Abstract
Objectives
Recent reports indicate the emergence of a new carbapenemase-producing Klebsiella pneumoniae clone, ST307. We sought to better understand the global epidemiology and evolution of this clone and evaluate its association with antimicrobial resistance (AMR) genes.
Methods
We collated information from the literature and public databases and performed a comparative analysis of 95 ST307 genomes (including 37 that were newly sequenced).
Results
We show that ST307 emerged in the mid-1990s (nearly 20 years prior to its first report), is already globally distributed and is intimately associated with a conserved plasmid harbouring the blaCTX-M-15 ESBL gene and several other AMR determinants.
Conclusions
Our findings support the need for enhanced surveillance of this widespread ESBL clone in which carbapenem resistance has occasionally emerged.
Introduction
Several reports have indicated the recent emergence of a new MDR Klebsiella pneumoniae (Kp) clone, ST307. We have recently generated data suggesting that ST307 is becoming an important cause of ESBL-producing Kp infections in Norway (M. A. K. Hetland, A. Fostervold, I. H. Löhr on behalf of The Norwegian Study Group on Klebsiella pneumoniae, unpublished data), and others have reported it as an emerging cause of KPC-producing Kp infections.1–3
Here we summarize what is known in the literature and investigate 95 geographically diverse ST307 whole-genome sequences from 11 countries to better understand the emergence and global molecular epidemiology of this clone and identify the antimicrobial resistance (AMR) genes with which it is associated.
Materials and methods
Ethics
The proposed collection of novel clinical Kp isolates from Norway was reviewed and approved by the Regional Committees for Medical and Health Research Ethics West (Norway, application ID: 2017/1185).
Literature search
We searched PubMed for abstracts containing the words ‘ST307’ with/without ‘Klebsiella pneumoniae’ as of April 2018. ST307-isolate genomes were identified from our collections and public databases using Kleborate.4 In total, 549 published and 37 newly sequenced genomes were identified, representing 11 countries on five continents. However, we were unable to find corresponding literature reports for 45 of the 58 genome assemblies identified among those deposited in GenBank (as of December 2017) and hence these genomes were excluded from comparative analyses.
Sequencing
Novel genomes were sequenced on the Illumina MiSeq platform generating 150 or 250 bp paired end (PE) reads as described previously.5 The oldest isolate (Kp616 from Iran, 2009) was also subjected to long-read Oxford Nanopore sequencing and hybrid genome assembly using Unicycler v0.4.4.6,7 The completed Kp616 genome comprised a 5246307 bp chromosome plus two plasmids (pKp616_1, 58 kbp; pKp616_2, 55 kbp; GenBank accession number GCA_003076555.1).
A core chromosomal single-nucleotide variant (SNV) alignment was generated using RedDog (reference: Kp616 chromosome) as described previously.5 Recombination was removed using Gubbins.8 A preliminary tree made using FastTree9 indicated that the majority of strains from Texas (n = 451/468, 96.4%) formed a distinct monophyletic clade; hence, we randomly selected one isolate per year to represent this clade in comparative analyses. The final recombination-free alignment of 1465 SNVs in 95 genomes was subjected to Bayesian phylogenetic analysis using BEAST 2 v2.4.710 as described previously.5 A GTR, relaxed clock, constant population size model was determined to be the best fit and we confirmed a strong temporal signal by date randomization and linear regression (Figure S1, available as Supplementary data at JAC Online).
