Abstract
Background and Aims
In recent years, increasing numbers of long non-coding RNAs (lncRNAs) have been identified in humans, animals and plants, and several of them have been shown to play important roles in diverse biological processes. However, little work has been performed on the regulation mechanism of lncRNA biogenesis and expression, especially in plants. Compared with studies of tomato MADS-box transcription factor RIPENING INHIBITOR (RIN) target coding genes, there are few reports on its relationship to non-coding RNAs. The aim of the present study was to identify and explore the specific role of RIN target lncRNAs in tomato fruit development and ripening.
Methods
lncRNA targets of RIN were identified by chromatin immunoprecipitation sequencing (ChIP-seq) combined with RNA deep sequencing analysis. Six selected lncRNA targets were validated by quantitative real-time PCR, ChIP and electrophoretic mobility shift assays, and we further confirmed differential expression between wild-type and ripening-deficient mutant fruit, and RIN direct binding in the promoter regions. By means of virus-induced gene silencing (VIGS) assays and a clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) genome editing strategy, the ripening-related function of a specific target lncRNA (lncRNA2155) was studied.
Key Results
We identified 187 lncRNAs as direct RIN targets, which exhibited RIN binding sites in their promoters and showed different expression between the wild-type and rin mutant. Six target lncRNAs were shown to bind with RIN directly in their promoters in vivo and in vitro. Moreover, using CRISPR/Cas9 technology to knock out the locus of the target lncRNA2155 indicated that it delayed fruit ripening in tomato.
Conclusions
Collectively, these findings provide new insight into RIN in the transcriptional regulation of lncRNAs and suggest that lncRNAs will contribute to a better understanding of the RIN regulatory network that controls fruit ripening.
Keywords: MADS box transcription factor RIN, long non-coding RNAs, chromatin immunoprecipitation, electrophoretic mobility shift assay, fruit ripening, Solanum lycopersicum
INTRODUCTION
Fruits have important nutritious and health roles in the human diet. Several characteristics of the flesh, including colour, flavour, aroma, texture and nutrition, have been studied for their dramatic changes during growth (Osorio et al., 2011). As a tightly coordinated and regulated set of physiological and biochemical processes, the molecular mechanisms of fruit ripening are worthy of study (Giovannoni, 2004). Several well-characterized tomato mutations with impressive effects on ripening can initiate transcription regulation of fruit ripening (Giovannoni, 2007), such as ripening inhibitor (rin) (Vrebalov et al., 2002), nonripening (nor) (Ng and Tigchelaar, 1977), colorless ripening (Cnr) (Manning et al., 2006), greenripe (Gr) (Barry and Giovannoni, 2006) and never ripe (Nr) (Lanahan et al., 1994). Similarly, transcription factors (TFs) have been identified as major contributory factors for understanding fruit development and ripening, including TOMATO AGAMOUS-LIKE1 (TAGL1) (Itkin et al., 2009), two FRUITFULL homologues (TDR4/FUL1 and MBP7/FUL2) (Bemer et al., 2012), ETHYLENE RESPONSE FACTOR 6 (ERF6) (Lee et al., 2012) and AP2a (Chung et al., 2010). The MADS-box TF RIPENING INHIBITOR (RIN) has a key role in regulating tomato fruit ripening and has been widely studied in tomato. The rin mutant, carrying a mutation in RIN, inhibits numerous ripening-related phenotypes, including loss of the respiratory climacteric and associated ethylene evolution, severely affecting carotenoid accumulation, softening and production of flavour compounds (Vrebalov et al., 2002). Recent work has begun to suggest that rin is not a null mutation, but rather it is a gain-of-function mutation that produces a protein that actively represses ripening (Ito et al., 2017). RIN mainly binds to the C-A/T-rich-G (the consensus CArG) motifs and interacts with the promoters of many ripening-related genes. In total, 241 direct RIN target genes have been identified by chromatin immunoprecipitation (ChIP)-chip and transcriptome analysis (Fujisawa et al., 2013). Transcriptome studies of wild-type and rin mutant fruit showed that MADS-RIN activity contributes to the expression of a great number of ripening-related genes, most of which have functionally defined roles, such as various cell-wall-integral and carbohydrate-modifying proteins which help to build the structure of ripening fruit (Zhong et al., 2013); SlUBC32 and PSMD2, which are involved in the ubiquitin-proteasome pathway, are identified as novel direct targets of RIN (Wang et al., 2014). RIN binds directly to the promoter region of MIR172a, implying the potential regulation of microRNA (miRNA) accumulation by RIN (Gao et al., 2015). However, most studies of RIN targets have focused on coding genes, and little is known about the role of RIN in long non-coding RNAs (lncRNAs).
