Abstract
Background: Type 2 (T2) inflammation drives airway dysfunction in many patients with asthma; yet, we lack a comprehensive understanding of the airway immune cell types and networks that sustain this inflammation. Moreover, defects in the airway immune system in patients with asthma without T2 inflammation are not established.
Objectives: To determine the gene networks that sustain T2 airway inflammation in T2-high asthma and to explore the gene networks that characterize T2-low asthma.
Methods: Network analysis of sputum cell transcriptome expression data from 84 subjects with asthma and 27 healthy control subjects was used to identify immune cell type–enriched networks that underlie asthma subgroups.
Results: Sputum T2 gene expression was characterized by an immune cell network derived from multiple innate immune cells, including eosinophils, mast cells/basophils, and inflammatory dendritic cells. Clustering of subjects within this network stratified subjects into T2-high and T2-low groups, but it also revealed a subgroup of T2-high subjects with uniformly higher expression of the T2 network. These “T2-ultrahigh subjects” were characterized clinically by older age and more severe airflow obstruction and pathologically by a second T2 network derived from T2-skewed, CD11b+/CD103−/IRF4+ classical dendritic cells. Subjects with T2-low asthma were differentiated from healthy control subjects by lower expression of a cytotoxic CD8+ T-cell network, which was negatively correlated with body mass index and plasma IL-6 concentrations.
Conclusions: Persistent airway T2 inflammation is a complex construct of innate and adaptive immunity gene expression networks that are variable across individuals with asthma and persist despite steroid treatment. Individuals with T2-low asthma exhibit an airway deficiency in cytotoxic T cells associated with obesity-driven inflammation.
Keywords: asthma, sputum gene expression, type 2, dendritic cells, CD8 cytotoxic T cells
At a Glance Commentary
Scientific Knowledge on the Subject
Increased airway type 2 (T2) inflammation is a key mechanism of asthma. Murine models have implicated multiple immune cell types in the initiation of airway T2 inflammation, but the immune cells and mechanisms that result in chronic activation of T2 inflammation in human T2-high asthma is unclear. Furthermore, airway immune system abnormalities in T2-low asthma are poorly understood.
What This Study Adds to the Field
We demonstrate how gene expression network analysis can be applied to sputum cell samples to identify abnormalities of immune function in asthma. In T2-high asthma, we show that the gene network at the core of persistent T2 airway inflammation is derived from innate immune cells that include eosinophils, mast cells/basophils, and inflammatory dendritic cells. Expression of this T2 gene network is variable across individuals with T2-high asthma, and a subgroup of individuals with T2-high asthma exhibit very high expression of genes in this network. These individuals with “T2-ultrahigh” asthma are characterized by older age and high expression of a gene signature specific for CD11b+/CD103−/IRF4+ classical dendritic cells. In T2-low asthma, we show that the gene expression profiles reveal a deficiency in airway cytotoxic CD8+ T cells and that this deficiency is associated with obesity-related systemic inflammation.
Of the 300 million patients with asthma worldwide, many respond well to available asthma treatments and experience minimal asthma-associated morbidity (1). However, 5–10% have more severe disease that is resistant to current asthma therapies. These patients experience more frequent asthma exacerbations and significant disability (2). Various pathobiologic mechanisms likely drive this heterogeneity in disease severity, and increased airway type 2 (T2) inflammation driven by the T2 cytokines (IL-4, IL-5, and IL-13) is the most established mechanism of asthma (3). The majority of the understanding of the initiation and maintenance of airway T2 inflammation comes from murine asthma models that demonstrate epithelium-derived cytokines, such as TSLP (thymic stromal lymphopoietin) and IL-33, as activators of tissue-resident immune cells, such as type 2 innate lymphoid cells (ILC2), to produce large amounts of the T2 cytokines (3–6). Alternatively, few human studies have investigated which airway immune cell types underlie the persistent production of T2 cytokines in the airways of individuals with T2-high asthma. Thus, very little is known regarding how airway T2 inflammation is maintained in human asthma. Furthermore, even less is known regarding immune dysfunction in the approximately 40% of individuals with asthma who do not have T2 airway inflammation (7).
One approach to advancing understanding of asthma biology is to study sputum cells, and sputum cells can be studied at both cellular and molecular levels. Indeed, we have shown the feasibility of extracting high-quality RNA from induced sputum cells (8), and we have reported that many individuals with asthma have persistent expression of T2 cytokines despite treatment with inhaled or systemic corticosteroids (7). These patients with steroid-refractory T2-high asthma are older and have more severe disease (7), but our prior sputum cell gene profiling work was PCR based, and the limited number of genes we studied did not reveal the gene expression networks that might explain disease severity in these patients. Furthermore, we were unable to determine the networks operating in patients with T2-low asthma.
Whole-transcriptome gene expression analyses have been conducted in sputum cell samples from individuals with asthma, and these studies have supported a role for eosinophilic, neutrophilic, and paucigranulocytic asthmatic disease (9, 10). However, the multiple immune cell types that compose sputum samples have hindered a more comprehensive understanding of sputum expression patterns. In the present study, we used gene expression network analysis to decipher the immune cell types and mechanisms that underlie both T2-high and T2-low asthma.
Methods
Cell Type–Specific Marker Gene Selection
We analyzed 12 flow-sorted immune cell gene expression profiles from the human Immunological Genome Project (Gene Expression Omnibus accession no. GSE3982) and the IRIS (Immune Response In Silico) Project (Gene Expression Omnibus accession no. GSE22886) (see Tables E1 and E2 in the online supplement). These data sets were combined using ComBat, a batch effect correcting tool in the sva package (11). We proceeded to perform differential expression between each cell type and all other cell types using the limma software package (12). For each cell type, we generated a random forest prediction model to identify the most important significantly upregulated genes that identify cell type; all genes with an importance value greater than 0 were kept (13, 14). Owing to high similarity in the gene expression profiles of several immune cell types, we combined T cells with T-helper cells and mast cells with basophils. This left us with gene expression signatures for 10 purified immune cell types (Table E2). To validate dendritic cell subtypes, we used gene expression data from purified murine lung dendritic cells obtained from the Immunological Genome Project database (15).
