Abstract
Background
As a result of the growing prevalence of the plasmid-mediated mobile colistin resistance gene mcr-1 among Gram-negative bacteria, the surveillance of mcr-1 has been globally applied. In our study, we aimed to shed light on the possibility of transmission of mcr-1-resistant isolates through market retail fruits.
Methods and results
Herein, 133 different fruit surface samples were collected and screened for the different MCR variants (mcr-1 to mcr-8) using PCR and confirmed with sequencing. We identify for the first time mcr-1-carrying Escherichia coli and Klebsiella pneumoniae from market retail fruits in Guangzhou, China. Minimum inhibitory concentrations were detected by the broth microdilution method. Liquid mating was performed to check the transferability of the mcr-1 gene. Pulsed field gel electrophoresis analysis of S1 nuclease-digested DNA and Southern blotting were performed to check the location of the mcr-1 gene. Then, whole-genome sequencing and in silico multilocus sequence typing analysis were performed.
Conclusion
We showed that E. coli GB110 can mediate the spreading of antibiotic resistance genes through the food chain, while K. pneumoniae GB015 was considered to be the progenitor of the most successful multidrug-resistant clone. Since fruits are usually consumed fresh, this may serve as a direct source of mcr-1-producing bacteria in humans that requires prompt surveillance and intervention to limit the spread of resistance.
Keywords: colistin, mcr-1, Escherichia coli, Klebsiella pneumoniae, fruit
Introduction
Colistin is a polymyxin antibiotic that has been used for many years in veterinary medicine. Nowadays, a need for using colistin in human medicine has evolved as the last resort drug for the treatment of infections caused by multidrug-resistant bacteria, especially carbapenem-resistant Enterobacteriaceae. However, the use of colistin as a last resort antibiotic is seriously threatened by the rise of plasmid-borne mobile colistin resistance (mcr family) genes (mcr-1, -2, -3, -4, -5, -6, -7 and -8),1–3 which can spread rapidly via horizontal gene transfer between bacterial strains and species.4 In August 2016, a survey reported detection of mcr-1-positive Enterobacteriaceae from farming soil in Shandong province, China, suggesting that Enterobacteriaceae harboring mcr-1 can contaminate agricultural products.5 Here, we report the identification of mcr-1 in Escherichia coli and Klebsiella pneumoniae isolated from market retail fruits in Guangzhou, China.
Methods and results
A total of 133 fruit surface samples were collected from market retail fruits in Guangzhou, China, from June to November 2016, at various intervals. Different fruits were collected, including apples (n=52), oranges (n=31), tangerines (n=16), pears (n=15), bananas (n=11), pomegranates (n=6) and grapes (n=2). Initially, surface samples from the fruits were wiped with sterile cotton swabs moistened with saline. Each swab contained one fruit surface sample and was transported to the laboratory immediately. The swabs were cultured on to lysogeny broth (LB) liquid medium (Oxoid, Basingstoke, UK), then DNA extraction was performed using the boiling method. All samples were screened for mcr-1 to mcr-8 using PCR and confirmed by Sanger sequencing. The primers and conditions used for the PCR assays are listed in Table S1.3
Out of 133 fruit samples, mcr-1 was detected in two samples (1.5%). No mcr-2 to mcr-8 genes were identified among these samples. To screen for colistin resistance, the two mcr-1-positive samples were directly seeded on LB agar plates containing 4 µg/mL colistin and incubated at 37°C for 24 hours. The species of selected colonies from the LB agar plates were identified using the API 20E system (bioMérieux, Marcy l’Etoile, France) and confirmed by 16S rRNA sequencing. As a result, mcr-1-harboring E. coli GB110 and K. pneumoniae GB015 were identified from apple and orange samples, respectively. Minimum inhibitory concentrations (MICs) were detected by the broth microdilution method and interpreted according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2017). It was found that E. coli GB110 was susceptible to all tested agents except colistin and polymyxin B; and K. pneumoniae GB015 was resistant to colistin, polymyxin B and ampicillin (Table 1).
Table 1.
