Abstract
Background and Aims
The genus Solanum includes important vegetable crops and their wild relatives. Introgression of their useful traits into elite cultivars requires effective recombination between hom(e)ologues, which is partially determined by genome sequence differentiation. In this study we compared the repetitive genome fractions of wild and cultivated species of the potato and tomato clades in a phylogenetic context.
Methods
Genome skimming followed by a clustering approach was used as implemented in the RepeatExplorer pipeline. Repeat classes were annotated and the sequences of their main domains were compared.
Key Results
Repeat abundance and genome size were correlated and the larger genomes of species in the tomato clade were found to contain a higher proportion of unclassified elements. Families and lineages of repetitive elements were largely conserved between the clades, but their relative proportions differed. The most abundant repeats were Ty3/Gypsy elements. Striking differences in abundance were found in the highly dynamic Ty3/Gypsy Chromoviruses and Ty1/Copia Tork elements. Within the potato clade, early branching Solanum cardiophyllum showed a divergent repeat profile. There were also contrasts between cultivated and wild potatoes, mostly due to satellite amplification in the cultivated species. Interspersed repeat profiles were very similar among potatoes. The repeat profile of Solanum etuberosum was more similar to that of the potato clade.
Conclusions
The repeat profiles in Solanum seem to be very similar despite genome differentiation at the level of collinearity. Removal of transposable elements by unequal recombination may have been responsible for structural rearrangements across the tomato clade. Sequence variability in the tomato clade is congruent with clade-specific amplification of repeats after its divergence from S. etuberosum and potatoes. The low differentiation among potato and its wild relatives at the level of interspersed repeats may explain the difficulty in discriminating their genomes by genomic in situ hybridization techniques.
Keywords: Solanum, transposable elements, repeat profiles, relative abundance, Solanum etuberosum, Solanum tuberosum, Solanum lycopersicum, crop wild relatives
INTRODUCTION
The genus Solanum includes various important vegetable crops, such as tomato and potato, and wild relatives that contain useful traits for introgression into elite crop cultivars (Bradshaw et al., 2006; Hajjar and Hodgkin, 2007; Grandillo et al., 2011; Ramsay and Bryan, 2011; Castañeda-Álvarez et al., 2016). However, genetic resources may not be directly usable due to limited crossability caused by pre- and post-zygotic hybridization barriers (Camadro et al., 2004; Jansky, 2009; Grandillo et al., 2011). Once these barriers have been overcome and a fertile hybrid progeny obtained, the next challenge is to ensure that homoeologous chromosomes pair and recombine. Even then, local loss of collinearity may cause linkage drag, where undesirable alien traits remain completely linked with the traits of interest. These difficulties are largely related to the degree of genome differentiation between the crop and its wild relative, which means that the higher the differentiation, the harder it is to introgress genes of interest from the donor to the recipient genome.
Divergence between two genomes can be explained in terms of large-scale structural differences and of nucleotide-level differences, particularly of repetitive DNA sequences. Structural differentiation of genomes with chromosome rearrangements, such as inversions or translocations, may hinder recombination and increase linkage drag or cause (semi-)sterility. In addition, rapid evolution of tandem and interspersed repetitive elements can be a major factor in reduced pairing between homoeologous chromosomes in hybrids between related species (Dvorak, 1983). Various aspects of genome differentiation between related species do not necessarily go hand in hand with their phylogenetic relationship.
Phylogenetic relationships within the genus Solanum have long been under debate. In particular, the tomato and potato clades, which diverged 7–8 Mya, are well defined (Rodriguez et al., 2009; Särkinen et al., 2013). The tomato clade started diversifying only 2 Mya, while the potato clade did so 7 Mya (Särkinen et al., 2013). Solanum etuberosum, which is frequently included in phylogenetic analyses of these groups, has a debated position with respect to these two clades: originally it was included within section Petota (Hawkes, 1990) but later it was included in section Etuberosum together with other non-tuber-bearing species (Spooner et al., 2014), a sister clade to both the tomato and the potato clades (Rodriguez et al., 2009).
Despite their relatedness, the genomes in the tomato and potato clade species have evolved in different directions. Tomato and its close relatives exhibit more macro- and micro-genomic rearrangements (Seah et al., 2004; Van Der Knaap et al., 2004; Tang et al., 2008; Anderson et al., 2010; Szinay et al., 2010, 2012; Verlaan et al., 2011), whereas the potatoes and some of their wild relatives have maintained higher chromosome collinearity (Lou et al., 2010; Gaiero et al., 2016). Potato species are more syntenic with species belonging to other sections in Solanum and other genera of the Solanaceae (Lou et al., 2010; Peters et al., 2012; Szinay et al., 2012), which suggests that the species in the tomato clade present a more derived state of genome organization. Large-scale chromosomal and small-scale DNA rearrangements are caused by active transposable elements (TEs), which promote chromosomal breakages and subsequent rearrangements (McClintock, 1946; Bennetzen, 1996, 2000; Kidwell and Holyoake, 2001; Raskina et al., 2008; Belyayev, 2014; Bennetzen and Wang, 2014), thus contributing to genome divergence. Their repeat profiles can give information on the phylogenetic relationships within and between clades.
For evolutionary studies of the repetitive fractions of the genome, two strategies can be used as proxies. One of them is the ability of genomic in situ hybridization (GISH) to discriminate parental genomes in hybrids, while the other is through differences in genome size. GISH has been successfully applied to hybrids between cultivated tomato and Solanum peruvianum or Solanum lycopersicoides (Parokonny et al., 1997; Ji and Chetelat, 2003; Ji et al., 2004). Among potatoes, this genome painting strategy permits the distinction of parental chromosomes in interspecific hybrids between Solanum tuberosum Group Tuberosum and non-tuber-bearing potato relatives carrying the E genome (Matsubayashi, 1991), such as Solanum brevidens (Dong et al., 2001, 2005; Gavrilenko et al., 2002; Tek et al., 2004) and S. etuberosum (Dong et al., 1999; Gavrilenko et al., 2003). GISH was not able to distinguish the parental chromosomes in hybrids between potato, S. tuberosum Group Phureja and its closer A-genome tuber-bearing wild relatives such as Solanum commersonii (Gaiero et al., 2017). The lack of GISH differentiation suggests that a major part of the repetitive sequences among their genomes has not differentiated enough, in spite of the estimated 7-Myr divergence in the potato clade (Särkinen et al., 2013). The second proxy for the evolution of repetitive sequences is genome size. Genome size values are on average slightly higher for species in the tomato clade than in the potato clade (see Table 1), although there is considerable variation among tomato species (Grandillo et al., 2011). The processes of genome size increase and reduction can be largely explained in terms of different dynamics of expansion/removal of repetitive elements (Feschotte et al., 2002; Leitch and Leitch, 2013; Belyayev, 2014). These dynamics may differ among related clades or individual species giving rise to variable degrees of divergence in the repeat composition of related genomes (Novák et al., 2014; Kelly et al., 2015; Macas et al., 2015).
Table 1.
Taxa sampled including taxonomic classification, three-letter code, accession details, genome size (http://data.kew.org/cvalues) and sequence data source
| Taxonomy | Species | Code | Accession | Genome size (1C, Mbp) | Sequence data source |
|---|---|---|---|---|---|
| Subgenus Potato Section PetotaSeries Tuberosa | Solanum tuberosum Group Phureja | phu | DM | 831 | http://solanaceae.plantbiology.msu.edu/ |
| Solanum tuberosum Group Tuberosum | tbr | RH | 860 | http://solanaceae.plantbiology.msu.edu/ | |
| Series Commersoniana | Solanum commersonii | cmm | 04.02.3 | 792* | Gaiero et al. unpublished |
| Solanum chacoense | chc | 07.01.7 | 617 | Gaiero et al. unpublished | |
| Series Bulbocastana | Solanum cardiophyllum | cph | 675* | Biosystematics, WUR | |
| Section Etuberosum | Solanum etuberosum | etu | 763 | Biosystematics, WUR | |
| Section Lycopersicon | Solanum lycopersicum | lyc | Heinz 1706 | 1002 | www.tomatogenome.net |
| Solanum pimpinelllifolium | pim | LA1584 | 831 | www.tomatogenome.net | |
| Section Arcanum | Solanum arcanum | arc | LA2157 | 1125 | www.tomatogenome.net |
| Solanum neorickii | neo | LA2133 | Not determined | www.tomatogenome.net | |
| Section Neolycopersicon | Solanum pennelli | pen | LA0716 | 1200 | www.tomatogenome.net |
| Section Eriopersicon | Solanum habrochaites | hab | LYC4 | 905 | www.tomatogenome.net |
| Solanum peruvianum | per | LA1954 | 1125 | www.tomatogenome.net |
* Genome size determined in this study.