Subjects and Sputum Sampling
The sputum RNA samples used in this study were acquired from the Airway Tissue Bank at the University of California, San Francisco. Subject characterization, sputum collection, and RNA extraction were performed using standardized and uniform protocols detailed in the online supplement (8, 16).
RNA Sequencing
Bar-coded whole-transcriptome libraries were generated with the Ion AmpliSeq Transcriptome Human Gene Expression Kit (catalogue no. A26325; Life Technologies) according to the manufacturer’s protocol. Sequencing data were generated with Ion Proton Sequencer (Life Technologies) using standard reagents and protocols. Read mapping was performed with TMAP (Torrent Mapping Alignment Program), and a read count table for each gene amplicon was generated using the Proton AmpliSeq plugin (Life Technologies). Sample sequencing depth was 10–14 million reads.
Statistical Methods
Using DESeq2 (17), gene expression data were variance stabilizing transformation normalized. Multidimensional scaling analysis of the top 500 most variant genes (12) removed 10 outlier samples (Figure E1). Genes with fewer than 10 counts in 15% of samples were excluded from downstream analyses. This approach resulted in a final analysis dataset of 13,536 genes across 84 subjects with asthma and 27 healthy control subjects. Weighted gene coexpression analysis (WGCNA) was performed using a soft-threshold power of 12 and a deep-split metric of 3, and we obtained 24 coexpression networks (Table E3). Eigengenes were calculated using the first principal component of all genes within each network. Using Spearman’s correlation, we determined the association of these gene network eigengene values with each other and with other clinical traits.
Results
Specific Immune Cell Type–enriched Gene Sets
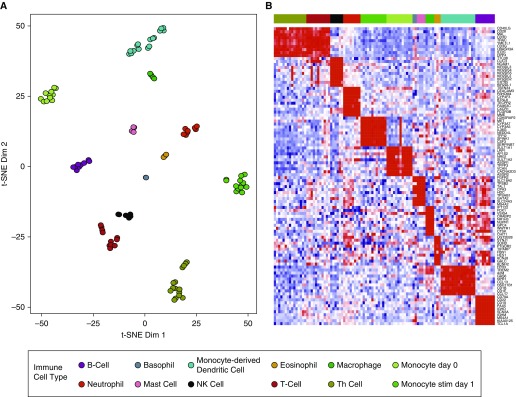
Gene expression data derived from induced sputum cells are a composite of expression from multiple immune cell types in the sample. To determine whether gene expression patterns identified from induced sputum are representative of changes in immune cell composition, we identified sets of genes enriched for specific immune cell types. These gene sets were developed by performing a combination of differential expression and feature selection analyses on published gene expression data (18) for 12 purified human immune cell types (Tables E1–E2) (19). Analysis of these gene sets by t-Distributed Stochastic Neighbor Embedding and plotting cells using two t-Distributed Stochastic Neighbor Embedding dimensions resulted in cell-type separation (Figure 1A). Moreover, ordering by cell types showed cellular specificity in gene expression profiles (Figure 1B).
Figure 1.
The development of gene sets enriched for specific human immune cell types. (A) Immune cell–enriched gene sets derived from analysis of the Garvan Institute for Medical Research and IRIS (Immune Response In Silico) human immune cell databases separate cell types into distinct clusters in a t-Distributed Stochastic Neighbor Embedding (t-SNE) plot (Tables E1 and E2). (B) Heat map of the top 10 genes from the immune cell type–enriched gene sets. Ordering by cell types shows cellular specificity in expression. Dim = dimension; NK = natural killer; Th = T helper.
Sputum RNA-Sequencing Networks Reflect Airway Immune Cell Diversity
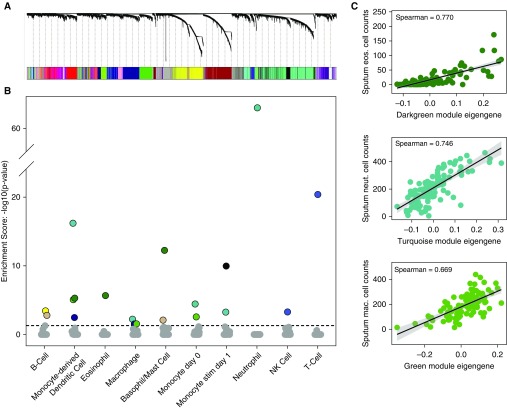
Whole-transcriptome sequencing performed on the induced sputum samples from 27 healthy control subjects and 84 subjects with asthma identified 13,536 expressed genes (Table E4). To identify coexpression networks, we performed WGCNA analysis on the subject–gene expression matrix, which identified 24 networks that we randomly assigned color labels (Figure 2A). Enrichment analysis for immune cell–type gene sets revealed that at least 1 network was enriched for each of the 10 immune cell–type signatures tested, and 9 of the 24 sputum networks were significantly enriched for at least 1 cell-type signature (Figure 2B). For example, the turquoise network was highly enriched in genes specific for neutrophils (Figure 2B), and the dark green network was highly enriched in genes specific for eosinophils, basophils/mast cells, and monocyte-derived dendritic cells (Figure 2B).
Figure 2.
Weighted gene coexpression analysis of sputum transcriptome data identifies gene networks reflective of specific immune cell types. (A) Weighted gene coexpression analysis hierarchical clustering dendrogram of the 13,536 genes that passed quality control filtering for induced sputum samples. Gene dissimilarity was calculated using topological overlap measure. The colors corresponding to network assignments are given in the bar below the dendrogram. (B) Plot of P values for gene set enrichment score of the immune cell–type gene sets in the sputum networks. Ten sputum networks were highly enriched with genes specific for different immune cells. For example, the dark green network was highly enriched with genes specific for eosinophils, basophils/mast cells, and dendritic cells. (C) Correlation plots between sputum cytospin cell counts and sputum network eigengene expression values. Specifically, correlations between the dark green module eigengene values and eosinophil cell counts, turquoise module eigengene values and neutrophil cell counts, and green module eigengene values and macrophage cell counts are shown. eos. = eosinophil; mac. = macrophage; neut. = neutrophil; NK = natural killer.