Characteristics of mcr-1-producing Escherichia coli GB110, Klebsiella pneumoniae GB015 and their transconjugants isolated from market retail fruits in China
| Isolate code | Source | Isolation site | MLST | Plasmid replicon type | Encoding genes of resistance | MICs |
|---|---|---|---|---|---|---|
|
| ||||||
| E. coli GB110 | Apple | Market retail fruits | ST189 | IncHI1,IncFIA | aadA2, aadA1, mcr-1, floR, cmlA1, sul2, sul3, tetA, tetM, dfrA12, mdfA | CL (16), PB (8), TGC (≤1), AMP (4), AMC (8), CTX (≤0.125), CAZ (≤0.125), FEP (0.25), GEN (≤1), AMK (4), ETP (≤0.125), IPM (≤0.125), MEM (≤0.125), FOS (≤8), NIT (≤8), CIP (≤0.03) |
| K. pneumoniae | Orange | Market | ST442 | IncHI1, | bla, mcr-1, SHV-110 | CL (64), PB (64), TGC (≤1), AMP (64), AMC (8), |
| GB015 | retail fruits | IncFIB | qnrS1, oqxA, oqxB, fosA6, sul1, tetA, dfrA1 | CTX (≤0.125), CAZ (0.5), FEP (≤0.125), GEN (≤1), AMK (4), ETP (≤0.125), IPM (0.5), MEM (≤0.125), FOS (16), NIT (64), CIP (1) | ||
| E. coli C600 (transconjugant of E. coli GB110) | – | – | – | IncFIA | mcr-1 | CL (16), PB (8), TGC (≤1), AMP (4), AMC (8), CTX (≤0.125), CAZ (≤0.125), FEP (≤0.125), GEN (≤1), AMK (4), ETP (≤0.125), IPM (≤0.125), MEM (≤0.125), FOS (≤8), NIT (8), CIP (≤0.03) |
| E. coli C600 (transconjugant of K. pneumoniae GB015) | – | – | – | IncHI1 | mcr-1 | CL (16), PB (16), TGC (≤1), AMP (4), AMC (8), CTX (≤0.125), CAZ (≤0.125), FEP (≤0.125), GEN (≤1), AMK (4), ETP (≤0.125), IPM (≤0.125), MEM (≤0.125), FOS (≤8), NIT (≤8), CIP (≤0.03) |
Abbreviations: AMC, amoxicillin–clavulanic acid; AMK, amikacin; AMP, ampicillin; CAZ, ceftazidime; CIP, ciprofloxacin; CL, colistin; CTX, cefotaxime; ETP, ertapenem; FEP, cefepime; FOS, fosfomycin; GEN, gentamicin; IPM, imipenem; MEM, meropenem; MIC, minimum inhibitory concentration; MLST, multilocus sequence typing; NIT, nitrofurantoin; PB, polymyxin B; TGC, tigecycline.
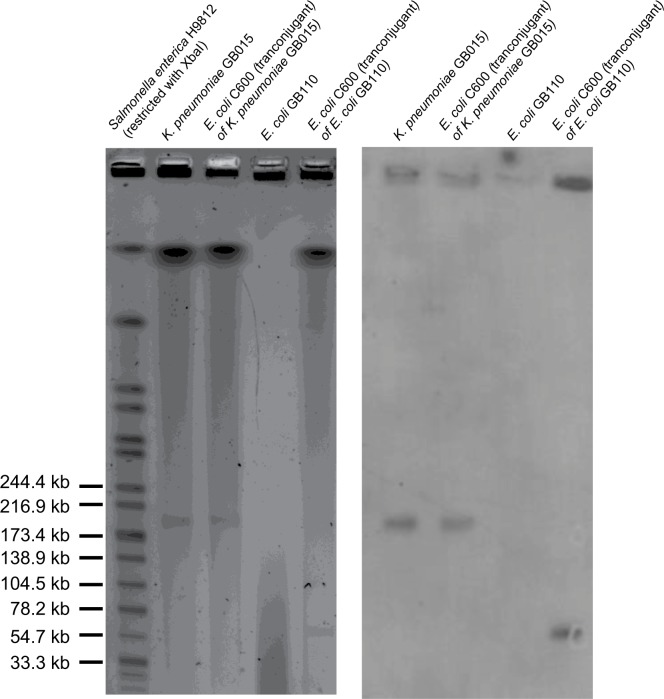
A conjugation experiment was performed using streptomycin-resistant E. coli C600 as the recipient. The mcr-1-producing isolates and recipient were mixed (ratio of 1:9) in LB and incubated overnight at 37°C. The mixture was then spread on LB agar plates containing colistin (2 µg/mL) and streptomycin (2,000 µg/mL). The transconjugants were confirmed for mcr-1 by PCR and Sanger sequencing. The results showed that mcr-1 was successfully transferred to streptomycin-resistant E. coli C600 through conjugation in both isolates, suggesting that mcr-1 was located on transferable plasmids. Plasmid incompatibility groups were identified by PCR assay as described previously.6 Pulsed field gel electrophoresis analysis of S1 nuclease-digested DNA (S1-PFGE), followed by Southern blotting, showed that mcr-1 was located on ~62.9 kb IncFIA and ~204.2 kb IncHI1 plasmids for the transconjugants of E. coli GB110 and K. pneumoniae GB015, respectively (Figure 1).
