Abstract
Background
Polymyxin B and E (colistin) have been pivotal in the treatment of XDR Gram-negative bacterial infections; however, resistance has emerged. A structurally related lipopeptide, octapeptin C4, has shown significant potency against XDR bacteria, including polymyxin-resistant strains, but its mode of action remains undefined.
Objectives
We sought to compare and contrast the acquisition of resistance in an XDR Klebsiella pneumoniae (ST258) clinical isolate in vitro with all three lipopeptides to potentially unveil variations in their mode of action.
Methods
The isolate was exposed to increasing concentrations of polymyxins and octapeptin C4 over 20 days. Day 20 strains underwent WGS, complementation assays, antimicrobial susceptibility testing and lipid A analysis.
Results
Twenty days of exposure to the polymyxins resulted in a 1000-fold increase in the MIC, whereas for octapeptin C4 a 4-fold increase was observed. There was no cross-resistance observed between the polymyxin- and octapeptin-resistant strains. Sequencing of polymyxin-resistant isolates revealed mutations in previously known resistance-associated genes, including crrB, mgrB, pmrB, phoPQ and yciM, along with novel mutations in qseC. Octapeptin C4-resistant isolates had mutations in mlaDF and pqiB, genes related to phospholipid transport. These genetic variations were reflected in distinct phenotypic changes to lipid A. Polymyxin-resistant isolates increased 4-amino-4-deoxyarabinose fortification of lipid A phosphate groups, whereas the lipid A of octapeptin C4-resistant strains harboured a higher abundance of hydroxymyristate and palmitoylate.
Conclusions
Octapeptin C4 has a distinct mode of action compared with the polymyxins, highlighting its potential as a future therapeutic agent to combat the increasing threat of XDR bacteria.
Introduction
Infections by XDR bacteria are an increasing concern due to the lack of effective antibiotics, thereby resulting in high mortality.1,2 Common therapeutic interventions include fosfomycin, tigecycline and polymyxins; however, resistance has emerged.2–9 Few new antibiotics or combinations are clinically available to combat XDR infections, hence it is desirable to discover novel classes.10
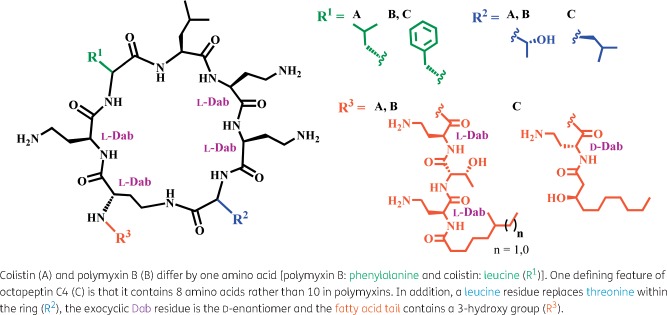
Octapeptins are structurally similar to the polymyxins, with both lipopeptide classes consisting of a cyclic heptapeptide ring and linear tail capped with a fatty acid, containing multiple positively charged diaminobutyric acid (Dab) residues (Figure 1).11–13 These Dab residues are critical for interactions of the polymyxins with the basal component of LPS, lipid A. Their mode of action involves the initial binding to lipid A, displacement of magnesium (Mg2+) and calcium (Ca2+), outer membrane permeabilization, leakage of cytoplasmic contents and subsequent cell death; however, the exact mechanism is yet to be discerned.14,15 Polymyxin resistance leads to modification of the phosphate groups on lipid A with 4-amino-4-deoxyarabinose (Ara4N) and/or phosphoethanolamine (pEtN). This reduces polymyxin binding by removing the negative phosphate that attracts the cationic Dab residues, stabilizing the outer membrane and negating the infiltration of this antibiotic class.16,17 Constitutive up-regulation of this pathway is achieved through chromosomal variations in the two-component regulatory systems (TCSs) crrAB, pmrAB and phoPQ and the negative regulator mgrB in Klebsiella pneumoniae.8,9,18 The structurally similar octapeptins retain most of the key binding motifs and might be expected to employ a similar mode of action. The most significant structural difference between the polymyxins and octapeptins is a truncated linear exocyclic peptide (one residue instead of three) linked to a β-hydroxy-fatty acid (instead of an alkyl fatty acid, a critical component in polymyxin activity19) in the octapeptins.11–13 More minor variations include l-Dab to d-Dab and l-Thr to l-Leu substitutions. Despite their similarity, prior research has revealed that octapeptins retain the ability to kill polymyxin-resistant (Pmx-R) bacteria and several exhibit broad-spectrum activity (against Gram-positive bacteria, fungi and protozoa).11,20,21 Interestingly, several octapeptin in vivo mouse studies have shown activity against Pmx-R infections and less toxicity compared with polymyxins.13,22–24
Figure 1.
Structural comparison between the three lipopeptide antibiotics used in this study. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
We have recently reported the first syntheses of octapeptin C411 and A3,22 followed by detailed biological characterization of octapeptin C4 that demonstrates its potential as a new ‘last-resort’ antibiotic.23 In view of the limited understanding of the mechanism by which octapeptins target bacteria, and to help advance their preclinical development, we sought to investigate the differences driving development of octapeptin C4 and polymyxin resistance at a genetic level. Two studies have previously investigated the acquisition of resistance to octapeptins. One was performed using EM49 (a mixture of octapeptin classes A and B), which exhibited no increase in resistance after 10 passages for Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus and Candida albicans.25 The other investigated lipid A modifications in octapeptin C4-resistant P. aeruginosa isolates obtained from a subculture surviving a single overnight treatment at 2 or 32 mg/L.23
The ST258 lineage of K. pneumoniae is commonly involved in outbreaks as it frequently harbours carbapenem resistance.26–30 We have previously used WGS to investigate the acquisition of polymyxin resistance in an epidemic lineage of K. pneumoniae ST258 isolated from a Greek hospital.26 An XDR polymyxin-susceptible isolate, 20_GR_12, was selected from this cohort and represents a strain in which a ‘last-resort’ antibiotic is employed. This isolate was passaged with an increasing concentration of polymyxins or octapeptin C4 for 20 days followed by antimicrobial susceptibility testing, WGS, complementation assays and analysis of lipid A composition to elucidate the potential mode of action.
Materials and methods
Bacterial strains and growth conditions
The clinical polymyxin-susceptible XDR K. pneumoniae ST258 (closely related to the NJST258_2 clade) isolate, 20_GR_12, was sourced through Hygeia General Hospital, Athens, Greece as previously described.26 Cultures were grown in LB and, for single colony isolation, cultures were grown on either LB or nutrient agar (NA) plates.
Antimicrobial susceptibility testing
MIC was determined by the broth microdilution method according to CLSI guidelines.31 Cultures were grown in CAMHB and, to assess cross-resistance of day 20 isolates, broth was supplemented with the concentration of antibiotic tolerated at that timepoint (Table S1, available as Supplementary data at JAC Online). Clinical breakpoints were determined in accordance with CLSI guidelines32 and, for tigecycline, as per EUCAST (Version 8.0, 2018) (see http://www.eucast.org). As no clinical breakpoint has been reported for octapeptin C4, an MIC ≥32 mg/L was defined as resistant.
