Abstract
Background
Gaucher disease is a rare pan-ethnic, lysosomal storage disorder resulting due to beta-Glucosidase (GBA1) gene defect. This leads to the glucocerebrosidase enzyme deficiency and an increased accumulation of undegraded glycolipid glucocerebroside inside the cells’ lysosomes. To date, nearly 460 mutations have been described in the GBA1 gene. With the aim to determine mutations spectrum and molecular pathology of Gaucher disease in India, the present study investigated one hundred unrelated patients (age range: 1 day to 31 years) having splenomegaly, with or without hepatomegaly, cytopenia and bone abnormality in some of the patients.
Methods
The biochemical investigation for the plasma chitotriosidase enzyme activity and β-Glucosidase enzyme activity confirmed the Gaucher disease. The mutations were identified by screening the patients’ whole GBA gene coding region using bidirectional Sanger sequencing.
Results
The biochemical analysis revealed a significant reduction in the β-Glucosidase activity in all patients. Sanger sequencing established 71 patients with homozygous mutation and 22 patients with compound heterozygous mutation in GBA1 gene. Lack of identification of mutations in three patients suggests the possibility of either large deletion/duplication or deep intronic variations in the GBA1 gene. In four cases, where the proband died due to confirmed Gaucher disease, the parents were found to be a carrier. Overall, the study identified 33 mutations in 100 patients that also covers four missense mutations (p.Ser136Leu, p.Leu279Val, p.Gly383Asp, p.Gly399Arg) not previously reported in Gaucher disease patients. The mutation p.Leu483Pro was identified as the most commonly occurring Gaucher disease mutation in the study (62% patients). The second common mutations identified were p.Arg535Cys (7% patients) and RecNcil (7% patients). Another complex mutation Complex C was identified in a compound heterozygous status (3% patients). The homology modeling of the novel mutations suggested the destabilization of the GBA protein structure due to conformational changes.
Conclusions
The study reports four novel and 29 known mutations identified in the GBA1 gene in one-hundred Gaucher patients. The given study establishes p.Leu483Pro as the most prevalent mutation in the Indian patients with type 1 Gaucher disease that provide new insight into the molecular basis of Gaucher Disease in India.
Electronic supplementary material
The online version of this article (10.1186/s12881-019-0759-1) contains supplementary material, which is available to authorized users.
Keywords: Gaucher disease, β-Glucosidase, Chitotriosidase, GBA1 gene, Glucocerebrosidase, Indian population, p.Leu483Pro most common mutation, novel mutations in GBA1 gene
Background
Gaucher disease (GD), an autosomal recessively inherited disease, is caused due to a defect in glucosylceramidase beta gene (GBA1; OMIM*606463) coding for the enzyme glucocerebrosidase (EC 3.2.1.45; alternate names: acid β-glucosidase and GCase). Its prevalence is 1:50000 in general population and 1:950 live births in Ashkenazi Jewish population [1, 2].
Since Philippe Charles Ernest Gaucher first described the GD phenotypes a hundred and thirty-six years ago, the eponym ‘Gaucher Disease’ was introduced. In 1934, a study by Aghion revealed that the distorted macrophages, also called Gaucher cells, resulted due to an accumulation of the lipid glucocerebroside [3]. In 1965, a study by Dr. Brady and Dr. Patrick uncovered the fact that the metabolic defect in GD was due to deficiency of the enzyme β-Glucocerebrosidase [4, 5]. In 1968, the GD was classified as a Lysosomal storage disorder since the Weinreb et.al. work on a rat model established the Lysosomal localization of β-Glucocerebrosidase [6].
The multi-systemic GD has heterogeneous phenotypes, however, based on the age of onset, presence/absence and progression of neurologic manifestation; the GD is clinically divided into three types. GD type 1 (non-neurological) is the most common form whereas neuronopathic GD i.e. type 2 (acute neuronopathic) and type 3 (chronic neuronopathic) occurs less frequently. The neuronal involvement in the GD is a grey area. Though, a study by Kinghorn et al. on an orthologs GBA1 gene knocked out Drosophila model, demonstrated that the blocking of autophagy flux results in the glucosylceramide accumulation in the brain and age-dependent locomotor defects [7]. The most common clinical features in GD include hepatosplenomegaly, anemia, thrombocytopenia, growth retardation, seizures, and radiological bone disease.
The loss-of-function mutations in the GBA1 gene prevent the GCase enzyme from cleaving the β-glucosyl linkage of Glucocerebroside, required to break down glycolipid glucocerebroside into glucose and ceramide [8]. The human GBA1 gene (12 exons and 11 introns), located on chromosome 1q22, has a highly homologous pseudogene sequence located 16 kb downstream which shares 96% exonic homology with the functional gene [9]. As per the Human Genome Mutation Database (HGMD), nearly 460 disease causing GBA1 mutations have been reported in GD. The first mutation reported in GD was p.Leu483Pro [10]. The most common mutation identified in the Romanian patients is p.Asn409Ser [11]. These mutations along with c.84dupG (84GG) and IVS2 + 1G>A accounts for the 96% of mutations in Ashkenazi Jew patients and approximately 50–60% of mutations in non-Jewish Gaucher’s disease patients [12, 13]. Our recent publication established that the carrier frequency of p.Leu483Pro in Indian population is 1:600 [14].
The given study intended to explore the molecular features of GD in Indian patients. Hence, 100 Indian Gaucher patients were screened by DNA sequencing of the GBA1 gene to identify the unknown and common Indian mutations for GD and understand the genotypic effect on the phenotype of these patients. The study also aimed at developing a common molecular screening test for GD in India.
Methods
Patients
The present study comprises the patients referred as clinical cases from Institute of Human Genetics after genetic counselling as well as from outside referring physicians. The Ethics committee of the Foundation for Research in Genetics and Endocrinology (FRIGE) at the Institute of Human Genetics approved the study and it was performed in accordance with the tenets of the Declaration of Helsinki. Irrespective of the case reference, a written informed consent for investigation and publication of the data was obtained from the patients or their guardian as per the institutional ethics committee guidelines.
The present study on GD was carried out in 100 unrelated patients (90 children, 8 adults, and 2 fetal tissues) referred in the time from 2011 to 2018 with a clinical suspicion of GD. The patients’ common clinical presentation included mild to severe liver/spleen enlargement, anaemia, thrombocytopenia and presence of Gaucher cells in bone marrow.
Biochemical investigations
The plasma, leukocyte, and genomic DNA (gDNA) was isolated from each patient using six millilitres of blood, drawn in ethylenediaminetetraacetic acid vacutainer.
Plasma chitotriosidase screening
A previously described protocol was followed for the estimation of chitotriosidase enzyme activity in blood plasma [15]. In brief, the plasma along with the substrate 4-MU (4-Methylumbelliferyl-β-D-N,N′,N″-triacetylchitotrioside) were incubated at 37 °C and the fluorescence was measured using photo fluorometer (360 nm primary and 465 nm secondary filter).
