Abstract
Background
Clinical characteristics (taxonomy, virulence genes and antimicrobial resistance ) of Aeromonas in isolated from extra-intestinal and intestinal infections were investigated to describe epidemiology, associated virulence factors and optimal therapy options.
Methods
Clinical samples (n = 115) of Aeromonas were collected from a general hospital in Beijing between the period 2015 and 2017. Taxonomy was investigate by Multilocus phylogenetic analysis (MLPA), 10 putative virulence factors by use of polymerase chain reaction (PCR) and antimicrobial resistance to 15 antibiotics by use of the microbroth dilution method.
Results
The most common species of Aeromonas detected in samples of intestinal tract included; A. caviae (43.9%), A. veronii (35.7%), and A. dhakensis (12.2%). Prevalent species of Aeromonas collected from extra-intestinal infections included; A. hydrophila (29.4%), A. caviae (29.4%), and A. dhakensis (23.5%). A. hydrophila were detected in 1% of stool samples and 29.4% (5/17) of extra-intestinal infections. A. hydrophila strains in extra-intestinal infections were related to malignancy. The most common medical conditions among patients with Aeromonas infections included malignancy and liver-transplant related cholecystitis. Multiple drug resistance (MDR) was prevalent in extra-intestinal isolates (82.3%, 14/17) and was greater than the prevalence in intestinal isolates (30.6%, 30/98) (P < 0.05). Resistant rates of extra-intestinal isolates were 70.6, 35.3, 23.5 and 5.9% for ceftriaxone, ciprofloxacin, gentamicin and imipenem, respectively, and were higher than found in previous studies. Despite differences in the number and type of virulence genes among samples of Aeromonas, no significant correlation was found between invasion and virulent genes in intestinal or extra-intestinal infections.
Conclusions
Overall results of this study support a role for Aeromonas spp. as a potential causative infectious agent of gastroenteritis, and malignancy, liver cirrhosis, post liver transplantation in immunocompromised patients. A. hydrophila was more prevalent in samples of extra-intestinal infections when compared to samples of intestinal infections, and was especially prominent in samples of patients presenting with malignancy. Aeromonas isolates from extra-intestinal samples had high rates of drug resistance but 3rd generation cephalosporins, fluoroquinolones and aminoglycosides remain as options to treat severe diarrhea. However, increasing MDR of extra-intestinal infection samples warrants monitoring.
Electronic supplementary material
The online version of this article (10.1186/s12879-019-3766-0) contains supplementary material, which is available to authorized users.
Keywords: Aeromonas, Multilocus phylogenetic analysis (MLPA), Virulence genes , Multi-drug resistance
Background
The genus Aeromonas is a common, gram-negative, facultative anaerobe, coccobacillary-to-bacillary bacteria that belongs to Aeromonadaceae [1].The genus Aeromonas is comprised of mesophiles and psychrophiles which can cause a number of diseases to warm and cold-blooded animals [2]. Recently, mesophilic Aeromonas have received increased attention as an emergent agent of foodborne illness [3]. In humans, Aeromonas can cause extra-intestinal diseases, especially in immunocompromised individuals, including septicemia, wound infections, urinary tract infections, hepatobiliary tract infections and necrotizing fasciitis [4].
Aeromonas have a complex taxonomy and the genus is comprised of over 30 species, however their identification has been limited by use of conventional biochemical identification methods such as matrix-assisted laser desorption/ionization time of flight masss spectrometry (MALDI-TOF MS), and 16S ribosomal ribonucleic acid (rRNA) sequencing [5–8]. To this end, the use of 5 or more housekeeping genes has been demonstrated as an effective approach for multilocus phylogenetic analysis (MLPA) and species identification of Aeromonas spp. [5, 9]. In addition, MLPA has been recommended for the verification of taxonomic affiliation by genome sequencing before being submitted to the NCBI database [10]. Current literature indicates that A. hydrophila, A. veronii bv sobria, and A. caviae are responsible for the majority of human infections and clinical isolations [11]. However, caution must be exercised as A. dhakensis can be misidentified as A. hydrophila by use of some phenotypic methods [12] and MLPA is suggeted for molecular subtyping [13, 14].
A. dhakensis was initially described as a A. hydrophila subspecies in 2002, and A. aquariorum described later, and was recommended to be reclassified as a separate species in 2012 [15].
The pathogenesis of Aeromonas spp. involves a series of virulence factors [16]. Haemolytic toxins include: aerolysin-related cytotoxic enterotoxin (Act) [17], heat-labile cytotonic enterotoxin (Alt), heat-stable cytotonic toxins (Ast) [18], hemolysin (HlyA) and aerolysin (AerA) [19]. In addition, the type III secretion system (TTSS) [20], polar flagellum (fla), lateral flagella (laf) [21, 22], elastase (Ela) [23] and lipase (Lip) [24] contribute to the pathogenicity of Aeromonas.
Most cases of diarrheal due to Aeromonads are self-limiting and treatment with oral or intravenous fluids is effective. However, patients with serious diarrhea or extra-intestinal infection should receive an antimicrobial treatment [2]. Previously, Aeromonas has been observed as resistant to ampicillin, while 3rd generation cephalosporin, fluoroquinolone and aminoglycosides demonstrated excellent antimicrobial activity to Aeromonas species isolated from clinical sources [14, 25–27]. However, extensive use of antibiotics in aquaculture and human treatment has led to increasing resistance in bacterium to antimicrobial drugs. Therefore it is prudent to monitor the development of antimicrobial resistance in species of Aeromonas to common clinical treatment options.
In the presented study, we investigated characteristics of strains of Aeromonas isolated from intestinal infections and extra-intestinal infection. Furthermore we evaluated virulence associated genes and antimicrobial resistance of species of Aeromonas.