Selection of resistance
A single colony of 20_GR_12 was selected and grown overnight at 37°C, shaking at 220 rpm. This culture was grown to log phase (OD600 = 0.4–0.6) and plated into three separate 96-well polystyrene, non-treated plates (Sigma–Aldrich) with colistin, polymyxin B or octapeptin C4 (n = 6). Following overnight incubation, the well that harboured the densest growth (OD600 ≥ 1) underwent a 1:1000 dilution and was transferred to a new plate with the concentration range adjusted accordingly. The highest concentration used for the polymyxins was 128 mg/L, and 32 mg/L for octapeptin C4. This process was performed for 20 days with the following 5 days of no antibiotic exposure. At day 20, the culture was further diluted (1:1000) and placed in non-supplemented broth to be incubated overnight, followed by an MIC test to evaluate resistance stability. Fold-change significance was determined via GraphPad Prism 7 with a one-way ANOVA with a Tukey’s multiple comparisons test where significance was P < 0.05.
Lipid A modifications
Lipid A was extracted using the ammonium hydroxide–isobutyric acid protocol as previously described.33 Day 20 cultures were grown overnight in LB supplemented with antibiotic (Table S1). Overnight inocula were subcultured (1:100) into 100 mL of LB broth and grown to an OD600 = 0.8–1. Cultures were pelleted (15008 g, 20 min, 4°C), washed with 1× PBS (15008 g, 15 min, 4°C) and freeze-dried.33 Ten milligrams of lyophilized cells was suspended in isobutyric acid:ammonium hydroxide (5:3 v/v) at 100°C for 4 h, supernatants isolated by centrifugation (12470 g, 15 min), diluted with water (1:1 v/v) and lyophilized. Extracts then underwent two methanol washes (1180 g, 15 min) and extracted lipid A (1 mg/L) was solubilized in methanol (5 mM ammonium acetate). Samples were infused at a low rate of 5 μL/min into a QSTAR Elite (Applied Biosystems) hybrid quadrupole Time-of-Flight (TOF) mass spectrometer. Data were exported from Analyst (SCIEX), normalized to the highest mass intensity and graphed in GraphPad Prism 7.
DNA extractions and library preparation
Glycerol stocks from day 20 isolates were grown on NA plates. Single colonies were grown in antibiotic supplemented broth (Table S1), incubated overnight and DNA was extracted using the DNeasy Blood and Tissue Kit (QIAGEN) according to the manufacturer’s guidelines. Two colonies were selected from day 0 and four colonies from four replicates per treatment group. Quantification of DNA was acquired using Qubit®3.0 (Thermo Fisher Scientific) and 1 ng of DNA underwent library preparation with the Nextera XT kit (Illumina) as per the manufacturer’s instructions. Quality control was checked with a 2100 Bioanalyzer (Agilent Technologies) and LabChip GX (PerkinElmer).
Sequencing and analysis
Libraries were sequenced on an Illumina NextSeq with 150 bp paired-end sequencing reads with ≥95× coverage with the exception of CST_2 (colony 1) (48×). Trimmomatic34 was used to trim paired-end reads and SPAdes v3.10.1 implemented for assembly.35 Annotation of assembled genomes was accomplished using the Rapid Annotation using Subsystem Technology (RAST).36 The Centre for Genomic Epidemiology (CGE) tools were implemented to delineate laterally acquired resistance genes (ResFinder 3.0)37 and plasmids (PlasmidFinder 1.3).38 Reads were aligned using BWA-MEM39, analysed through FreeBayes40 and impact of change determined through SnpEff.41 Nucleotide sequences have been deposited under NCBI BioProject PRJNA415530 (www.ncbi.nlm.nih.gov/bioproject/415530).
Complementation assays
Genes speculated to cause resistance underwent complementation as previously described.26,42 Briefly, genes harbouring a potential variation contributing to resistance were amplified using the 2X Phusion HF master mix (Thermo Fisher) with the primers listed in Table S2. The gene was cloned into the pCR-BluntII-TOPO using the Zero Blunt TOPO PCR cloning kit (Invitrogen). The plasmid was transformed in electrocompetent E. coli TOP10 via electroporation, grown on Mueller-Hinton agar (MHA) (kanamycin: 50 mg/L) at 37°C and plasmids extracted using the QIAprep Spin Miniprep Column kit (QIAGEN). Plasmids were transformed into the initial susceptible strain (20_GR_12) and incubated on MHA (zeocin: 1500 mg/L). Furthermore, the WT gene was amplified from the initial strain and placed into the resistant day 20 isolates followed by MIC determination.
Results
Rapid resistance acquisition for polymyxins dissimilar to octapeptin C4
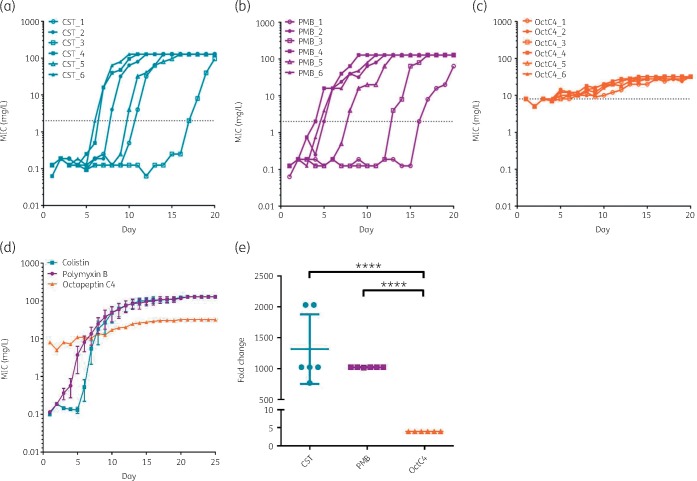
Selected resistance over the 20 day time course revealed significant variability between polymyxins and octapeptin C4 (Figure 2d). Initially, an MIC of 0.125 mg/L was measured for both colistin and polymyxin B. The majority of replicates treated with the polymyxins had a clinical resistance phenotype of >2 mg/L by day 10 (Figure 2a and b). Every replicate had a rapid escalation in MIC to >64 mg/L, generally within 5 days from the point when the MIC reached 0.5 mg/L. In contrast, octapeptin C4 resistance progressed steadily amongst replicates (Figure 2c) with only a 4-fold increase (from an initial MIC of 8 mg/L) compared with a ≥1000-fold increase for the polymyxins (Figure 2e). Although octapeptin C4 resistance remained stable, the extent of growth started to diminish during the last passages in the presence of 32 or 16 mg/L octapeptin C4.
Figure 2.
