously classified as Desulfomicrobium, which was recently found to be increased in the presence of low levels of lead in environmental samples, but nearly absent in the presence of high levels of lead.63 Given the negative association observed in our study, it is possible Ruminococcaceae NK4214 experiences a similar effect with high PBDE-99 exposure, but this hypothesis must be confirmed by other studies. Other genera with species reclassified to Ruminococcaceae NK4A214 include Oscillibacter, a genus that previously was found to be more abundant in toddlers whose mothers were obese, particularly among high income individuals.64 According to the SILVA database, Lachnospiraceae UCG-003, which had higher abundance with higher PCBE-100 exposure, contains only uncultured bacteria, and no studies have been conducted indicating potential health effects. Finally, although Σ4PBDE was associated with changes in the order Clostridiales (which contains the Ruminococcaceae and Lachnospiraceae genera affected by individual PBDEs), it is a large and diverse order with no consistent health effects. For example, the most notorious member of the order, Clostridium difficile, causes severe gastrointestinal intestinal distress and is directly responsible for 1.3%–9.3% of deaths in infected individuals in the United States depending on the source of the infection.65 However, at least one family in the order, the Eubacteriaceae, is more common in controls and patients with inactive inflammatory bowel disease (IBD) than in patients with active IBD.66
It is of note that our results indicate the importance of early pregnancy exposures over later pregnancy (delivery) exposures. While the exposures were positively correlated between the time points, there was not a 1:1 correspondence, and so it is possible that we detected an importance of timing of exposure. This could be an indication of how PBDEs/PCBs modify bacterial community structure (i.e., associations found with early pregnancy concentrations may indicate that the exposures alter the maternal microbiome which is then transferred, whereas associations found with concentrations measured at delivery may reflect the environment in the infant gut to which early colonizing bacteria are exposed). However, the discord in our findings may be an artifact of the limited size of our pilot population. Extensive in vitro research is needed to explore the mechanisms by which PBDEs and PCBs could affect the microbiome. Further, studies in larger populations should assess the relative importance of exposures during different windows of development.
Our pilot study has many strengths. It is one of the first prospective studies examining associations of environmental exposures in pregnancy with the development of the child microbiome. The homogeneous nature of our study participants reduces confounding by factors such as race and socioeconomic position. In addition, we were able to adjust for delivery mode and family income. Because being in a higher income bracket had a significant deleterious association with Faith’s PD, inclusion of this variable was particularly important. In addition to these covariates, our primary analysis explored the associations between common congeners of two classes of chemicals with nearly ubiquitous exposure (PBDEs and PCBs) and the microbiome. Our results indicate the potential of the microbiome as a target of chemicals and suggest more research should be done on the public health implications of differences in bacterial composition induced by environmental exposures. Further, we explored several ways in which the microbiome may be affected by environmental exposures, including large effects on within- and between-subject diversity and smaller effects on individual taxa. Lastly, the statistical methods used in this study are uniquely suited for environmental epidemiological microbiome analyses. In addition to testing for differences at each taxonomic level, MiCAM allows for OTUs within higher-level taxa to respond to exposure differently (i.e., some may increase while others decrease). This flexibility allows for the detection of associations that may not be discernable by running individual regressions for higher-level taxa. Further, the permutation testing and multiple-testing correction incorporated into the OMiAT and MiCAM algorithms ensures the robustness of our results. Finally, OMiAT and MiCAM allow for continuous exposures, which is important for environmental epidemiological situations where there is no unexposed population.
Our study also has limitations. Because this pilot study had a limited sample size, it is possible our findings are the result of small sample bias and random error.67 However, the methods for taxon-level microbiome analyses do not estimate effect sizes, and so our study does not exaggerate the associations of PBDEs and PCBs with the developing microbiome. Further, the consistent findings among PCBs despite different levels of exposure for each congener support their validity. Regardless, our results should be replicated in a larger cohort to confirm the taxa affected by PCBs. Additionally, we cannot rule out unmeasured confounding by factors such as dust ingestion, which is associated with PBDE concentration in human tissues and could affect the microbiome similarly to ingested air pollution, as found by Kish and colleagues.68,69 Another limitation related to the pilot nature of our study is the limited generalizability of the subjects selected for the pilot. GESTE subjects are primarily French-Canadian and upper-middle class. While this reduces confounding by these factors in our study, it limits the generalizability of our findings. In addition, we restricted our study to children who were ever breastfed because early nutrition (breastfeeding/formula) is known to influence the development of the microbiome.70 Therefore, our results should not be generalized to exclusively formula-fed infants. Further, we were not able to adjust our analyses for duration of breastfeeding/childhood diet or supplemental nutritional intake and medication use owing to the limited sample size. However, these variables could not affect PBDE/PCB exposure during pregnancy temporally, and therefore their inclusion in the model would improve precision but their exclusion does not bias effect estimates. Lastly, the temporal distance between exposure and microbiome measurement may be the cause for concern because of the relative instability of the infant microbiome.70 Ideally, we could compare the microbiome in mid-childhood to maternal samples as well as child samples from infancy and toddlerhood to determine whether the observed effects can be detected earlier, which would indicate that our observed associations were persistent perturbations. Additionally, it would have been optimal to measure repeated PBDE and PCB exposure in childhood as it may be correlated with maternal exposure in pregnancy. However, this pilot was conducted using available samples, and future studies will need to examine earlier samples in other cohorts.