Materials and methods
Isolates of Aeromonas
Overall, 1286 stool samples were collected from adults over 14 years old presenting with acute diarrhea at a general hospital in Beijng, China, between June and July 2015, 2017. Epidemiology related medical records were completed to assess clinical history and physical fitness of patients (Additional file 1). Samples of Stool were enriched in alkaline peptone water broth (Beijing landbrige, China) for 8 h at 37 °C, and a loop of the resulting mixture was subcultured on a blood agar plate (Oxoid, UK) supplemented with 20% ampicillin (Sigma, USA) for 16–24 h at 37 °C [28]. An oxidase test (BioMerieuX, France) was performed to select the colonies which were different from Enterobacteriaceae. Microorganisms were identified by use of an automatic bacteriologic analyzer (VITEK2 Compact, BioMerieuX, France). Salmonella spp, Shigella spp and Vibrio spp were also detected on a routine basis. simultaneously.
Extra-intestinal infections due to Aeromonas were monitored and the strains were isolated between 2015 and 2017. Clinical samples of blood or bile were cultured in a BACTEC FX400 (BD Diagnostic Instrument Systems, USA). Samples positive for Aeromonas were simultaneously subcultured on a blood agar plate and a Maconkey agar plate (BioMerieuX, France). Identification of the isolated microorganisms was completed by use of an automatic bacteriologic analyzer (VITEK2 Compact, BioMerieuX, France). Concurrently, medical records of the patients with extra-intestinal infections due to species of Aeromonas were reviewed and age, gender, underlying conditions, microbiological findings and outcome were assembled.
Strains were stored in a Luria broth: glycerol mixture (80:20) at − 80 °C until identification was performed.
Molecular identification and subtyping of Aeromonas isolates
Molecular identification and subtyping of Aeromonas isolates was completed by use of 16S rRNA sequencing and MLPA. Total chromosomal DNA from Aeromonas was prepared by use of the DNA purification kit (Tiangen Biotech, China) as specified by the manufacturer. PCR amplification was performed by use of 2 × Taq PCR MasterMix (Tiangen Biotech, China). Primers synthesis and sequencing of PCR products were conducted (Shanghai Sangon Biotech, China). Due to the limitations of molecular identification by 16S rRNA sequencing, phylogenetic analysis of the seven selected housekeeping genes gyrB, rpoD, recA, dnaJ, gyrA, dnaX and atpD was completed to identify strains of Aeromonas. Primers [5] used for PCR amplification are provided in Additional file 2. Concatenated 7-gene phylogenetic trees were constructed and compared with representative species by use of MLPA as previously described [5]. Unrooted neighbour-joining phylogenetic trees were prepared by use of MEGA 5.0 software with Bootstrap values calculated by use of 1000 replicates.
Detection of virulence-associated genes
The presence of 10 genes encoding virulence factors was determined by use of PCR. Primers are listed in Additional file 2, including alt [29], ast [30], hlyA, aerA, act, ascF-G of TTSS, laf [14], lip, fla, and ela [31]. PCR amplification reactions were performed at a final volume of 40 μl, containing 20 μl of Taq PCR MasterMix (2×), 1 μl 10 μM primer, 1 μl DNA template (~ 30-40 ng), and 17 μl ddH2O. Cycling conditions consisted of an initial single cycle at 95 °C for 5 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing was completed at 55 °C–60 °C for 30 s, elongation was completed at 72 °C for 1 min and followed by a final cycle at 72 °C for 7 min. The PCR products were sequenced for further confirmation.
Antibiotic susceptibility test
Antibiotic susceptibility tests were performed by use of the microbroth dilution method according to guidelines of the current Clinical and Laboratory Standards Institute (CLSI). Minimum inhibitory concentrations (MIC) of strains of Aeromonas strains to 15 antibiotics were determined and included; gentamycin (GEN), imipenem (IPM), ampicillin (AMP), cefoxitin (FOX), ceftriaxone (CRO), amoxicillin-clavulanate (AMC), nalidixic acid (NAL), ciprofloxacin (CIP), chloramphenicol (CHL), tetracycline (TCY), doxycycline (DOX), azithromycin (AZM), cefepime (FEP), sulfonamides (Sas) and trimethoprim-sulfamethoxazole (SXT). E. coli ATCC 25922 was used as the quality-control strain for susceptibility testing.
Definitions
Multiple drug resistance (MDR) was was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories, according the criteria for defining MDR, XDR and PDR in Enterobacteriaceae [32].
Intestinal infections related to a strain of Aeromonas were diagnosed as patients presenting with acute diarrhea and a sample culture positive for a strain of Aeromonas. Extra-intestinal infections related to a strain of Aeromonas were diagnosed as patients presenting with inflammation in a region not identified as intestinal and a sample culture positive for a strain of Aeromonas.
Statistical methods
Data were analyzed by use of the x2 test and Fisher’s exact test (SPSS 15.0), When P < 0.05 results were considered statistically significant.
Results
Clinical features
Aeromonas spp. were identified as the causative agent of diarrhea in 98 (7.6%) of 1286 patients. Clinical and epidemiological characteristics were shown in Additional file 3. Gender ratio (male: female) was 0.94 (46/49) among 98 patiens presenting with diarrhea caused by strains of Aeromonas. Sources of infections were largely unknown and likely originate from contaminated food. However, 3% of patients identified seafood, cooked food or frozen drinks as likely sources. Twenty percent of patients presented with vomiting, 35.8% abdominal pain, 11.6% fever (body temperature ≥ 37.7 °C), and 9.1% had mild dehydration. Approximately 70% of patients presenting with diarrhea caused by a strain of Aeromonas had loose stools for ≥3 times per day, 29.4% had watery stools, and 1.0% had mucus-like stool. Erythrocytes and leukocytes were present in 28.4 and 11.6% of samples of stool collected from patients infected by strains of Aeromonas when observed by use of high magnification (HP, × 40). In addition, 6.3% of stool samples presented with erythrocytes and leukocytes. Infection of patients by other enteropathogens was observed in three patients (3/98, 3.1%). Combinations of infectious species included; Salmonella typhimurium with A.caviae, Vibrio fluvialis with A. veronii and Vibrio parahaemolyticus with A. veronii.