β-Glucosidase enzyme assay
The substrate 4-methylumbelliferyl-β-D-glucopyranoside was used to measure the Lysosomal hydrolase enzyme (β-Glucosidase) activity from leukocyte using fluorometric assay [16].
Molecular genetics investigations
DNA extraction and purification
DNA, isolated from whole blood using the standard salting-out method, was quantified using a QIAxpert (Cat. No: 9002340) from Qiagen [17]. The DNA samples were purified using The Genomic DNA Clean & Concentrator™-25 (DCC™) Kit, from Zymo Research, Irvine, California, U.S.A (Cat. No. D4064) and were stored at − 20 °C until investigated.
Primary screening of the common Gaucher mutations (p.Leu483Pro)
Our earlier Polymerase Chain Reaction- Restriction Fragment Length Polymorphism (PCR-RFLP) based protocol for p.Leu483Pro screening was applied [18]. The Thermal Cycler-2720 (Applied Biosystems, Inc. India) amplified the DNA samples and the restriction endonucleases MspI (New England Biolabs) digested the PCR product. In brief, 10 μl of PCR product along with 0.5 μL of MspI (10 U/μl) enzyme was incubated at 37 °C for 3 h. The digested DNA fragments were separated on 2.5% agarose gel.
The above protocol also covers the mutation p.Arg463Cys that can be distinguished from p.Leu483Pro based on the different band pattern of the DNA fragments on the agarose gel. The codon numbering of the mutations described in this study uses the current nomenclature for GBA1 mutations that includes the first 39 amino acids of the leader sequence.
Single-gene sequencing (GBA1 gene)
The GBA1 gene containing 12 exons was amplified using primer sets standardized by nested PCR for specific amplification of the functional gene (see Additional file 1). For exons 1–3, 35 cycles of amplification; each consisting of initial denaturation (94 °C; 4 min), denaturation (94 °C; 30 s), annealing (65.5 °C; 30 s), elongation (72 °C; 30 s), and final elongation (72 °C; 10 min) were run. Amplification for exon 4–5 involved initial denaturation (96 °C; 2 min), denaturation (96 °C; 30 s), annealing (61 °C; 30 s), elongation (74 °C; 60 s), and final elongation (74 °C; 5 min) were run. Exon 6–12 included initial denaturation (96 °C; 2 min) followed by 33 cycles each consisting of denaturation (96 °C; 30 s), annealing (58 °C to 61 °C; 30 s), elongation (74 °C; 60 s), and final elongation (74 °C; 5 min).
The GBA1 gene Sanger sequencing was performed on the Applied Biosystems™ SeqStudio™ Genetic Analyzer with SeqStudio™ Data Collection Software using our earlier described protocol [14]. In brief the cycle sequencing products (samples) were purified, resuspended in Hi-Di Formamide and transferred to 96 wells plate, where the samples were denatured at 95 °C for 2 min and snap chilled at -20 °C for 2–3 min before proceeding with the sequencing. The sequences obtained were aligned to the available reference sequence (NM_001005741.2) in The National Centre for Biotechnology Information (NCBI) GeneBank database to detect variation.
In Silico analysis
Prediction of the functional effect of the variants
Six in silico tools identifying the effect of DNA mutations (MutationTaster2), coding non-synonymous variants (SIFT), coding and non-coding variants (FATHMM), and amino acid substitution (PolyPhen2, PROVEAN, and Mutation Assessor) were employed.
Orthologous conservation of the GBA residues harbouring the novel variant
A previously described protocol to check the inter-species conservation of the GBA residues incorporating novel variants was employed [19]. In brief, the Clastal Omega; an online multiple sequence alignment program was used to align the Homo sapiens (NP_001005741) protein sequence along other species.
Homology modeling, structure validation and protein stability due to novel variants
The mutated protein structure of the four novel missense variants was modeled using our previously described protocol [19]. In brief, the mutant protein model was build using the template β-Glucosidase crystallographic structure (PDB ID: 1OGS) and the Root Mean Square Deviation (RMSD) of the mutant structures with respect to the wild-type structure was calculated.
Results
The patients included in the study were in the age range 1 day to 31 years at the time of investigations. The study comprises 62 male and 36 female patients presented with unexplained hepatomegaly, moderate splenomegaly, anaemia, and thrombocytopenia with or without any bone abnormality. Bone marrow examination in the majority of the patients showed classical Gaucher cells. Table 1 covers the demographic profile of the patients. Out of 100 patients investigated, 75 patients were diagnosed with GD type I, 12 patients were diagnosed with GD type II, and 11 patients were diagnosed with GD type III. Two fetuses were diagnosed with GD by enzymatic study however, their GD type remains indistinct. Parental consanguinity was observed in 26 (26%) patients. The parents in consanguineous marriage were first-degree relatives and hence the average inbreeding coefficient of the 26 patients is 3.1%. Majority of the patients were observed from the western India (43%; Gujarat and Maharashtra), followed by the northern zone (32%; Rajasthan, Punjab, New Delhi, Himachal Pradesh, Jammu and Kashmir, and Uttar Pradesh), southern zone (21%; Karnataka, Kerala, Telangana, and Tamil Nadu), and eastern zone (4%; Bihar, and West Bengal).
Table 1.
Demographic profile of the patients with Gaucher disease
| Total patients n = 100 | GDI n = 77 | GDII n = 12 | GDIII n = 11 | |
|---|---|---|---|---|
| Gender | ||||
| Male | 62 (62%) | 47 (61.03%) | 7 (58.33%) | 8 (72.72%) |
| Female | 36 (36%) | 28 (36.36%) | 5 (41.66%) | 3 (27.27%) |
| Fetus | 2 (2%) | – | – | – |
| Regional distribution | ||||
| East India | 4 (4%) | 3 (3.89%) | 1 (8.33%) | – |
| West India | 43 (43%) | 32 (41.55%) | 7 (58.33%) | 4 (36.36%) |
| North India | 32 (32%) | 24 (31.16%) | 3 (25%) | 5 (45.45%) |
| South India | 21 (21%) | 18 (23.37%) | 1 (8.33%) | 2 (18.18%) |
Abbreviations: GDI, Gaucher disease type I, GDII Gaucher disease type II, GDIII Gaucher disease type III
Data are represented as n (%)
Biochemical analysis
Out of 100 patients, plasma chitotriosidase was elevated (172–72,000 nmolh−1ml−1plasma) in 71 patients, undetectable in 5 patients, and normal (1.0–102.4 nmolh−1ml−1plasma) in 13 patients. However, no information about the plasma chitotriosidase activity in 11 patients is available. The β-Glucosidase enzyme assay confirmed the GD. Significantly reduced activity (< 10% of normal mean) of the β-Glucosidase enzyme was observed in the patients (see Additional file 2). The β-Glucosidase enzyme activity results were not available in five patients.