The processes shaping the repeat composition of related plant genomes can be inferred by conducting a detailed study of the repetitive DNA in various species within a clade and across related clades. The availability of high-thoughput sequencing (HTS) data for tomato, potato and their wild relatives allows us to compare their repetitive fractions. There are two classes of TEs: class I or retrotransposons with an RNA intermediate and a ‘copy-and-paste’ mechanism and class II or DNA transposons, with DNA as intermediate and with a ‘cut-and-paste’ transposition mechanism. Class I is divided into two subclasses, those flanked by long terminal repeats (LTR retrotransposons) and those without or with short terminal repeats (non-LTR retrotransposons) (Finnegan, 1989). The classes and subclasses are further divided hierarchically into order, superfamily, family and subfamily (Wicker et al., 2007; Piégu et al., 2015). TEs can thus be annotated and their relative abundances in related genomes determined.
TE classification and abundances carry phylogenetic signal (Dodsworthet al., 2015a) and have successfully been used to answer phylogenetic questions in the tomato clade (Dodsworth et al., 2016). From the structural point of view, the genome of S. etuberosum shows many rearrangements with respect to both potato and tomato (Lou et al., 2010; Szinay et al., 2012), but a recent analysis shows much greater genome collinearity with the potato lineage than with the tomato lineage (M. E. Schranz, unpubl. res.). We expect TE analysis to provide further evidence of the relationship of this species to the tomato and potato clades.
The aim of this study is to elucidate differentiation of major repetitive sequence classes between and among species belonging to the tomato and potato clades of the genus Solanum in terms of their dynamics and evolutionary processes. We compared the classes of repetitive sequences of cultivated and wild species belonging to those clades and we assessed whether the composition of this genome fraction in S. etuberosum is more similar to that found in the tomato or in the potato clade.
MATERIALS AND METHODS
Taxa sampled, genome size determinations, DNA isolation and sequencing
We included 13 taxa from three sections of the genus Solanum including seven taxa from the tomato clade (section Lycopersicon), five from the potato clade (section Petota) and S. etuberosum (section Etuberosum). For some of the taxa we obtained sequence data from the 100 Tomato Genome Sequencing Consortium, www.tomatogenome.net (Aflitos et al., 2014), or from various research groups (Table 1). Genomic DNA of S. commersonii and Solanum chacoense was extracted from approximately 2.5 g of fresh etiolated leaf tissue samples using the nuclei enrichment protocol described by Bernatzky and Tanksley (1986), slightly modified. Libraries were prepared using the Nextera Library Preparation Kit (Illumina) and were sequenced on an Illumina HiSeq2000 sequencer at Applied Bioinformatics, Wageningen University and Research, for S. commersonii (100-bp paired-end reads) and on an Illumina HiSeq4000 sequencer at The Beijing Genomics Institute (BGI) for S. chacoense (150-bp paired-end reads). Nuclear DNA measurements were performed according to Doležel and Göhde (1995). Propidium iodide (PI, 50 mg mL–1) was used to stain nuclei and tomato (2C = 1.96; Doležel et al., 1992) was used as an internal standard. Three DNA estimations were carried out for each plant (5000 nuclei per analysis) on three different days. Nuclear DNA content (2C value) was calculated as sample peak mean/standard peak mean × 2C DNA content of the standard (pg).
Repeat identification from sequence data
We sampled the raw sequence data using the SEQTK command (https://github.com/lh3/seqtk) with a seed of 10 to reduce the genome coverage to 0.1× for all species, and different numbers of paired-end reads sampled depending on the genome size. All sequence reads were then trimmed to the same length (75 bp) and filtered by quality with 95 % of bases equal to or above the quality cut-off value of 10. We employed the similarity-based read clustering method for reads from each species compared to themselves as described by Novák et al. (2010) as implemented in the RepeatExplorer pipeline (https://repeatexplorer-elixir.cerit-sc.cz/; Novák et al., 2013). We used the pipeline default parameters and included a database of Solanum repeats which was available at that moment from The Plant Repeat Database (currently out of service; http://plantrepeats.plantbiology.msu.edu/index.html). The clustering was performed using the default settings of 90 % similarity over 55 % of the read length. This analysis resulted in the clustering of overlapping reads, and these clusters represented different families of repetitive sequences. Reads within individual clusters were also assembled to form contigs, representing sequence variants of corresponding repeats. For the comparative analyses we performed an all-to-all similarity comparison across all species following the same approach. Each set of reads was downsampled to represent 1 % of each genome (i.e. coverage of 0.01) based on 1C values (Table 1). Samples from each species were identified with the three-letter prefixes mentioned above (Table 1), and concatenated to produce datasets as input for RepeatExplorer (Novák et al., 2013) for graph-based clustering. From these data sets, the pipeline retrieved 5 757 692 reads.
Repeat classification
We performed basic repeat classification using a combined approach that involved similarity searches with DNA and protein databases, as implemented in the RepeatExplorer pipeline (Novák et al., 2013), and improved it by including a custom Solanum repeats database. Clusters that were not classified in that way could be annotated by the examination of cluster graph shape and by similarity searches using BLASTN and BLASTX against public databases (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The detection of subrepeats in assembled contigs was performed by similarity dot-plot analysis using a sliding window of 100 bp and similarity cut-off of 40 %. All these sources were combined and used for final manual annotation and quantification of repeats from clusters that represented at least 0.01 % of the investigated genomes. Overall repeat composition was calculated excluding clusters of organelle DNA representing contamination of nuclear DNA preparations by chloroplast and mitochondrial DNA.
Sequence conservation across repeats
We compared the relative abundance of the largest clusters and we also investigated the graph representations of individual clusters with the SeqGrapheR program (Novák et al., 2010) in order to identify protein domains and sequence variants derived from each species or clades as parallel paths in the graph.
RESULTS
Repeat proportion across all species
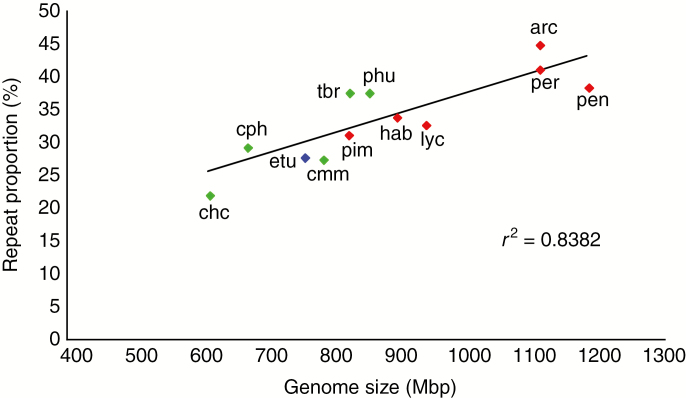
We estimated repeat proportions in the genomes of all species through comparative clustering in RepeatExplorer. Combined, the repeats identified for each species represent between 22.24 % (Solanum cardiophyllum) and 45.12 % (Solanum arcanum) of the total genome for each species (Table 2). There is a high correlation (r2 = 0.84) between repeat proportion and genome size (Fig. 1). There is also a clear grouping of potato clade species with lower genome sizes and tomato clade species with larger genomes.
Table 2.