To validate that our enrichment analyses correctly assigned cell types to their respective enriched networks, we correlated subject network expression with subject sputum cytospin cell counts. For example, we found strong correlations between the dark green network expression and eosinophil cell counts (ρ = 0.77), turquoise network expression and neutrophil cell counts (ρ = 0.746), and green network expression and macrophage cell counts (ρ = 0.669) (Figure 2C). These results reveal the diversity of immune cell types in the human airway and the utility of sputum gene expression network analysis to identify and quantify these immune cells.
A Sputum Network Identifies Patients with T2-High Asthma and a New Subgroup of Patients with T2-UltraHigh Asthma
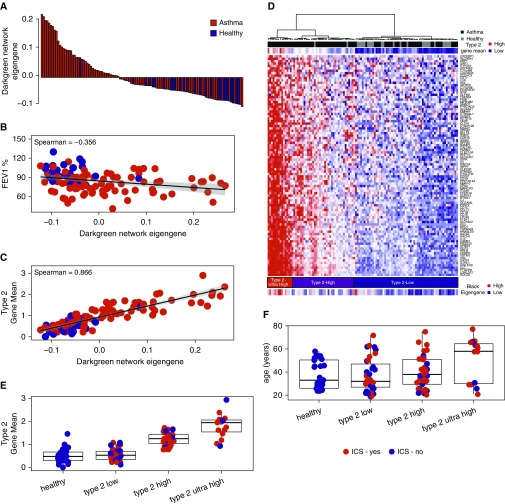
We tested each network’s expression for association with asthma status and asthma-related demographic variables (Figure E2). The dark green network was most strongly associated with asthma, and expression levels of this network were 2.42-fold higher among subjects with asthma (P = 9.82 × 10−9) (Figure 3A). Moreover, dark green network expression negatively correlated with FEV1 percent predicted (ρ = −0.35; P = 2.4 × 10−3) (Figure 3B) and positively correlated with biomarkers of airway T2 inflammation, including blood eosinophil cell counts (ρ = 0.67; P = 9 × 10−15) and fractional exhaled nitric oxide (FeNO) concentrations (ρ = 0.52; P = 5 × 10−8) (Figure E2). We found that all three T2 cytokine genes—IL4, IL5, and IL13—were present in this network and that the T2 cytokine gene mean (8) strongly correlated to dark green network expression (ρ = 0.86; P = 5 × 10−33) (Figure 3C). Therefore, we used the 92 genes in the dark green network to hierarchically cluster subjects into T2-high and T2-low asthma subgroups (Figure 3D). T2-high and T2-low clusters were identified on the basis of the first branching point in the dendrogram (Figure 3D). The T2-high cluster consisted of predominately subjects with asthma (n = 49) and few healthy control subjects (n = 2). The T2-low cluster consisted of nearly equal numbers of subjects with asthma (n = 35) and healthy control subjects (n = 25). Subjects with T2-high asthma had higher blood and sputum eosinophil concentrations, IgE concentrations, and FeNO concentrations, validating the cluster-based T2 status assignment (Table 1). Hierarchical clustering also identified a subgroup of subjects with T2-high asthma with uniformly higher expression of genes in the T2 network (Figure 3D); we classified them as subjects with “T2-ultrahigh” asthma (n = 15) (Figure 3D). This T2-ultrahigh subcluster was characterized by higher blood and sputum eosinophil concentrations, FeNO concentrations, and T2 cytokine expression, but their IgE concentrations were not especially high (Table 1 and Figure 3E). Furthermore, we found that subjects with T2-ultrahigh asthma were older (Table 1 and Figure 3F) and had more severe airflow obstruction (Table 1), despite the majority (11 of 15 [73%]) receiving inhaled corticosteroids (Table 1).
Figure 3.
Sputum transcriptome network analysis identifies a type 2 (T2) inflammation network that clusters subjects with T2-high, T2-ultrahigh, and T2-low asthma. (A, B) Eigengene values of the dark green network were robustly increased in subjects with asthma (black) compared with healthy control subjects (light gray) and were associated with lower FEV1 values. (C) A summary metric of IL-4, IL-5, and IL-13 airway T2 gene expression (T2 gene mean) was strongly associated with the dark green network eigengene values. (D) Hierarchical clustering of subjects based on expression of genes in the dark green network resulted in three primary clusters. Cluster 1 was characterized by low T2 gene expression and contained an equal distribution of subjects with asthma (n = 35) and healthy control subjects (n = 25). Cluster 2 was characterized by high T2 gene expression and consisted predominately of subjects with asthma (n = 34) and few healthy control subjects (n = 2). Cluster 3 was characterized by extremely high T2 gene expression (T2-ultrahigh) and was comprised exclusively of subjects with asthma (n = 15). Black network eigengene values strongly correlated with dark green network eigengene values. (E) The T2 gene mean was robustly higher in subjects with T2-ultrahigh asthma. (F) Subjects with T2-ultrahigh asthma are characterized by older age. ICS = inhaled corticosteroids.
Table 1.