Molecular analysis
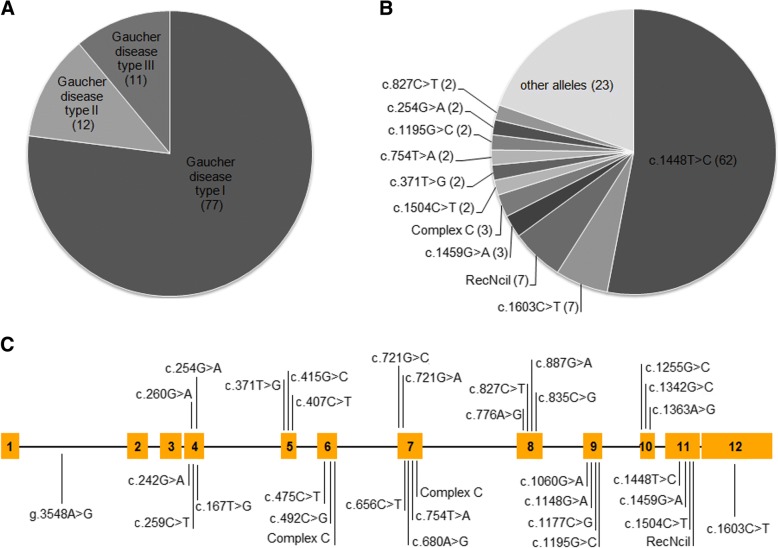
The bidirectional sequencing of the coding GBA1 gene revealed a total 33 mutations in 93 patients. Figure 1 depicts an illustrative representation of the variants identified through Sanger sequencing.
Fig. 1.
Illustrative representation of the distributions of the variants identified in Indian Gaucher patients investigated in this study. a Majority of the patients were affected with Gaucher disease type I (77 patients), followed by Gaucher disease type II (12 patients) and Gaucher disease type III (11 patients). b The most common mutation c.1448T>C was observed in 62 patients (including homozygotes and compound heterozygotes). The second most common mutations identified were c.1603C>T and RecNcil. c Variations on exon 4–12 were observed on GBA1 gene. Also a mutation g.3548A>G was observed in the intron 1. The mutations were more clustered on exon 4 and exon 7 to exon 11
The identification of the genetic cause of GD in all the patients involved an initial screening for the common mutations c.1448 T>C (p.Leu483Pro) observed in Indian patients with GD [18, 20]. Patient P1 to P51 were found to be homozygous for p.Leu483Pro mutation in exon 11 of GBA1 gene (Table 2). Patient P52 to P62 harbour a heterozygous copy of the same mutation with the second disease-causing mutant allele. Hence, the mutation p.Leu483Pro was observed as a most common mutation in the patients affected with GD (including homozygous and compound heterozygous). However, during the initial screening, no molecular output was obtained in the remaining patients. As a single variant or no variant was detected in the initial screening, the further investigation involved the sequencing of the complete coding region of GBA1 gene in these patients.
Table 2.
Molecular analysis of the patients with Gaucher disease
| Patient ID | Variant location (GBA1 geneb) | Zygosity | Allele frequency | dbSNP reference number | |
|---|---|---|---|---|---|
| 1000 Genomes | ExAC | ||||
| P1-P51 | Ex11:c.1448T>C/p.L483P | Hom | 0.0034 | 0.0031 | rs421016 |
| P52 | Ex11:c.1448T>C/p.L483P | Com Hetz | 0.0034 | 0.0031 | rs421016 |
| Ex5:c.407C>T/p.S136 La | NR | NR | rs878853316 | ||
| P53 | Ex11:c.1448T>C/p.L483P | Com Hetz | 0.0034 | 0.0031 | rs421016 |
| Ex4:c.167T>G/p.V56G | NR | NR | rs878853318 | ||
| P54 | Ex11:c.1448T>C/p.L483P | Com Hetz | 0.0034 | 0.0031 | rs421016 |
| Ex10:c.1363A>G/p.M455 V | NR | NR | NR | ||
| P55 | Ex11:c.1448T>C/p.L483P | Com Hetz | 0.0034 | 0.0031 | rs421016 |
| Ex7:c.656C>T/p.T219I | NR | NR | NR | ||
| P56-P57 | Ex11:c.1448T>C/p.L483P | Com Hetz | 0.0034 | 0.0031 | rs421016 |
| Ex12:c.1603C>T/p.R535C | NR | 0.00004357 | rs747506979 | ||
| P58-P59 | Ex11:c.1448T>C/p.L483P | Com Hetz | 0.0034 | 0.0031 | rs421016 |
| Ex5:c.371T>G/p.M124R | NR | 0.000008237 | NR | ||
| P60 | Ex11:c.1448T>C/p.L483P | Com Hetz | 0.0034 | 0.0031 | rs421016 |
| Ex10:c.1255G>C/p.D419H | NR | NR | NR | ||
| P61-P62 | Ex11:c.1448T>C/p.L483P | Com Hetz | 0.0034 | 0.0031 | rs421016 |
| Ex11:RecNcil | – | ||||
| P63-P64 | Ex12:c.1603C>T/p.R535C | Hom | NR | 0.00004357 | rs747506979 |
| P65 | Ex12:c.1603C>T/p.R535C | Com Hetz | NR | 0.00004357 | rs747506979 |
| Ex6,7:Complex C | – | ||||
| P66-P67 | Ex11:c.1504C>T/p.R502C | Hom | NR | 0.00006 | rs80356771 |
| P68-P70 | Ex11:c.1459G>A/p.A487T | Hom | NR | NR | rs878853317 |
| P71 | Ex6:c.492C>G/p.S164R | Com Hetz | NR | 0.00003295 | NR |
| Ex4:c.254G>A/p.G85E | NR | 0.000008 | rs77829017 | ||
| P72 | Ex4:c.254G>A/p.G85E | Com Hetz | NR | 0.000008 | rs77829017 |
| Ex11:RecNcil | – | ||||
| P73-P74 | Ex7:c.754T>A/p.F252I | Hom | NR | 0.00002 | rs381737 |
| P75-P76 | Ex9:c.1195G>C/p.G399Ra | Com Hetz | NR | NR | NR |
| Ex12:c.1603C>T/ p.R535C | NR | 0.00004357 | rs747506979 | ||
| P77 | Ex7:c.721G>A/p.G241R | Hom | NR | NR | rs409652 |