Genome proportions of different repeat classes and superfamilies in Solanum species
| cph | cmm | chc | phu | tbr | etu | neo | per | arc | pen | pim | hab | lyc | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Repeats | Lineage/class | ||||||||||||||
| LTR retroelements | 0.118 | 0.135 | 0.123 | 0.087 | 0.072 | 0.106 | 2.127 | 1.913 | 1.819 | 2.310 | 1.872 | 1.967 | 1.993 | ||
| Ty1/Copia | 0.069 | 0.124 | 0.139 | 0.067 | 0.068 | 0.108 | 0.170 | 0.180 | 0.177 | 0.215 | 0.232 | 0.210 | 0.225 | ||
| Maximus-SIRE | 0.075 | 0.128 | 0.123 | 0.131 | 0.152 | 0.087 | 0.576 | 0.530 | 0.535 | 0.544 | 0.547 | 0.627 | 0.605 | ||
| Angela | 0.048 | 0.019 | 0.014 | 0.005 | 0.010 | 0.005 | 0.009 | 0.008 | 0.007 | 0.015 | 0.011 | 0.007 | 0.008 | ||
| Tork | 0.185 | 0.247 | 0.248 | 0.175 | 0.231 | 0.190 | 1.153 | 1.261 | 1.188 | 1.004 | 0.983 | 1.077 | 1.104 | ||
| Ale | 0.177 | 0.152 | 0.172 | 0.186 | 0.209 | 0.080 | 0.156 | 0.137 | 0.158 | 0.167 | 0.149 | 0.141 | 0.155 | ||
| Ivana | 0.014 | 0.017 | 0.010 | 0.011 | 0.010 | 0.021 | 0.115 | 0.092 | 0.090 | 0.101 | 0.125 | 0.097 | 0.107 | ||
| Bianca | 0.201 | 0.274 | 0.250 | 0.114 | 0.181 | 0.244 | 0.600 | 0.588 | 0.512 | 0.646 | 0.686 | 0.688 | 0.708 | ||
| TAR | 0.647 | 0.871 | 0.772 | 0.526 | 0.609 | 0.676 | 1.253 | 1.166 | 1.166 | 1.044 | 1.273 | 1.264 | 1.229 | ||
| Total Ty1/Copia | 1.416 | 1.834 | 1.727 | 1.214 | 1.470 | 1.411 | 4.031 | 3.961 | 3.834 | 3.736 | 4.007 | 4.112 | 4.141 | ||
| Ty3/Gypsy | 3.627 | 4.536 | 4.414 | 5.146 | 5.412 | 3.748 | 3.151 | 2.948 | 2.760 | 3.731 | 2.733 | 3.111 | 2.900 | ||
| Chromoviruses | 8.392 | 13.29 | 13.45 | 16.22 | 17.02 | 14.94 | 9.558 | 16.34 | 12.48 | 10.70 | 10.09 | 10.61 | 11.39 | ||
| Ogre | 0.386 | 0.505 | 0.523 | 0.918 | 0.840 | 0.214 | 0.513 | 0.522 | 0.506 | 0.415 | 0.456 | 0.540 | 0.504 | ||
| Athila | 1.260 | 2.922 | 3.403 | 4.412 | 4.418 | 1.980 | 4.746 | 3.946 | 4.846 | 4.099 | 3.397 | 4.794 | 3.868 | ||
| Total Ty3/Gypsy | 13.67 | 21.26 | 21.79 | 26.70 | 27.45 | 20.88 | 17.97 | 23.76 | 20.59 | 18.94 | 16.67 | 19.06 | 18.66 | ||
| Other | Caulimovirus | (Pararetrovirus) | 1.072 | 1.121 | 1.017 | 0.170 | 0.190 | 1.991 | 0.723 | 0.484 | 0.576 | 0.794 | 0.727 | 0.529 | 0.514 |
| LINE | 0.168 | 0.552 | 0.458 | 0.888 | 1.130 | 0.033 | 0.496 | 0.469 | 0.394 | 0.577 | 0.382 | 0.440 | 0.432 | ||
| SINE | 0.016 | 0.027 | 0.020 | 0.051 | 0.030 | 0.016 | 0.013 | 0.010 | 0.011 | 0.008 | 0.014 | 0.012 | 0.011 | ||
| Helitron | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.021 | 0.024 | 0.019 | 0.031 | 0.015 | 0.025 | 0.017 | ||
| DNA transposons | 0.353 | 0.390 | 0.370 | 0.180 | 0.160 | 0.306 | 0.252 | 0.265 | 0.266 | 0.336 | 0.393 | 0.317 | 0.308 | ||
| hAT | 0.200 | 0.237 | 0.238 | 0.107 | 0.109 | 0.051 | 0.121 | 0.130 | 0.119 | 0.144 | 0.175 | 0.159 | 0.143 | ||
| CACTA | 0.134 | 0.121 | 0.103 | 0.098 | 0.106 | 0.055 | 0.091 | 0.089 | 0.074 | 0.109 | 0.099 | 0.088 | 0.092 | ||
| Mutator | 0.039 | 0.030 | 0.032 | 0.012 | 0.009 | 0.022 | 0.093 | 0.091 | 0.090 | 0.135 | 0.135 | 0.106 | 0.131 | ||
| Harbinger | 0.022 | 0.029 | 0.022 | 0.013 | 0.016 | 0.016 | 0.036 | 0.043 | 0.036 | 0.029 | 0.052 | 0.022 | 0.040 | ||
| Total DNA transp | 0.747 | 0.807 | 0.765 | 0.411 | 0.399 | 0.450 | 0.593 | 0.618 | 0.585 | 0.753 | 0.854 | 0.691 | 0.714 | ||
| Tandem repeats | rDNA | 3.342 | 0.602 | 1.837 | 2.712 | 2.705 | 0.892 | 3.891 | 1.982 | 3.090 | 2.448 | 1.864 | 2.476 | 2.458 | |
| Satellites | 1.255 | 1.056 | 1.143 | 5.104 | 3.741 | 1.928 | 5.406 | 4.864 | 8.804 | 5.682 | 2.959 | 3.601 | 2.801 | ||
| Total tandem repeats | 4.597 | 1.658 | 2.980 | 7.816 | 6.446 | 2.820 | 9.927 | 6.846 | 11.894 | 8.130 | 4.823 | 6.077 | 5.259 | ||
| Annotated repetitive total | 21.80 | 27.40 | 28.88 | 37.33 | 37.19 | 27.71 | 35.27 | 38.08 | 39.72 | 35.28 | 29.37 | 32.91 | 31.74 | ||
| Unclassified repetitive | 0.443 | 0.591 | 0.638 | 0.491 | 0.626 | 0.514 | 3.500 | 3.291 | 5.404 | 3.383 | 2.037 | 1.185 | 1.213 | ||
| All repetitive total | 22.24 | 27.99 | 29.52 | 37.83 | 37.81 | 28.23 | 38.77 | 41.37 | 45.12 | 38.67 | 31.41 | 34.10 | 32.95 | ||
Fig. 1.
Correlation between repeat proportion and genome size in potato species (green), tomato species (red) and Solanum etuberosum (blue).