Clinical Characteristics of Type 2 Clusters
| Characteristic | Healthy Control Subjects (n = 27) | T2-Low Asthma (n = 35) | T2-High Asthma (n = 34) | T2-Ultrahigh Asthma (n = 15) | P Value* |
|---|---|---|---|---|---|
| Age, yr | 37.9 (12.1) | 37.4 (12.8) | 40.1 (14.3) | 51.5 (18.5) | 0.009 |
| ICS, n (%) | N/A | 15 (43) | 23 (68) | 11 (73) | 0.02 |
| Female, n (%) | 17 (63) | 21 (60) | 19 (56) | 11 (73) | 0.59 |
| BMI, kg/m2 | 25.4 (6.5) | 29.6 (6.1) | 30.7(7.6) | 26.0 (5.4) | 0.31 |
| FEV1, % predicted | 98.0 (12.7) | 83.0 (15.4) | 79.3 (15.1) | 73.5 (12.5) | <0.001 |
| FVC, % predicted | 98.8 (15.0) | 96.6 (16.1) | 94.2 (17.0) | 85.9 (15.2) | 0.06 |
| Blood cell counts, ×106/L, median (IQR) | |||||
| Neutrophils | 3.3 (2.9–4.5) | 4.2 (3.4–5.0) | 3.8 (3.3–4.8) | 3.6 (2.7–4.0) | 0.05 |
| Eosinophils | 90 (60–170) | 160 (110–250) | 290 (250–350) | 470 (390–550) | <0.001 |
| Sputum cells, %, median (IQR) | |||||
| Neutrophils | 29 (18–36) | 23 (11–31) | 24 (15–35) | 36 (24–48) | 0.05 |
| Eosinophils | 0 (0–0.1) | 0.2 (0–0.6) | 2.0 (0.9–4.3) | 7.3 (4.0–13.2) | <0.001 |
| FeNO† | 15.9 (11.9) | 32.0 (35.1) | 40.6 (29.7) | 52.4 (33.2) | 0.09 |
| Blood IgE, median (IQR)‡ | 16 (7–57) | 65 (37–319) | 318 (96–741) | 113 (63–248) | 0.3 |
| T2GM | −0.58 (0.33) | −0.54 (0.27) | 0.31 (0.43) | 1.60 (1.30) | <0.001 |
| ACT score, median (IQR)§ | N/A | 20 (17–22) | 19.5 (16–22) | 18 (16–21) | 0.71 |
Definition of abbreviations: ACT = Asthma Control Test; BMI = body mass index; FeNO = fractional exhaled nitric oxide; ICS = inhaled corticosteriods; IQR = interquartile range; N/A = not applicable; T2 = type 2; T2GM = type 2 gene mean.
Data are reported as mean (SD) unless otherwise indicated.
P values for association based on univariate ordinal logistic regression modeling in the subjects with asthma.
FeNO was available for 55 subjects with asthma.
Blood IgE was available for 67 subjects with asthma.
ACT score was available for 71 subjects with asthma.
Sputum T2 Inflammation Is Characterized by the Activity of Multiple Innate Immune Cells
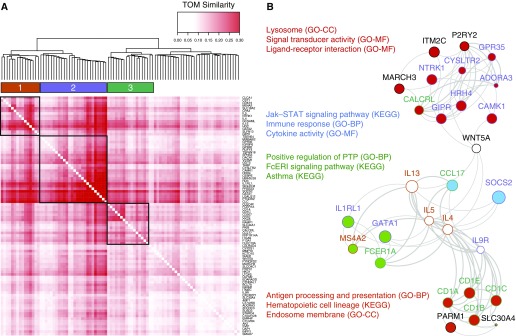
Our immune cell enrichment analysis indicated that the sputum T2 network is derived from multiple innate immune cells, including eosinophils, mast cells/basophils, and monocyte-derived dendritic cells. To better understand the mechanisms underlying the genes in these immune cell enrichments, we examined the connectivity of the 92 T2 network genes using the WGCNA topological overlap matrix. Hierarchical clustering of these genes identified three highly correlated subsets of genes (labeled subnetworks 1–3) within the T2 network (Figure 4). Subnetwork 1 contained all three T2 cytokine genes (IL4, IL5, IL13), together with three genes highly specific to mast cells/basophils (CPA3 [carboxypeptidase A3], HDC [histidine decarboxylase], MS4A2 [membrane spanning 4-domains A2]) and GATA2 (GATA-binding protein 2), a critical transcription factor in basophil and mast cell differentiation (20, 21). We also observed three genes—NTRK1 (neurotrophic receptor tyrosine kinase 1), CST1 (cystatin SN), and CLCA1 (chloride channel accessory 1)—highly induced from airway epithelial cells by IL-13 stimulation (21, 22).
Figure 4.
A multicellular innate immunity network underlies asthmatic persistent type 2 inflammation. (A) Topological overlap measure (TOM) plot showing the subclusters of dark green network genes with highly correlated expression. The heat map is based on correlations of dissimilarity between genes. (B) FGNet enrichment results for the genes within the dark green module. The four enrichment metagroups are denoted by the fill of the node (gene) colors, where genes involved in multiple metagroups are filled in white. The gene subclusters identified in A are denoted by the node outline colors, where a black outline indicates that the gene was not within any of the three subclusters in A. FCeRI = high-affinity IgE receptor; GO-BP = Gene Ontology–Biological Process; GO-CC = Gene Ontology–Cellular Component; GO-MF = Gene Ontology–Molecular Function; KEGG = Kyoto Encyclopedia of Genes and Genomes; PTP = protein tyrosine phosphatase; STAT = signal transducer and activator of transcription.
Subnetwork 2 contained the most interconnected genes in the T2 network (Figure 4). CEBPE (CCAAT enhancer–binding protein epsilon) and GATA1, two genes with critical roles in eosinophil differentiation and maturation, were within this network (23). Moreover, 17 of the 26 genes in this network were upregulated in sputum eosinophils after allergen challenge (P = 3.65 × 10−21) (24). In addition, a hub gene in this network was IL1RL1 (IL-1 receptor-like 1), encoding the ST2 receptor for IL-33 cytokine signaling, a key signaling mechanism in T2 inflammation (25).
Consistent with our enrichment, the genes in subnetwork 3 were highly representative of inflammatory dendritic cells (IDECs), including CD1A, CD1B, CD1C, CD1E, RAMP1 (receptor activity modifying protein 1), CD207, and F13A1 (coagulation factor XIII A chain) (Figure 4). We also noted the presence of other IDEC markers in this subnetwork, such as the T2 chemokine CCL17 (C-C motif chemokine ligand 17) and FCER1A (Fc fragment of IgE receptor Ia) subunit of the high-affinity IgE receptor. Collectively, these data suggest that the T2 cytokine signal in the airways of patients with T2-high asthma is derived from the interaction of multiple innate immune cells, including eosinophils, mast cells/basophils, and IDECs.