| P78 | Ex10:c.1342G>C/p.D448H | Hom | NR | 0.0001 | rs1064651 |
| P79−P80 | Ex8:c.827C>T/p.S276F | Hom | NR | 0.00000837 | NR |
| P81 | Ex9:c.1060G>A/p.D354N | Hom | NR | 0.000008 | rs398123526 |
| P82 | Ex8:c.776A>G/p.Y259C | Hom | NR | NR | NR |
| P83 | Ex5:c.415G>C/p.A139P | Hom | NR | NR | rs878853314 |
| P84 | Ex9:c.1177C>G/p.L393 V | Hom | NR | NR | rs878853315 |
| P85 | Ex7:c.721G>C/p.G241R | Hom | NR | NR | NR |
| P86 | Ex4:c.260G>A/p.R87Q | Hom | NR | NR | rs78769774 |
| P87 | Ex8:c.835C>G/p.L279 V† | Hom | NR | NR | NR |
| P88 | In1:g.3548A>G/g.3548A > G | Com Hetz | 0.0078 | NR | rs18897815 |
| Ex11:RecNcil | – | ||||
| P89 | Ex4:c.259C>T/p.R87W | Com Hetz | NR | 0.00002 | rs1141814 |
| Ex11:RecNcil | – | ||||
| P90 | Ex9:c.1148G>A/p.G383D† | Com Hetz | NR | NR | NR |
| Ex11:RecNcil | – | ||||
| P91 | Ex6:c.475C>T/p.R159W | Com Hetz | NR | NR | rs439898 |
| Ex11:RecNcil | – | ||||
| P92 | Ex7:c.680A>G/p.N227S | Com Hetz | 0.0002 | 0.00007 | rs364897 |
| Ex6,7:Complex C | – | ||||
| P93 | Ex8:c.887G>A/p.R296Q | Com Hetz | NR | 0.00003 | rs78973108 |
| Ex6,7:Complex C | – | ||||
| P94 | Patients’DNA is not available Mother- Ex11:c.1448T> C/p.L483P Father- Ex11:c.1448T> C/p.L483P |
Single Hetz | 0.0034 | 0.0031 | rs421016 |
| P95 | Patients’ DNA is not available Mother- Ex11:c.1448T>C/p.L483P |
Single Hetz | 0.0034 | 0.0031 | rs421016 |
| Father- Ex4:c.242G>A/p.S81 N | NR | NR | NR | ||
| P96 | Patients’ DNA is not available Mother- Ex6:c.475C>T/p.R159W |
Single Hetz | NR | NR | rs439898 |
| Father- Ex11:c.1448T>C/p.L483P | 0.0034 | 0.0031 | rs421016 | ||
| P97 | Patients’ DNA is not available Mother- Ex6,7:Complex C Father- Ex6,7:Complex C |
Single Hetz | – | ||
| P98 | Mutation not found | NA | NA | NA | NA |
| P99 | Mutation not found | NA | NA | NA | NA |
| P100 | No amplification from Exon 1 to Exon 7 | NA | NA | NA | NA |
Abbreviations: Com Hetz Compound Heterozygous, dbSNP The Single Nucleotide Polymorphism database, ExAC The Exome Aggregation Consortium, Ex Exon, Homozygous (Hom), NR Not Reported
RecNcil: [c.1448T>C (p.L483P), c.1483G>C (p.A495P), c.1497G>C (p.V499 V)]]
Complex C: [c.475C>T (p.R159W), c.667T>C (p. W223R), c.681T>G (p.N227K), c.689 T>G (p.V230G), c.703T>C (p.S235P), c.721G>A (p.G241R), c.754T>A (p.F252I)]
aNovel variants identified in the given study
bThe above variants refers to the GBA1 gene with transcript ID ENST00000327247.5 and reference sequence number NM_001005741.2
The second most common mutations identified in the given study are c.1603C>T (p.Arg535Cys) in exon 12 and the complex mutation RecNcil [c.1448 T>C (p.L483P), c.1483G>C (p.A495P), c.1497G>C (p.V499 V)] in exon 11. These mutations existed in both homozygous and compound heterozygous status in seven patients each. Another complex mutation Complex C [c.475C>T (p.R159W), c.667T>C (p.W223R), c.681T>G (p.N227K), c.689T>G (p.V230G), c.703T>C (p.S235P), c.721G>A (p.G241R), c.754T>A (p.F252I)] was identified in a compound heterozygous status in three patients. Also, the mutation c.1459G>A (p.Ala487Thr) was revealed in three patients in homozygous form.
The given study uncovered the known mutations like c.1504C>T (p.Arg502Cys), c.371T>G (p.Met124Arg), c.754T>A (p.Phe252Ile), c.827C>T (p.Ser276Phe), and c.254G>A (p.Gly85Glu); each in different sets of two patients. These mutations were comparatively less commonly observed.
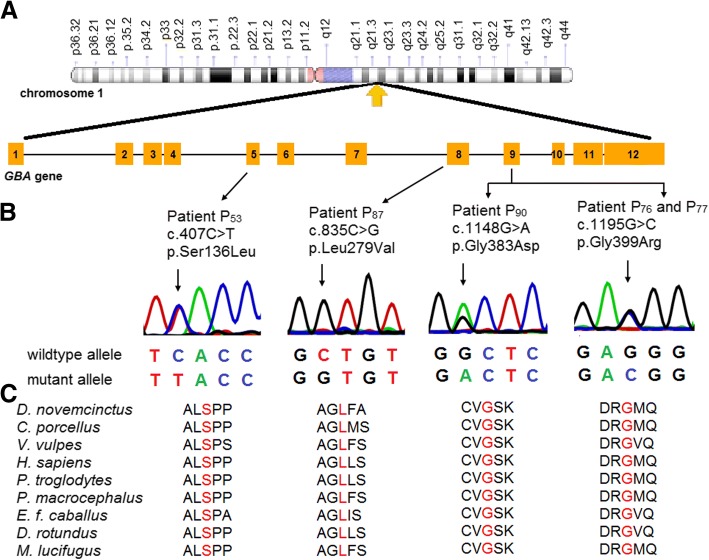
The sequencing of the coding region of GBA1 gene revealed four novel variants in the given study (Fig. 2a and b). The novel variant c.835C>G (p.Leu279Val) was identified in the homozygous state in exon 8. The remaining three novel variants, c.407C>T (p.Ser136Leu), c.1195G>C (p.Gly399Arg), and c.1148G>A (p.Gly383Asp) were identified as compound heterozygous along with other known mutations (Table 2).
Fig. 2.