Comparative analysis of major groups of interspersed repeats across and within clades
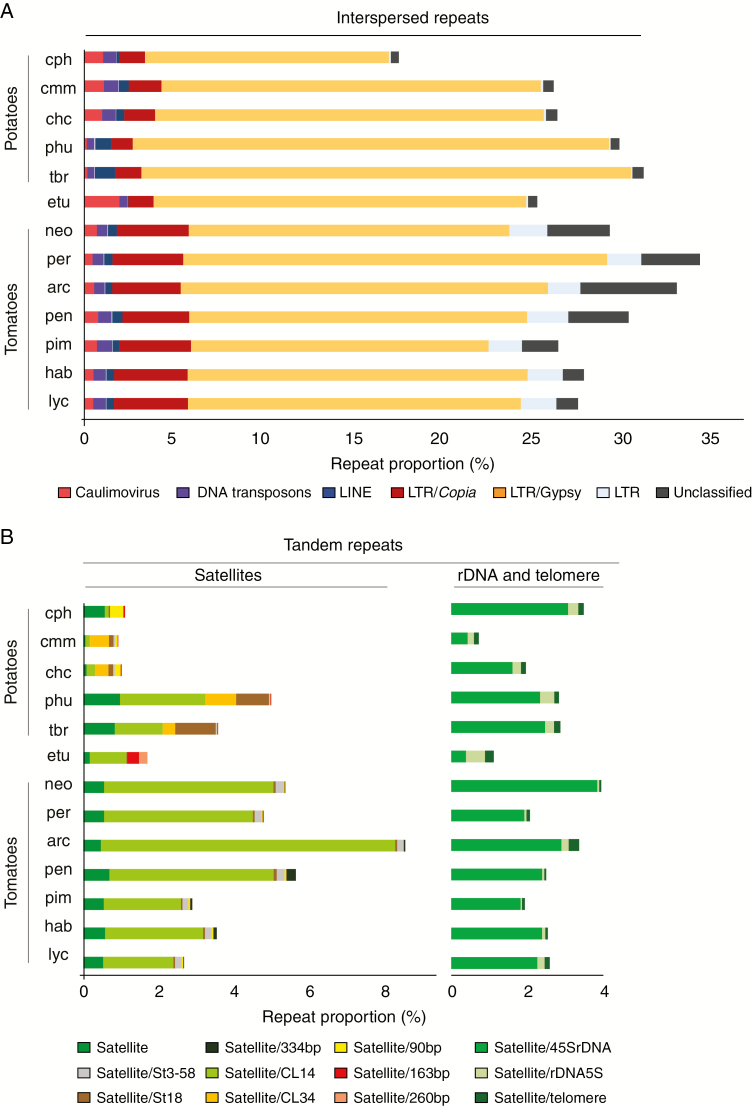
The repetitive fractions of the genomes of all species are composed mainly of LTR retrotransposons. A high proportion of these LTR elements remained unclassified in the tomato clade. Among those that we could annotate, Ty3/Gypsy elements were the most abundant (Fig. 2A), mostly those belonging to the Chromovirus lineage (Table 2). Although these elements are highly prolific in all species, they show significant variation in abundance, with some species having as much as twice the relative content as the others such as S. tuberosum Group Tuberosum vs. S. cardiophyllum (Table 2). In the case of tomato and its relatives, the most frequently observed were Jinling elements (Supplementary Data Table S1), which were almost undetectable in the potato clade.
Fig. 2.
All-to-all similarity comparison of sequence reads from Solanum species based on cluster composition across all 13 species included in this study. The bar plots show the size of the repetitive fraction of the genome, represented as a percentage of each genome. Different colours represent different repeat families. (A) Relative abundance of interspersed repeats. (B) Relative abundance of tandem repeats. See Table 1 for species codes.
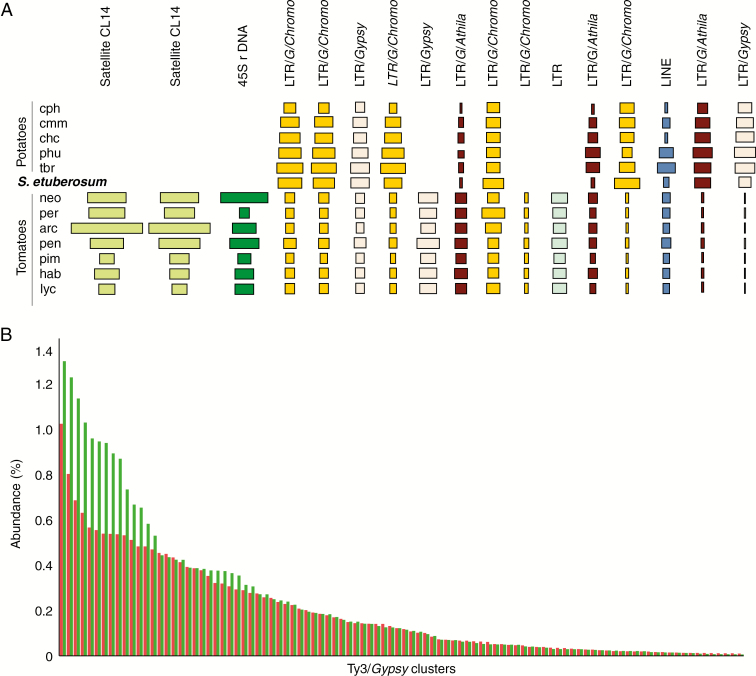
When analysing individual sequence clusters, some of the largest represent Ty3/Gypsy elements, three of them belong to the Chromovirus lineage (Fig. 3A). Several of the most abundant LTR elements including Chromoviruses occur in higher numbers in the potato than in the tomato clade; however several variants (clusters 8, 11 and 12) appear only in the tomato clade (Fig. 3A). We plotted the relative abundance of all clusters annotated as Ty3/Gypsy in descending order from the largest to the smallest for potato species compared to tomato and its wild relatives (Fig. 3B). We found that in potato species a higher proportion of Ty3/Gypsy repeats belong in a few large clusters, while in the tomato clade repeat sequences are more evenly distributed among smaller clusters (Fig. 3B).
Fig. 3.
(A) Sequence composition of the largest 17 clusters derived from the comparative analysis across all 13 species of Solanum included in this study. The size of the rectangle is proportional to the number of reads in a cluster for each species. Colours of the rectangles correspond to repeat type. See Table 1 for species codes. (B) Relative abundance of clusters containing LTR elements of the Ty3/Gypsy type, arranged from the largest to the smallest clusters in potato species (green bars) and tomato species (red bars).
Some types of repeats are more variable among potato species, such as the Caulimovirus type of Pararetroviruses (Fig. 2A). In terms of abundance Caulimoviruses represent only 0.17 and 0.19 % of the genome of cultivated S. tuberosum Group Phureja and Tuberosum, respectively, but their occurrence is almost ten times more prevalent in the wild potatoes (S. cardiophyllum, S. commersonii and S. chacoense, Fig. 2A and Table 2). On the other hand, they show comparable levels of abundance ranging from 0.50 to 0.70 % of the genome across all tomato species (Fig. 2A and Table 2).
Comparative analysis of major groups of tandem repeats across and within clades
Wild potatoes have three- to six-fold lower proportions of tandem repeats than the cultivated potatoes (including satellites, rDNA and telomeric repeats). This discrepancy is mostly caused by one satellite repeat that shows high homology with the satellite CL14 (Torres et al., 2011) when compared to the Solanum repeats database. This satellite is virtually absent in wild potatoes but it is conspicuously abundant in the tomato clade. The two largest clusters in our comparative analysis represent variants of the satellite element CL14 that are only present in the tomato clade (Fig. 3A). Cluster 3 is a variant of the rDNA 45S tandem repeats only present in the tomato clade. Satellite St18 is far more abundant in cultivated potatoes than in the remaining species (Fig. 2B), while St3-58 has a much higher genomic proportion in tomatoes than in potatoes and notably is absent in S. etuberosum and S. cardiophyllum. We also found some lineage-specific tandem repeats, such as a 334-bp satellite that is only present among tomato and its wild relatives, a 90-bp satellite that is more prevalent in wild potatoes, and satellite element CL34 which is present in the potato clade except for S. cardiophyllum and the outgroup species S. etuberosum (Fig. 2B and Table S1).
The relative abundances and the patterns of presence/absence of different repeat elements in the genome of S. etuberosum are more similar to those found in the potato clade. However, S. etuberosum does show some species-specific elements, such as two satellites with 163- and 260-bp repeat units representing 0.32 and 0.22 % of the total genome, respectively (Fig. 2B and Table S1).
Taxon-specific repeats
We identified a total of 58 clusters present in the tomato but not in the potato clade with a maximum genomic abundance of 4.2 %. Among these, the single cluster classified as Helitron was only found among tomato species (Table 2). Tomato-clade-specific repeats include many Chromoviruses belonging to supercluster 7 and among the Ty1/Copia, many Tork elements (Table S1). Twelve clusters found in the potato species could not be detected in tomato species; however, among these, the maximum genomic abundance was only 0.8 %. Solanum cardiophyllum lacked some repeat types that were found in low abundances in other potato species. None of the species-specific repeats identified among the rest of the potato species was significantly abundant (Table S1).