Between 2015 and 2017, 17 strains of Aeromonas causing extra-intestinal infections were identified (Table 1 and Additional file 4). With the excepted for 3 children accepting a liver transplant (age < 4 years), the average age of the 14 patients was 58.5 years old. The gender ratio (male: female) was 1.83 (11/6). None of the 17 patients were ICU admissions nor was there any acute respiratory failure or mortality. Six (35.3%) patients suffered from Aeromonas-related cholecystitis following a liver transplant and 6 (35.3%) pantients presented with malignant tumors. Overall, the most common underlying conditions of patients presenting with Aeromonas infections were liver transplantation and malignancy (12/17), In addition, patients presenting with Aeromonas related infections were associated with increased prevalence of lung cancer in our study.
Table 1.
Clinical characteristics of 17 patients presenting with extra-intestinal infections likely caused by species of Aeromonas
| Strains | Species | Disease spectrum | Underlying condition | Polymicrobial infection |
|---|---|---|---|---|
| BJ127 | A. dhakensis | Wound infection after cholecystectomy | Liver cirrhosi and Gallbladder cyst | – |
| BJ069 | A. dhakensis | Bacteremia | Pneumonia | Klebsiella pneumoniae |
| BJ123 | A. media | Bacteremia | Pneumonia and uremia | – |
| BJ015 | A. dhakensis | Cholecystitis | Post Liver transplantation | Stenotrophomonas maltophilia |
| BJ022 | A. dhakensis | Cholecystitis | Post Liver transplantation | Klebsiella pneumoniae |
| BJ016 | A. caviae | Cholecystitis | Post Liver transplantation | Klebsiella pneumoniae |
| BJ093 | A. caviae | Cholecystitis | Post Liver transplantation | – |
| BJ128 | A. caviae | Chronic Cholecystitis | Post Liver transplantation | Klebsiella pneumoniae |
| BJ124 | A. sanarellii | Cholecystitis | Post Liver transplantation | – |
| BJ042 | A. caviae | Urinary infection | Renal tuberculosis | – |
| BJ043 | A. caviae | Urinary infection | Renal insufficiency | – |
| BJ126 | A. veronii | Wound infection after Rectal cancer radical resection | Rectal cancer and Esophagus cancer | Proteus mirabilis |
| BJ014 | A. hydrophila | Hydrothorax | Lung cancer | – |
| BJ017 | A. hydrophila | Hydrothorax | Lung cancer | – |
| BJ018 | A. hydrophila | Wrapped empyema | Lung cancer | – |
| BJ054 | A. hydrophila | Hydrothorax | Esophagus cancer | – |
| BJ125 | A. hydrophila | Wound infection after mastectomy | Breast cancer | – |
Eleven patients presented with monomicrobial related Aeromonas infections, and 6 patients presented with polymicrobial Aeromonas infections (Table 1). Of polymicrobial infections, two consisted of A.caviae and Klebsiella pneumoniae; and two were A.aquariorum with Klebsiella pneumoniae. One patient presented with A.aquariorum and Stenotrophomonas maltophilia and one patient with A.veronii and Proteus mirabilis. Klebsiella pneumoniae was the most common combined pathogen (66.7%, 4/6).
Genotyping of species of Aeromonas
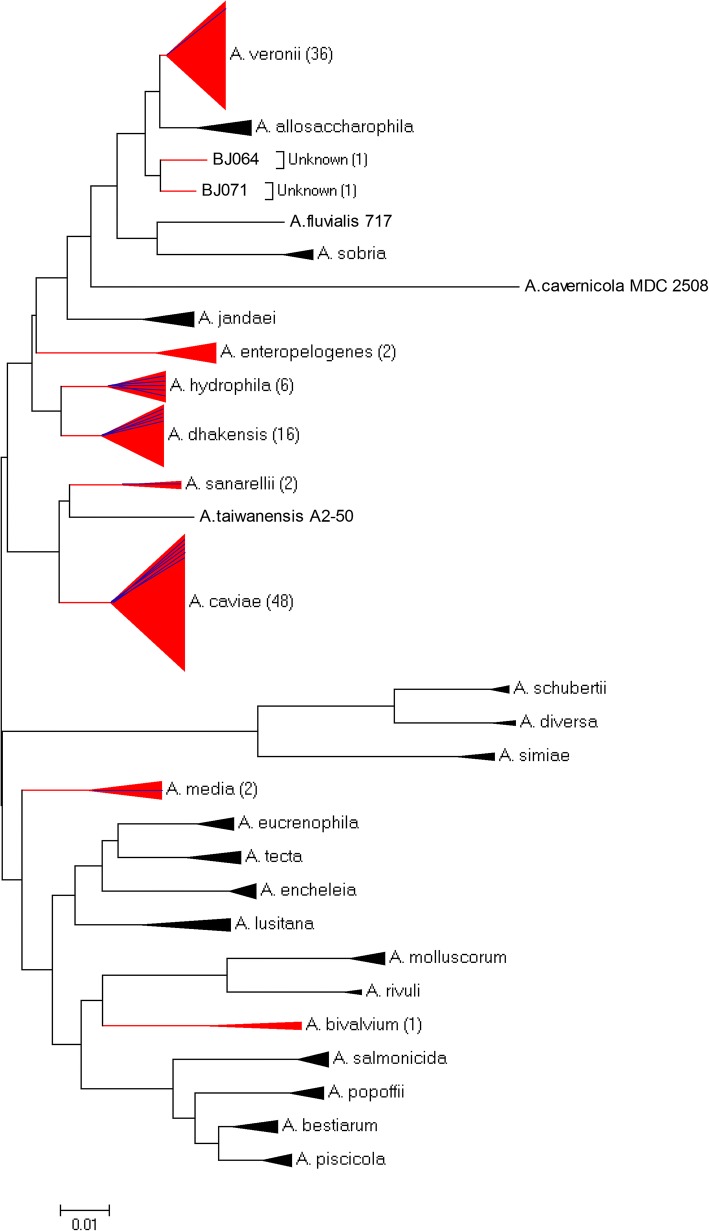
Results of MLPA performed with the concatenated 7-gene phylogenetic tree analysis classified 113 of 115 (98.3%) Aeromonas isolates to 8 different species (Fig. 1). The four most prevalent species of Aeromonas were A. caviae (41.7%), A. veronii (31.3%), A. dhakensis (13.9%), and A. hydrophila (5.2%). As presented in Table 2, comparative analysis of genotyping demonstrated differences between intestinal and extra-intestinal isolates was completed. Overall, there was a significant difference in the assemblage of isolates as intestinal isolates generally contained species of A. caviae (43.9%), A. veronii (35.7%), and A. dhakensis (12.2%). In contrast, extra-intestinal isolates generally contained A. hydrophila (29.4%), A. caviae (29.4%), and A. dhakensis (23.5%). There was significant difference between intestinal and extra-intestinal isolates for the species A. veronii and A. hydrophila (P < 0.05, x2 test). Five of 6 strains of A. hydrophila were isolated from patients with solid tumors, while only 1 strain of A. hydrophila was associated with an intestinal infection.