Identification of novel variants in GBA1 gene. a Illustrative representation of the distributions of the novel variants identified in Indian Gaucher patients investigated in this study. b Sanger sequencing discovered four missense variants in GBA1 gene. The variants p.Ser136Leu, p.Gly383Asp, and p.Gly399Arg, were identified, as compound heterozygotes along with another know mutant allele. The variant p.Leu279Val existed in homozygous form. An arrow indicates the point of variation. c The multiple alignment of the protein sequence surrounding the novel variants against various orthologous sequence revealed the conservative status of the wildtype residues (marked red)
Apart from this, the study also found 18 known mutations; each in one patient. These mutations are g.3548A>G, c.167T>G (p.Val56Gly), c.1363A>G (p.Met455Val), c.656C>T (p.Thr219Ile), c.1255G>C (p.Asp419His), c.492C>G (p.Ser164Arg), c.721G>A (p.Gly241Arg), c.1342G>C (p.Asp448His), c.1060G>A (p.Asp354Asn), c.776A>G (p.Tyr259Cys), c.415G>C (p.Ala139Pro), c.1177C>G (p.Leu393Val), c.721G>C (p.Gly241Arg), c.260G>A (p.Arg87Gln), c.259C>T (p.Arg87Trp), c.475C>T (p.Arg159Trp), c.680A>G (p.Asn227Ser), c.887G>A (p.Arg296Gln).
The carrier analysis was carried out in four couples where the proband died due to confirmed GD. In case of the patient P94, the parents were found to be the carrier for the most commonly identified mutation p.Leu483Pro. While one parent in case of patients P95 and P96 was heterozygous for p.Leu483Pro. The following compound heterozygous mutation identified in the second parent of the patient P95 and P96 were c.242G>A (p.Ser81Asn) and c.475C>T (p.Arg159Trp) respectively. For the patient P97, the parents were identified as a carrier for the Complex C.
However, no variation in the exonic or exon-intronic boundaries of GBA1 gene was detected in three patients (P98, P99, and P100). These patients presented common clinical indications like hepatosplenomegaly, unexplained hepatomegaly, moderate splenomegaly, anemia, and thrombocytopenia with or without any bone abnormality as also indicated in GD patients. Along with these common clinical indications, patient P99 and P100 manifested neurological regression evident at the age of 8 years and one and half years respectively. Hence, patient P99 and P100 were classified as GD type 3 and GD type 2 respectively. Their significant decrease in the β-Glucosidase enzyme activity confirmed the GD diagnosis. In such cases of the uninformative Sanger sequencing results, the possibility of either large deletion/duplication or deep intronic variations cannot be ruled out.
Overall, the mutation p.Leu483Pro was observed as a most common mutation with its prevalence in 62% patients affected with GD. Its frequency was 60.75% of the total mutations detected.
In silico analysis of the novel variants
The in silico tools described above established the functional effects of the variants identified (see Additional file 3). The novel variants were found to be disease causing. These predicting tools suggest the probably damaging and deleterious effect of the novel variants on protein function. These variants were found neither in the 1000 Genomes database nor in the Exome Aggregation Consortium (ExAc).
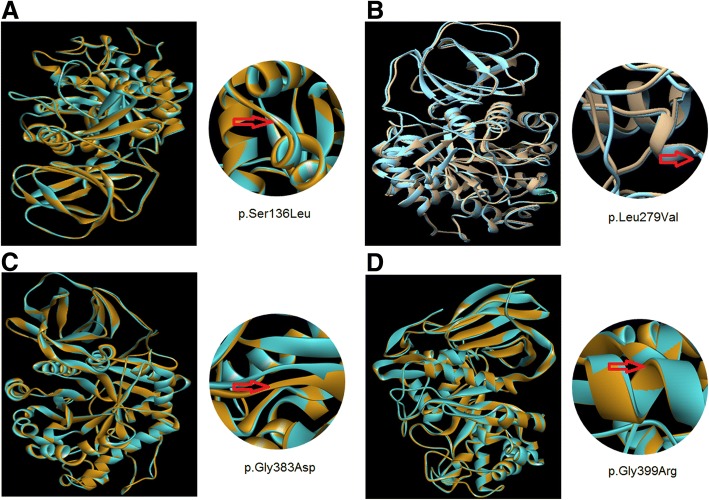
The protein sequence alignment of Homo sapiens along with other species using Clastal Omega suggests that these variations occurred at highly evolutionarily conserved and functionally active residual domain in the protein (Fig. 2c). The protein homology modeling of the novel missense point variants in the GBA1 gene suggest their damaging effect at highly evolutionarily conserved and functionally active domain residues in the protein leading to conformational changes or destabilization of the protein structure (Fig. 3). The variant p.Ser136Leu caused changes in the loop region joining α1 and α1a helix. The variant p.Leu279Val caused destabilization of the normal shape of the active site cavity. The variant p.Gly383Asp caused conformational changes in the loop regions and the variant p.Gly399Arg caused conformational changes in the alpha helix.
Fig. 3.
Homology modeling of novel missense variants identified in the GBA1 gene. The native structure (blue) and mutant structure (brown) are superimposed. a The model of the variant p.Ser136Leu depicting the changes in the loop region joining α1 and α1a; at the codon number 136 (TCA-TTA). b The model of the variant p.Leu279Val depicting the destabilization of the normal shape of the active site cavity at the codon number 279 (CTG-GTG). c The model of the variant p.Gly383Asp depicting the conformational changes in the loop regions at the codon number 383 (GGC-GAC). d The model of the variant p.Gly399Arg depicting the conformational changes in the alpha helix at the codon number 399 (GGG-CGG). All the models reveal the conformational changes in the GBA protein structure
Discussion
The given study demonstrates the mutation spectrum of GD in a large Indian cohort. It reveals 100 cases of GD (77 with GD type 1, 12 with GD type 2, and 11 with GD type 3) with maximum GD type 1 cases.
The literature reports p.Leu483Pro, p.Asn409Ser, c.84dupG (84GG) and IVS2 + 1G>A as the most common mutations in GD. A study by Bisariya et al. in 24 Indian GD patients reported the mutant alleles p.Leu483Pro, p.Asn409Ser, IVS2+1G>A, p.Asp448His and c.1263_1317del (55Del) collectively in approximately 50% of the patients; p.Leu483Pro was the most common mutation, followed by p.Asp448His [21]. However, in the given study only p.Leu483Pro was observed as the most common mutation. The other mutations mentioned above were not detected.
Variation in these observations can be likely due to different ethnic group involved in these studies. Endogamous marriage practice has clustered specific community in different zones of India. The majority of the patients, in the given study, are from western and northern zone that provides an idea that GD mutations spectrum may vary in different parts of India. However, the mutation p.Leu483Pro remains as the most commonly observed mutation in Indian GD patients [18, 20, 21]. Considering these results along with our recently published data on carrier frequency of p.Leu483Pro (1:600), it is justifiable to consider p.Leu483Pro as the most prevalent mutation in the Indian population; irrespective of the ethnic group [14].
Several ethnicity specific mutations have been observed in the GD. For instance, the mutation p.Gly416Ser is observed in high frequency in Brazilian GD type 3 patients [22]. The mutations p.Leu483Pro and p.Phe252Ile are relatively common in Japanese GD patients [23, 24]. The most prevalent mutations in Turkish, Romanian, and Czech and Slovak GD patients are p.Leu483Pro and p.Asn409Ser [11, 25, 26]. Based on these, the mutation p.Leu483Pro can be considered Pan-ethnic.