Sequence divergence of the repeats across clades
We compared the sequences appearing in the tomato and potato clades and S. etuberosum in two of the most abundant shared clusters for which we could identify coding domains. Variants were evidenced by alternative paths in the cluster graph layouts (Novák et al., 2010). Cluster 5 (Fig. 3A) was the largest Ty3/Gypsy Chromovirus cluster for which we were able to identify the reverse transcriptase (RT), RNase H (RH) and integrase (INT) domains in the graph layout (Fig. 4A). These domains were conserved across clades; however, we observed alternative narrow paths for the linking sequences in species belonging to the potato and tomato clades (Fig. 4B). For the largest Ty1/Copia cluster (CL25) we identified reads coding for the RT and RH domains (Fig. 4C), but no alternative clade-specific paths were observed in this case (Fig. 4D). In both cases, the paths observed in S. etuberosum (blue dots in Fig. 4) coincided with those of the potato species.
Fig. 4.
Repeat sequence differentiation across clades for LTR elements. (A,C) Cluster graph layout (Novák et al., 2010) representation of (A) Ty3/Gypsy Chromovirus cluster CL005 and (C) of Ty1/Copia cluster CL025 from the comparative analysis across all species. Sequence reads are represented by nodes of the graph and reads with identity of at least 90 % with minimal overlap of 55 % are connected by lines. Reads are coloured based on their similarity to conserved coding domains of LTR retrotransposons. (B,D) Nodes of the graph are coloured based on their species of origin in the comparative analysis across all species. The parts of the graphs that represent the most variable sequence regions in (B) the CL5 element and (C) the CL25 element, which can differentiate between clades, are indicated by black arrows. These variants are evident as narrow parallel paths on the graph representation.
DISCUSSION
In this work we compared the classes, families and lineages of repetitive elements across representative wild and cultivated species of the tomato and potato clades using consistent sequence sampling strategies in order to generate equivalent data sets for each taxon. The combined data set allowed us to interpret the different evolutionary dynamics that have shaped the present composition of the repetitive fraction of the genomes of these groups of species in the current phylogenetic context. The lack of abundant species-specific TEs among the potato species probably explains the difficulty in discriminating their genomes using genome painting techniques such as GISH (Gaiero et al., 2017); however, our analysis has identified unique clusters in some tandem repeats across these clades which can be useful as cytogenetic markers.
Genome size variation and repeat content
The similarity between the genome sizes of species in the potato clade to the modal value of 600 Mbp for angiosperms (Dodsworth et al., 2015b) was in sharp contrast to the values found in the tomato clade ranging from 905 to 1200 Mbp. The correlation between repeat content and genome size shown in Fig. 1 is comparable to correlations published for other genera (Uozu et al., 1997; Neumann et al., 2006; Piégu et al., 2006; Zedek et al., 2010), tribes such as Fabae (Macas et al., 2015) and across the angiosperms (Kidwell, 2002; Vitte and Bennetzen, 2006; Bainard and Gregory, 2013; Lee and Kim, 2014). The observed differences in repeat proportions indicate that the genomes in the tomato and potato clades must have reached a different balance between TE insertion and removal processes since their divergence from their common ancestor.
Tomato species contain more degraded or truncated elements (e.g. solo LTRs) that were identified as LTRs without further classification or that remained simply unclassified. The resulting degraded repeats constitute what is sometimes called genomic ‘dark matter’ and are the result of sequence removal from full-length elements by ectopic recombination (Lee and Schatz, 2012). For the species that had been analysed previously (S. arcanum, S. habrochaites and S. pennellii), Aflitos et al. (2014) suggested that the unique portion of their genomes is roughly the same. Our results show that different abundances of some satellite repeats and a significantly higher proportion of unclassified elements largely explain the rest of the genome size increase in the tomato species.
Interspersed repeats
The most abundant repeats in our study were the LTR-type retrotransposons, particularly the Ty3/Gypsy elements. This higher abundance has already been reported using very different approaches for potato and tomato BAC-end sequences (Datema et al., 2008), tomato chromosome 6 (Peters et al., 2009) and the assembled genomes of tomato (The Tomato Genome Consortium, 2012), S. pennellii (Bolger et al., 2014), potato (Xu et al., 2011) S. commersonii (Aversano et al., 2015) and S. chacoense (Leisner et al., 2018). Tomato and potato LTRs are hypothesized to be the product of large-scale amplification events that took place about 2.8 Mya (The Tomato Genome Consortium, 2012), possibly as a result a large-scale epigenetic change and massive bursts of transposable element activity (Belyayev, 2014).
Ty3/Gypsy elements were, on average, more frequent in the potato species than in the tomato species with the exception of the Jinling elements. These were the most abundant classified Ty3/Gypsy elements found in tomatoes. The presence and distribution of these TEs in tomatoes has already been described. Jinling elements are located in the pericentromere heterochromatin where they are thought to have spread 5 Mya (Wang et al., 2006), during the radiation of the tomato clade after its divergence from the potato clade (Wang et al., 2006; Särkinen et al., 2013). The largest clusters classified as Ty3/Gypsy in potatoes were 30–50 % more prolific than in the tomato clade. In tomatoes, they were more evenly distributed across sequence clusters than in potatoes. This higher sequence divergence across Ty3/Gypsy elements in tomato species as a whole probably reflects different dynamics of this type of TE in the two clades and independent amplification events of different sequence variants within each clade, as shown for Chromovirus CL5.
The Ty1/Copia elements were more abundant in tomato species than in potato species. Manetti et al. (2009) proposed that the Copia element insertion frequency, but not their abundance, may be correlated with the mating system. In the potato clade, diploid species are self-incompatible (Hawkes, 1958). Within the tomato clade, although we did not find a clear relationship between mating system and repeat content, selfing species such as S. lycopersicum, S. habrochaites or S. pimpinellifolium had the lowest repeat abundances and consequently their genome sizes were the lowest among tomatoes and similar to those of potatoes.
Our study used unassembled sequences because we focused on building deliberately equivalent datasets for all the species analysed to compare the relative abundance of repeats. For several of these species, information is available about repetitive sequence distribution and insertion site preferences in those genomes that have been assembled and thoroughly studied cytogenetically. In potato pachytene chromosome complements, there is a large number of chromomeres in the euchromatin, while in tomato, euchromatin is relatively free of such chromomeres in most of the chromosomes (cf. Ramanna and Prakken, 1967; Ramanna and Wagenvoort, 1976; Wagenvoort, 1988; our own obervations). Chromomeres correspond to repeat-rich regions in the genome assemblies of potato (Xu et al., 2011) and tomato (The Tomato Genome Consortium, 2012). In the tomato chromosome 6, Ty1/Copia elements are more abundant in the gene-rich short-arm euchromatin and Ty3/Gypsy repeats are preferentially localized in the heterochromatin, both in the pericentromere and in small-sized chromomeres (Peters et al., 2009).
Interspersed repeats may have caused chromosomal breakages leading to structural rearrangements (Gaut et al., 2007; Belyayev, 2014) in the genomes of the tomato clade. Our approach did not allow us to associate chromosome rearrangements with repeat localization, whereas large-scale changes followed by removal of repeats by unequal recombination (Gaut et al., 2007; Xu and Du, 2014) in the tomato clade might have produced the large amounts of truncated and unclassified LTR elements we found. Peters et al. (2012) described such mobile elements at the synteny breakpoints with the potato and pepper genomes. Rearrangements have occurred between chromosomal fragments located in the pericentromere heterochromatin (Verlaan et al., 2011) and other repeat-rich regions (Seah et al., 2004). Lineage-specific transpositional bursts and ectopic recombination might have been responsible for the chromosome rearrangements found among tomato species but which have not taken place in potatoes and their wild relatives.