Figure 1.
S1-PFGE and Southern hybridization with the mcr-1 probe.
Abbreviations: E. coli, Escherichia coli; K. pneumoniae, Klebsiella pneumoniae; S1-PFGE, pulsed field gel electrophoresis analysis of S1 nuclease-digested DNA.
The genome DNA of E. coli GB110 and K. pneumoniae GB015 were extracted using a Qiagen Blood & Tissue kit (Qiagen, Hilden, Germany) and DNA libraries were constructed with 350 bp paired-end fragments. In total, 9,367,660 and 10,515,584 paired-end 150 bp reads were produced by the Illumina Hiseq2000 platform for E. coli GB110 and K. pneumoniae GB015, respectively. Reads were assembled using SPAdes v3.12.0.7 This Whole Genome Shotgun project has been deposited at GenBank under the accession number PRJNA482733. The genome annotation was performed using PATRICK (https://www.patricbrc.org/) and ISFinder (https://www-is.biotoul. fr/). The assembled genomes showed that ISApl1 was located upstream of mcr-1 for E. coli GB110 and downstream of mcr-1 for K. pneumoniae GB015, consistent with reported variability in the location of ISAba1, which was probably involved in the original mobilization of mcr-1 from Moraxella spp.4,8
Multilocus sequence typing (MLST), serotyping, antimicrobial resistance genes and virulence factors were annotated by the Center for Genomic Epidemiology website (http://www.genomicepidemiology.org/).
MLST analysis of E. coli GB110 (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) showed that it belonged to sequence type 189 (165 cplx), which was reported from poultry retail meat mediating the spread of extended-spectrum β-lactamase (ESBL) genes in Colombia,9 while K. pneumoniae GB015 belonged to ST442. ST442 and ST11, the most common carbapenem-resistant clones in China, are considered to be the progenitors of K. pneumoniae ST258, which is widespread worldwide as the most successful multidrug-resistant clone of K. pneumoniae.10
The serotype of E. coli GB110 was identified as O38:H26, which can cause diarrhea in humans.11 The E. coli heat-stable enterotoxin-1 (EAST-1)-encoding gene astA was found in E. coli GB110, which is associated with human diseases. These results indicate the risk that eating contaminated fruits could lead to diarrheal diseases.
The E. coli GB110 was found to harbor ten resistance genes, including mcr-1 and aminoglycoside resistance genes, aadA2 and aadA1, while K. pneumoniae GB015 was found to possess nine resistance genes, including mcr-1, blaSHV-110, qnrS1 and fosA6 (Table 1).
Conclusion
In our study, we reported the detection of mcr-1 from market retail fruits, which is significant since, unlike meat and vegetables, fruits are usually consumed without cooking or processing, making them a potentially high-risk source of mcr-1 acquisition and infection in humans. This work sheds light on the urgent need for continued surveillance and prompt intervention in China to guard against the worldwide distribution of mcr-1, which threatens the use of colistin as a last resort antibiotic.
Supplementary material
Table S1.