The mutation c.84dupG (84GG), along with other common Jewish GD mutation, accounts for the 50–60% of mutations in non-Jewish GD patients. However, it was observed neither in the given study nor in our previously published data on GD [18]. This may account for the low prevalence of bone disease in the patients investigated [27]. Several studies on non-Jewish population have also failed to identify this mutation [11, 18, 21, 26, 28].
The given study has also identified the complex allele RecNcil in seven patients and Complex C in three patients. These mutations usually result either due to non-homologous recombination between the functional GBA1 gene and the highly homologous, non-functional GBA1 pseudogene, gene conversion, fusion or duplication [28, 29]. A study by Horowitz et al. reported RecNcil in 7.8% of alleles of non-Jewish patients [28]. The given study reports two patients (GD type 1 and GD type 2) with p.Leu483Pro/RecNcil. Lee et al. also reported the same mutant complex in a patient with GD type 2 [30]. Nevertheless, the phenotypic heterogeneity prevails amongst these patients.
A study by Koprivica et al. suggests the association of p.Leu483Pro with GD type 3 [31]. However, the given study demonstrates that, though associated with all types of GD, the mutation p.Leu483Pro is most commonly observed in GD type 1. A study on Romanian GD patience was in accordance to our given study [11]. A study by Wan et al. in Taiwanese population have reported the occurrence of p.Leu483Pro in 53.5% of type 1 and type 2 GD patients collectively [32]. Similarly, the p.Leu483Pro mutation was identified in 60% of the total Thai patients with GD type 1 and type 2 [33]. These phenotypic variations in different population, due to p.Leu483Pro mutant allele could be because of the effect of the modifier gene on mutant allele. A Genome-wide Association Study by Zhang et al. identified CLN8 as a potential modifier gene responsible for the phenotypic variation in the type 1 GD patients with p.Asn409Ser homozygotes [34]. However, the impact of modifier genes on GD is a grey area. It is also likely that many of our type 1 GD patients with p.Leu483Pro genotype may develop neurological symptoms at the later stage as observed in the Spanish population [35]. Owing to the high cost of enzyme replacement therapy and poor financial background of the patients, only two patients with type 1 GD are provided the therapy. However, the long term follows up of our remining patients with type 1 GD will help to understand the heterogenic effect of the said genotype on the phenotype. Those with p.Leu483Pro carriers are also at an increased risk of Parkinson disease (PD) and extended study of these families will help to understand the natural history of progression from carrier to PD. A study by Wang et al. in Chinese population established that patients with p.Leu483Pro had 3.4% chances of developing PD compared to the 0.3% of the controls and hence a long-term follow would be essential to understand the impact of PD on Indian population [36].
The majority of mutations found in the given study were clustered in exons 4, 7, 8, 9, 10, and 11. Around 45% of the mutations have been observed in exons 8 to 11. The expression studies have shown that certain residues substitution in exon 8, 9, 10, and 11 are highly disruptive and can produce compromised protein [37].
Conclusion
In conclusion, our results confirmed the remarkable heterogeneity of the mutational spectrum of the GBA1 gene and provided new insight into molecular pathology of GD. The mutation c.1448T>C (p.Leu483Pro) was identified in 62% patients and hence can be used for mass screening. The same mutation was also observed in majority of the patients with GD type I. This is likely to provide the base to understand the progression of some of these patients in type 3 GD and possible effect on the development of Parkinson disease. Since the mutations were clustered in the exons 4, 7, 8, 9, 10, and 11, this region can be considered as hot spot for mutation analysis.
Additional files
List of primers used for GBA1 gene sequencing. The exons and the exon-intron boundaries of both the genes were bidirectionally sequenced using the given set of primers. (DOC 32 kb)
Biochemical analysis of the Gaucher Disease patients. The plasma chitotriosidase enzyme activity and β-Glucosidase enzyme activity were checked using the standard protocol. (DOCX 17 kb)
In silico analysis of the functional effect of the variants identified in the patients with Gaucher disease. The in silico tools predicting the effect of DNA variants, amino acid substitution, non-coding variants, and coding non-synonymous variants were employed to predict the functional effect of the variants identified in the given study. (DOCX 20 kb)
ClinVar Accession ID of the variants generated in the given study. The variants identified through Sanger sequencing are reported in NCBI ClinVar database. The file provides accession ID and the links to an individual variant. (DOCX 14 kb)
Acknowledgements
We are grateful to the coordinators and all the members of Task force on Lysosomal storage Disorders: Dr. Roli Mathur and Dr. Babbanjee. We thank Dr. Madhulika Kabra, Dr. Neeraj Gupta, and Dr. Shankar VH for referring patients. We show our gratitude to the patients for their support and without their consent this study would not have been possible.
Funding
This work is supported by Department of Health Research/Indian Council of Medical Research [grant no.: GIA/31(ii)/2014-DHR] and Department of Biotechnology [BT/PR4112/MED/12/654/2014], Government of India. The funding agency was not directly or indirectly involved in the study design, specimen collection, analysis, interpretation of the data, and preparation of the manuscript.
Availability of data and materials
The dataset generated and/or analyzed during the current study is available in the ClinVar repository (see Additional file 4).
Abbreviations
- 4-MU
4-Methylumbelliferyl-β-D-N,N′,N″-triacetylchitotrioside
- dbSNP
The Single Nucleotide Polymorphism database
- FATHMM
The Functional Analysis Through Hidden Markov Models
- GBA1
beta-Glucosidase
- GD
Gaucher disease
- gDNA
genomic DNA
- NCBI
National Center for Biotechnology Information
- PCR
Polymerase Chain Reaction
- PolyPhen-2
Polymorphism Phenotyping version 2
- PROVEAN
Protein Variation Effect Analyzer
- RFLP
Restriction Fragment Length Polymorphism
- RMSD
Root Mean Square Deviation
- SIFT
The Sorting Intolerant from Tolerant
Authors’ contributions
Conceived the experiments: JS and FS. Designed the experiments: JS, RB, MM (Mehul Mistri). Clinical analysis: JS, AB, AD, PR, GK, AS, SP, RP, IP, AK, MM (Mamta Muranja), MG, RR, RS, SN, SD, CD, SK, SB. Laboratory workup: RB and MM (Mehul Mistri). Data analysis: RB, DP, and MM (Mehul Mistri). Manuscript writing: DP, JS, and MM (Mehul Mistri). Critical revisions and approval of final manuscript: JS, DP and FS. All authors reviewed and approved the final manuscript.
Ethics approval and consent to participate
The given study has been approved by the institutional ethics committee [FRIGE’s Institute of Human Genetics] with approval number FRIGE/IEC/14/2016 dated 19 November 2016. This process is in accordance with the declaration of Helsinki. An informed consent for investigation and publication of their clinical details and/or clinical images was obtained from the patient or an accompanying guardian at the time of enrolment for the study [This was in accordance with the requirement of the institutional ethics committee].