An Activated T2-skewed CD11b+IRF4+CD103− Dendritic Cell Network Is Increased in T2-High Asthma
The T2 network (dark green) results showed the centrality of innate immune cells in persistent airway T2 inflammation, but we also found a strong correlation between the black network and the T2 gene network (ρ = 0.67; P = 1.3 × 10−15) (Figure 3D). Notably, the black network contained CD11B (a dendritic cell marker), ZBTB46 (zinc finger and BTB domain containing 46) (26), and FLT3 (fms-related tyrosine kinase 3), three genes highly selective for classical dendritic cells (cDCs) (27, 28). In fact, expression of ZBTB46 and FLT3 was highly correlated with black network expression (Figure 5A). Prior work in mouse lungs has established CD11b+/CD103− and CD11b−/CD103+ subpopulations of cDCs (6, 29) driven by IRF4 (IFN-regulatory factor 4) and IRF8 (30), respectively. Both CD11B and IRF4 are part of the black network, and expression of these genes was highly correlated with black network expression (Figure 5B). In contrast, neither CD103 nor IRF8 was part of the black network, and expression of both genes was negatively correlated with black network expression (Figure 5C). To confirm these findings in a global analysis, we examined transcriptome-wide expression data for purified mouse lung CD11b+/CD103− and CD11b−/CD103+ dendritic cell populations. These murine gene sets were obtained from the Immunological Genome Project database (30). Using gene set enrichment analysis (31), we found that black network genes were strongly enriched among the genes most highly expressed in CD11b+/CD103−, as compared with CD11b−/CD103+ cells (P < 1.0 × 10−5) (Figure 5D).
Figure 5.
A type 2 (T2) inflammation–skewed CD11b+IRF4+CD103− classical dendritic cell (DC) sputum network is increased in T2-high asthma. (A) ZBTB46 (zinc finger and BTB domain containing 46) and FLT3 (fms-related tyrosine kinase 3), two genes highly specific for classical DCs, are tightly correlated with the eigengene values of the black module. (B) Gene expression for the DC surface marker CD11B and the CD11B DC transcription factor IRF4 (IFN-regulatory factor 4) are tightly correlated with the eigengene values of the black module. (C) The cell surface marker CD103 and the CD103+ DC transcription factor IRF8 are negatively correlated to the eigengene values of the black network. Circles represent each subject and are shaded according to the expression of the T2 gene mean (red = high expression; blue = low expression). (D) Gene set enrichment analysis. The green curve displays the running enrichment score for the black network genes as the analysis walks down the ranked distribution of genes ordered by fold change in expression between CD11b+CD103− DCs versus CD11b−C103+ DCs. Genes are represented by the vertical black bars. P < 0.001.
Functional enrichment analyses (32, 33) of black network genes identified Gene Ontology categories involved in LPS-mediated signaling and its primary activation target, NF-κB (nuclear factor κB) (Figure E3). In fact, all five NF-κB family members were located in the black network. LPS is a strong maturation stimulus for dendritic cells, and NF-κB signaling is required for dendritic cell development (34). Enrichment analysis of black network genes also identified positive regulation of IL-2 production. IL-2 is produced by mature dendritic cells and stimulates T-cell expansion (35). Taken together, our data suggest that this black network is tracking mature CD11b+/CD103−/IRF4+ dendritic cell stimulation of T cells. This conclusion is further supported by the observation that the OX40 ligand (TNFSF4 [TNF superfamily member 4]) and the OX40 receptor (TNFRSF4 [TNF receptor superfamily member 4]) genes are in this network. The OX40 receptor is expressed only on activated rather than naive T cells, and likewise, OX40L is expressed only by activated dendritic cells (36). Moreover, OX40L is expressed by a specific type of dendritic cell driving T-helper type 2 cell (Th2) differentiation (37). This complements recent data showing that CD11b+/IRF4+ dendritic cells drive T2 inflammation in the mouse lung (6, 30). Furthermore, the CRLF2 (cytokine receptor like factor 2) gene, which serves as the coreceptor for the master T2 cytokine TSLP, and CCL22, a T2 chemokine produced by dendritic cells, were both present in the black network. Taken together, our data support a role for CD11b+/CD103−/IRF4+ and CD103+/CD11b−/IRF8+ dendritic cell populations in the human airway that are increased and decreased, respectively, in individuals with T2-high asthma. Moreover, our data support a role for CD11b+/CD103−/IRF4+ dendritic cells as key activators of T cells in a T2-skewed manner to drive persistent airway T2 inflammation.
Sputum CD8+ Cytotoxic T-Cell Network Is Decreased in T2-Low Asthma and Negatively Associates with Obesity
To identify immune mechanisms of T2-low asthma, we compared eigengene expression of each of the immune cell–enriched WGCNA networks between subjects with T2-low asthma (n = 35) and healthy control subjects (n = 27). We found that expression of the royal blue network was robustly lower in subjects with T2-low asthma than in healthy control subjects (false discovery rate, 0.03) (Table E5). Immune cell–type enrichment analysis identified the royal blue network as enriched for T-cell and natural killer cell genes (Figure 2B). Because CD8+ cytotoxic T cells (CD8+ T cells) express many of the same genes as natural killer cells, we analyzed the expression data for CD8+ T-cell genes. Of the CD8+ cell surface markers, we found that CD3E, CD3G, CD3D, CD8A, and CD8B (38) were all within the royal blue network and that their expression was strongly correlated with royal blue network expression (Figure 6A and Table E3). Furthermore, many CD8+ T-cell genes involved in cytotoxic function, including killer cell lectin-like receptors, PRF1 (perforin 1), and the granzymes, were identified as highly correlated members of the royal blue network (Figures 6B and 6C and Table E3). Finally, we identified many additional CD8+ T cell–associated genes in the royal blue network, including the CD8+ T-cell transcription factors EOMES (eomesodermin) and TBX21 (T-box 21) (39, 40) as well as the cell surface markers CD40LG, CD96, NKG7 (natural killer cell granule protein 7), CD28, and IL12RB2 (IL-12 receptor subunit β2) (Table E3 and Figure 6D). To confirm that the royal blue module was representing CD8+ T cells, we used gene set enrichment analysis (31) to test overrepresentation of genes from the royal blue module in the most differentially expressed genes between flow-sorted CD8+ T cells and all other immune cell subtypes (Table E2). This analysis confirmed that genes in the royal blue network were strongly enriched among the genes most highly expressed in CD8+ T cells (Figure 6E).