De novo genome assemblies were generated with Unicycler v0.4.4.6 AMR genes and virulence loci were detected using Kleborate.4,11,12 Capsule synthesis (K) and lipopolysaccharide (O) loci were typed using Kaptive.13
Sample information accession numbers, citations and genotyping results for the 95 genomes included in the final analyses are listed in Table S1. Notably, this collection represents the most diverse sample of ST307 Kp genomes to date, including isolates (i) from diverse sources, e.g. human infections [urine (n = 34), blood (n = 31), respiratory (n = 10) and unknown/other (n = 17)], two human rectal carriage isolates and one isolate from canal water; and (ii) from diverse geographies, including 47 genomes (49.5%) from seven countries not represented in previous comparative analyses.1,2
Results and discussion
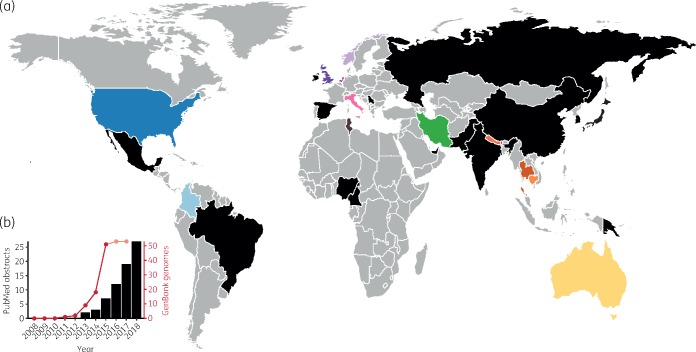
A total of 26 literature reports were identified by systematic search and a further 6 papers were identified by association with published genome sequences (Table S2). The oldest recorded ST307 isolate was collected in The Netherlands in 2008 (Kp MLST database; https://bigsdb.pasteur.fr/klebsiella/klebsiella.html) but the earliest clinical strain reported in the literature was collected in Pakistan in 2009.14 This was followed by sporadic isolations across Europe, Asia, Africa and the Americas, the majority (>98%) from a variety of human clinical specimens plus a minority from other sources, e.g. human rectal samples, companion animals and environmental/sewage water samples (summarized in Table S2 and Figure 1). Several reports indicated local dissemination of ST307 harbouring KPC genes, blaKPC-2 (Columbia, USA, South Korea) and blaKPC-3 (Italy)3,15–18 while an analysis of >1700 ESBL-producing Kp from a hospital network in Texas, USA found high prevalence of blaCTX-M-15-positive ST307 strains, ∼1/3 of which also carried blaKPC-2 genes and three carried blaKPC-3.1 This was consistent with other reports that blaCTX-M-15 is common in ST3071–3,14,19 (also see Table S2). Aside from blaKPC genes, carbapenem resistance conferred by NDM-1 or OXA-48 carbapenemases has been reported,1–3,19,20 as has resistance to the novel β-lactam inhibitor combination ceftazidime/avibactam21 and colistin.20,22
Figure 1.
Geographical distribution and increasing reports of ST307. (a) Countries of collection of ST307 isolates reported in the literature, the international K. pneumoniae MLST database and/or for which genome data are available (also see Table S2). Countries for which isolate genomes were included in the current analysis are coloured as shown in Figure 2. All other countries where ST307 has been reported are coloured black. (b) Reports of ST307 in the literature and among genome assemblies deposited in GenBank. Black bars show the cumulative number of PubMed abstracts as of April 2018, identified using the search criteria ‘ST307’ with/without ‘Klebsiella pneumoniae’ (Table S2). The red line shows the cumulative number of isolates for which genome assemblies are deposited in GenBank as of December 2017. Dates indicate year of isolate collection, not date of deposition, hence recent values will likely increase as further genomes are deposited. The first ST307 isolate reported in the international MLST database was collected in 2008.
We performed the first molecular dating analysis for ST307 using the 95 genomes in our final subsampled collection. This indicated that ST307 emerged in 1994 [95% highest posterior density (HPD), 1974–2006], close to the emergence date estimated previously for ST258,23 despite the fact that the latter was reported in the literature and recognized as a disseminated clone more than a decade earlier than ST307.24 The estimated mutation rate for ST307 (1.18 × 10−6 substitutions/site/year, 95% HPD, 8.01 × 10−7–1.58 × 10−6; Figure S1) was remarkably similar to that estimated previously for ST258 (1.03 × 10−6 substitutions/site/year, 95% HPD 8.09 × 10−7–1.24 × 10−6)23 but faster than that of hypervirulent Kp ST23 (3.40 × 10−7 substitutions/site/year, 95% HPD 2.43 × 10−7–4.38 × 10−7),5 which represent the only Kp clones for which comparable analyses have been published to date.