Consent for publication
Informed written consent was obtained from all the participants or the accompanying guardian for publication of the participants’ clinical details and/or clinical images. A copy of the written consent is available for review by the editor of this journal.
Competing interests
Dr. Ashwin Dalal is a member of the editorial board (Associate Editor) of the journal BMC Medical Genetics. The authors declare that they have no competing interests (financial or non-financial) in the present study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jayesh Sheth, Phone: +91-79-26921414, Email: jshethad1@gmail.com.
Riddhi Bhavsar, Email: riddhib4@gmail.com.
Mehul Mistri, Email: mistrimehul@yahoo.co.in.
Dhairya Pancholi, Email: riahp7261@gmail.com.
Ashish Bavdekar, Email: ashish.bavdekar1@gmail.com.
Ashwin Dalal, Email: ashwindalal@gmail.com.
Prajnya Ranganath, Email: pragnya4@gmail.com.
Katta M Girisha, Email: girish.katta@manipal.edu.
Anju Shukla, Email: anju.shukla@manipal.edu.
Shubha Phadke, Email: shubharaophadke@gmail.com.
Ratna Puri, Email: ratnadpuri@yahoo.com.
Inusha Panigrahi, Email: inupan@yahoo.com.
Anupriya Kaur, Email: anukaur.genetics@gmail.com.
Mamta Muranjan, Email: muranjanmamta@gmail.com.
Manisha Goyal, Email: manidr2000@gmail.com.
Radha Ramadevi, Email: radharamadevi@gmail.com.
Raju Shah, Email: rajucshah@gmail.com.
Sheela Nampoothiri, Email: sheeladr@gmail.com.
Sumita Danda, Email: sumita_danda@hotmail.com.
Chaitanya Datar, Email: dr.cdatar@gmail.com.
Seema Kapoor, Email: drseemakapoor@gmail.com.
Seema Bhatwadekar, Email: ssbkar16@gmail.com.
Frenny Sheth, fshethad1@googlemail.com.
References
- 1.Stirnemann J, Vigan M, Hamroun D, Heraoui D, Rossi-Semerano L, Berger MG, et al. The French Gaucher’s disease registry: clinical characteristics, complications and treatment of 562 patients. Orphanet J Rare Dis. 2012;9(7):77. doi: 10.1186/1750-1172-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ron I, Horowitz M. ER retention and degradation as the molecular basis underlying Gaucher disease heterogeneity. Hum Mol Genet. 2005;14(16):2387–2398. doi: 10.1093/hmg/ddi240. [DOI] [PubMed] [Google Scholar]
- 3.AE. La maladie de Gaucher dans l’enfance. Faculté de Médicine. 1934.
- 4.Brady RO, Kanfer JN, Shapiro D. Metabolism of glucocerebrosides. II. Evidence of an enzymatic deficiency in gaucher’s disease. Biochem Biophys Res Commun. 1965;18(18):221–225. doi: 10.1016/0006-291X(65)90743-6. [DOI] [PubMed] [Google Scholar]
- 5.Patrick AD. A Deficiency of Glucocerebrosidase in Gaucher’s Disease. BiochemJ. 1965;97(2):17C–24C. doi: 10.1042/bj0970017C. [DOI] [Google Scholar]
- 6.Weinreb NJ, Brady RO, Tappel AL. The lysosomal localization of sphingolipid hydrolases. Biochim Biophys Acta. 1968;159(1):141–146. doi: 10.1016/0005-2744(68)90251-9. [DOI] [PubMed] [Google Scholar]
- 7.Kinghorn KJ, Grönke S, Castillo-Quan JI, Woodling NS, Li L, Sirka E, et al. A Drosophila model of neuronopathic Gaucher disease demonstrates lysosomal-autophagic defects and altered mTOR signalling and is functionally rescued by rapamycin. J Neurosci. 2016;36(46):11654–11670. doi: 10.1523/JNEUROSCI.4527-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DL S, Tayebi N, Orvisky E, Stubblefield B, Madike V, Sidransky E. Glucocerebrosidase gene mutations in patients with type 2 Gaucher disease. Hum Mutat. 2000;15(2):181–188. doi: 10.1002/(SICI)1098-1004(200002)15:2<181::AID-HUMU7>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 9.Horowitz M, Wilder S, Horowitz Z, Reiner O, Gelbart T, Beutler E. The human glucocerebrosidase gene and pseudogene: structure and evolution. Genomics. 1989;4(1):87–96. doi: 10.1016/0888-7543(89)90319-4. [DOI] [PubMed] [Google Scholar]
- 10.Tsuji S, Choudary PV, Martin BM, Stubblefield BK, Mayor JA, Barranger JA, et al. A mutation in the human glucocerebrosidase gene in neuronopathic Gaucher’s disease. N Engl J Med. 1987;316(10):570–575. doi: 10.1056/NEJM198703053161002. [DOI] [PubMed] [Google Scholar]
- 11.Drugan C, Procopciuc L, Jebeleanu G, Grigorescu-Sido P, Dussau J, Poenaru L, et al. Gaucher disease in Romanian patients: incidence of the most common mutations and phenotypic manifestations. Eur J Hum Genet. 2002;10(9):511–515. doi: 10.1038/sj.ejhg.5200845. [DOI] [PubMed] [Google Scholar]
- 12.Hruska KS, LaMarca ME, Scott CR, Sidransky E. Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Hum Mutat. 2008;29(5):567–583. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- 13.Pastores GM, Hughes DA. Gaucher Disease. 2018. https://www.ncbi.nlm.nih.gov/books/NBK1269/. 2018. Assessed 18 Sept 2018.