Fig. 1.
Unrooted neighbor -joined phylogenetic tree of species of Aeromonas isolated patients presenting with intestinal or extra-intestinal infections (n = 1000 bootstrap replicates). The tree was constructed by use of MLPA of seven housekeeping genes (gyrB, rpoD, recA, dnaJ, gyrA, dnaX and atpD). Red tree lines and triangles represent species detected in this study, blue lines represent the numbers of the extra-intestinal infections due to species of Aeromonas. The number of identified strains is indicated in brackets. Black tree lines and triangles represent other representative species
Table 2.
Distribution of species of Aeromonas spp. in isolates collected from intestinal and extra-intestinal samples
| Species | No. total strains (%) | No. intestinal isolates (%) | No. extra-intestinal isolates (%) |
|---|---|---|---|
| A. veronii | 36 (31.3) | 35 (35.7) | 1 (5.9) |
| A. caviae | 48 (41.7) | 43 (43.9) | 5 (29.4) |
| A. dhakensis | 16 (13.9) | 12 (12.2) | 4 (23.5) |
| A. media | 2 (1.7) | 1 (1.0) | 1 (5.9) |
| A. hydrophila | 6 (5.2) | 1 (1.0) | 5 (29.4) |
| A. sanarellii | 2(1.7) | 1 (1.0) | 1 (5.9) |
| A. enteropelogenes | 2 (1.7) | 2 (2.0) | 0 (0.0) |
| A. bivalvium | 1 (0.9) | 1 (1.0) | 0 (0.0) |
| Unknown | 2 (1.7) | 2 (2.0) | 0 (0.0) |
| Total | 115 | 98 | 17 |
Distribution of virulence genes in strains of Aeromonas
Presence of multiple virulence genes was common among isolates of Aeromonas and 40 virulence combinations of 10 putative virulence genes were identified. The predominant combination (i.e. pattern) of virulence genes was alt/ela/lip/fla (pattern1), which presented in 27.0% of patients presenting with an infection related to a isolate of Aeromonas. In addition, the patterns of act/fla (pattern 2), alt/ela/lip (pattern 3) and act/ascF-G/fla (pattern 4) were prevalent among patients.. As presented in Table 3, the pattern of virulence genes varied among genus’s. Of the four most prevalent species, the haemolytic genes act was prevalent in A. veronii and A. dhakensis. The haemolytic genes hlyA was prevalent in A. hydrophila and A. dhakensis, and the haemolytic genes aerA was more prevalent in A. dhakensis. The enterotoxin gene ast was identified primarily in A. hydrophila. The enterotoxin gene alt, extracellular protease genes ela and lip were less prevalent in A. veronii. The TTSS genes (ascF-G) was prevalent in A. hydrophila. A. veronii carried pattern 2 and 4 and A. caviae carried pattern1 and 3. The species A. dhakensis and A. hydrophila had diverse virulence patterns, 93.3% A. dhakensis and 100% A. hydrophila had 5 or more virulence genes.
Table 3.
Virulence related genes identified in isolates of species of Aeromonas
| Gene | A.veronii | A. caviae | A. dhakensis | A. hydrophila |
|---|---|---|---|---|
| No. strains (%) | No. strains (%) | No. strains (%) | No. strains (%) | |
| ast | 2 (5.6) | 0 (0.0) | 3 (18.8) | 6 (100.0) |
| act | 36 (100.0) | 1 (2.1) | 10 (62.5) | 1 (16.7) |
| alt | 0 (0.0) | 43 (89.6) | 14 (87.5) | 4 (66.7) |
| hlyA | 0 (0.0) | 0 (0.0) | 14 (87.5) | 5 (83.3) |
| aerA | 0 (0.0) | 0 (0.0) | 10 (62.5) | 1 (16.7) |
| ascF-G | 14 (38.9) | 2 (4.2) | 4 (25.0) | 4 (66.7) |
| ela | 6 (16.7) | 48 (100.0) | 16 (100.0) | 6 (100.0) |
| lip | 3 (8.3) | 48 (100.0) | 15 (93.8) | 6 (100.0) |
| fla | 29 (80.6) | 38 (79.2) | 16 (100.0) | 6 (100.0) |
| laf | 2 (5.6) | 2 (4.2) | 1 (6.3) | 2 (33.3) |
Susceptibility to antimicrobials
Resistance profiles of the 115 Aeromonas isolates to 15 antimicrobial agents were shown in Table 4. High resistance to ampicillin (93.9%) and Nalidixic acid (54.8%) was observed in Aeromonas isolates. The majority of isolates (≥80%) were susceptible to chloramphenicol, gentamicin and the new generation antibiotics ciprofloxacin, ceftriaxone, cefepime, imipenem, sulfonamides, trimethoprim- sulfamethoxazole, doxycycline and azithromycin. Resistance was most prevalent for ciprofloxacin, amoxicillin-clavulanate, cefoxitin, ceftriaxone, sulfonamides, gentamicin and azithromycin in A. hydrophila, as a resistance of 66.7, 100.0, 66.7, 66.7, 50.0, 50.0, and 66.7% was observed, respectively. Higher rates of resistance to cefoxitin was also observed in A. dhakensis (87.5%). Only 3 strains exhibited resistance to imipenem, all of which were identified as belonging to A. dhakensis. Significantly higher rates of resistance to 10 antibiotics (ciprofloxacin, nalidixic acid, amoxicillin-clavulanate, cefoxitin, ceftriaxone, cefepime, sulfonamides, trimethoprim-sulfamethoxazole, gentamicin and azithromycin) were found among extra-intestinal isolates when compared with intestinal isolates (P < 0.05, x2 test).