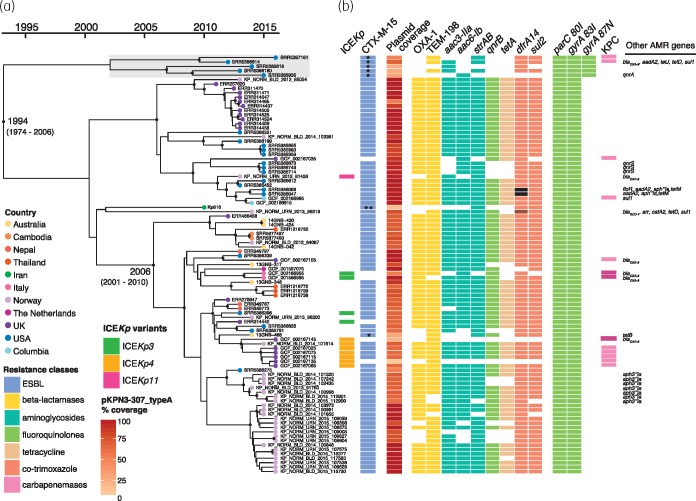
The phylogeny revealed two deep-branching lineages, one of which has become globally distributed, comprising genomes from the Americas, Asia, Australia, the Middle East and Europe (including the 12 genomes reported by Villa et al.2; see Figure 2 and also available for interactive viewing at https://microreact.org/project/ryiY_FlfQ). Within this lineage there was evidence of transfer of ST307 between countries, and for all countries with three or more genomes there were multiple clusters within the global lineage. The countries with the highest representation were distributed most broadly (Norway, n = 30; USA, n = 22; UK, n = 22), suggesting that the same patterns would likely be detected for most countries if sampling was increased (Figure 2). The second lineage included only subsampled strains from Texas (2011–15, shaded grey in Figure 2), indicating that the majority of the hundreds of infections attributed to ST307 in the report by Long et al.1 resulted from prolonged local transmission of this clade, a finding that was not previously evident because the genomes were not compared with those from other geographies. Texan isolates were also found in the global lineage, suggesting that the USA may be a potential origin for ST307, as most of its genetic diversity was present in that location.
Figure 2.
Bayesian phylogeny of 95 ST307 isolates. (a) Dated phylogeny of ST307 isolates (n = 95), with tips coloured by country of isolation (as shown in the inset legend and Figure 1). Black dots on internal nodes indicate ≥95% posterior probability. Light grey shading shows subsampled Texan-specific clade. (b) Presence of the yersiniabactin-carrying ICEKp elements (variants coloured as shown in the inset legend), antimicrobial resistance genes (blocks coloured by drug class), and coverage of blaCTX-M-15 plasmid pKPN3-307_typeA. In the CTX-M-15 column: * indicates blaCTX-M-15 is inserted in chromosome; ** blaCTX-M-15 inserted in IncN plasmid. Brown in the dfrA14 column indicates dfrA27 allele and black the dfrA12 allele. In the KPC column, light pink indicates blaKPC-2 and dark pink indicates blaKPC-3. These data are available for interactive viewing at https://microreact.org/project/ryiY_FlfQ.
We used Kleborate4 to detect Kp virulence determinants that are positively associated with invasive infections.11,12,25 While these determinants are most commonly detected among drug-susceptible ‘hypervirulent’ Kp, they are occasionally found among MDR strains,11,12 posing a risk of severe, difficult-to-treat infections. We detected no evidence of the Kp virulence plasmid (encoding the salmochelin and aerobactin siderophores plus RmpA/RmpA2),26 but a minority of genomes (n = 12, 12.6%) harboured the yersiniabactin siderophore locus located within three distinct chromosomally integrated ICEKp variants (ICEKp3, ICEKp4 and ICEKp11). Hence our data indicate a lower prevalence of ICEKp than that previously reported for ST307 (66%, 8 of 12 genomes investigated2). The distribution of ICEKp insertions on the ST307 core genome tree indicated more than four independent acquisitions with limited expansion of recipient sub-lineages (Figure 2), consistent with the patterns recently reported for ST258 and other common MDR Kp clones.11 Unlike these other clones,13,23 all ST307 shared the same K and O loci (KL102, associated with wzi allele 173, and O2v2), both of which have also been identified among various other Kp STs, including those from other disseminated MDR clones, e.g. ST105, ST152 and ST1583.12,13 An additional putative capsule synthesis locus (Kp616 genes C2861_20465 to C2861_205202), which was previously reported in ST3072 and is unlike any known K locus, was also conserved (present at ≥95% nucleotide identity and ≥90% coverage in 95/95 genomes) but is rare among the broader Kp population (just 10 Kp nucleotide blast matches were identified in the GenBank nucleotide database).