lncRNAs are broadly defined as non-coding RNAs longer than 200 nucleotides in length (Jin et al., 2013). Recent advances suggest that lncRNAs play critical roles in transcriptional and post-transcriptional regulation (Vanwerven et al., 2012), as well as in epigenetic modification, cell differentiation and development (Zofall et al., 2012). Although lncRNAs have received more attention in recent years, research has mainly concerned functional analysis (Bai et al., 2015), and knowledge of the transcriptional regulation mechanism of lncRNAs remains scant.
Here, we report the genome-wide identification of lncRNA targets of RIN using ChIP sequencing (ChIP-seq) combined with transcriptome analysis. A total of 627 lncRNAs were identified to have RIN binding sites and 187 of these lncRNAs were significantly differentially expressed in wild-type and rin mutant fruits. Further analysis indicated that RIN binds directly to the promoter regions of several target lncRNAs in vivo and in vitro. Using a clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-mediated knockout approach, we explored the functional role of a specific target lncRNA2155 in fruit ripening. Overall, our results shed new light on the transcriptional regulation of lncRNAs, and help to better enrich the network of fruit ripening.
MATERIAL AND METHODS
Plant materials and growth conditions
Wild-type tomato (Solanum lycopersicum ‘Ailsa Craig’) and rin mutant (‘Ailsa Craig’ background) were grown in a glasshouse under standard conditions (16 h under light at 26 °C, 8 h in the dark at 20 °C), with regular addition of fertilizer and lighting. To collect fruits, they were tagged at anthesis, and harvested at the immature green (IM), mature green (MG), breaker (BR), BR+3, pink (PK) and red-ripe (RR) stages based on days post-anthesis (dpa), respectively. Immediately upon harvest, the pericarp was manually dissected, frozen in liquid nitrogen, and stored at −80 °C until use. Seeds of 35S-driven overexpression of RIN in tomato were kindly provided by Prof. Guozheng Qin (Institute of Botany, CAS, Beijing, China). Wild-type MicroTom (S. lycopersicum ‘Micro Tom’) were also planted for virus-induced gene silencing (VIGS) and CRISPR/Cas9 transgenic lines.
ChIP-seq data analysis
Sequencing reads were mapped to the tomato genome available at the Genome Project (Tomato Gene Consortium, 2012) using Bowtie2 (http://solgenomics.net/organism/ Solanum_lycopersicum/genome) (Langmead et al., 2009). The raw sequencing data were deposited in the National Center for Biotechnology Information Sequence Read Archive, which are publicly available (http://www.ncbi.nlm.nih.gov/sra/) under accession number SRA053345. To estimate the number of uniquely mapped reads, we used SAMtools (Sequence Alignment Map format) for mapping score as a filter (samtools view –bq 1). For peak calling, data were analysed using the program MACS2 (Zhang et al., 2008), and 32 243 peaks were obtained. MACS2 provided the false-discovery rate for each peak, and we used 0.01 as the cut-off threshold. Using these criteria, we obtained 23 594 significantly enriched and overlapping peaks from two RIN ChIP-seq replicates. RIN binding motifs were then predicted by using MEME-ChIP. We extracted the sequences 250 bp upstream and downstream of the peak summits for MEME-ChIP enrichment analysis, which used the programs MEME and DREME to identify sequence motifs (Machanick and Bailey, 2011).
RNA-seq analysis
Clean reads were obtained by RNA-seq and deposited in the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under accession number SRP106775. The program FastQC (v.0.11.3) was used to check whether the quality of reads was credible for further analyses. All clean reads were then mapped to the tomato reference genomes (SGN release SL2.50; ftp://ftp.sgn.cornell.edu/tomato_genome) combined with tomato lncRNAs from our data using TopHat (v.1.4.6), and fragments were assigned to genes by the feature count and count program. Reads per kilobase per million mapped reads (RPKM) was used to express the expression value. Fold changes were calculated by using log2 (RPKM RIN RNAi /RPKM AC). lncRNAs were considered as differentially expressed under the threshold of |log2 fold change| > 1, P < 0.05.