Acquired resistance in XDR K. pneumoniae over time for polymyxins and octapeptin C4. (a) Colistin. (b) Polymyxin B. (c) Octapeptin C4. (d) Overall comparison of acquired resistance for 20 day antibiotic exposure and the following 5 days without exposure (mean ± SEM, n = 6). (e) Fold change of colistin, polymyxin B and octapeptin C4 day 0 and day 20 MICs (mean ± SD) (****P < 0.001). The broken lines represent breakpoints (2 mg/L for polymyxins, 8 mg/L set for octapeptin C4 to highlight divergence from day 0). Highest concentration used was 128 mg/L for polymyxins and 32 mg/L for octapeptin C4. CST, colistin; PMB, polymyxin B; OctC4, octapeptin C4. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Lack of cross-reactivity and reduction of resistance in octapeptin C4-selected isolates
Day 20 isolate MICs of a broad array of antibiotic classes were determined to evaluate whether acquired resistance conferred cross-resistance or resulted in regained susceptibility (Table 1). No cross-reactivity was apparent between polymyxins and octapeptin C4. Non-susceptibility to amoxicillin, aztreonam, ceftriaxone, ciprofloxacin, piperacillin and trimethoprim was ubiquitous amongst treatment groups. Chloramphenicol resistance was observed in the initial isolate but normally diminished over the time course across treatment groups. In some instances, cefepime susceptibility was restored (replicates OctC4_3, OctC4_4 and OctC4_6). These replicates also regained susceptibility to meropenem, as did PMB_2. Replicate OctC4_2 also exhibited susceptibility to tetracycline and tigecycline but replicate OctC4_4 showed variability towards these antibiotics, with both resistant and susceptible MICs depending on the colonies selected.
Table 1.
MICs of several antibiotic classes for day 20 replicates compared with the initial isolate
| Straina | MIC (mg/L)b |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CST | PMB | OctC4 | AMX | ATM | FEP | CRO | CHL | CIP | GEN | MEM | PIP | TET | TGC | TMP | |
| Initial | ≤0.25S | ≤0.125S | 8 | >64R | >64R | ≥16R | >64R | ≥32R | >64R | ≤4S | ≥32R | >64I | >64R | ≤2S,I | >64R |
| CST_1 | >128R | >128R | ≤8 | >64R | >64R | ≥16R | >64R | ≤8S | ≥32R | 4S | ≥32R | >64I | >64R | ≤4S,R | >64R |
| CST_2 | >128R | >128R | ≤4 | >64R | >64R | >64R | >64R | 4S | ≥32R | ≤4S | ≥32R | >64I | >64R | 2I | >64R |
| CST_3 | ≥128R | ≥128R | ≤4 | >64R | >64R | >64R | >64R | 8S | >64R | ≤4S | >64R | >64I | >64R | ≤4I,R | >64R |
| CST_4 | >128R | >128R | ≤8 | >64R | >64R | >64R | >64R | ≤8S | >64R | ≤4S | >64R | >64I | >64R | ≤4I,R | >64R |
| CST_5 | >128R | >128R | 2 | >64R | >64R | >64R | >64R | ≤8S | >64R | ≤4S | >64R | >64I | >64R | ≤4I,R | >64R |
| CST_6 | >128R | >128R | ≤8 | >64R | >64R | >64R | >64R | 8S | >64R | ≤4S | >64R | >64I | >64R | ≥4R | >64R |
| PMB_1 | 128R | 128R | ≤4 | >64R | >64R | >64R | 32R | 4S | 32R | ≤4S | >64R | >64I | >64R | ≤4I,R | >64R |
| PMB_2 | >128R | >128R | 8 | >64R | >64R | ≤8I | >64R | 4S | >64R | ≤4S | ≤0.25S | >64I | >64R | ≤2S,I | >64R |
| PMB_3 | >128R | >128R | 4 | >64R | >64R | ≥32R | >64R | 8S | >64R | 2S | ≥32R | >64I | >64R | 2I | >64R |
| PMB_4 | >128R | >128R | 4 | >64R | >64R | ≥8I,R | ≥32R | ≤2S | 48R | 2S | ≥8R | >64I | >64R | 2I | >64R |
| PMB_5 | >128R | >128R | ≤8 | >64R | >64R | ≥8I,R | ≥32R | ≤8S | >64R | ≤4S | ≥2I,R | >64I | >64R | ≤2S,I | >64R |
| PMB_6 | >128R | >128R | ≤8 | >64R | >64R | ≥16R | >64R | 8S | >64R | ≤4S | ≥32R | >64I | >64R | ≤4I,R | >64R |
| OctC4_1 | 0.25S | ≤0.25S | 32 | >64R | >64R | ≥16R | >64R | 8S | >64R | ≤2S | ≥32R | >64I | >64R | ≤2S,I | >64R |
| OctC4_2 | 0.25S | 0.25S | 32 | >64R | >64R | ≥8I,R | >64R | 8S | >64R | ≤2S | ≥32R | >64I | ≤2S | ≤1S | ≥8R |
| OctC4_3 | ≤0.5S | 0.25S | 32 | >64R | >64R | ≤4S,I | 32R | 8S | >64R | ≤4S | ≤0.25S | >64I | >64R | 2I | >64R |
| OctC4_4 | ≤0.5S | 0.25S | >32 | >64R | >64R | ≤4S,I | 32R | 8S | >64R | 1S | ≤0.25S | >64I | ≥2S,R | ≤2S,I | ≥4R |
| OctC4_5 | 0.5S | 0.5S | 32 | >64R | >64R | >64R | >64R | ≤8S | >64R | 2S | >64R | >64I | >64R | 2I | >64R |
| OctC4_6 | ≤0.5S | ≤0.5S | 32 | >64R | >64R | ≤4S,I | ≥32R | ≤16S, I | >64R | 2S | ≤0.25S | >64I | >64R | 2I | >64R |
Resistance determined as per CLSI guidelines, except EUCAST guideline used for tigecycline (S, susceptible; I, intermediate; R, resistant). Fluctuations in MIC values (n = 4) are displayed by two letters defining the resistance level. Grey shading indicates resistant MIC values. Resistance to octapeptin C4 defined at an MIC ≥32 mg/L, as no clinical breakpoint has been reported for this compound.
The initial polymyxin-susceptible isolate (20_GR_12) and this strain subjected to 20 days of treatment with colistin (CST), polymyxin B (PMB) or octapeptin C4 (OctC4) (_1, _2, _3, _4, _5 and _6 indicate replicate numbers).
MIC determined for: CST, colistin; PMB, polymyxin B; OctC4, octapeptin C4; AMX, amoxicillin; ATM, aztreonam; FEP, cefepime; CRO, ceftriaxone; CHL, chloramphenicol; CIP, ciprofloxacin; GEN, gentamicin; MEM, meropenem; PIP, piperacillin; TET, tetracycline; TGC, tigecycline; and TMP, trimethoprim.