- 14.Sheth J, Pancholi D, Mistry M, Nath P, Ankleshwaria C, Bhavsar R. Biochemical and molecular characterization of adult patients with type I Gaucher disease and carrier frequency analysis of Leu444Pro- a common Gaucher disease mutation in India. BMC Med Genet. 2018. 10.1186/s12881-018-0687-5. [DOI] [PMC free article] [PubMed]
- 15.Sheth JJ, Sheth FJ, Oza NJ, Gambhir PS, Dave UP, Shah RC. Plasma chitotriosidase activity in children with lysosomal storage disorders. Indian J Pediatr. 2010;77(2):203–205. doi: 10.1007/s12098-009-0249-0. [DOI] [PubMed] [Google Scholar]
- 16.Shapria E, Blitzer MG, Miller JB, et al. Fluorometric assays in Biochemical Genetics: A laboratory Manual. Oxford: Oxford University press; 1989. [Google Scholar]
- 17.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ankleshwaria C, Mistri M, Bavdekar A, Muranjan M, Dave U, Tamhankar P, et al. Novel mutations in the glucocerebrosidase gene of Indian patients with Gaucher disease. J Hum Genet. 2015;60(5):285. doi: 10.1038/jhg.2015.27. [DOI] [PubMed] [Google Scholar]
- 19.Sheth J, Bhavsar R, Gandhi A, Sheth F, Pancholi D. A case of Raine syndrome presenting with facial dysmorphy and review of literature. BMC Med Genet. 2018;19(1):76. doi: 10.1186/s12881-018-0593-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muranjan M. Gaucher Disease. In: Gupta P, Menon PS, Ramji S, Lodha R, editors. PG textbook of pediatrics. New Delhi: Jaypee Brothers Medical Publishers (P) Ltd; 2015. pp. 109–118. [Google Scholar]
- 21.Bisariya V, Mistry PK, Liu J, Chaudhari MR, Gupta N, Kabra M. The mutation spectrum in Indian patients with Gaucher disease. Genome Biol. 2011;12(Suppl 1):26. doi: 10.1186/gb-2011-12-s1-p26. [DOI] [Google Scholar]
- 22.Rozenberg R, Araújo FT, Fox DC, Aranda P, Nonino A, Micheletti C, et al. High frequency of mutation G377S in Brazilian type 3 Gaucher disease patients. Braz J Med Biol Res. 2006;39(9):1171–1179. doi: 10.1590/S0100-879X2006000900004. [DOI] [PubMed] [Google Scholar]
- 23.Kawame H, Maekawa K, Eto Y. Molecular screening of Japanese patients with Gaucher disease: phenotypic variability in the same genotypes. Hum Mutat. 1993;2(5):362–367. doi: 10.1002/humu.1380020507. [DOI] [PubMed] [Google Scholar]
- 24.Eto Y, Ida H. Clinical and molecular characteristics of Japanese Gaucher disease. Neurochem Res. 1999;24(2):207–211. doi: 10.1023/A:1022553819241. [DOI] [PubMed] [Google Scholar]
- 25.Emre S, Gürakan F, Yüce A, Rolf A, Scott R, Ozen H. Molecular analysis of Turkish Gaucher disease patients: identification of novel mutations in glucocerebrosidase (GBA) gene. Eur J Med Genet. 2008;51(4):315–321. doi: 10.1016/j.ejmg.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Hodanová K, Hrebícek M, Cervenková M, Mrázová L, Vepreková L, Zemen J. Analysis of the β-glucocerebrosidase gene in Czech and Slovak Gaucher patients: mutation profile and description of six novel mutant alleles. Blood Cells Mol Dis. 1999;25(5–6):287–298. doi: 10.1006/bcmd.1999.0256. [DOI] [PubMed] [Google Scholar]
- 27.Sibille A, Eng CM, Kim SJ, Pastores G, Grabowski GA. Phenotype/genotype correlations in Gaucher disease type I: clinical and therapeutic implications. Am J Hum Genet. 1993;52(6):1094–1101. [PMC free article] [PubMed] [Google Scholar]
- 28.Horowitz M1, Tzuri G, Eyal N, Berebi A, Kolodny EH, Brady RO, et al. Prevalence of nine mutations among Jewish and non-Jewish Gaucher disease patients. Am J Hum Genet. 1993;53(4):921–930. [PMC free article] [PubMed] [Google Scholar]
- 29.Eyal N, Wilder S, Horowitz M. Prevalent and rare mutations among Gaucher patients. Gene. 1990;96:277–283. doi: 10.1016/0378-1119(90)90264-R. [DOI] [PubMed] [Google Scholar]
- 30.Lee YS, Poh LK, Ida H, Loke KY. Type II Gaucher disease: compound heterozygote with RecNciI and L444P mutations. J Trop Pediatr. 2001;47(2):115–117. doi: 10.1093/tropej/47.2.115. [DOI] [PubMed] [Google Scholar]
- 31.Koprivica V, Stone DL, Park JK, Callahan M, Frisch A, Cohen IJ, et al. Analysis and classification of 304 mutant alleles in patients with type 1 and type 3 Gaucher disease. Am J Hum Genet. 2000;66(6):1777–1786. doi: 10.1086/302925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan L, Hsu CM, Tsai CH, Lee CC, Hwu WL, Tsai FJ. Mutation analysis of Gaucher disease patients in Taiwan: high prevalence of the RecNciI and L444P mutations. Blood Cells Mol Dis. 2006;36(3):422–425. doi: 10.1016/j.bcmd.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Tammachote R, Tongkobpetch S, Srichomthong C, Phipatthanananti K, Pungkanon S, Wattanasirichaigoon D, et al. A common and two novel GBA mutations in Thai patients with Gaucher disease. J Hum Genet. 2013;58(9):594–599. doi: 10.1038/jhg.2013.60. [DOI] [PubMed] [Google Scholar]
- 34.Zhang CK, Stein PB, Liu J, Pastores GM, Wang Z, Yang R, et al. Genome-wide Association Study of N370S Homozygous Gaucher Disease Reveals the Candidacy of CLN8 gene as a Genetic Modifier Contributing to Extreme Phenotypic Variation. Am J Hematol. 2012;87(4):377–383. doi: 10.1002/ajh.23118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alfonso P, Aznarez S, Giralt M, Pocovi M, Giraldo P, Gaucher's Disease Registry S. Mutation analysis and genotype/phenotype relationships of Gaucher disease patients in Spain. J Hum Genet. 2007;52(5):391–396. doi: 10.1007/s10038-007-0135-4. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Liu L, Xiong J, Zhang X, Chen Z, Yu L, et al. Glucocerebrosidase L444P mutation confers genetic risk for Parkinson’s disease in central China. Behav Brain Funct. 2012;8:57. doi: 10.1186/1744-9081-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grace ME, Newman KM, Scheinker V, Berg-Fussman A, Grabowski GA. Analysis of human acid beta-glucosidase by site-directed mutagenesis and heterologous expression. J Biol Chem. 1994;269(3):2283–2291. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used for GBA1 gene sequencing. The exons and the exon-intron boundaries of both the genes were bidirectionally sequenced using the given set of primers. (DOC 32 kb)
Biochemical analysis of the Gaucher Disease patients. The plasma chitotriosidase enzyme activity and β-Glucosidase enzyme activity were checked using the standard protocol. (DOCX 17 kb)
In silico analysis of the functional effect of the variants identified in the patients with Gaucher disease. The in silico tools predicting the effect of DNA variants, amino acid substitution, non-coding variants, and coding non-synonymous variants were employed to predict the functional effect of the variants identified in the given study. (DOCX 20 kb)
ClinVar Accession ID of the variants generated in the given study. The variants identified through Sanger sequencing are reported in NCBI ClinVar database. The file provides accession ID and the links to an individual variant. (DOCX 14 kb)
Data Availability Statement
The dataset generated and/or analyzed during the current study is available in the ClinVar repository (see Additional file 4).