RNA extraction and quantitative real-time PCR
Total RNA was isolated from fruit samples using Trizol reagent prepared in our lab (Zhu et al., 2015). RNA integrity was validated by 1.5 % (v/v) agarose gel electrophoresis. Genomic DNA was removed from the extracted total RNA by DNase I (TaKaRa, Shiga, Japan). RNA concentration and purity were measured using a NAS-99 spectrophotometer (ATC Gene, NJ, USA). A 2-μg aliquot of total RNA was reverse transcribed via cDNA synthesis using a TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kit (Trans, Beijing, China) with random primer. Quantitative real-time PCR (qRT-PCR) was performed with a Bio-Rad CFX96 real-time PCR detection system in standard mode using SYBR Green Supermix (Bio-Rad, CA, USA). The following PCR programme was used: 95 °C for 2 min, followed by 39 cycles of 95 °C for 15 s and 60 °C for 30 s. Samples were normalized against the Actin gene (Solyc03g078400), and relative gene expression values were measured using the cycle threshold (Ct) 2−ΔΔCt method. Three biological replicates were included and each independent sample was performed in triplicate. Oligonucleotide primers used are listed in Supplementary Data Table S1.
Chromatin immunoprecipitation
The ChIP assay was performed as described by Wang et al. (2014). The pericarp of the fruit tissue was sliced and fixed with 1 % formaldehyde under vacuum, and then subjected to nuclear isolation. The chromatin was sheared to an average length of approximately 500 bp by sonication. A small aliquot of the sonicated chromatin was reversibly cross-linked, and was used as the input DNA control. The remaining chromatin sample was centrifuged and the supernatant was diluted 10-fold in ChIP dilution buffer and pre-cleared by incubation with Dynabeads (Millipore, Waltham, MA, USA) for 1 h at 4 °C. The monoclonal anti-FLAG antibody (Sigma, St Louis, MO, USA) and pre-immune serum IgG (negative control) were used. DNA fragments were purified using the QIAquick PCR Purification Kit (Qiagen, Dusseldorf, Germany). ChIP assays were repeated with three biological replicates. The immunoprecipitated DNA was analysed by qPCR using the primers detailed in Table S1.
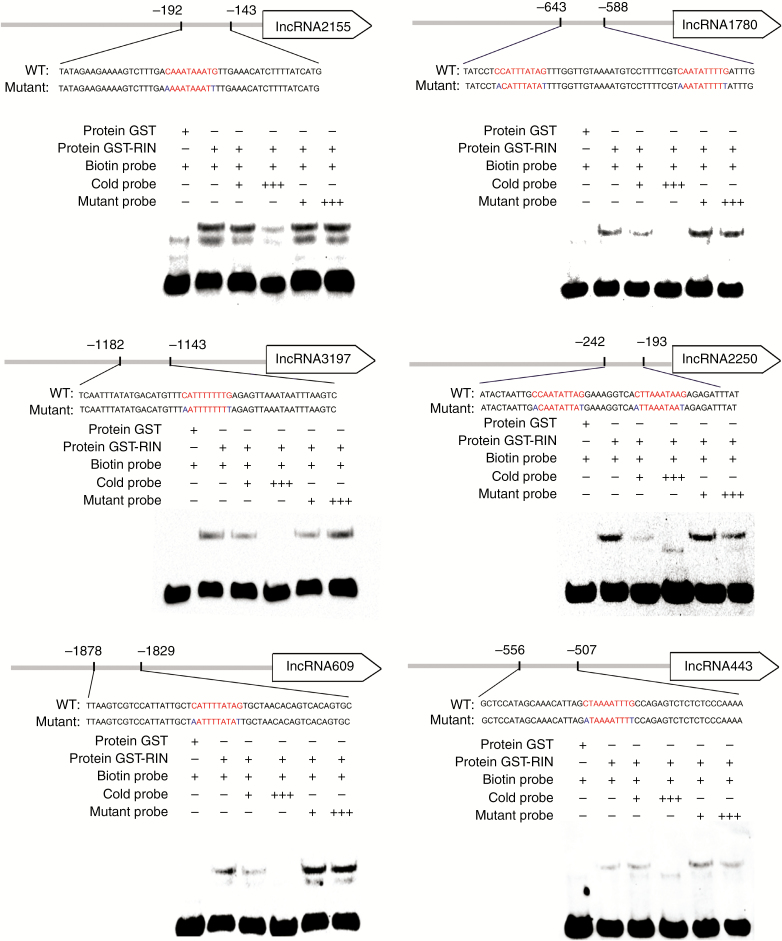
Electrophoretic mobility shift assay
An electrophoretic mobility shift assay (EMSA) was carried out according to the method of Han et al. (2016) with some modifications. The full-length RIN was amplified from tomato cDNA, fused in frame to glutathione S-transferase (GST) and expressed in Escherichia coil BM Rosetta (DE3) cells by induction with 0.5 mm IPTG (isopropyl β-d-thiogalactoside) for 4 h at 16 °C. The recombinant protein was purified with Glutathione-Sepharose 4B (GE Healthcare, NJ, USA). An EMSA was performed using the Electrophoretic Mobility Shift Assay kit from Thermo Fisher Scientific (Waltham, MA, USA). Probes containing one or more CArG boxes which were derived from the promoter regions of lncRNAs were labelled with biotin using the DNA 3′ End Biotinylation kit (Thermo Fisher Scientific). The competitor was the same unlabelled DNA fragment, and the mutant control was the probe with an ANNNNNNNNT sequence instead of CArG box. The probes were incubated with the fusion protein for 25 min. After cross-linking, the biotin-labelled probes on the membrane were detected by using the chemiluminescence reagents provided with the EMSA kit. The probes are listed in Table S1.