Octapeptin C4-resistant isolates harbour an increase in hydroxymyristate and palmitoylate dissimilar to Ara4N lipid A modifications in Pmx-R strains
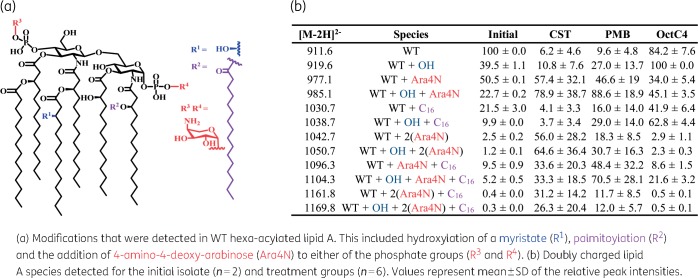
In the initial isolate, MS/MS analysis of extracted lipid A fractions showed that the major singly charged peak was at m/z 1824.2, which corresponded to a hexa-acylated lipid A species composed of two phosphate groups, two glucosamines and four 3-hydroxy-myristoyl groups (3-OH-C14), with two of these further acylated with myristate (C14) (Figure 3a and Figure S1). This mass correlated with a doubly charged species of m/z 911.6 with greater intensity, herein designated as the WT lipid A. Lesser quantities of various modifications accompanied the WT lipid A, including a hydroxyl modification of a myristate (m/z 919.6, WT + C14:OH), palmitoylation (m/z 1030.7, WT + C16) and Ara4N (m/z 977.1, WT + Ara4N) (Figure 3b). The detection of Ara4N species may indicate the initial strain is heteroresistant, with a resistant subpopulation existing within a phenotypically susceptible isolate. Pmx-R isolates showed near complete loss of WT lipid A and fortification of Ara4N on phosphate groups, mainly in hydroxymyristate species [m/z 985.1, WT + C14:OH + Ara4N; m/z 1042.7, WT + 2(Ara4N); m/z 1050.7, WT + C14:OH + 2(Ara4N)] (Figure 3b, Figure S2 and Figure S3). The other commonly reported resistance modification to lipid A, pEtN (m/z 973.2), was never observed. Lipid A from the octapeptin C4-selected isolates differed from the Pmx-R isolates and was similar to the WT profile (major peak of the hydroxymyristate derivative), but with a significant 5-fold increase in representation of palmitoylation (Figure 3b and Figure S4). The Ara4N modification was enhanced compared with WT, but not to the extent seen with Pmx-R isolates.
Figure 3.
Lipid A modifications identified after 20 days of exposure to colistin, polymyxin B or octapeptin C4. CST, colistin; PMB, polymyxin B; OctC4, octapeptin C4. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Plasmid loss associated with octapeptin C4 resistance
To ascertain the genetic basis for resistance and subsequent phenotypic traits, four day 20 replicates were selected from each treatment group. Clonal expansion of genomic variations was monitored by selecting four colonies per replicate along with two colonies from the initial isolate. The initial isolate harboured resistance genes targeting aminoglycosides, β-lactams, fosfomycin, quinolones, sulphonamides, tetracycline and trimethoprim (Table 2). Five plasmid replicons were identified including ColRNAI, IncFIB(K)-Kpn3, IncFII(K), IncN and IncX3. The only replicates with other resistance gene alterations in polymyxin-treated groups were PMB_2 [loss of aph(3′)-Ia, blaKPC-2, blaOXA-9 and IncX3 replicon, n = 4] and PMB_3 (loss of IncX3, n = 1) (Table 2). High variability was observed for octapeptin C4-exposed replicates including the absence of aph(3′)-Ia, aph(3′′)-Ib, aph(6)-Id, blaKPC-2, blaOXA-9, blaTEM-1B, sul2, tet(A) and dfrA14. Furthermore, plasmid replicon loss was apparent in three of the four replicates including IncFIB(K)-Kpn3, IncFII(K) and IncN.
Table 2.
Detection of acquired resistance genes and plasmid replicons compared with the initial isolate
| Straina | Antibiotic class impactedb |
Plasmidc |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A |
B |
F |
Q |
S |
T |
Tr |
|||||||||||||||||
| aac(6')- Ib | aph(3')- Ia | aph(3′′)- Ib | aph(6)- Id | blaKPC-2 | blaKPC-9 | blaLEN-12 | blaOXA-9 | blaSHV-12 | blaTEM-1A | blaTEM-1B | fosA | aac(6')- Ib-cr | oqxA | oqxB | sul2 | tet(A) | dfrA14 | I | II | III | IV | V | |
| Initial | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |||
| CST_1 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 1 | 3 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | ||
| CST_2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | |||
| CST_3 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | |||
| CST_4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | |||
| PMB_1 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 1 | 3 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | ||
| PMB_2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | |||||||
| PMB_3 | 4 | 3 | 4 | 4 | 3 | 3 | 3 | 1 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 3 | ||
| PMB_4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 1 | 3 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | ||
| OctC4_1 | 4 | 4 | 4 | 4 | 1 | 3 | 4 | 1 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | ||
| OctC4_2 | 4 | 4 | 3 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | |||||||||
| OctC4_3 | 4 | 4 | 2 | 2 | 4 | 3 | 4 | 1 | 2 | 2 | 4 | 4 | 4 | 4 | 2 | 2 | 2 | 4 | 4 | 4 | 2 | 4 | |
| OctC4_4 | 4 | 1 | 1 | 4 | 1 | 4 | 4 | 4 | 4 | 1 | 1 | 1 | 4 | 1 | 4 | ||||||||
Number represents the presence of the gene or plasmid, of four colonies, harbouring this attribute.
The initial polymyxin-susceptible isolate (20_GR_12) and this strain subjected to 20 days of treatment with colistin (CST), polymyxin B (PMB) or octapeptin C4 (OctC4) (_1, _2, _3 and _4 indicate replicate numbers).
Acquired resistance genes as determined by ResFinder 3.0. (≥90% identity and ≥60% length) for: A, aminoglycoside; B, β-lactam; F, fosfomycin; Q, quinolone; S, sulphonamide; T, tetracycline; and Tr, trimethoprim.
Plasmid replicons detected by PlasmidFinder 1.3 (≥95% identity): I, ColRNAI; II, IncFIB(K)-Kpn3; III, IncFII(K); IV, IncN; and V, IncX3.