Conclusions
In conclusion, our work suggests the potential importance of environmental factors for the developing human gut microbiome. Larger studies should be conducted to confirm our results and to explore whether other environmental contaminants have similar associations. Further, it is vital that the clinical implications of changes in gut microbial structure be examined, particularly when these changes occur in childhood and may have life-long effects.
Conflict of interest statement
The authors declare that they have no financial conflict of interest with regard to the content of this report.
Supplementary Material
Footnotes
Published online 7 February 2019
Supplemental digital content is available through direct URL citations in the HTML and PDF versions of this article (www.environepidem.com).
The results reported herein correspond to specific aims of grant R01ES027845 to investigators Andrea Baccarelli and Larissa Takser from the National Institute of Environmental Health Sciences (NIEHS). This work was supported by grants R21ES024841, P30ES009089, and K23ES022242 from the NIEHS and MOP-84551 from the Canadian Institutes of Health Research.
Data Access: Use of the data may be possible under certain conditions by contacting Larissa Takser (Larissa.Takser@USherbrooke.ca).
References
- 1.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet 201213260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang D, Leung RK, Guan W, Au WW. Involvement of gut microbiome in human health and disease: brief overview, knowledge gaps and research opportunities. Gut Pathog 2018103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoen AG, Madan JC, Li Z, et al. Sex-specific associations of infants’ gut microbiome with arsenic exposure in a US population. Sci Rep 2018812627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ribado JV, Ley C, Haggerty TD, Tkachenko E, Bhatt AS, Parsonnet J. Household triclosan and triclocarban effects on the infant and maternal microbiome. EMBO Mol Med 201791732–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nash MJ, Frank DN, Friedman JE. Early Microbes Modify Immune System Development and Metabolic Homeostasis-The “Restaurant” Hypothesis Revisited. Front Endocrinol (Lausanne) 20178349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Song X, Wei W, et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun 20178875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med 20146237237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiGiulio DB, Romero R, Amogan HP, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One 20083e3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim ES, Rodriguez C, Holtz LR. Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community. Microbiome 2018687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauder AP, Roche AM, Sherrill-Mix S, et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 2016429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amato KR, Yeoman CJ, Cerda G, et al. Variable responses of human and non-human primate gut microbiomes to a Western diet. Microbiome 2015353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gohir W, Whelan FJ, Surette MG, Moore C, Schertzer JD, Sloboda DM. Pregnancy-related changes in the maternal gut microbiota are dependent upon the mother’s periconceptional diet. Gut Microbes 20156310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol 20153169–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young VB. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ 2017356j831. [DOI] [PubMed] [Google Scholar]
- 15.Rosenfeld CS. Gut dysbiosis in animals due to environmental chemical exposures. Front Cell Infect Microbiol 20177396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. In: Services USDoHaH. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention; 2018. Fourth National Report on Human Exposure to Environmental Chemicals: Updated Tables, March 2018. https://www.cdc.gov/exposurereport/ [Google Scholar]
- 17.Lam J, Lanphear BP, Bellinger D, et al. Developmental PBDE exposure and IQ/ADHD in childhood: a systematic review and meta-analysis. Environ Health Perspect 2017125086001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vuong AM, Webster GM, Romano ME, et al. Maternal Polybrominated Diphenyl Ether (PBDE) Exposure and Thyroid Hormones in Maternal and Cord Sera: The HOME Study, Cincinnati, USA. Environ Health Perspect 20151231079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dishaw LV, Macaulay LJ, Roberts SC, Stapleton HM. Exposures, mechanisms, and impacts of endocrine-active flame retardants. Curr Opin Pharmacol 201419125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowell WJ, Lederman SA, Sjödin A, et al. Prenatal exposure to polybrominated diphenyl ethers and child attention problems at 3-7 years. Neurotoxicol Teratol 201552Pt