Primers and PCR conditions used in this study
| Purpose | Primer | Nucleotide sequence (5′→3′) | Amplicon size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
|
| ||||
| Amplification of mcr-1 | MCR-1F | ATCAGCCAAACCTATCCCATCG | 1,257 | 55 |
| MCR-1R | GCAGACGCACAGCAATGCCTAT | |||
| Amplification of mcr-2 | MCR-2F | GCGATGGCGGTCTATCCTGTAT | 378 | 55 |
| MCR-2R | TGCGATGACATGGGGTGTCAGC | |||
| Amplification of mcr-3 | MCR-3F | TATGGGTTACTATTGCTGG | 814 | 55 |
| MCR-3R | CTACCCTGATGCTCATCG | |||
| Amplification of mcr-4 | MCR-4F | GTCATAGTGGTATAAAAGTACAG | 669 | 55 |
| MCR-4R | CCACCGTCTATCAGAGCCAAC | |||
| Amplification of mcr-5 | MCR-5F | GCGGTTGTCTGCATTTATCAC | 1,042 | 50 |
| MCR-5R | CTTTGAAAACCTGTCTTCGGCA | |||
| Amplification of mcr-6 | MCR-6F | GTCCGGTCAATCCCTATCTGT | 556 | 55 |
| MCR-6R | ATCACGGGATTGACATAGCTAC | |||
| Amplification of mcr-7 | MCR-7F | TGCTCAAGCCCTTCTTTTCGT | 892 | 55 |
| MCR-7R | TTCATCTGCGCCACCTCGT | |||
| Amplification of mcr-8 | MCR-8F | AACCGCCAGAGCACAGAATT | 667 | 60 |
| MCR-8R | TTCCCCCAGCGATTCTCCAT | |||
Acknowledgments
This work was supported by the Science and Technology Research Project of Henan Province (grant 182102310553) and the National Natural Science Foundation of China (grant numbers 81722030 and 81830103).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Mendes AC, Novais Â, Campos J, et al. mcr-1 in carbapenemase-producing Klebsiella pneumoniae with hospitalized patients, Portugal, 2016-2017. Emerg Infect Dis. 2018;24(4):762–766. doi: 10.3201/eid2404.171787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Wang Y, Zhou Y, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7(1):122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R, van Dorp L, Shaw LP, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. 2018;9(1):1179. doi: 10.1038/s41467-018-03205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng B, Huang C, Xu H, et al. Occurrence and Genomic Characterization of ESBL-Producing, MCR-1-Harboring Escherichia coli in Farming Soil. Front Microbiol. 2017;8:2510. doi: 10.3389/fmicb.2017.02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong LL, Zhang YF, Doi Y, et al. Coproduction of MCR-1 and NDM-1 by colistin-resistant Escherichia coli isolated from a healthy individual. Antimicrob Agents Chemother. 2016;61(1) doi: 10.1128/AAC.01962-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nurk S, Bankevich A, Antipov D, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20(10):714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snesrud E, Mcgann P, Chandler M. The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. MBio. 2018;9(1):e02381–e02317. doi: 10.1128/mBio.02381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castellanos LR, Donado-Godoy P, León M, et al. High heterogeneity of Escherichia coli sequence types harbouring ESBL/AmpC genes on IncI1 plasmids in the Colombian Poultry Chain. PLoS One. 2017;12(1):e0170777. doi: 10.1371/journal.pone.0170777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–5884. doi: 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buvens G, Lauwers S, Piérard D. Prevalence of subtilase cytotoxin in verocytotoxin-producing Escherichia coli isolated from humans and raw meats in Belgium. Eur J Clin Microbiol Infect Dis. 2010;29(11):1395–1399. doi: 10.1007/s10096-010-1014-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Primers and PCR conditions used in this study
| Purpose | Primer | Nucleotide sequence (5′→3′) | Amplicon size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
|
| ||||
| Amplification of mcr-1 | MCR-1F | ATCAGCCAAACCTATCCCATCG | 1,257 | 55 |
| MCR-1R | GCAGACGCACAGCAATGCCTAT | |||
| Amplification of mcr-2 | MCR-2F | GCGATGGCGGTCTATCCTGTAT | 378 | 55 |
| MCR-2R | TGCGATGACATGGGGTGTCAGC | |||
| Amplification of mcr-3 | MCR-3F | TATGGGTTACTATTGCTGG | 814 | 55 |
| MCR-3R | CTACCCTGATGCTCATCG | |||
| Amplification of mcr-4 | MCR-4F | GTCATAGTGGTATAAAAGTACAG | 669 | 55 |
| MCR-4R | CCACCGTCTATCAGAGCCAAC | |||
| Amplification of mcr-5 | MCR-5F | GCGGTTGTCTGCATTTATCAC | 1,042 | 50 |
| MCR-5R | CTTTGAAAACCTGTCTTCGGCA | |||
| Amplification of mcr-6 | MCR-6F | GTCCGGTCAATCCCTATCTGT | 556 | 55 |
| MCR-6R | ATCACGGGATTGACATAGCTAC | |||
| Amplification of mcr-7 | MCR-7F | TGCTCAAGCCCTTCTTTTCGT | 892 | 55 |
| MCR-7R | TTCATCTGCGCCACCTCGT | |||
| Amplification of mcr-8 | MCR-8F | AACCGCCAGAGCACAGAATT | 667 | 60 |
| MCR-8R | TTCCCCCAGCGATTCTCCAT | |||