Chromosomal variations in LPS pathways associated with polymyxin resistance whilst phospholipid (PL) transport associated with octapeptin C4 resistance
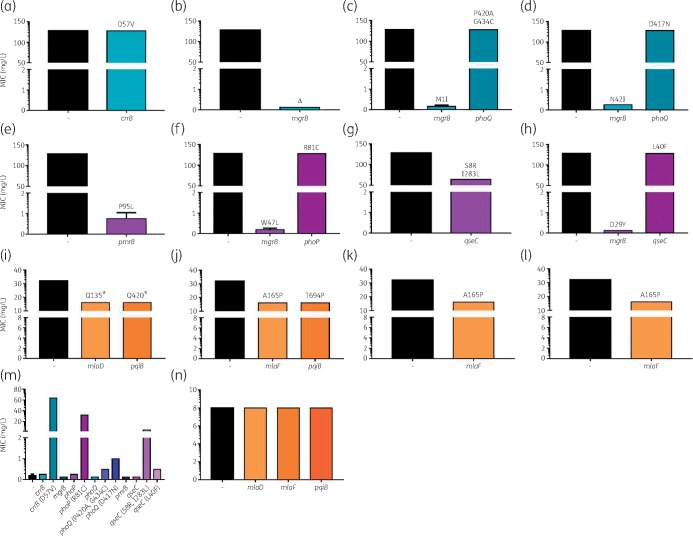
Genomic alterations identified in replicates treated with polymyxin or octapeptin C4 differed significantly. In Pmx-R replicates, affected genes were predominantly associated with LPS processing and lipid A modifications, including crrB, hepIII, lptC, mgrB, pmrB, phoPQ and yciM (Table 3). An additional TCS gene, qseC, was also disrupted in PMB_3 (S8R, I283L) and PMB_4 (L40F). Although similar genes were impacted across replicates, the mutation positions differed. Additionally, an accumulation of variations in LPS pathways was apparent within a single replicate. Variants were observed in all four colonies indicating clonal expansion. Complementation assays were conducted to unveil these genes’ contributions to resistance (Figure 4). Polymyxin susceptibility was restored in CST_2 (complete deletion of mgrB), CST_3 (M1I), CST_4 (N42I), PMB_2 (W47L) and PMB_4 (D29Y) once complemented with pTOPO-mgrB (Figure 4b–d, f and h). The PmrB (P95L) variant in PMB_1 was validated to contribute to resistance (Figure 4e). Alterations in CrrB (D57V), PhoP (R81C) and QseC (S8R, I283L) were confirmed to cause resistance once these mutated genes were introduced into the initial strain (Figure 4m). Subtle increases in polymyxin MIC were detected for PhoQ (P420A, G434C), PhoQ (D417N) and QseC (L40F) but did not surpass the breakpoint MIC (Figure 4m). This confirms the presence of multiple resistance-conferring mutations in a single isolate with several contributing to polymyxin tolerance.
Table 3.
Genomic alterations detected in day 20 resistant isolates
| Straina | Gene | Gene description | Nt changeb | Amino acid changec |
|---|---|---|---|---|
| CST_1 (4) | crrB | two-component hybrid sensor and regulator | A170T | D57V |
| CST_2 (4) | hepIII | LPS heptosyltransferase III | A238Δfs | R80Gtr |
| CST_2 (4) | mgrB | putative inner membrane protein | 1-144Δ | 1-47Δ |
| CST_3 (4) | mgrB | putative inner membrane protein | G3A | M1I |
| CST_3 (4) | phoQ | sensor protein | C1258G, G1300T | P420A, G434C |
| CST_4 (3) | epsJ | glycosyltransferase | T932G | L310STOP |
| CST_4 (3) | lptC | LPS export system protein | Δ498Afs | N166Ktr |
| CST_4 (4) | mgrB | putative inner membrane protein | G-41T, A125T | N42I |
| CST_4 (4) | phoQ | sensor protein | G1249A | D417N |
| PMB_1 (4) | pmrB | sensor protein | C284T | P95L |
| PMB_2 (4) | dnaJ | chaperone protein | A892C | T298P |
| PMB_2 (4) | mgrB | putative inner membrane protein | G140T | W47L |
| PMB_2 (4) | phoP | transcriptional regulatory protein | C241T | R81C |
| PMB_2 (4) | hepIII | LPS heptosyltransferase III | TGAAGAGACCCG153Δ | Y51STOP |
| PMB_3 (4) | qseC | sensory histidine kinase | GCCTGAGCCTGC17Δfs, A847C | S8R, I283L |
| PMB_4 (4) | mgrB | putative inner membrane protein | G85T | D29Y |
| PMB_4 (4) | qseC | sensory histidine kinase | CTGGATAAGCTG118Δfs | L40F |
| PMB_4 (4) | yciM | LPS regulatory protein | T128G | V43G |
| OctC4_1 (4) | mlaD | uncharacterized ATP-binding cassette (ABC) transporter, periplasmic component | C403T | Q135STOP |
| OctC4_1 (4) | pqiB | paraquat-inducible protein B | C1258T | Q420STOP |
| OctC4_1 (4) | traH | conjugal transfer protein | G417T | M139I |
| OctC4_2 (4) | pqiB | paraquat-inducible protein B | A2080C | T694P |
| OctC4_2 (2) | rpsA | small subunit (SSU) ribosomal protein S1p | T1031A | L344Q |
| OctC4_3 (2) | hinT | YcfF/hinT protein: purine nucleoside phosphoramidase | Δ240Cfs | D81Rtr |
| OctC4_4 (2) | azoR | flavin mononucleotide (FMN)-dependent NADH-azoreductase | T152A | L51Q |
| OctC4_2 (4), OctC4_3 (4), OctC4_4 (4) | mlaF | uncharacterized ABC transporter, ATP-binding protein | GCCGC493Δfs | A165Ptr |
Strain represented as: treatment group (colistin, CST; polymyxin B, PMB; octapeptin C4, OctC4)_replicate number (number of colonies impacted from the four selected).
Nt variations present in ≥90% of reads and ≥50× coverage compared with the initial strain, 20_GR_12. Δ symbolizes a deletion, – in front of the nt position indicates an alteration upstream and fs represents a frameshift mutation.
The introduction of a truncation in the protein downstream of the alteration is noted as tr.
Figure 4.
Complementation assays to delineate contribution to resistance of variations detected in day 20 treated strains. (a–d) Colistin treatment groups complemented with WT gene. (a) CST_1 with pTOPO-crrB. (b) CST_2 with pTOPO-mgrB. (c) CST_3 with pTOPO-mgrB or pTOPO-phoQ. (d) CST_4 with pTOPO-mgrB or pTOPO-phoQ. (e–h) Polymyxin B treatment groups complemented with WT gene. (e) PMB_1 with pTOPO-pmrB. (f) PMB_2 with pTOPO-mgrB or pTOPO-phoP. (g) PMB_3 with pTOPO-qseC. (h) PMB_4 with pTOPO-mgrB or pTOPO-qseC. (i–l) Octapeptin C4 treatment groups complemented with WT gene. (i) OctC4_1 with pTOPO-mlaD or pTOPO-pqiB. (j) OctC4_2 with pTOPO-mlaF or pTOPO-pqiB. (k) OctC4_3 with pTOPO-mlaF. (l) OctC4_4 with pTOPO-mlaF. (m) 20_GR_12, the initial strain, complemented with WT genes and genes harbouring mutations potentially causing polymyxin resistance. (n) Complementation of octapeptin C4 resistance-associated WT genes in 20_GR_12. A - symbol indicates that no complementation was conducted and represents the initial MIC. The y-axis split signifies the breakpoint for polymyxins (2 mg/L) and initial MIC for octapeptin C4 (8 mg/L). Values represent mean ± SD (n = 4). Text above bars (a–l) indicates the amino acid change(s) in the selected resistant isolate. Δ represents a complete gene deletion and * is a stop codon. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Several octapeptin C4 replicates harboured changes in mlaDF, pqiB and traH in all four colonies. Additional genes that were altered in two colonies per replicate included azoR, hinT and rpsA. Strikingly, mlaF (A165P) was impacted in three different octapeptin C4 replicates at the same position (Table 3). Complementation assays that introduced pTOPO-mlaD, -mlaF or -pqiB into octapeptin C4-resistant replicates reduced the MIC by 2-fold; however, consistently only partial growth was observed at 8 mg/L (Figure 4i–l). Introduction of WT genes into the initial isolate revealed that the vector and gene did not influence the MIC and confirmed that these alterations were responsible for the resistance observed (Figure 4m and n).