B143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sagiv SK, Kogut K, Gaspar FW, et al. Prenatal and childhood polybrominated diphenyl ether (PBDE) exposure and attention and executive function at 9-12 years of age. Neurotoxicol Teratol 201552Pt B151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vuong AM, Braun JM, Sjödin A, et al. Prenatal polybrominated diphenyl ether exposure and body mass index in children up to 8 years of age. Environ Health Perspect 20161241891–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erkin-Cakmak A, Harley KG, Chevrier J, et al. In utero and childhood polybrominated diphenyl ether exposures and body mass at age 7 years: the CHAMACOS study. Environ Health Perspect 2015123636–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee LK, Ding C, Yang KL, He J. Complete debromination of tetra- and penta-brominated diphenyl ethers by a coculture consisting of dehalococcoides and desulfovibrio species. Environ Sci Technol 2011458475–8482 [DOI] [PubMed] [Google Scholar]
- 25.Yang CW, Huang HW, Chang BV. Microbial communities associated with anaerobic degradation of polybrominated diphenyl ethers in river sediment. J Microbiol Immunol Infect 20175032–39 [DOI] [PubMed] [Google Scholar]
- 26.Hertz-Picciotto I, Charles MJ, James RA, Keller JA, Willman E, Teplin S. In utero polychlorinated biphenyl exposures in relation to fetal and early childhood growth. Epidemiology 200516648–656 [DOI] [PubMed] [Google Scholar]
- 27.Yamashita F, Hayashi M. Fetal PCB syndrome: clinical features, intrauterine growth retardation and possible alteration in calcium metabolism. Environ Health Perspect 19855941–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Errico MN, De Tullio G, Di Gioacchino M, et al. Immune effects of polychlorinated biphenyls, smoking and alcohol. Int J Immunopathol Pharmacol 2012251041–1054 [DOI] [PubMed] [Google Scholar]
- 29.Heilmann C, Grandjean P, Weihe P, Nielsen F, Budtz-Jørgensen E. Reduced antibody responses to vaccinations in children exposed to polychlorinated biphenyls. PLoS Med 20063e311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hertz-Picciotto I, Park HY, Dostal M, Kocan A, Trnovec T, Sram R. Prenatal exposures to persistent and non-persistent organic compounds and effects on immune system development. Basic Clin Pharmacol Toxicol 2008102146–154 [DOI] [PubMed] [Google Scholar]
- 31.Boas M, Feldt-Rasmussen U, Main KM. Thyroid effects of endocrine disrupting chemicals. Mol Cell Endocrinol 2012355240–248 [DOI] [PubMed] [Google Scholar]
- 32.Knerr S, Schrenk D. Carcinogenicity of “non-dioxinlike” polychlorinated biphenyls. Crit Rev Toxicol 200636663–694 [DOI] [PubMed] [Google Scholar]
- 33.Choi JJ, Eum SY, Rampersaud E, Daunert S, Abreu MT, Toborek M. Exercise attenuates PCB-induced changes in the mouse gut microbiome. Environ Health Perspect 2013121725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Zhang W, Hua J, et al. Dysregulation of intestinal health by environmental pollutants: involvement of the estrogen receptor and aryl hydrocarbon receptor. Environ Sci Technol 2018522323–2330 [DOI] [PubMed] [Google Scholar]
- 35.Whittaker RH. Evolution and measurement of species diversity. Taxon 1972212/3213–251 [Google Scholar]
- 36.Abdelouahab N, Langlois MF, Lavoie L, Corbin F, Pasquier JC, Takser L. Maternal and cord-blood thyroid hormone levels and exposure to polybrominated diphenyl ethers and polychlorinated biphenyls during early pregnancy. Am J Epidemiol 2013178701–713 [DOI] [PubMed] [Google Scholar]
- 37.Frings CS, Fendley TW, Dunn RT, Queen CA. Improved determination of total serum lipids by the sulfo-phospho-vanillin reaction. Clin Chem 197218673–674 [PubMed] [Google Scholar]
- 38.LaA Guardia MJ, Hale RC, Harvey E. Detailed polybrominated diphenyl ether (PBDE) congener composition of the widely used penta-, octa-, and deca-PBDE technical flame-retardant mixtures. Environ Sci Technol 2006406247–6254 [DOI] [PubMed] [Google Scholar]
- 39.Longnecker MP, Ryan JJ, Gladen BC, Schecter AJ. Correlations among human plasma levels of dioxin-like compounds and polychlorinated biphenyls (PCBs) and implications for epidemiologic studies. Arch Environ Health 200055195–200 [DOI] [PubMed] [Google Scholar]
- 40.Song SJ, Amir A, Metcalf JL, et al. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems 201613pii–e0002116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One 20127e33865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson CR, Langner HW, Donahoe-Christiansen J, Inskeep WP, McDermott TR. Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ Microbiol 200838532–542 [DOI] [PubMed] [Google Scholar]