Figure 6.
The sputum gene expression network enriched for cytotoxic CD8+ T-cell genes is decreased in type 2 inflammation–low asthma and associated with body mass index (BMI). (A) The cell surface markers of CD8+ T cells, CD3 and CD8, are strongly correlated to eigengene values for the royal blue module. (B) KLRB1 (killer cell lectin-like receptor B1) gene expression is strongly correlated to eigengene values for the royal blue module. (C) Genes for cytotoxic function, PRF1 (perforin 1) and GZMB (granzyme B), are strongly correlated to eigengene values for the royal blue module. (D) CD8+ T-cell transcription factors EOMES (eomesodermin) and TBX21 (T-box 21) are strongly correlated to eigengene values for the royal blue module. (E) Gene set enrichment analysis. The green curve displays the enrichment score for the royal blue network genes as the analysis walks down the ranked distribution of genes ordered by fold change in expression between CD8+ T cells relative to all other immune cell types. Genes are represented by the vertical black bars. (F) Eigengene values of the royal blue module are strongly negatively correlated to BMI and plasma IL-6 concentration. Red dots represent subjects with asthma, and blue dots represent healthy control subjects. KLRK1 = killer cell lectin-like receptor K1.
Interestingly, we found that the royal blue network was negatively associated with body mass index (ρ = −0.44; P = 1.58 × 10−6) (Figure 6F), with lower network expression levels in obese subjects with asthma than in nonobese subjects with asthma (P = 0.005) (Figure E4). Obesity induces a state of chronic low-grade inflammation characterized by elevated plasma concentrations of inflammatory cytokines such as IL-6, and we recently determined that this inflammation strongly associates with disease severity in asthma (41). Thus, we assessed the relationship between the CD8+ T-cell network and plasma IL-6 concentrations. We found a strong negative correlation between plasma IL-6 concentrations and expression of the CD8+ T-cell network (Figure 6F). As a sensitivity analysis to determine if this observation was secondary to inhaled corticosteroids, we stratified subjects into those receiving or not receiving inhaled corticosteroids. We found that subjects with asthma who were not receiving inhaled corticosteroids demonstrated a similar negative correlation between CD8+ T-cell gene expression and body mass index (Figure E5). In concert, these findings suggest that expression of the CD8+ cytotoxic T-cell network is decreased in individuals with T2-low asthma compared with healthy control individuals and that this decrease tracks with obesity and systemic IL-6 inflammation.
Discussion
The mechanism by which the airway immune system sustains persistent T2 cytokine production is poorly understood, and limited data exists for whether this inflammatory process is heterogeneous across individuals with T2-high asthma. To address this deficiency, we performed a complete characterization of the airway immune system by analysis of whole-transcriptome gene expression patterns generated from the sputum cell pellets of subjects with asthma and healthy control subjects. We found that the core T2 inflammation network exhibited gene expression profiles characteristic of multiple innate immune cells, including mast cells/basophils, eosinophils, and IDECs. Further dissecting this network into subnetworks, we found that one such subnetwork was clearly derived from the gene expression of eosinophils, reaffirming the centrality of eosinophils to airway T2 inflammation and the dependence of airway eosinophilia on T2 cytokines. Interestingly, the subnetwork containing IL4, IL5, and IL13 also contained canonical mast cell/basophil genes, suggesting these cells as candidate cellular sources of T2 cytokines in the airways. In support of this possibility, we recently reported that ex vivo IL-33 stimulation of sputum cell samples resulted in IL13 and IL5 expression only in sputum samples that had a strong mast cell/basophil gene signature before stimulation (21). Most surprisingly, a third subnetwork was observed that contained genes characteristic of IDECs. Although this is one of the first reports, to our knowledge, of an IDEC gene signature in airway T2 inflammation, these cells are a dominant feature in skin tissue in atopic dermatitis (42, 43). The IDEC signature includes the CD1 family genes that are involved in antigen presentation and the FCER1A gene subunit for the high-affinity IgE receptor, suggesting roles for these cells in both adaptive immune and IgE responses.
Our workflow of sputum collection and RNA sequencing, coupled with unsupervised clustering of our core T2 network, represents a robust and unbiased method for classifying individuals with asthma as “T2 high” and “T2 low.” Expression of these T2 network genes varied widely among the T2-high subgroup, and we found a subgroup of subjects with nearly uniformly high expression of all T2 network genes. These subjects with “T2-ultrahigh” asthma were older and had more severe airflow obstruction (7) despite the fact that they were usually receiving inhaled corticosteroids. These data suggest that not all airway T2 inflammation responds to inhaled corticosteroids, a finding that is consistent with our recent report of individuals with T2-high asthma who are older and whose asthma is nonresponsive to intramuscular triamcinolone (7). Our findings for a distinct T2-ultrahigh subgroup require confirmation in future studies of airway T2 inflammation and severe asthma.
Our analysis also revealed a cDC network that was increased in subjects with T2-high asthma, but especially in the T2-ultrahigh subgroup. This human airway cDC network clearly mirrored the well-described murine airway CD11b+/CD103− cDC population, which was shown to be a critical driver of the T2 immune response in mice (6, 30, 44). Functional enrichment analysis of genes in this network demonstrated evidence of cDC activation via NF-κB signaling and upregulation of IL-2 production. Notably, the Gene Ontology signature for production of T cell–activating IL-2 and the presence of the dendritic cell ligand OX40 (TNFSF4) and the T-cell OX40 receptor (TNFRSF4) genes in this network leads us to conclude that this network captures key molecular events in the dendritic cell activation of Th2 cells (6, 30, 37). The especially high expression of this network in the subjects with T2-ultrahigh asthma provides a potential mechanism underlying this severe disease subtype and a rationale for targeting CD11b+ cDCs and/or the associated pathways in patients with T2-ultrahigh asthma.