Discussion
Polymyxins can lead to high levels of resistance during therapeutic use, likely driven by suboptimal exposure in the clinic due to the risk of nephrotoxicity.43 Resistance in K. pneumoniae appears to be stable and incurs a minimal fitness cost.44,45 These clinical characteristics were reflected in our study whereby once the isolate could tolerate 0.5 mg/L of either colistin or polymyxin B, the clinical breakpoint was vastly exceeded within 48 hours, well within the duration of clinical antibiotic therapy. This rapid progression of resistance was not observed for octapeptin C4, in which only comparatively minor increases in MIC were observed. The slow progression in resistance profile could be an advantageous characteristic of octapeptin C4 as a potential clinical intervention.
Following 20 days of increasing sublethal antibiotic exposure, no cross-resistance was apparent between polymyxins and octapeptin C4. Colistin and polymyxin B resulted in similar resistance profiles with the only deviation seen in sample PMB_2, in which susceptibility to meropenem was regained. This was due to the absence of blaKPC-2 and blaOXA-9, along with the homogeneous loss of IncX. Clinically, meropenem is being used in combination with polymyxins, and these results suggest that, in some cases, meropenem may overcome polymyxin resistance.46,47 Furthermore, previous research has identified the loss of blaKPC plasmids in Pmx-R clinical isolates and suggests that this loss is due to a potential fitness cost.48 Our results also show plasmid loss in octapeptin C4-exposed replicates, and this corresponded to a reduction in resistance to cefepime, meropenem and tetracycline. However, these findings are preliminary and whether this resembles a fitness cost associated with octapeptin C4 exposure or results from repeated passaging under no selective pressure warrants further investigation. Similarly, chloramphenicol susceptibility was restored in day 20 isolates exposed to polymyxins or octapeptin C4. Whether this is the result of repeated passaging or a novel loss of chloramphenicol resistance via gaining resistance to the three lipopeptides is yet to be discerned.
The mutations observed in selected Pmx-R ST258 strains can be compared to those we have previously identified in closely related Pmx-R clinical ST258 isolates (2_GR_12, 4_GR_12, 10_GR_13, 13_GR_14 and 14_GR_14).26 Similarly, the vast majority of resistance was attributed to mgrB (60%), albeit not via an IS element disruption. Additional mutations were identified in phoPQ accompanying the mgrB disruption, which was also apparent in this study [CST_3 (mgrB: M1I, phoQ: P420A, G434C), CST_4 (mgrB: N42I, phoQ: D417N), PMB_2 (mgrB: W47L, phoP: R81C)]. Other mutations in crrB, mgrB, pmrB, phoPQ and yciM have previously been described in Pmx-R strains.17,18,49 Overall, this indicates that in vitro experiments can give rise to genomic changes similar to those observed in the clinic. The notion that one alteration in a TCS drives resistance, the circumstance for the majority of clinical isolates, is well accepted.50 However, our findings contradict this concept although the high concentration of polymyxin used for our in vitro resistance selection could be influencing this finding.
We also identified alterations in another TCS, QseBC, which is known to facilitate cross-talk with PmrAB in E. coli.51 In E. coli, PmrB acts as a non-cognate partner to the QseBC TCS and has the ability to not only phosphorylate PmrA, but also QseB. The absence of QseC was shown to impact virulence due to the accumulation of phosphorylated QseB and, in particular, alterations in the histidine kinase domain attenuates its ability to dephosphorylate QseB.51,52 Furthermore, the deletion of qseC and pmrA, promoting phosphorylation of QseB by PmrB, stimulated tolerance to polymyxin B.53 This signalling pathway is not well-characterized in K. pneumoniae. We observed partial tolerance to PMB when a frameshift mutation was apparent at nt 118; however, full resistance in PMB_4 was promoted by alterations in mgrB (D29Y) and yciM (V43G), which has recently been identified to cause resistance.49 Similarly, PMB_3 harboured a frameshift in qseC (GCCTGAGCCTGC17Δfs), although an additional I283L change in the histidine kinase region resulted in an MIC of 4 mg/L. This did not explain the full resistance profile exhibited by PMB_3 and due to the presence of both alleles during complementation the true extent of resistance cannot be deduced. Considering PMB_3 still resulted in the addition of Ara4N to lipid A, we speculate that due to the perturbation in the QseC kinase this is increasing the accumulation of phosphorylated QseB and allows for the up-regulation of transcriptional targets. Subsequent transcription could be activating PmrA, similar to other TCSs in K. pneumoniae, allowing for the expression of the pmrHFIJKLM operon (Figure 5a).
Figure 5.
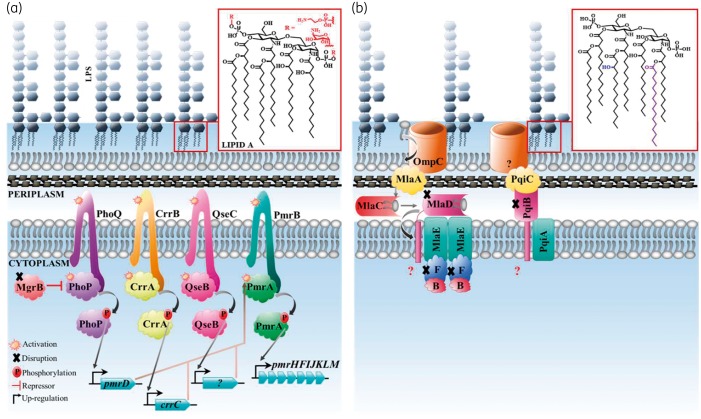
Proposed pathway associated with K. pneumoniae polymyxin and octapeptin C4 resistance observed in this study. (a) To facilitate resistance to polymyxins, genomic variations are acquired in TCSs. These encompass CrrAB, QseBC, PmrAB and PhoPQ with MgrB acting as a negative repressor. Once this pathway is activated during resistance, sensor histidine kinases (CrrB, QseC, PmrB and PhoQ) will phosphorylate response regulators (CrrA, QseB, PmrA and PhoP) and allow for the expression of target genes [crrC, unknown (?), pmrD and pmrHFIJKLM]. Disruptions in MgrB allow for the up-regulation of this pathway resulting in the expression of pmrHFIJKLM, which allows for Ara4N to be attached to phosphate groups on lipid A. The pEtN lipid A modification, facilitated via the pmrCAB operon, was not observed in this study. (b) The major disruptions identified during octapeptin C4 resistance were in the Mla and Pqi pathway. OmpC removes PLs from the outer membrane and transfers these to MlaA. PLs are transported across the periplasm via MlaC and transported to the MlaBDEF complex where the subsequent fate of PLs is unknown. An unknown porin complexes with PqiC to transport metabolites and potentially PLs across the periplasm via the PqiAB complex. Mutations in these pathways elevate the octapeptin C4 MIC and subsequently hydroxymyristate and palmitoylate are added to lipid A to potentially stabilize the outer membrane. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