- 43.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 201613581–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navas-Molina JA, Peralta-Sánchez JM, González A, et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol 2013531371–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 201341Database issueD590–D596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koh H, Blaser MJ, Li H. A powerful microbiome-based association test and a microbial taxa discovery framework for comprehensive association mapping. Microbiome 2017545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacobson JL, Fein GG, Jacobson SW, Schwartz PM, Dowler JK. The transfer of polychlorinated biphenyls (PCBs) and polybrominated biphenyls (PBBs) across the human placenta and into maternal milk. Am J Public Health 198474378–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frederiksen M, Vorkamp K, Mathiesen L, Mose T, Knudsen LE. Placental transfer of the polybrominated diphenyl ethers BDE-47, BDE-99 and BDE-209 in a human placenta perfusion system: an experimental study. Environ Health 2010932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol 20119244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stackebrandt E, Cummins CS, Johnson JL. Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. Family propionibacteriaceae: The genus Propionibacterium. In: The Prokaryotes: Volume 3: Archaea. Bacteria: Firmicutes, Actinomycetes 2006New York, NY: Springer New York;400–418 [Google Scholar]
- 51.Plé C, Breton J, Richoux R, et al. Combining selected immunomodulatory Propionibacterium freudenreichii and Lactobacillus delbrueckii strains: reverse engineering development of an anti-inflammatory cheese. Mol Nutr Food Res 201660935–948 [DOI] [PubMed] [Google Scholar]
- 52.Altieri C. Dairy propionibacteria as probiotics: recent evidences. World J Microbiol Biotechnol 201632172. [DOI] [PubMed] [Google Scholar]
- 53.Rogers MAM, Aronoff DM. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect 201622178.e1–178.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sipka S, Eum SY, Son KW, et al. oral administration of PCBs induces proinflammatory and prometastatic responses. Environ Toxicol Pharmacol 200825251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allali I, Delgado S, Marron PI, et al. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microbes 20156161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maffeis C, Martina A, Corradi M, et al. Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes. Diabetes Metab Res Rev 201632700–709 [DOI] [PubMed] [Google Scholar]
- 57.Said HS, Suda W, Nakagome S, et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res 20142115–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klinder A, Shen Q, Heppel S, Lovegrove JA, Rowland I, Tuohy KM. Impact of increasing fruit and vegetables and flavonoid intake on the human gut microbiota. Food Funct 201671788–1796 [DOI] [PubMed] [Google Scholar]
- 59.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014505559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petrov VA, Saltykova IV, Zhukova IA, et al. Analysis of gut microbiota in patients with Parkinson’s disease. Bull Exp Biol Med 2017162734–737 [DOI] [PubMed] [Google Scholar]
- 61.Rehman A, Rausch P, Wang J, et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut 201665238–248 [DOI] [PubMed] [Google Scholar]
- 62.Pan X, Xue F, Nan X, et al. Illumina sequencing approach to characterize thiamine metabolism related bacteria and the impacts of thiamine supplementation on ruminal microbiota in dairy cows fed high-grain diets. Front Microbiol 201781818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen YT, Kieu HT, West S, Dang YT, Horn H. Community structure of a sulfate-reducing consortium in lead-contaminated wastewater treatment process. World J Microbiol Biotechnol 20173310. [DOI] [PubMed] [Google Scholar]
- 64.Galley JD, Bailey M, Kamp Dush C, Schoppe-Sullivan S, Christian LM. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PLoS One 20149e113026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lessa FC, Mu Y, Bamberg WM, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med 2015372825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Papa E, Docktor M, Smillie C, et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 20127e39242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Button KS, Ioannidis JP, Mokrysz C, et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci 201314365–376 [DOI] [PubMed] [Google Scholar]
- 68.Wu N, Herrmann T, Paepke O, et al. Human exposure to PBDEs: associations of PBDE body burdens with food consumption and house dust concentrations. Environ Sci Technol 2007411584–1589 [DOI] [PubMed] [Google Scholar]
- 69.Kish L, Hotte N, Kaplan GG, et al. Environmental particulate matter induces murine intestinal inflammatory responses and alters the gut microbiome. PLoS One 20138e62220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bäckhed F, Roswall J, Peng Y, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 201517690–703 [DOI] [PubMed] [Google Scholar]