Table 4.
Antibiotic susceptibility patterns of species of Aeromonas
| Total | Intestinal infection | Extra-intestinal infection | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Antibiotic | Ra [n (%)] | I [n (%)] | S [n (%)] | R [n (%)] | I [n (%)] | S [n (%)] | R [n (%)] | I [n (%)] | S [n (%)] |
| Ampicillinc | 108(93.9) | 3(2.6) | 4(3.5) | 92(93.9) | 2(2.0) | 4(4.1) | 16(94.1) | 1(5.9) | 0(0.0) |
| Amoxicillin-clavulanatec | 15(13.0) | 76(66.1) | 24(20.9) | 8(8.2) | 68(69.4) | 22(22.4) | 7(41.2) | 8(47.1) | 2(11.8) |
| Imipenemb | 3(2.6) | 10(8.7) | 102(88.7) | 2(2.0) | 7(7.1) | 88(89.8) | 1(5.9) | 3(17.6) | 14(82.4) |
| Ceftriaxoneb | 17(14.8) | 3(2.6) | 95(82.6) | 5(5.1) | 3(3.1) | 90(91.8) | 12(70.6) | 0(0.0) | 5(29.4) |
| Cefepimeb | 5(4.3) | 3(2.6) | 107(93.0) | 1(1.0) | 0(0.0) | 97(99.0) | 4(23.5) | 3(17.6) | 10(58.8) |
| Cefoxitinb | 28(24.3) | 6(5.2) | 81(70.4) | 17(17.3) | 5(5.1) | 76(77.6) | 11(64.7) | 1(5.9) | 5(29.4) |
| Gentamicinb | 6(5.2) | 4(3.5) | 105(91.3) | 2(2.0) | 2(2.0) | 94(95.9) | 4(23.5) | 2(11.8) | 11(64.7) |
| Nalidixic acidc | 63(54.8) | – | 52(45.2) | 49(50.0) | 0(0.0) | 49(50.0) | 14(82.4) | 0(0.0) | 3(17.6) |
| Ciprofloxacinb | 7(6.1) | 0(0.0) | 108(93.9) | 1(1.0) | 0(0.0) | 97(99.0) | 6(35.3) | 0(0.0) | 11(64.7) |
| Chloramphenicolb | 10(8.7) | 2(1.7) | 103(89.6) | 8(8.2) | 2(2.0) | 88(89.8) | 2(11.8) | 0(0.0) | 15(88.2) |
| Tetracyclineb | 21(18.3) | 5(4.3) | 89(77.4) | 15(15.3) | 3(3.1) | 80(81.6) | 6(35.3) | 2(11.8) | 9(52.9) |
| Doxycyclinec | 4(3.5) | 4(3.5) | 107(93.0) | 2(2.0) | 2(2.0) | 94(95.9) | 2(11.8) | 2(11.8) | 13(76.5) |
| Azithromycinc | 5(4.3) | – | 110(95.7) | 0(0.0) | – | 98(100.0) | 5(29.4) | – | 12(70.6) |
| Trimethoprim- Sulfamethoxazoleb | 6(5.2) | – | 109(94.8) | 1(1.0) | 0(0.0) | 97(99.0) | 5(29.4) | 0(0.0) | 12(70.6) |
| Sulfonamidesc | 19 (16.5) | – | 96(83.5) | 12(12.2) | 0(0.0) | 86(87.8) | 7(41.2) | 0(0.0) | 10(58.8) |
aR: Resistant; I: Intermediate; S: Sensitive
bBreakpoints are based on the CLSI M45-A3 standards for Aeromonas spp.
cOther breakpoints refer to the CLSI M100-S26E criteria for Enterobacteriaceae
Of the 115 strains, 33 strains (28.7%) exhibited 35 multiple-drug resistance (MDR) patterns to 15 antimicrobial agents. Eighty-three percent (5/6) of strains of A. hydrophila and 81.2% (13/16) of strains of A. dhakensis presented with MDR, while less MDR isolates were found in A.caviae (39.6%, 19/48) and A.veronii (16.7%, 6/36). Intestinal strains (30.6%, 30/98) presented with significantly less rates of MDR when compare to isolates from extra-intestinal strains (82.3%, 14/17), indicating acquisition of MDR was likely from in the hospital.
Discussion
In the presented study, 115 isolates of Aeromonas were collected from a general hospital in Beijing between 2015 and 2017. Overall, the abundance and prevalence of strains of Aeromonas were different between intestinal and extra-intestinal infections. In our study, 1% of samples isolated from samples of stool of patients with intestinal infection were positive for A. hydrophila, while in 29.4% of extra-intestinal infections. Thus, results of this study indicated that the A. hydrophila was not the primary pathogen contributing to acute gastroenteritis, however it was more prevalent in extra-intestinal infections when compared to samples from patients with intestinal infections. Interestingly, 5 strains of A. hydrophila strains from extra-intestinal infections were present in patients presented with malignant tumor. These results might indicate a preference of strains of A. hydrophila and other Aeromonas spp. to colonize differently. A. veronii was more common in samples of patients presenting with acute gastroenteritis (35.7%) but was rare in patients with extra-intestinal infections (5.9%), which was similar to previous results [14, 33].
In addition, results of our study demonstrate a potential relationship between Aeromonas and clinical cirrhosis or malignancy as previously reported [34, 35] and liver-transplant related cholecystitis. These results might be related to bacterial translocation, use of antacids [35] or immunosuppressive agents following liver transplantation.