The Mla pathway was impacted in octapeptin C4-exposed replicates. These genes are responsible for PL importation from the outer membrane.54 Removal of mlaC in E. coli was previously identified to increase the abundance of palmitoylated lipid A to stabilize the outer membrane, which correlated with the phenotype in our study. Similarly, prior research exposing P. aeruginosa to octapeptin C4 (32 mg/L) revealed an increase in palmitoylated lipid A.23 Literature reports have demonstrated that octapeptins have the capacity to bind to PLs55 and it is likely that octapeptin C4 utilizes this pathway in order to traverse to the outer membrane (Figure 5b). The involvement of PqiB in membrane integrity has only recently been characterized in E. coli.56 PqiB was identified to connect to PqiC and potentially deliver substrate(s) from the outer to inner membrane. The contribution of the Pqi and Mla pathway appeared to be additive when evaluating the MIC reduction in OctC4_1 and OctC4_2. Further genes impacted, though not homogeneously amongst the colonies, included rpsA (40S ribosomal protein), azoR (quinone reductase), traH (plasmid conjugal transfer protein) and hinT (purine nucleoside phosphoramidase), which may indicate several intracellular targets.57–60 The lack of mutations associated with Ara4N modifications to lipid A is consistent with the lipid A profile of the octapeptin C4-resistant isolates. This observation supports the hypothesis that the octapeptins work by a different mode of action compared with the polymyxins, one that does not require an initial binding to lipid A and explains the lack of cross-resistance between the two classes of lipopeptides. However, further studies are required to determine if this occurs ubiquitously for K. pneumoniae and if the same phenomenon is observed for other Gram-negative pathogens. The development of resistance in in vitro experiments entails several caveats compared with the clinical in vivo condition, including a limited regulation of cell growth and antibiotic concentration and exposure to a concentration of antibiotic that may not reflect an in vivo scenario.61,62 Hence, it would be of interest to discern the development of octapeptin resistance in vivo. Nonetheless, the slow progression of resistance, potential fitness cost if resistance develops and the alternative mechanism of infiltration of octapeptin C4 highlight the potential for octapeptins as future antibiotics.
Supplementary Material
Acknowledgements
We would like to acknowledge Dr Ilias Karaiskos and Dr Helen Giamarellou for supplying the clinical isolate and Dr Alysha Elliott for the initial characterization of the strain. We thank: Dr Alejandra Gallardo-Godoy, David Edwards and Ruby Pelingon for the synthesis and purification of octapeptin C4; Dr Tim Bruxner and Angelika Christ for their support with the sequencing; Alun Jones for his assistance with MS (IMB Mass Spectrometry Facility at the University of Queensland); and Dr Hannah Sidjabat for her assistance with the initial quality control of the strains.
Funding
This research was supported by National Health and Medical Research Council (NHMRC) grants APP1005350 and APP1045326, and National Institutes of Health grant R21AI098731/R33AI098731-03. M. A. C. is an NHMRC Principal Research Fellow (APP1059354). L. J. M. C. is an Australian Research Council Future Fellow (FT110100972). M. E. P. is an Australian Postgraduate Award scholar. M. A. T. B. is supported in part by a Wellcome Trust Strategic Award (104797/Z/14/Z).
Transparency declarations
M. A. C. currently holds a fractional Professorial Research Fellow appointment at the University of Queensland with his remaining time as CEO of Inflazome Ltd, a company with headquarters in Dublin, Ireland that is developing drugs to address clinical unmet needs in inflammatory disease by targeting the inflammasome. All other authors: none to declare.
Author contributions
M. E. P., M. A. C., L. J. M. C. and M. A. T. B. conceived this study. M. E. P., M. D. C. and D. G. performed the sequencing analysis and M. E. P., M. S. B. and S. R. performed the experiments. M. E. P. wrote the paper with input from the other authors.
References
- 1. Bhatt P, Tandel K, Shete V. et al. Burden of extensively drug-resistant and pandrug-resistant Gram-negative bacteria at a tertiary-care centre. New Microbes New Infect 2015; 8: 166–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karaiskos I, Giamarellou H.. Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin Pharmacother 2014; 15: 1351–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Falagas ME, Maraki S, Karageorgopoulos DE. et al. Antimicrobial susceptibility of multidrug-resistant (MDR) and extensively drug-resistant (XDR) Enterobacteriaceae isolates to fosfomycin. J Antimicrob Agents 2009; 35: 240–3. [DOI] [PubMed] [Google Scholar]
- 4. Arca P, Reguera G, Hardisson C.. Plasmid-encoded fosfomycin resistance in bacteria isolated from the urinary tract in a multicentre survey. J Antimicrob Chemother 1997; 40: 393–9. [DOI] [PubMed] [Google Scholar]
- 5. Liu YY, Wang Y, Walsh TR. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161–8. [DOI] [PubMed] [Google Scholar]
- 6. Tsuruoka T, Miyata A, Yamada Y.. Two kinds of mutants defective in multiple carbohydrate utilization isolated from in vitro fosfomycin-resistant strains of Escherichia coli K-12. J Antibiot (Tokyo) 1978; 31: 192–201. [DOI] [PubMed] [Google Scholar]
- 7. Sun Y, Cai Y, Liu X. et al. The emergence of clinical resistance to tigecycline. Int J Antimicrob Agents 2013; 41: 110–6. [DOI] [PubMed] [Google Scholar]
- 8. Cheng HY, Chen YF, Peng HL.. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J Biomed Sci 2010; 17: 60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cannatelli A, Giani T, D’Andrea MM. et al. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 2014; 58: 5696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bassetti M, Russo A, Carnelutti A. et al. Antimicrobial resistance and treatment: an unmet clinical safety need. Expert Opin Drug Saf 2018; 17: 669–80. [DOI] [PubMed] [Google Scholar]
- 11. Becker B, Butler MS, Hansford KA. et al. Synthesis of octapeptin C4 and biological profiling against NDM-1 and polymyxin-resistant bacteria. Bioorg Med Chem Lett 2017; 27: 2407–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Velkov T, Roberts KD, Li J.. Rediscovering the octapeptins. Nat Prod Rep 2017; 34: 295–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qian CD, Wu XC, Teng Y. et al. Battacin (octapeptin B5), a new cyclic lipopeptide antibiotic from Paenibacillus tianmuensis active against multidrug-resistant Gram-negative bacteria. Antimicrob Agents Chemother 2012; 56: 1458–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koike M, Iida K, Matsuo T.. Electron microscopic studies on mode of action of polymyxin. J Bacteriol 1969; 97: 448–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clausell A, Garcia-Subirats M, Pujol M. et al. Gram-negative outer and inner membrane models: insertion of cyclic cationic lipopeptides. J Phys Chem B 2007; 111: 551–63. [DOI] [PubMed] [Google Scholar]
- 16. Raetz CR, Reynolds CM, Trent MS. et al. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem 2007; 76: 295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Olaitan AO, Morand S, Rolain JM.. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 2014; 5: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng YH, Lin TL, Lin YT. et al. Amino acid substitutions of CrrB responsible for resistance to colistin through CrrC in Klebsiella pneumoniae. Antimicrob Agents Chemother 2016; 60: 3709–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vaara M, Siikanen O, Apajalahti J. et al. A novel polymyxin derivative that lacks the fatty acid tail and carries only three positive charges has strong synergism with agents excluded by the intact outer membrane. Antimicrob Agents Chemother 2010; 54: 3341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meyers E, Parker WL, Brown WE. et al. EM49: a new polypeptide antibiotic active against cell membranes. Ann N Y Acad Sci 1974; 235: 493–501. [DOI] [PubMed] [Google Scholar]