Protein extraction and western blot
Protein extraction was performed according to Ma et al. (2016). In brief, fruit samples were placed in extraction buffer containing 10 % TCA/acetone and centrifuged at 4 °C, the supernatant was removed, and the precipitate was then mixed with 80 % MeOH and 0.1 m ammonium acetate, and centrifuged. The supernatant was discarded and the tubes were washed with 80 % acetone, centrifuged and the supernatant was again discarded. Then phenol/SDS solution [Tris-phenol, pH 8.0; SDS buffer (30 % sucrose, 2 % SDS, 0.2 m Tris pH 8.0, 5 % β-mercaptoethanol)] was added (1: 1, v/v) to extract protein. After washing in MeOH and 80 % acetone, the protein was finally suspended in SDS buffer (0.5 m Tris pH 7.0, 1.4 % SDS) and stored at −80 °C until use. Protein concentration was determined by the Bradford method (Solarbio, Beijing, China) using bovine serum albumin as standard. For immunoblotting, proteins were separated by SDS-PAGE (12 % acrylamide gels) and electrotransferred to Immobilon-P PVDF membranes (Millipore, MMAS, USA). The membrane was blocked in 5 % skimmed milk for 2 h at room temperature. An anti-FLAG monoclonal antibody (Sigma, MO, USA) was used. Membranes were washed with 0.05 % Tween 20 in Tris-buffered saline three times and then reacted with the corresponding secondary antibodies conjugated to horseradish peroxidase (HRP) at a dilution of 1: 10 000. An enhanced chemiluminescence kit (Absin, Beijing, China) was used for detection after incubation with the HRP-conjugated secondary antibody (B&M, Beijing, China).
Virus-induced gene silencing
VIGS assays were carried out on MicroTom fruit using tobacco rattle virus (TRV) following the method of Fu et al. (2005). The primers used for the lncRNA and phytoene desaturase (PDS) gene fragments were designed by the VIGS tool (http://solgenomics.net/tools/vigs). Specific cDNA fragments were amplified and inserted into pTRV2 vectors which were transferred to Agrobacterium strain GV3101, with pTRV1, pTRV2 and pTRV2 fragments respectively (PDS and lncRNA cDNA). After culturing the bacteria at 28 °C overnight, cultures were harvested and resuspended in infiltration buffer (10 mm MgCl2, 200 μm acetosyringone) to a final OD600 of 1.0. Vectors pTRV1 and pTRV2 (empty or with insert) were mixed at a 1: 1 ratio and incubated at room temperature for 3 h. Agrobacterium was infiltrated into the carpopodium of fruits with a 1-mL syringe. Tomato fruits infiltrated with pTRV1 and pTRV2 were used as controls. Each inoculation was carried out three times, and on each occasion six different plants were infiltrated. When the VIGS phenotype was visible, different sections of tomato fruits were collected and stored at −80 °C. Primers for this analysis are listed in Table S1.