Prevalence of antimicrobial-resistance was greater in extra-intestinal isolates when compared to the previous study. In our study, rates of resistance to ceftriaxone, ciprofloxacin, gentamicin and imipenem was 70.6, 35.3, 23.5 and 5.9%, while a study completed in Taiwan was 7.7, 6, 3.3 and 1.1%, respectively [36] . Additionally, a study completed in Korea the rates of resistance were 15.5, 10.1, 7.1 and 9.8%, respectively [11]. When compared with the rates of intestinal isolates, the rate of MDR in extra-intestinal isolates was greater. These findings indicate selective pressures in hospitals on strains of infectious bacteria due to the extensive use of antimicrobial agents and warrants more attention in the future.
In our study, two bacteremia-related Aeromonas species were identified; A. media and A. dhakensis. These results were different from previous results where A. caviae was identified as bacteremia-related Aeromonas species in Japan, A. hydrophila and A. veronii biovar sobria in Taiwan, and A. hydrophila and A. caviae in Korea and Taiwan [11, 36].
A study completed in Southern India reported a resistance rate to ceftriaxone resistant of 31% (9/29) for isolates of Aeromonas from samples of stool [37]. In our study resistance rates for ceftriaxone, ciprofloxacin and gentamicin and imipenem were 5.1, 1.0, 2.0 and 2.0% in Aeromonas isolates of patients presenting with diarrhea and were similar to rates in Shanghai (5.7, 3.6, 0.5, and 2.6%, respectively) [14]. These results along with results of the study competed in Shanghai indicate that 3rd generation cephalosporins, fluoroquinolones and aminoglycosides are a treatment option for severe diarrhea but not for extra-intestinal infections originating in Eastern China.
It is important to note that only 3 strains exhibited resistance to imipenem, all of which belonged to the genus A. dhakensis. The genus A. dhakensis should be the focus of future research as they harbored high numbers of virulence genes, high rates of drug resistance and a high degree of infection in intestinal and extra-intestinal samples. In addition, A. hydrophila presented with a high number of virulence genes and high rates of drug resistance. A. hydrophila have previously been isolated from wounds in two cases as reported by Christopher J. Grim et al. [38], and were classified as having MDR and multiple virulence genes.
In the presented study, Klebsiella pneumoniae was the most common combined pathogen. These results demonstrate that cholecystitis post Liver transplant predisposed patients to polymicrobial Aeromonas infections, while malignant cancers, such as rectal cancer, might predispose patients to monomicrobial Aeromonas infection. A previous study in Taiwan found that E. coli was the most common pathogen (42%) in polymicrobial infections, then Klebsiella spp. (24%) [35]. Conversely, cirrhosis predisposed patients to monomicrobial Aeromonas bacteremia while malignant cancer predisposed patients to polymicrobial Aeromonas bacteremia [35]. This difference indicates a high degree of heterogeneity in the distribution of intestinal bacteria, and region specific presence of Aeromonas infections.
The pathogenic mechanism of Aeromonas was multifactorial and complex, and likely involves a series of virulence genes involved in this process. Despite Aeromonas harboring different numbers and types of virulence genes, there was no significant correlation found between infection and virulent genes of Aeromonas in intestinal infections and extra-intestinal infections. For example, intestinal infections, alt have been reported as associated with loose stool, alt plus ast with watery stools, and act with bloody diarrhea [39]. In the presented study, 3 watery stool samples were associated with ast, however 25 samples of watery stool were not. In addition, a relationship between infection and presence of virulent genes was not observed and might be related to the limited number of strains isolated in extra-intestinal infections. Similarly, a study completed by Wu et al. found no association between the presence of the genes aerA, hlyA, alt, ast, ascFG in isolates of Aeromonas and development of extra-intestinal infections or bacterium [34].
In conclusion, Aeromonas spp. should be considered as a causative infectious agent in immunocompromised patients especially those presenting with malignancy, liver cirrhosis and following a liver transplant. In addition, A. hydrophila was more prevalent in extra-intestinal infections when compared to intestinal infections, especially for patients presenting with a malignancy. Extra-intestinal Aeromonas isolates possessed higher rates of drug resistance. However, 3rd generation cephalosporins, fluoroquinolones and aminoglycosides remain as effective treatments for patients presenting with severe diarrhea but not for extra-intestinal infections. In addition, increasing prevalence of drug resistance and MDR in extra-intestinal isolates of Aeromonas requires attention and further monitoring.
Additional files
Medical Record table upload. (DOCX 14 kb)
Sequence of primers used for amplification of housekeeping genes and virulence factor genes. (DOC 58 kb)
Clinical data, genes of toxins and drug resistant patterns for 95 diarrhea patients related to Aeromonas spp. (XLS 578 kb)
Clinical data, genes of toxins and drug resistant patterns for 17 patients related to Aeromonas spp. in extra-intestinal infections. (XLS 49 kb)
Acknowledgements
We thank the medical staff from the intestinal clinic of Beijing Friendship Hospital.
for registering the information of diarrhea patients. We also would like to thank Dr. Duochun Wang for the helpful expert technical assistance.
Funding
This work was supported by Capital’s Funds for Health Improvement and Research (No.2016–4-1101) and Beijing Natural Science Foundation (No.7164249). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Abbreviations
- Act
aerolysin-related cytotoxic enterotoxin
- AerA
aerolysin
- Alt
heat-labile cytotonic enterotoxin
- AMC
amoxicillin-clavulanate
- AMP
ampicillin
- Ast
heat-stable cytotonic toxins
- AZM
azithromycin
- CHL
chloramphenicol
- CIP
ciprofloxacin
- CLSI
Clinical and Laboratory Standards Institute
- CRO
ceftriaxone
- DOX
doxycycline
- Ela
elastase
- FEP
cefepime
- FOX
cefoxitin
- GEN
gentamycin
- HlyA
hemolysin
- HP
high magnification
- IPM
imipenem
- Lip
lipase
- MDR
multiple-drug resistance
- MIC
minimum inhibitory concentrations
- MLPA
multilocus phylogenetic analysis
- NAL
nalidixic acid
- PCR
Polymerase chain reaction
- Sas
sulfonamides
- SXT
trimethoprim-sulfamethoxazole
- TCY
tetracycline
- TTSS
type III secretion system
Authors’ contributions
YD and SJ designed the study. ZY, YL, NZ, ZP performed experiments. KB revised it critically for important intellectual content. All authors contributed to the data analysis and writing. All authors read and approved the manuscript.