- 21. Chitty JL, Butler MS, Suboh A. et al. Antimicrobial octapeptin C4 analogues active against Cryptococcus species. Antimicrob Agents Chemother 2018; 62: pii=e00986-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han M-L, Shen HH, Hansford KA. et al. Investigating the interaction of octapeptin A3 with model bacterial membranes. ACS Infect Dis 2017; 3: 606–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Velkov T, Gallardo-Godoy A, Swarbrick JD. et al. Structure, function, and biosynthetic origin of octapeptin antibiotics active against extensively drug-resistant Gram-negative bacteria. Cell Chem Biol 2018; 25: 380–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blaskovich MAT, Pitt ME, Elliott AG. et al. Can octapeptin antibiotics combat extensively drug-resistant (XDR) bacteria? Expert Rev Anti Infect Ther 2018; 16: 485–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meyers E, Pansy FE, Basch HI. et al. EM49, a new peptide antibiotic. III. Biological characterization in vitro and in vivo. J Antibiot (Tokyo) 1973; 26: 457–62. [DOI] [PubMed] [Google Scholar]
- 26. Pitt ME, Elliott AG, Cao MD. et al. Multifactorial chromosomal variants regulate polymyxin resistance in extensively drug-resistant Klebsiella pneumoniae. Microb Genom 2018; doi:10.1099/mgen.0.000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Navon-Venezia S, Kondratyeva K, Carattoli A.. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 2017; 41: 252–75. [DOI] [PubMed] [Google Scholar]
- 28. Bowers JR, Kitchel B, Driebe EM. et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One 2015; 10: e0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kitchel B, Rasheed JK, Patel JB. et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 2009; 53: 3365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Munoz-Price LS, Poirel L, Bonomo RA. et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 2013; 13: 785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—Eleventh Edition: M07. CLSI, Wayne, PA, USA, 2018. [Google Scholar]
- 32.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing—Twenty-Sixth Edition: M100. CLSI, Wayne, PA, USA, 2016. [Google Scholar]
- 33. Choi MJ, Ko KS.. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother 2015; 59: 6763–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bankevich A, Nurk S, Antipov D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aziz RK, Bartels D, Best AA. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008; 9: 75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zankari E, Hasman H, Cosentino S. et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67: 2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carattoli A, Zankari E, García-Fernández A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014; 58: 3895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li H, Durbin R.. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009; 25: 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garrison E, Marth G.. Haplotype-based Variant Detection from Short-read Sequencing. arXiv 2012:1207.3907. [q-bio.GN]. [Google Scholar]
- 41. Cingolani P, Platts A, Wang Le L. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012; 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jayol A, Poirel L, Brink A. et al. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother 2014; 58: 4762–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pogue JM, Lee J, Marchaim D. et al. Incidence of and risk factors for colistin-associated nephrotoxicity in a large academic health system. Clin Infect Dis 2011; 53: 879–84. [DOI] [PubMed] [Google Scholar]
- 44. Arena F, Henrici De Angelis L, Cannatelli A. et al. Colistin resistance caused by inactivation of the MgrB regulator is not associated with decreased virulence of sequence type 258 KPC carbapenemase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 2016; 60: 2509–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee JY, Choi MJ, Choi HJ. et al. Preservation of acquired colistin resistance in Gram-negative bacteria. Antimicrob Agents Chemother 2015; 60: 609–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee GC, Burgess DS.. Treatment of Klebsiella pneumoniae carbapenemase (KPC) infections: a review of published case series and case reports. Ann Clin Microbiol Antimicrob 2012; 11: 32.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dickstein Y, Leibovici L, Yahav D. et al. Multicentre open-label randomised controlled trial to compare colistin alone with colistin plus meropenem for the treatment of severe infections caused by carbapenem-resistant Gram-negative infections (AIDA): a study protocol. BMJ Open 2016; 6: e009956.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wright MS, Suzuki Y, Jones MB. et al. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother 2015; 59: 536–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Halaby T, Kucukkose E, Janssen AB. et al. Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob Agents Chemother 2016; 60: 6837–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Baron S, Hadjadj L, Rolain J-M. et al. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents 2016; 48: 583–91. [DOI] [PubMed] [Google Scholar]
- 51. Guckes KR, Kostakioti M, Breland EJ. et al. Strong cross-system interactions drive the activation of the QseB response regulator in the absence of its cognate sensor. Proc Natl Acad Sci USA 2013; 110: 16592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Breland EJ, Zhang EW, Bermudez T. et al. The histidine residue of QseC is required for canonical signaling between QseB and PmrB in uropathogenic Escherichia coli. J Bacteriol 2017; 199: pii=e00060-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guckes KR, Breland EJ, Zhang EW. et al. Signaling by two-component system noncognate partners promotes intrinsic tolerance to polymyxin B in uropathogenic Escherichia coli. Sci Signal 2017; 10: pii=eaag1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Malinverni JC, Silhavy TJ.. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci USA 2009; 106: 8009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Swanson PE, Paddy MR, Dahlquist FW. et al. Characterization of octapeptin-membrane interactions using spin-labeled octapeptin. Biochemistry 1980; 19: 3307–14. [DOI] [PubMed] [Google Scholar]
- 56. Nakayama T, Zhang-Akiyama Q-M. . pqiABC and yebST, putative mce operons of Escherichia coli, encode transport pathways and contribute to membrane integrity. J Bacteriol 2016; 199: pii=e00606-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Duval M, Korepanov A, Fuchsbauer O. et al. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol 2013; 11: e1001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu G, Zhou J, Fu QS. et al. The Escherichia coli azoreductase AzoR is involved in resistance to thiol-specific stress caused by electrophilic quinones. J Bacteriol 2009; 191: 6394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arutyunov D, Arenson B, Manchak J. et al. F plasmid TraF and TraH are components of an outer membrane complex involved in conjugation. J Bacteriol 2010; 192: 1730–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chou TF, Bieganowski P, Shilinski K. et al. 31P NMR and genetic analysis establish hinT as the only Escherichia coli purine nucleoside phosphoramidase and as essential for growth under high salt conditions. J Biol Chem 2005; 280: 15356–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rosenthal AZ, Elowitz MB.. Following evolution of bacterial antibiotic resistance in real time. Nat Genet 2011; 27: 11–3. [DOI] [PubMed] [Google Scholar]
- 62. Hughes D, Andersson DI.. Evolutionary trajectories to antibiotic resistance. Annu Rev Microbiol 2017; 8: 579–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.