Selection of target sequences and vector construction of pYLCRISPR/Cas9
CRISPR-P (http://cbi.hzau.edu.cn/crispr/) was used to select three specific single guide RNAs (sgRNAs) for targeting lncRNA2155 (Table S2). Selection criteria aimed to maximize editing efficiency: GC content in target sites was > 40 % and target sequences avoided more than four consecutive T nucleotides as RNA polymerase III would recognize such regions as a termination signal. The RNA Folding program (http://mfold.rna.albany.edu /?q= mfold /RNA -Folding-Form2.3) was used to ensure that no more than 5 bp were between the target and sgRNA sequences because sgRNA secondary structure greatly influenced editing efficiency. The primers used for this analysis are listed in Table S1.
DNA extraction and mutation analysis of transgenic lines
DNA was extracted from 100 mg fresh frozen leaves using a hi-DNA secure plant kit (Tiangen, Beijing, China). Primers were designed to contain the target sites and were used for PCR amplification of T0 and T1 transgenic lines. PCR products were directly sequenced or cloned into the pEasy-T1 (TransGen Biotech, China) vector and mutations were then identified. The sequences of the mutants were analysed by DSDecode (http://dsdecode.scgene.com/). Primers for this analysis are listed in Table S1.
Statistical analysis
GraphPad Prism 6.0 was used for all statistical analyses. Statistical significance was calculated with Student’s t test, and P values less than 0.05 were considered significantly different, while P values less than 0.01 were considered highly significant. For three or more data sets, statistical significance was calculated by Duncan’s test. Statistically significant differences (P < 0.05) are indicated by different lowercase letters in figures.
RESULTS
Mapping of binding sites of RIN in the promoter regions of lncRNAs
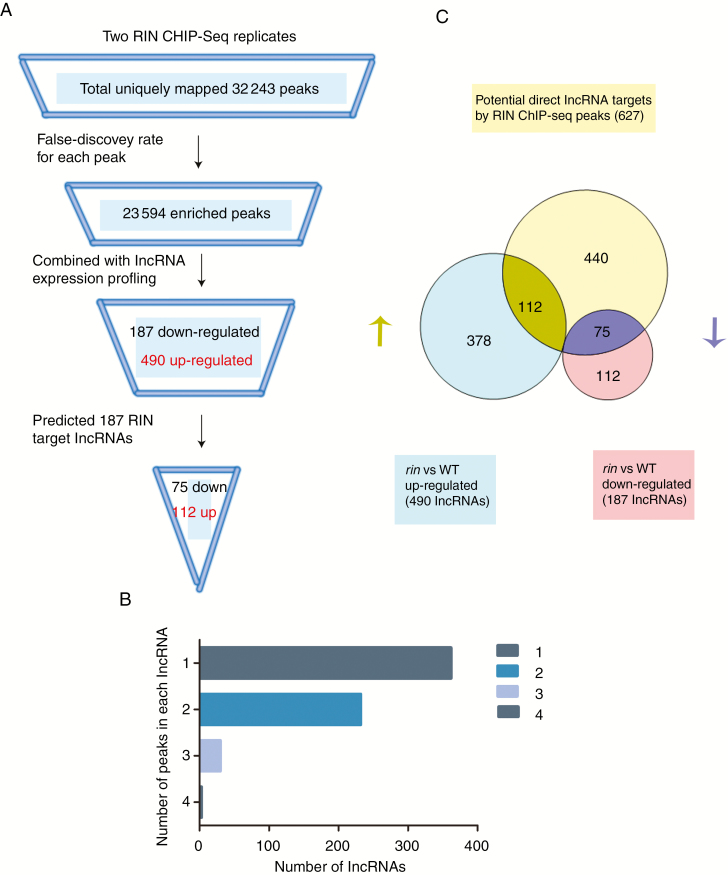
The whole genome of binding sites of RIN in tomato lncRNA promoter regions were detected by using the ChIP-seq approach (Zhong et al., 2013). ChIP-seq of transcripts from ‘Ailsa Craig’ (AC) fruits was performed in two biological replicates. A total of 32 243 peaks were obtained (Fig. 1A). To identify immuno-enriched regions, we made use of the MACS2 software program (Zhang et al., 2008). MACS2 generated a list of 23 594 enriched peaks from high-throughput ChIP-seq data (Fig. 1A). We extracted the sequence of 250 bp up- and downstream of the peak summits to perform MEME-ChIP enrichment analysis, which used the programs MEME and DREME to identify sequence motifs. The ChIP-seq expression score (ES) of the ChIPed DNA relative to the input DNA, defined as signal enrichment for the region, localized to the upstream regulatory region of the corresponding lncRNAs, was used to discard RIN binding sites with very low enrichment using a threshold ES > 3. We defined the promoter region as 2 kb upstream of the lncRNA start site. Using these criteria, we identified 928 of the 23 594 enriched peaks which localized to a transcriptional regulatory region. The 928 binding sites were mapped to 627 lncRNAs. Of these lncRNAs, 362 had one binding site in the promoter region, 232 had two, 30 had three and three had four (Fig. 1B). Overall, on the basis of the presence of information on the genomic positions of the RIN binding sites, we identified 627 candidate target lncRNAs that carried one or more RIN binding sites in the transcriptional regulatory region (Table S3).