Ethics approval and consent to participate
The study obtained ethical approval (2017-P2–095-01) from the Medical Ethics Committee of Beijing Friendship Hospital, Capital Medical University, Beijing, P.R China. Given that the study was retrospective without any interventions in treatment and all the data were collected and analyzed anonymously, the requirement for informed consent was waived. Permissions for obtaining related medical records were approved by hosipital.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yanyan Zhou, Email: zhouyanyan1216@163.com.
Li Yu, Email: wersdu@163.com.
Zheng Nan, Email: nanzheng0127@163.com.
Pingping Zhang, Email: applezhang890804@163.com.
Biao Kan, Email: kanbiao@icdc.cn.
Donghui Yan, Email: n66ydh@163.com.
Jianrong Su, Email: youyilab@163.com.
References
- 1.Sawabe T, Oliver JD. International committee on systematics of prokaryotes subcommittee on the taxonomy of Aeromonadaceae, Vibrionaceae and related organisms minutes of the meetings, 31 march 2016, Roscoff, France. Int J Syst Evol Microbiol. 2017;67(3):759–760. doi: 10.1099/ijsem.0.001675. [DOI] [PubMed] [Google Scholar]
- 2.Igbinosa IH, Igumbor EU, Aghdasi F, et al. Emerging Aeromonas species infections and their significance in public health. ScientificWorldJournal 2012; 2012: 625023. [DOI] [PMC free article] [PubMed]
- 3.Pablos M, Remacha MA, Rodriguez-Calleja JM, et al. Identity, virulence genes, and clonal relatedness of Aeromonas isolates from patients with diarrhea and drinking water. Eur J Clin Microbiol Infect Dis. 2010;29(9):1163–1172. doi: 10.1007/s10096-010-0982-3. [DOI] [PubMed] [Google Scholar]
- 4.Janda JM, Abbott SL. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010;23(1):35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Murcia AJ, Monera A. Saavedra MJ, et al.Multilocus phylogenetic analysis of the genus Aeromonas. Syst Appl Microbio. 2011;34(3):189–199. doi: 10.1016/j.syapm.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Murcia A, Beaz-Hidalgo R, Svec P, et al. Aeromonas cavernicola sp. nov., isolated from fresh water of a brook in a cavern. Curr Microbiol. 2013;66(2):197–204. doi: 10.1007/s00284-012-0253-x. [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Murcia A, Beaz-Hidalgo R, Navarro A, et al. Aeromonas lusitana sp. nov., isolated from untreated water and vegetables. Curr Microbiol. 2016;72(6):795–803. doi: 10.1007/s00284-016-0997-9. [DOI] [PubMed] [Google Scholar]
- 8.Figueras MJ, Latif-Eugenin F, Ballester F, et al. ‘Aeromonas intestinalis’ and ‘Aeromonas enterica’ isolated from human faeces, ‘Aeromonas crassostreae’ from oyster and ‘Aeromonas aquatilis’ isolated from lake water represent novel species. New Microbes New Infect. 2017;15:74–76. doi: 10.1016/j.nmni.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schleifer KH. Classification of Bacteria and archaea: past, present and future. Syst Appl Microbiol. 2009;32(8):533–542. doi: 10.1016/j.syapm.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Beaz-Hidalgo R, Hossain MJ, Liles MR, et al. Strategies to avoid wrongly labelled genomes using as example the detected wrong taxonomic affiliation for aeromonas genomes in the GenBank database. PLoS One. 2015;10(1):e0115813. doi: 10.1371/journal.pone.0115813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhee JY, Jung DS, Peck KR. Clinical and therapeutic implications of Aeromonas bacteremia: 14 years nation-wide experiences in Korea. Infect Chemother. 2016;48(4):274–284. doi: 10.3947/ic.2016.48.4.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aravena-Roman M, Harnett GB, Riley TV, et al. Aeromonas aquariorum is widely distributed in clinical and environmental specimens and can be misidentified as Aeromonas hydrophila. J Clin Microbiol. 2011;49(8):3006–3008. doi: 10.1128/JCM.00472-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yano Y, Hamano K, Tsutsui I, et al. Occurrence, molecular characterization, and antimicrobial susceptibility of Aeromonas spp. in marine species of shrimps cultured at inland low salinity ponds. Food Microbiol. 2015;47:21–27. doi: 10.1016/j.fm.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Li F, Wang W, Zhu Z, et al. Distribution, virulence-associated genes and antimicrobial resistance of Aeromonas isolates from diarrheal patients and water, China. J Inf Secur. 2015;70(6):600–608. doi: 10.1016/j.jinf.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Beaz-Hidalgo R, Martinez-Murcia A, Figueras MJ. Reclassification of Aeromonas hydrophila subsp. dhakensis Huys et al. 2002 and Aeromonas aquariorum Martinez-Murcia et al. 2008 as Aeromonas dhakensis sp. nov. comb nov. and emendation of the species Aeromonas hydrophila. Syst Appl Microbiol. 2013;36(3):171–176. doi: 10.1016/j.syapm.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Tomas JM. The main Aeromonas pathogenic factors. ISRN Microbiol. 2012;2012:256261. doi: 10.5402/2012/256261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chopra AK, Houston CW, Peterson JW, et al. Cloning, expression, and sequence analysis of a cytolytic enterotoxin gene from Aeromonas hydrophila. Can J Microbiol. 1993;39(5):513–523. doi: 10.1139/m93-073. [DOI] [PubMed] [Google Scholar]
- 18.Sha J, Kozlova EV, Chopra AK. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect Immun. 2002;70(4):1924–1935. doi: 10.1128/IAI.70.4.1924-1935.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heuzenroeder MW, Wong CY, Flower RL. Distribution of two hemolytic toxin genes in clinical and environmental isolates of Aeromonas spp.: correlation with virulence in a suckling mouse model. FEMS Microbiol Lett. 1999;174(1):131–136. doi: 10.1111/j.1574-6968.1999.tb13559.x. [DOI] [PubMed] [Google Scholar]