The molecular mechanisms underlying T2-low asthma remain poorly understood. We found decreased expression of a network containing genes highly characteristic of CD8+ cytotoxic T cells in T2-low asthma. A primary function of CD8+ T cells is host defense against viral infection, and our data lead us to speculate that CD8+ T-cell deficiency and an impaired immune response to viral infections could be a mechanism of exacerbations in T2-low asthma. Interestingly, we found that expression of this CD8+ cytotoxic T-cell network was strongly related to obesity and systemic inflammation, and it is possible that obesity-related systemic inflammation could cause CD8+ cytotoxic T-cell dysfunction, possibly via T-cell exhaustion (45). Supporting this hypothesis, IL-6 concentrations were negatively associated with expression of the CD8+ T-cell network, and genes specific for T-cell exhaustion, including PDCD1 (programmed cell death 1), LAG3 (lymphocyte-activating 3), and CTLA4 (cytotoxic T-lymphocyte–associated protein 4) (46), were in this CD8+ cytotoxic T-cell network (Table E3). The cross-sectional nature of our study opens the possibility that our findings in T2-low asthma could be driven by the treatment effect of inhaled corticosteroids on airway inflammation. However, the strong negative association between body mass index and the CD8+ T-cell gene network remained robust even in patients not receiving inhaled corticosteroids, suggesting that inhaled corticosteroid treatment was not responsible for this relationship.
We acknowledge limits to our use of gene expression proxies to infer the presence and quantity of specific cell types. Specifically, other cell types that were not represented in our analysis may also play a role in T2-high and T2-low disease. For example, mouse and human studies suggest that ILC2 cells are major producers of T2 cytokines. Using available mouse ILC2 cell data, we were unable to identify genes specifically enriched in ILC2 cells, preventing us from determining their contribution to the sputum T2 inflammatory signature. Moreover, gene profiles enriched in blood-derived immune cells used in our study may differ from these cells in the airway. Future studies employing single-cell sequencing of sputum cells will be needed to confirm and extend our results.
In conclusion, we demonstrate how gene coexpression network analysis in sputum cells can reveal airway immune cell dysfunction in asthma. Our work advances understanding of airway immune dysfunction in asthma and provides mechanistic targets for drug development in steroid-resistant T2-high and T2-low phenotypes of asthma.
Supplementary Material
Footnotes
Supported by NIH grants P01 HL107201, R01 HL080414, K23 HL138303, R01 HL135156-01, R01 MD010443, R01 HL128439, P01 HL132821, P30DK098722, and R01 MD010443 and by grants from the Parker B. Francis Foundation and the Sandler Asthma Basic Research Center at the University of California San Francisco.
Author Contributions: M.C.P. and M.A.S. are the guarantors of the paper, taking responsibility for the integrity of the work as a whole from inception to published article; M.C.P., J.V.F., and M.A.S. conceived of and designed the study; L.R. and N.D. did the primary analysis and made substantial contributions to the design of the study and interpretation of data; R.H. and C.R. made substantial contributions to the design of the study and data analysis; B.O’C. and P.G.W. made substantial contributions to the interpretation of data for the work; and M.C.P., L.R., N.D., and M.A.S. prepared the first draft of the manuscript. All authors revised the draft critically for important intellectual content. All authors gave final approval of the manuscript version to be published.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201807-1291OC on October 29, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.GBD 2015 Chronic Respiratory Disease Collaborators. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir Med. 2017;5:691–706. doi: 10.1016/S2213-2600(17)30293-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43:343–373. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
- 3.Fahy JV. Type 2 inflammation in asthma—present in most, absent in many. Nat Rev Immunol. 2015;15:57–65. doi: 10.1038/nri3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohapatra A, Van Dyken SJ, Schneider C, Nussbaum JC, Liang HE, Locksley RM. Group 2 innate lymphoid cells utilize the IRF4-IL-9 module to coordinate epithelial cell maintenance of lung homeostasis. Mucosal Immunol. 2016;9:275–286. doi: 10.1038/mi.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, Molofsky AB, et al. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 2013;502:245–248. doi: 10.1038/nature12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plantinga M, Guilliams M, Vanheerswynghels M, Deswarte K, Branco-Madeira F, Toussaint W, et al. Conventional and monocyte-derived CD11b+ dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38:322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Peters MC, Kerr S, Dunican EM, Woodruff PG, Fajt ML, Levy BD, et al. Refractory airway type-2 inflammation in a large subgroup of asthmatics treated with inhaled corticosteroids. J Allergy Clin Immunol. 2019;143:104–113. doi: 10.1016/j.jaci.2017.12.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters MC, Mekonnen ZK, Yuan S, Bhakta NR, Woodruff PG, Fahy JV. Measures of gene expression in sputum cells can identify TH2-high and TH2-low subtypes of asthma. J Allergy Clin Immunol. 2014;133:388–394. doi: 10.1016/j.jaci.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol. 2011;127:153–160.e9. doi: 10.1016/j.jaci.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 10.Baines KJ, Simpson JL, Wood LG, Scott RJ, Fibbens NL, Powell H, et al. Sputum gene expression signature of 6 biomarkers discriminates asthma inflammatory phenotypes. J Allergy Clin Immunol. 2014;133:997–1007. doi: 10.1016/j.jaci.2013.12.1091. [DOI] [PubMed] [Google Scholar]
- 11.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn M, Wing J, Weston S, Williams A, Keefer C, Engelhardt A, et al. caret: Classification and regression training 2017[accessed 2018 Jan 25]. Available from: https://cran.r-project.org/web/packages/caret/index.html.