Fig. 1.
Genome-wide identification of RIN target lncRNAs. (A) Schematic of identification target lncRNAs by RIN. The sequencing reads were mapped to the tomato genome and estimated 32 243 uniquely mapped reads. Then 23 594 significantly enriched and overlapping peaks were identified based on a 0.01 cut off threshold criterion. Combining these peaks with lncRNAs significant up- and down-regulated in ripening mutant tomato, RIN target lncRNAs were identified (112 up-regulated and 75 down-regulated). (B) The number of ChIP-seq peaks detected in each target lncRNA. (C) Venn diagram of potential direct RIN target lncRNAs selected based on ChIP-seq peaks and lncRNAs positively and negatively regulated by RIN. The overlapping area indicates the number of lncRNAs that are direct targets by RIN.
ChIP-seq combined with transcriptome analysis to identify target lncRNAs regulated by RIN
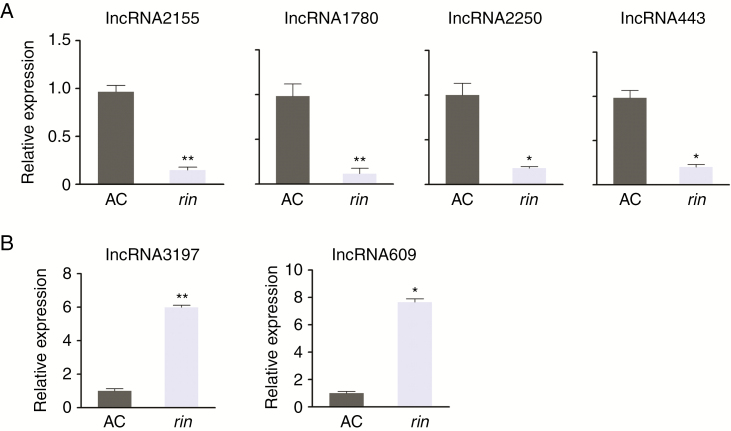
To further identify target lncRNAs regulated by RIN directly, we screened the candidate target lncRNAs together with lncRNA expression profiling (Zhu et al., 2015), using the threshold |log2 fold change| > 1, q < 0.05. Of the 627 potential target lncRNAs, 75 were included in the set of down-regulated lncRNAs in the rin mutant compared with AC (breaker stage of fruits). Similarly, 112 lncRNAs were up-regulated (Fig. 1A, C; Table S4). The remaining 112 down-regulated and 378 up-regulated lncRNAs were not selected as direct RIN targets which suggested that they were not directly regulated by RIN at the transcriptional level. Six lncRNAs were chosen based on their high ChIP-seq enrichment score for further analysis, and qRT-PCR validation of these selected target lncRNAs indicated that four were down-regulated and two were up-regulated in the rin mutant compared with AC (WT) (Fig. 2). The rin mutation caused fusion of adjacent truncated RIN and MC genes (RIN-MC), and the functions of the RIN-MC fusion gene in the rin mutant are largely unknown (Zhong et al., 2013). To identify the functions of RIN, we reanalysed ChIP-seq data and RNA-seq data of the rin knockout mutant (AC 35S::RIN RNAi in AC background) compared with AC (breaker+5 stage) (Li et al., 2018). In total, 56 positively and 78 negatively regulated target lncRNAs were identified in comparison with the wild-type (AC) with AC 35S::RIN RNAi (Fig. S1, Table S5). For our selected six lncRNAs, four (lncRNA2155, lncRNA1780, lncRNA2250, lncRNA609) were in the overlap group of RIN positively regulated targets (Fig. S1). The other two (lncRNA443, lncRNA3197) had very low expression at the Brearker+5 stage, so they could not be detected by RNA-seq of AC and AC 35S::RIN RNAi.