In contrast to the virulence loci, acquired AMR genes were highly prevalent; 93 (97.9%) isolates carried acquired resistance determinants associated with three or more drug classes (Table S1). The ParC 80I and GyrA 83I fluoroquinolone resistance-associated mutations were conserved in all genomes. The blaCTX-M-15 ESBL gene was found in 89 (93.7%) genomes, and 81 (85.3%) harboured it in combination with sul2, dfrA14 and strAB with/without aac(3)-IIa, which were all linked to an MDR plasmid (see below). blaKPCs and other AMR genes were occasionally identified (Figure 2 and Table S1). For the majority of genomes carrying blaCTX-M-15 (n = 88/89), BLASTn confirmed that this gene was located downstream of ISEcp1, which forms a transposon to mobilize blaCTX-M-15 and promotes its expression.27 Four complete ST307 IncFIIK/IncFIBKblaCTX-M-15 plasmids have been published,2 and share an insertion of the ISEcp1/blaCTX-M-15 transposon within Tn3. Read-mapping to the largest of these plasmids, pKPN3-307_typeA (accession KY271404) and assembly graph inspections of our blaCTX-M-15-positive genomes showed that all carried the same ISEcp1/blaCTX-M-15 transposon. In 82/89 cases, the same pKPN3-307_typeA IncFIIK/IncFIBK plasmid backbone was present and ISEcp1/blaCTX-M-15 was located in the same site within Tn3, consistent with conservation of the same IncFIIK/IncFIBK ESBL plasmid (including various deletion variants; Figure S2). The exceptions were as follows: (i) an Australian isolate (13GNB-468) had the ISEcp1/blaCTX-M-15 transposon inserted in the chromosomal gene feoB; (ii) Kp616 carried no pKPN3-307_typeA-like plasmid but harboured blaCTX-M-15 on an IncN plasmid; and (iii) the five representatives of the Texas-specific lineage carried two chromosomal insertions of the ISEcp1/blaCTX-M-15 transposon (within Kp616 loci C2861_02545 and C2861_22795, not detailed in the previous study1). This, coupled with the additional GyrA 87N fluoroquinolone resistance mutation in the Texan lineage, may have contributed to its prolonged transmission in the hospital setting by facilitating enhanced resistance to antimicrobials without any burden of plasmid maintenance. Regardless of these exceptions, the level of plasmid conservation in ST307 is remarkable, mirroring the association of ST258 with the blaKPC pKpQIL plasmid23 and suggesting that the plasmid confers limited fitness cost to the host (although plasmid-positive, blaCTX-M-15-negative genomes were observed; Table S1).
Complementing the increasing reports in the literature, our analyses reveal that ST307 is a highly successful MDR clone that shares many traits with ST258 (i.e. date of emergence, evolutionary rate, high prevalence of AMR genes, low prevalence of ICEKps but multiple independent acquisitions, and high conservation of a single plasmid), but is closely associated with blaCTX-M-15 rather than blaKPCs. With sufficient exposure ST307 can acquire and disseminate carbapenemases,1–3 and likely other clinically important AMR determinants. Our analyses show for the first time that ST307 is readily transferred between countries and has already become globally disseminated but remained largely unnoticed for almost 20 years. These findings indicate an urgent need for enhanced surveillance of MDR Kp to monitor ST307 alongside other well-known clones and detect emerging MDR threats.
Supplementary Material
Acknowledgements
We thank The Norwegian Surveillance System for Antimicrobial Drug Resistance (NORM) for data sharing; the Norwegian Klebsiella pneumoniae study group for collection of Norwegian isolates; Dr Mohammad Ali Boroumand for collection of the Iranian isolate; the Australian Group on Antimicrobial Resistance (AGAR), in particular Jan Bell, for providing isolates from Australia; and the staff at MDU Public Health Laboratory for sequencing Australian isolates.
Funding
This work was supported by: the Viertel Foundation of Australia; the NHMRC of Australia (fellowship number GNT1105905 to B. P. H.); The Western Norway Regional Health Authority (fellowship numbers 912037, 912119 and grant number 912050); and the Bill and Melinda Gates Foundation, Seattle.
Transparency declarations
The authors declare no conflicts of interest. K. L. W., J. H., I. H. L. and K. E. H. conceived the study and performed data analyses. M. A. K. H. performed the systematic literature review. A. F., I. H. L., M. H. and B. P. H. provided isolates and/or genome data. R. R. W. and L. M. J. generated the completed reference genome. K. L. W., J. H. and K. E. H. wrote the paper. All authors contributed to data interpretation, read and commented on the manuscript.