- 14.Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, Feuston BP. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003;43:1947–1958. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 15.Heng TSP, Painter MW, Elpek K, Lukacs-Kornek V, Mauermann N, Turley SJ, et al. Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 16.Gershman NH, Wong HH, Liu JT, Mahlmeister MJ, Fahy JV. Comparison of two methods of collecting induced sputum in asthmatic subjects. Eur Respir J. 1996;9:2448–2453. doi: 10.1183/09031936.96.09122448. [DOI] [PubMed] [Google Scholar]
- 17.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeffrey KL, Brummer T, Rolph MS, Liu SM, Callejas NA, Grumont RJ, et al. Positive regulation of immune cell function and inflammatory responses by phosphatase PAC-1. Nat Immunol. 2006;7:274–283. doi: 10.1038/ni1310. [DOI] [PubMed] [Google Scholar]
- 19.Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, et al. Immune Response In Silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6:319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Qi X, Liu B, Huang H. The STAT5-GATA2 pathway is critical in basophil and mast cell differentiation and maintenance. J Immunol. 2015;194:4328–4338. doi: 10.4049/jimmunol.1500018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon ED, Simpson LJ, Rios CL, Ringel L, Lachowicz-Scroggins ME, Peters MC, et al. Alternative splicing of interleukin-33 and type 2 inflammation in asthma. Proc Natl Acad Sci USA. 2016;113:8765–8770. doi: 10.1073/pnas.1601914113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rochman M, Kartashov AV, Caldwell JM, Collins MH, Stucke EM, KC K, et al. Neurotrophic tyrosine kinase receptor 1 is a direct transcriptional and epigenetic target of IL-13 involved in allergic inflammation. Mucosal Immunol. 2015;8:785–798. doi: 10.1038/mi.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du J, Stankiewicz MJ, Liu Y, Xi Q, Schmitz JE, Lekstrom-Himes JA, et al. Novel combinatorial interactions of GATA-1, PU.1, and C/EBPε isoforms regulate transcription of the gene encoding eosinophil granule major basic protein. J Biol Chem. 2002;277:43481–43494. doi: 10.1074/jbc.M204777200. [DOI] [PubMed] [Google Scholar]
- 24.Esnault S, Kelly EA, Schwantes EA, Liu LY, DeLain LP, Hauer JA, et al. Identification of genes expressed by human airway eosinophils after an in vivo allergen challenge. PLoS One. 2013;8:e67560. doi: 10.1371/journal.pone.0067560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon ED, Palandra J, Wesolowska-Andersen A, Ringel L, Rios CL, Lachowicz-Scroggins ME, et al. IL1RL1 asthma risk variants regulate airway type 2 inflammation. JCI Insight. 2016;1:e87871. doi: 10.1172/jci.insight.87871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satpathy AT, KC W, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med. 2012;209:1135–1152. doi: 10.1084/jem.20120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karsunky H, Merad M, Cozzio A, Weissman IL, Manz MG. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J Exp Med. 2003;198:305–313. doi: 10.1084/jem.20030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waskow C, Liu K, Darrasse-Jèze G, Guermonprez P, Ginhoux F, Merad M, et al. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat Immunol. 2008;9:676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer JU, Demiri M, Agace WW, MacDonald AS, Svensson-Frej M, Milling SW. Different populations of CD11b+ dendritic cells drive Th2 responses in the small intestine and colon. Nat Commun. 2017;8:15820. doi: 10.1038/ncomms15820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams JW, Tjota MY, Clay BS, Vander Lugt B, Bandukwala HS, Hrusch CL, et al. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat Commun. 2013;4:2990. doi: 10.1038/ncomms3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouaaz F, Arron J, Zheng Y, Choi Y, Beg AA. Dendritic cell development and survival require distinct NF-κB subunits. Immunity. 2002;16:257–270. doi: 10.1016/s1074-7613(02)00272-8. [DOI] [PubMed] [Google Scholar]
- 35.Sallusto F, Lanzavecchia A. The instructive role of dendritic cells on T-cell responses. Arthritis Res. 2002;4(Suppl 3):S127–S132. doi: 10.1186/ar567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croft M, So T, Duan W, Soroosh P. The significance of OX40 and OX40L to T-cell biology and immune disease. Immunol Rev. 2009;229:173–191. doi: 10.1111/j.1600-065X.2009.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenkins SJ, Perona-Wright G, Worsley AGF, Ishii N, MacDonald AS. Dendritic cell expression of OX40 ligand acts as a costimulatory, not polarizing, signal for optimal Th2 priming and memory induction in vivo. J Immunol. 2007;179:3515–3523. doi: 10.4049/jimmunol.179.6.3515. [DOI] [PubMed] [Google Scholar]
- 38.Bevan MJ. Helping the CD8+ T-cell response. Nat Rev Immunol. 2004;4:595–602. doi: 10.1038/nri1413. [DOI] [PubMed] [Google Scholar]
- 39.Knox JJ, Cosma GL, Betts MR, McLane LM.Characterization of T-bet and eomes in peripheral human immune cells Front Immunol 20145217[Published erratum appears in Front Immunol 2016;7:337.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li G, Yang Q, Zhu Y, Wang HR, Chen X, Zhang X, et al. T-bet and eomes regulate the balance between the effector/central memory T cells versus memory stem like T cells. PLoS One. 2013;8:e67401. doi: 10.1371/journal.pone.0067401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters MC, McGrath KW, Hawkins GA, Hastie AT, Levy BD, Israel E, et al. National Heart, Lung, and Blood Institute Severe Asthma Research Program. Plasma interleukin-6 concentrations, metabolic dysfunction, and asthma severity: a cross-sectional analysis of two cohorts. Lancet Respir Med. 2016;4:574–584. doi: 10.1016/S2213-2600(16)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dyjack N, Goleva E, Rios C, Kim BE, Bin L, Taylor P, et al. Minimally invasive skin tape strip RNA sequencing identifies novel characteristics of the type 2-high atopic dermatitis disease endotype. J Allergy Clin Immunol. 2018;141:1298–1309. doi: 10.1016/j.jaci.2017.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujita H, Shemer A, Suárez-Fariñas M, Johnson-Huang LM, Tintle S, Cardinale I, et al. Lesional dendritic cells in patients with chronic atopic dermatitis and psoriasis exhibit parallel ability to activate T-cell subsets. J Allergy Clin Immunol. 2011;128:574–582.e12. doi: 10.1016/j.jaci.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 44.Neyt K, Lambrecht BN. The role of lung dendritic cell subsets in immunity to respiratory viruses. Immunol Rev. 2013;255:57–67. doi: 10.1111/imr.12100. [DOI] [PubMed] [Google Scholar]
- 45.Peters MC, Fahy JV. Metabolic consequences of obesity as an “outside in” mechanism of disease severity in asthma. Eur Respir J. 2016;48:291–293. doi: 10.1183/13993003.01132-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.