- 20.Yu HB, Rao PS, Lee HC, et al. A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect Immun. 2004;72(3):1248–1256. doi: 10.1128/IAI.72.3.1248-1256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabaan AA, Gryllos I, Tomas JM, et al. Motility and the polar flagellum are required for Aeromonas caviae adherence to HEp-2 cells. Infect Immun. 2001;69(7):4257–4267. doi: 10.1128/IAI.69.7.4257-4267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gavin R, Merino S, Altarriba M, et al. Lateral flagella are required for increased cell adherence, invasion and biofilm formation by Aeromonas spp. FEMS Microbiol Lett. 2003;224(1):77–83. doi: 10.1016/S0378-1097(03)00418-X. [DOI] [PubMed] [Google Scholar]
- 23.Cascon A, Yugueros J, Temprano A, et al. A major secreted elastase is essential for pathogenicity of Aeromonas hydrophila. Infect Immun. 2000;68(6):3233–3241. doi: 10.1128/IAI.68.6.3233-3241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuang YC, Chiou SF, Su JH, et al. Molecular analysis and expression of the extracellular lipase of Aeromonas hydrophila MCC-2. Microbiology. 1997;143(Pt 3):803–812. doi: 10.1099/00221287-143-3-803. [DOI] [PubMed] [Google Scholar]
- 25.Vila J, Marco F, Soler L, et al. In vitro antimicrobial susceptibility of clinical isolates of Aeromonas caviae, Aeromonas hydrophila and Aeromonas veronii biotype sobria. J Antimicrob Chemother. 2002;49(4):701–702. doi: 10.1093/jac/49.4.701. [DOI] [PubMed] [Google Scholar]
- 26.Tena D, Gonzalez-Praetorius A, Gimeno C, et al. Extraintestinal infection due to Aeromonas spp.: review of 38 cases. Enferm Infecc Microbiol Clin. 2007;25(4):235–241. doi: 10.1157/13100463. [DOI] [PubMed] [Google Scholar]
- 27.Ghenghesh KS, Rahouma A, Zorgani A, et al. Aeromonas in Arab countries: 1995-2014. Comp Immunol Microbiol Infect Dis. 2015;42:8–14. doi: 10.1016/j.cimid.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Soltan Dallal MM, Mazaheri Nezhad Fard R, Kavan Talkhabi M, et al. Prevalence, virulence and antimicrobial resistance patterns of Aeromonas spp. isolated from children with diarrhea. Germs. 2016;6(3):91–96. doi: 10.11599/germs.2016.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rather MA, Willayat MM, Wani SA, et al. A multiplex PCR for detection of enterotoxin genes in Aeromonas species isolated from foods of animal origin and human diarrhoeal samples. J Appl Microbiol. 2014;117(6):1721–1729. doi: 10.1111/jam.12641. [DOI] [PubMed] [Google Scholar]
- 30.Martinez O, Rodriguez-Calleja JM, Santos JA, et al. Foodborne and indicator bacteria in farmed molluscan shellfish before and after depuration. J Food Prot. 2009;72(7):1443–1449. doi: 10.4315/0362-028X-72.7.1443. [DOI] [PubMed] [Google Scholar]
- 31.Sen K, Rodgers M. Distribution of six virulence factors in Aeromonas species isolated from US drinking water utilities: a PCR identification. J Appl Microbiol. 2004;97(5):1077–1086. doi: 10.1111/j.1365-2672.2004.02398.x. [DOI] [PubMed] [Google Scholar]
- 32.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 33.Puthucheary SD, Puah SM, Chua KH. Molecular characterization of clinical isolates of Aeromonas species from Malaysia. PLoS One. 2012;7(2):e30205. doi: 10.1371/journal.pone.0030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu CJ, Wu JJ, Yan JJ, et al. Clinical significance and distribution of putative virulence markers of 116 consecutive clinical Aeromonas isolates in southern Taiwan. J Inf Secur. 2007;54(2):151–158. doi: 10.1016/j.jinf.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Lay CJ, Zhuang HJ, Ho YH, et al. Different clinical characteristics between polymicrobial and monomicrobial Aeromonas bacteremia--a study of 216 cases. Intern Med. 2010;49(22):2415–2421. doi: 10.2169/internalmedicine.49.4117. [DOI] [PubMed] [Google Scholar]
- 36.Tang HJ, Lai CC, Lin HL, et al. Clinical manifestations of bacteremia caused by Aeromonas species in southern Taiwan. PLoS One. 2014;9(3):e91642. doi: 10.1371/journal.pone.0091642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhaskar M, Dinoop KP, Mandal J. Characterization of ceftriaxone-resistant Aeromonas spp. isolates from stool samples of both children and adults in southern India. J Health Popul Nutr. 2015;33:26. doi: 10.1186/s41043-015-0036-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grim CJ, Kozlova EV, Sha J, et al. Characterization of Aeromonas hydrophila wound pathotypes by comparative genomic and functional analyses of virulence genes. MBio. 2013;4(2):e00064–e00013. doi: 10.1128/mBio.00064-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albert MJ, Ansaruzzaman M, Talukder KA, et al. Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J Clin Microbiol. 2000;38(10):3785–3790. doi: 10.1128/jcm.38.10.3785-3790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Medical Record table upload. (DOCX 14 kb)
Sequence of primers used for amplification of housekeeping genes and virulence factor genes. (DOC 58 kb)
Clinical data, genes of toxins and drug resistant patterns for 95 diarrhea patients related to Aeromonas spp. (XLS 578 kb)
Clinical data, genes of toxins and drug resistant patterns for 17 patients related to Aeromonas spp. in extra-intestinal infections. (XLS 49 kb)
Data Availability Statement
The datasets used during the current study are available from the corresponding author on reasonable request.