References
- 1. Long SW, Olsen RJ, Eagar TN. et al. Population genomic analysis of 1,777 extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates, Houston, Texas: unexpected abundance of clonal group 307. MBio 2017; 8: e00489–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Villa L, Feudi C, Fortini D. et al. Diversity, virulence and antimicrobial resistance of the KPC- producing Klebsiella pneumoniae ST307 clone. MGen 2017; 3: e000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ocampo AM, Chen L, Cienfuegos AV. et al. A two-year surveillance in five Colombian tertiary care hospitals reveals high frequency of non-CG258 clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics. Antimicrob Agents Chemother 2016; 60: 332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lam MCC, Wick RR, Wyres KL. et al. Kleborate: comprehensive genotyping of Klebsiella pneumoniae genome assemblies. 2018. https://github.com/katholt/Kleborate.
- 5. Lam MMC, Wyres KL, Duchêne S. et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal group 23 reveals early emergence and rapid global dissemination. Nat Commun 2018; 9: 2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wick RR, Judd LM, Gorrie CL. et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017; 13: e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wick RR, Judd LM, Gorrie CL. et al. Completing bacterial genome assemblies with multiplex MinION sequencing. MGen 2017; 3: e000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Croucher NJ, Page AJ, Connor TR. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43: e15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Price MN, Dehal PS, Arkin AP.. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 2009; 26: 1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bouckaert R, Heled J, Kühnert D. et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comp Biol 2014; 10: e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lam MMC, Wick RR, Wyres KL. et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. MGen 2018; 4. doi:10.1099/mgen.0.000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lam MCC, Wyres KL, Judd LM. et al. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. bioRxiv 2018; doi:10.1101/376236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wick RR, Heinz E, Holt KE. et al. Kaptive Web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J Clin Microbiol 2018; 56: e00197-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Habeeb MA, Haque A, Nematzadeh S. et al. High prevalence of 16S rRNA methylase RmtB among CTX-M extended-spectrum β-lactamase-producing Klebsiella pneumoniae from Islamabad, Pakistan. Int J Antimicrob Agents 2013; 41: 524–6. [DOI] [PubMed] [Google Scholar]
- 15. Bonura C, Giuffrè M, Aleo A. et al. An update of the evolving epidemic of blaKPC carrying Klebsiella pneumoniae in Sicily, Italy, 2014: emergence of multiple non-ST258 clones. PLoS One 2015; 10: e0132936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoon E, Yang JW, Kim JO. et al. Carbapenemase-producing Enterobacteriaceae in South Korea: a report from the National Laboratory Surveillance System. Future Microbiol 2018; 13: 771–83. [DOI] [PubMed] [Google Scholar]
- 17. Castanheira M, Farrell SE, Wanger A. et al. Rapid expansion of KPC-2-producing Klebsiella pneumoniae isolates in two Texas hospitals due to clonal spread of ST258 and ST307 lineages. Microb Drug Resist 2013; 19: 295–7. [DOI] [PubMed] [Google Scholar]
- 18. Kim JO, Song SA, Yoon EJ. et al. Outbreak of KPC-2-producing Enterobacteriaceae caused by clonal dissemination of Klebsiella pneumoniae ST307 carrying an IncX3-type plasmid harboring a truncated Tn4401a. Diagn Microbiol Infect Dis 2017; 87: 343–8. [DOI] [PubMed] [Google Scholar]
- 19. Yoon E, Kang DY, Yang JW. et al. New Delhi metallo-β-lactamase-producing Enterobacteriaceae in South Korea between 2010 and 2015. Front Genet 2018; 9: 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Novovic K, Trudic A, Brkic S. et al. Molecular epidemiology of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae in Serbia from 2013 to 2016. Antimicrob Agents Chemother 2017; 61: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Giddins MJ, Macesic N, Annavajhala MK. et al. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC2-harbouring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother 2018; 62: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saavedra S, Diaz L, Wiesner M. et al. Genomic and molecular characterization of clinical isolates of Enterobacteriaceae harboring mcr-1 in Colombia, 2002 to 2016. Antimicrob Agents Chemother 2017; 61: e00841-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bowers JR, Kitchel B, Driebe EM. et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One 2015; 10: e0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kitchel B, Rasheed JK, Patel JB. et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 2009; 53: 3365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holt KE, Wertheim H, Zadoks RN. et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA 2015; 112: E3574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramirez MS, Traglia GM, Lin DL. et al. Plasmid-mediated antibiotic resistance and virulence in gram-negatives: the Klebsiella pneumoniae paradigm. Microbiol Spectr 2014; 2: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bonnet R. Growing group of extended spectrum β-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother 2004; 48: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.