Tandem repeats
Tandem repeats, including satellites, occurred in the tomato clade at a higher abundance than in the potato clade (Fig. 2B, Table 2). This class of repeats has been thoroughly described in both clades of Solanum, showing variation in location and abundance (Rokka et al., 1998; Tek and Jiang, 2004; Tek et al., 2005; Chang et al., 2008; Zhu et al., 2008; Brasileiro-Vidal et al., 2009; Szinay, 2010; Torres et al., 2011; Gong et al., 2012; He et al., 2013; Sharma et al., 2013; Tang et al., 2014). The patterns of occurrence are more evident when looking at the largest clusters, particularly satellite DNAs. The most abundant satellite in the tomatoes, CL14 (Torres et al., 2011), was originally described for potato and its relatives and has 99 % sequence identity with the PGR1 repeat (Tang et al., 2014). In our results, the CL14 elements were much more frequent in the tomato clade and displayed a sequence variant that is not present in the potato clade. Although our analysis does not reveal major dissimilarities in the types of tandem repeats described across clades, the quantitative differences produced specific profiles for each clade consistent with the notion of an ancestral ‘library’ of satellite sequences, which were differentially amplified in each clade, as proposed by Fry and Salser (1977).
Phylogenetic context
Among potatoes, the distantly related S. cardiophyllum showed the most obvious divergence within the potato clade, which is coherent with its position as an early branching species in the 1EBN group. However, we found a sharp contrast between cultivated and wild potatoes. The most striking difference is the overall much higher proportion of tandem repeats in cultivated potatoes. Interspersed repeats also showed differences, with 5–6 % more Ty3/Gypsy elements in cultivated potatoes and twice as much Caulimoviruses in wild potatoes. Amplification of a certain type of repeat can occur rapidly and even in a few marginal populations within a species (Belyayev, 2014). It is possible that Caulimoviruses underwent amplification after the divergence of cultivated and wild potatoes, or that a selective bias against them (Kidwell and Lisch, 2001) arose in domesticated potatoes. It remains to be tested whether domestication processes themselves underlie these differences.
The repeat profile of S. etuberosum was more similar to that of potato than to tomato species although a few TE types show unique patterns, particularly Caulimoviruses. The ten-fold higher abundance of this type of TE in S. etuberosum and possible sequence variants in other elements probably explain why GISH results have allowed discrimination between S. tuberosum and S. etuberosum chromosomes in hybrids (Dong et al., 1999; Gavrilenko et al., 2003). In terms of structural genome differentiation, S. etuberosum sometimes shares collinearity with potato species and sometimes with tomato species, while certain chromosome arms are entirely rearranged with respect to both clades (Lou et al., 2010; Szinay et al., 2012). Here we showed that the relative abundance and the patterns of presence/absence of repeats in S. etuberosum were more similar to those found in the potato clade than in the tomato clade. Moreover, S. etuberosum sequences were also more similar to those of potato species in the analysed TE clusters. Given the phylogenetic relationships among these clades, sequence similarity between TEs in potato and S. etuberosum is probably plesiomorphic. Tomato clade-specific sequence variants may have propagated by independent transposition after its divergence from the common ancestor of both clades and S. etuberosum.
Our results are congruent with the current phylogenetic hypotheses for these clades within the genus Solanum. At this point, we cannot establish causal relationships between the constitution of the repetitive fraction of the genome and the different paths that genome evolution has taken in the tomato and potato clades. In spite of this, the patterns we observed and our current understanding support the notion that the dynamics of repetitive elements may be related to the underlying mechanisms that have driven tomato and potato genomes in different directions.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: Output from the annotation of all the repeat classes and lineages across all species in the potato and tomato clade, plus S. etuberosum. The relative genome abundance for each cluster was calculated and the length of the cluster (in Mbp) was estimated where nuclear DNA content was known. All relative abundances for the same repeat type were added up for further comparisons across species and clades.
ACKNOWLEDGEMENTS
We are grateful to Dr Francisco Vilaró, Dr Alicia Castillo and technical staff at INIA for kindly providing in vitro plant material. We are also thankful to Bertus van der Laan and technical staff of the glasshouse facility of Wageningen University for kindly providing and taking care of the plants. We wish to thank Dr Gustavo Folle, Federico Santiñaque and Beatriz López-Carro for their assistance with nuclear DNA content measurements. We also thank Dr Lidjia Berke, Henri van de Geest and Dr Theo Borm for their help with sequence data transfer. We thank Dr Saulo A. Aflitos and Henri van de Geest for patiently assisting with bioinformatics questions. This work was supported by Comisión Sectorial de Investigación Científica, Universidad de la República Uruguay (grant code CSIC I + D 2012 383), and Instituto Nacional de Investigaciones Agropecuarias, Uruguay (grant Línea 4). P.G. was supported by travel grants from Universidad de la República and Comisión Sectorial de Investigación Científica.
LITERATURE CITED
- Aflitos SA, Schijlen E, de Jong H, et al. . 2014. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. The Plant Journal 80: 136–148. [DOI] [PubMed] [Google Scholar]
- Anderson LK, Covey PA, Larsen LR, Bedinger P, Stack SM.. 2010. Structural differences in chromosomes distinguish species in the tomato clade. Cytogenetic and Genome Research 129: 24–34. [DOI] [PubMed] [Google Scholar]
- Aversano R, Contaldi F, Ercolano MR, et al. . 2015. The Solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. The Plant Cell 27: 954–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainard JD, Gregory TR.. 2013. Genome size evolution: patterns, mechanisms, and methodological advances. Genome 56: vii–viii. [DOI] [PubMed] [Google Scholar]
- Belyayev A. 2014. Bursts of transposable elements as an evolutionary driving force. Journal of Evolutionary Biology 27: 2573–2584. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. 1996. The contributions of retroelements to plant genome organization, function and evolution. Trends in Microbiology 4: 347–353. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. 2000. Transposable element contributions to plant gene and genome evolution. Plant Molecular Biology 42: 251–269. [PubMed] [Google Scholar]
- Bennetzen JL, Wang H.. 2014. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annual Review of Plant Biology 65: 505–530. [DOI] [PubMed] [Google Scholar]
- Bernatzky R, Tanksley SD.. 1986. Toward a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics 112: 887–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A, Scossa F, Bolger ME, et al. . 2014. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nature Genetics 46: 1034–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw JE, Bryan GJ, Ramsay G.. 2006. Genetic resources (including wild and cultivated Solanum species) and progress in their utilisation in potato breeding. Potato Research 49: 49–65. [Google Scholar]
- Brasileiro-Vidal AC, Melo-Oliveira MB, Carvalheira GMG, Guerra M.. 2009. Different chromatin fractions of tomato (Solanum lycopersicum L.) and related species. Micron (Oxford, England : 1993) 40: 851–859. [DOI] [PubMed] [Google Scholar]
- Camadro EL, Carputo D, Peloquin SJ.. 2004. Substitutes for genome differentiation in tuber-bearing Solanum: interspecific pollen-pistil incompatibility, nuclear-cytoplasmic male sterility, and endosperm. Theoretical and Applied Genetics 109: 1369–76. [DOI] [PubMed] [Google Scholar]
- Castañeda-Álvarez NP, Khoury CK, Achicanoy HA, et al. . 2016. Global conservation priorities for crop wild relatives. Nature Plants 2: 16022. [DOI] [PubMed] [Google Scholar]
- Chang S-B, Yang T-J, Datema E, et al. . 2008. FISH mapping and molecular organization of the major repetitive sequences of tomato. Chromosome Research 16: 919–933. [DOI] [PubMed] [Google Scholar]
- Datema E, Mueller LA, Buels R, et al. . 2008. Comparative BAC end sequence analysis of tomato and potato reveals overrepresentation of specific gene families in potato. BMC Plant Biology 8: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodsworth S, Chase MW, Kelly LJ, et al. . 2015. a Genomic repeat abundances contain phylogenetic signal. Systematic Biology 64: 112–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodsworth S, Leitch AR, Leitch IJ.. 2015. b Genome size diversity in angiosperms and its influence on gene space. Current Opinion in Genetics and Development 35: 73–78. [DOI] [PubMed] [Google Scholar]
- Dodsworth S, Chase MW, Särkinen T, Knapp S, Leitch AR.. 2016. Using genomic repeats for phylogenomics: a case study in wild tomatoes (Solanum section Lycopersicon : Solanaceae). Biological Journal of the Linnean Society 117: 96–105. [Google Scholar]
- Doležel J, Göhde W.. 1995. Sex determination in dioecious plants Melandrium album and M. rubrum using high-resolution flow cytometry. Cytometry 19: 103–106. [DOI] [PubMed] [Google Scholar]
- Doležel J, Sgorbati S, Lucretti S.. 1992. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiologia Plantarum 85: 625–631. [Google Scholar]
- Dong F, Novy RG, Helgeson JP, Jiang J.. 1999. Cytological characterization of potato - Solanum etuberosum somatic hybrids and their backcross progenies by genomic in situ hybridization. Genome 42: 987–992. [Google Scholar]
- Dong F, McGrath JM, Helgeson JP, Jiang J.. 2001. The genetic identity of alien chromosomes in potato breeding lines revealed by sequential GISH and FISH analyses using chromosome-specific cytogenetic DNA markers. Genome 44: 729–734. [PubMed] [Google Scholar]
- Dong F, Tek AL, Frasca ABL, et al. . 2005. Development and characterization of potato-Solanum brevidens chromosomal addition/substitution lines. Cytogenetic and Genome Research 109: 368–372. [DOI] [PubMed] [Google Scholar]
- Dvorak J. 1983. Evidence of genetic suppression of heterogenetic chromosome pairing in polyploid species of Solanum, sect. Petota. Canadian Journal of Genetics and Cytology25: 530–539. [Google Scholar]
- Feschotte C, Jiang N, Wessler SR.. 2002. Plant transposable elements: where genetics meets genomics. Nature Reviews Genetics 3: 329–341. [DOI] [PubMed] [Google Scholar]
- Finnegan DJ. 1989. Eukaryotic transposable elements and genome evolution. Trends in Genetics 5: 103–107. [DOI] [PubMed] [Google Scholar]
- Fry K, Salser W.. 1977. Nucleotide sequences of HS-α satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell 12: 1069–1084. [DOI] [PubMed] [Google Scholar]
- Gaiero P, van de Belt J, Vilaró F, Schranz ME, Speranza P, de Jong H.. 2016. Collinearity between potato (Solanum tuberosum L.) and wild relatives assessed by comparative cytogenetic mapping. Genome 60: 228–240. [DOI] [PubMed] [Google Scholar]
- Gaiero P, Mazzella C, Vilaró F, Speranza P, de Jong H.. 2017. Pairing analysis and in situ hybridisation reveal autopolyploid-like behaviour in Solanum commersonii × S. tuberosum (potato) interspecific hybrids. Euphytica 213: 137. [Google Scholar]
- Gaut BS, Wright SI, Rizzon C, Dvorak J, Anderson LK.. 2007. Recombination: an underappreciated factor in the evolution of plant genomes. Nature Reviews Genetics 8: 77–84. [DOI] [PubMed] [Google Scholar]
- Gavrilenko T, Larkka J, Pehu E, Rokka V-M.. 2002. Identification of mitotic chromosomes of tuberous and non-tuberous Solanum species (Solanum tuberosum and Solanum brevidens) by GISH in their interspecific hybrids. Genome 45: 442–449. [DOI] [PubMed] [Google Scholar]
- Gavrilenko T, Thieme R, Heimbach U, Thieme T.. 2003. Somatic hybrids of Solanum etuberosum (+) dihaploid Solanum tuberosum and their backcrossing progenies: relationships of genome dosage with tuber development. Euphytica 131: 323–332. [Google Scholar]
- Gong Z, Wu Y, Koblízková A, et al. . 2012. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. The Plant Cell 24: 3559–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandillo S, Chetelat RT, Knapp S, et al. . 2011. Solanum section Lycopersicon. In: Kole, C, ed. Wild crop relatives: genomic and breeding resources: temperate fruits. Berlin: Springer-Verlag, 129–215. [Google Scholar]
- Hajjar R, Hodgkin T.. 2007. The use of wild relatives in crop improvement: a survey of developments over the last 20 years. Euphytica 156: 1–13. [Google Scholar]
- Hawkes JG. 1958. Significance of wild species and primitive forms for potato breeding. Euphytica 7: 257–270. [Google Scholar]
- Hawkes JG. 1990. The potato: evolution, biodiversity and genetic resources. London: Belhaven. [Google Scholar]
- He L, Liu J, Torres GA, Zhang H, Jiang J, Xie C.. 2013. Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species. Chromosome Research 21: 5–13. [DOI] [PubMed] [Google Scholar]
- Jansky SH. 2009. Breeding, genetics, and cultivar development. In: Singh J, Kaur L, eds. Advances in potato chemistry and technology. Amsterdam: Elsevier Ltd, 27–62. [Google Scholar]
- Ji Y, Chetelat RT.. 2003. Homoeologous pairing and recombination in Solanum lycopersicoides monosomic addition and substitution lines of tomato. Theoretical and Applied Genetics 106: 979–789. [DOI] [PubMed] [Google Scholar]
- Ji Y, Pertuzé R, Chetelat RT.. 2004. Genome differentiation by GISH in interspecific and intergeneric hybrids of tomato and related nightshades. Chromosome Research 12: 107–116. [DOI] [PubMed] [Google Scholar]
- Kelly LJ, Renny-Byfield S, Pellicer J, et al. . 2015. Analysis of the giant genomes of Fritillaria (Liliaceae) indicates that a lack of DNA removal characterizes extreme expansions in genome size. New Phytologist 208: 596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG. 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115: 49–63. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Holyoake AJ.. 2001. Transposon-induced hotspots for genomic instability. Genome Research 11: 1321–1322. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR.. 2001. Transposable elements, parasitic DNA, and genome evolution. Evolution 55: 1–24. [DOI] [PubMed] [Google Scholar]
- Lee H, Schatz MC.. 2012. Genomic dark matter: the reliability of short read mapping illustrated by the genome mappability score. Bioinformatics 28: 2097–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-I, Kim N-S.. 2014. Transposable elements and genome size variations in plants. Genomics & Informatics 12: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner CP, Hamilton JP, Crisovan E, et al. . 2018. Genome sequence of M6, a diploid inbred clone of the high glycoalkaloid-producing tuber-bearing potato species Solanum chacoense, reveals residual heterozygosity. The Plant Journal 94: 562–570. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Leitch AR.. 2013. Genome size diversity and evolution in land plants. In: Greilhuber J, Dolezel J, Wendel JF, eds. Plant genome diversity Volume 2 Vienna: Springer Vienna, 307–322. [Google Scholar]
- Lou Q, Iovene M, Spooner DM, Buell CR, Jiang J.. 2010. Evolution of chromosome 6 of Solanum species revealed by comparative fluorescence in situ hybridization mapping. Chromosoma 119: 435–442. [DOI] [PubMed] [Google Scholar]
- Macas J, Novak P, Pellicer J, et al. . 2015. In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe Fabeae. PLoS ONE 10: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetti ME, Rossi M, Nakabashi M, Grandbastien MA, Van Sluys MA.. 2009. The Tnt1 family member Retrosol copy number and structure disclose retrotransposon diversification in different Solanum species. Molecular Genetics and Genomics 281: 261–271. [DOI] [PubMed] [Google Scholar]
- Matsubayashi M. 1991. Phylogenetic relationships in the potato and its related species. In: Tsuchiya T, Gupta P, eds. Chromosome engineering in plants: genetics, breeding, evolution, part B. Amsterdam: Elsevier, 93–118. [Google Scholar]
- McClintock B. 1946. Maize genetics. In: Carnegie Institution of Washington Year Book No. 45. Washington, DC: Carnegie Institution of Washington, 176–186. [PubMed] [Google Scholar]
- Neumann P, Koblížková A, Navrátilová A, Macas J.. 2006. Significant expansion of Vicia pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics 173: 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Macas J.. 2010. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Pech J, Steinhaisl J, Macas J.. 2013. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29: 792–793. [DOI] [PubMed] [Google Scholar]
- Novák P, Hřibová E, Neumann P, Koblížková A, Doležel J, Macas J.. 2014. Genome-wide analysis of repeat diversity across the family Musaceae. PLoS ONE 9: e98918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parokonny A, Marshall J, Bennett MD, Cocking EC, Davey MR, Power JB.. 1997. Homoeologous pairing and recombination in backcross derivatives of tomato somatic hybrids (Lycopersicon esculentum (+) L. peruvianum). Theoretical and Applied Genetics 94: 713–723. [Google Scholar]
- Peters SA, Datema E, Szinay D, et al. . 2009. Solanum lycopersicum cv. Heinz 1706 chromosome 6: distribution and abundance of genes and retrotransposable elements. The Plant Journal 58: 857–869. [DOI] [PubMed] [Google Scholar]
- Peters SA, Bargsten JW, Szinay D, et al. . 2012. Structural homology in the Solanaceae: analysis of genomic regions in support of synteny studies in tomato, potato and pepper. The Plant Journal 71: 602–614. [DOI] [PubMed] [Google Scholar]
- Piégu B, Guyot R, Picault N, et al. . 2006. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Research 16: 1262–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piégu B, Bire S, Arensburger P, Bigot Y.. 2015. A survey of transposable element classification systems - A call for a fundamental update to meet the challenge of their diversity and complexity. Molecular Phylogenetics and Evolution 86: 90–109. [DOI] [PubMed] [Google Scholar]
- Ramanna MS, Prakken R.. 1967. Structure of and homology between pachytene and somatic metaphase chromosomes of the tomato. Genetica 38: 115–133. [Google Scholar]
- Ramanna M, Wagenvoort M.. 1976. Identification of the trisomic series in diploid Solanum tuberosum L., Group Tuberosum. I. Chromosome identification. Euphytica 25: 233–240. [Google Scholar]
- Ramsay G, Bryan G.. 2011. Solanum. In: Kole, C, ed. Wild crop relatives: genomic and breeding resources, vegetables. Berlin: Springer, 259–271. [Google Scholar]
- Raskina O, Barber JC, Nevo E, Belyayev A.. 2008. Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenetic and Genome Research 120: 351–357. [DOI] [PubMed] [Google Scholar]
- Rodriguez F, Wu F, Ané C, Tanksley S, Spooner DM.. 2009. Do potatoes and tomatoes have a single evolutionary history, and what proportion of the genome supports this history?BMC Evolutionary Biology 9: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokka VM, Clark MS, Knudson DL, et al. . 1998. Cytological and molecular characterization of repetitive DNA sequences of Solanum brevidens and Solanum tuberosum. Genome 41: 487–494. [DOI] [PubMed] [Google Scholar]
- Särkinen T, Bohs L, Olmstead RG, Knapp S.. 2013. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. BMC Evolutionary Biology 13: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seah S, Yaghoobi J, Rossi M, Gleason CA, Williamson VM.. 2004. The nematode-resistance gene, Mi-1, is associated with an inverted chromosomal segment in susceptible compared to resistant tomato. Theoretical and Applied Genetics 108: 1635–1642. [DOI] [PubMed] [Google Scholar]
- Sharma SK, Bolser D, de Boer J, et al. . 2013. Construction of reference chromosome-scale pseudomolecules for potato: integrating the potato genome with genetic and physical maps. G3: Genes, Genomes, Genetics 3: 2031–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner DM, Ghislain M, Simon R, Jansky SH, Gavrilenko T.. 2014. Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Botanical Review 80: 283–383. [Google Scholar]
- Szinay D. 2010. The development of FISH tools for genetic, phylogenetic and breeding studies in tomato (Solanum lycopersicum). PhD thesis, Wageningen University and Research, The Netherlands; ISBN:9789085856351. [Google Scholar]
- Szinay D, Bai Y, Visser R, de Jong H.. 2010. FISH applications for genomics and plant breeding strategies in tomato and other solanaceous crops. Cytogenetic and Genome Research 129: 199–210. [DOI] [PubMed] [Google Scholar]
- Szinay D, Wijnker E, van den Berg R, Visser RGF, de Jong H, Bai Y.. 2012. Chromosome evolution in Solanum traced by cross-species BAC-FISH. The New Phytologist 195: 688–698. [DOI] [PubMed] [Google Scholar]
- Tang X, Szinay D, Lang C, et al. . 2008. Cross-species bacterial artificial chromosome-fluorescence in situ hybridization painting of the tomato and potato chromosome 6 reveals undescribed chromosomal rearrangements. Genetics 180: 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Datema E, Guzman MO, et al. . 2014. Chromosomal organizations of major repeat families on potato (Solanum tuberosum) and further exploring in its sequenced genome. Molecular Genetics and Genomics 289: 1307–1319. [DOI] [PubMed] [Google Scholar]
- Tek AL, Jiang J.. 2004. The centromeric regions of potato chromosomes contain megabase-sized tandem arrays of telomere-similar sequence. Chromosoma 113: 77–83. [DOI] [PubMed] [Google Scholar]
- Tek AL, Stevenson WR, Helgeson JP, Jiang J.. 2004. Transfer of tuber soft rot and early blight resistances from Solanum brevidens into cultivated potato. Theoretical and Applied Genetics 109: 249–254. [DOI] [PubMed] [Google Scholar]
- Tek AL, Song J, Macas J, Jiang J.. 2005. Sobo, a recently amplified satellite repeat of potato, and its implications for the origin of tandemly repeated sequences. Genetics 170: 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tomato Genome Consortium 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres GA, Gong Z, Iovene M, et al. . 2011. Organization and evolution of subtelomeric satellite repeats in the potato genome. G3: Genes, Genomes, Genetics 1: 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozu S, Ikehashi H, Ohmido N, Ohtsubo H, Ohtsubo E, Fukui K.. 1997. Repetitive sequences: cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Molecular Biology 35: 791–799. [DOI] [PubMed] [Google Scholar]
- Van Der Knaap E, Sanyal A, Jackson SA, Tanksley SD.. 2004. High-resolution fine mapping and fluorescence in situ hybridization analysis of sun, a locus controlling tomato fruit shape, reveals a region of the tomato genome prone to DNA rearrangements. Genetics 168: 2127–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaan MG, Szinay D, Hutton SF, et al. . 2011. Chromosomal rearrangements between tomato and Solanum chilense hamper mapping and breeding of the TYLCV resistance gene Ty-1. The Plant Journal 68: 1093–1103. [DOI] [PubMed] [Google Scholar]
- Vitte C, Bennetzen JL.. 2006. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proceedings of the National Academy of Sciences of the United States of America 103: 17638–17643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenvoort M. 1988. Spontaneous structural rearrangements in Solanum L. Chromosome identification at pachytene stage. Euphytica 9: 159–167. [Google Scholar]
- Wang Y, Tang X, Cheng Z, Mueller L, Giovannoni J, Tanksley SD.. 2006. Euchromatin and pericentromeric heterochromatin: comparative composition in the tomato genome. Genetics 172: 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, et al. . 2007. A unified classification system for eukaryotic transposable elements. Nature Reviews Genetics 8: 973–982. [DOI] [PubMed] [Google Scholar]
- Xu X, Pan S, Cheng S, et al. . 2011. Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195. [DOI] [PubMed] [Google Scholar]
- Xu Y, Du J.. 2014. Young but not relatively old retrotransposons are preferentially located in gene-rich euchromatic regions in tomato (Solanum lycopersicum) plants. Plant Journal 80: 582–591. [DOI] [PubMed] [Google Scholar]
- Zedek F, Smerda J, Smarda P, Bureš P.. 2010. Correlated evolution of LTR retrotransposons and genome size in the genus Eleocharis. BMC Plant Biology 10: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Ouyang S, Iovene M, et al. . 2008. Analysis of 90 Mb of the potato genome reveals conservation of gene structures and order with tomato but divergence in repetitive sequence composition. BMC Genomics 9: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.