Fig. 2.
qRT-PCR validation of six target lncRNAs. lncRNAs down-regulated (A) and up-regulated (B) in rin according to RNA-seq data were quantified. Actin expression values were used as the internal reference. The relative level of lncRNA transcripts was normalized to that in ‘Ailsa Craig’ (AC) fruits where the amount was arbitrarily assigned a value of 1. Error bars indicate ±s.d. of three biological replicates, each measured in triplicate. Asterisks indicate a significant difference as determined by Student’s t-test (*P < 0.05; **P < 0.01).
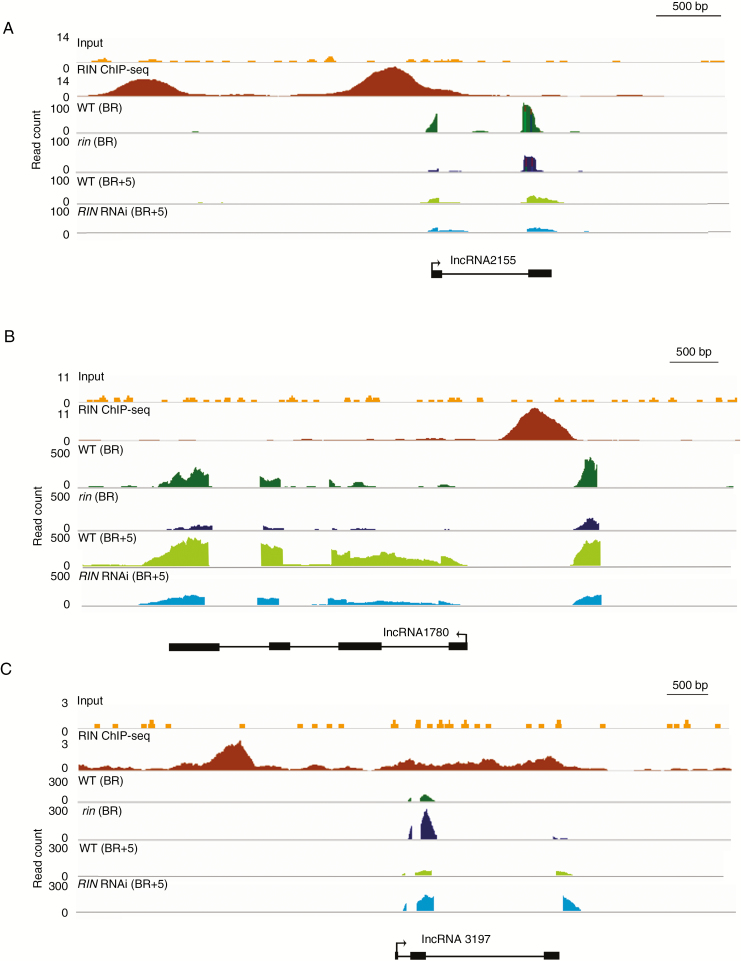
To better detect RIN target lncRNAs, the overlaps between RIN binding sites (ChIP peaks) and differential expression of transcriptome analysis in the promoter regions of lncRNA2155, lncRNA1780 and lncRNA3197 were manually visualized using the Integrative Genomics Viewer (IGV) genome browser (Fig. 3). The reliability of these RNA-seq analyses was verified by differential expression of RIN in AC and AC 35S::RIN RNAi, rin and rin 35S::RIN-MC RNA fruits (Fig. S2).
Fig. 3.
Visual verification of the ChIP-seq peaks and transcriptome analysis. A histogram of three selected genomic regions of lncRNA2155 (A), lncRNA1780 (B) and lncRNA3197 (C). The three tracks represent calls for significantly enriched regions of RIN public ChIP-seq data, RNA-seq in comparison with the breaker (BR) stage of wild type and rin fruits, and the BR+5 stage of wild type and AC RIN RNAi fruits; statistically significant peaks were observed. The corresponding gene organization in exons (boxes) and introns (lines) is also shown. The arrow indicates the sense of transcription.
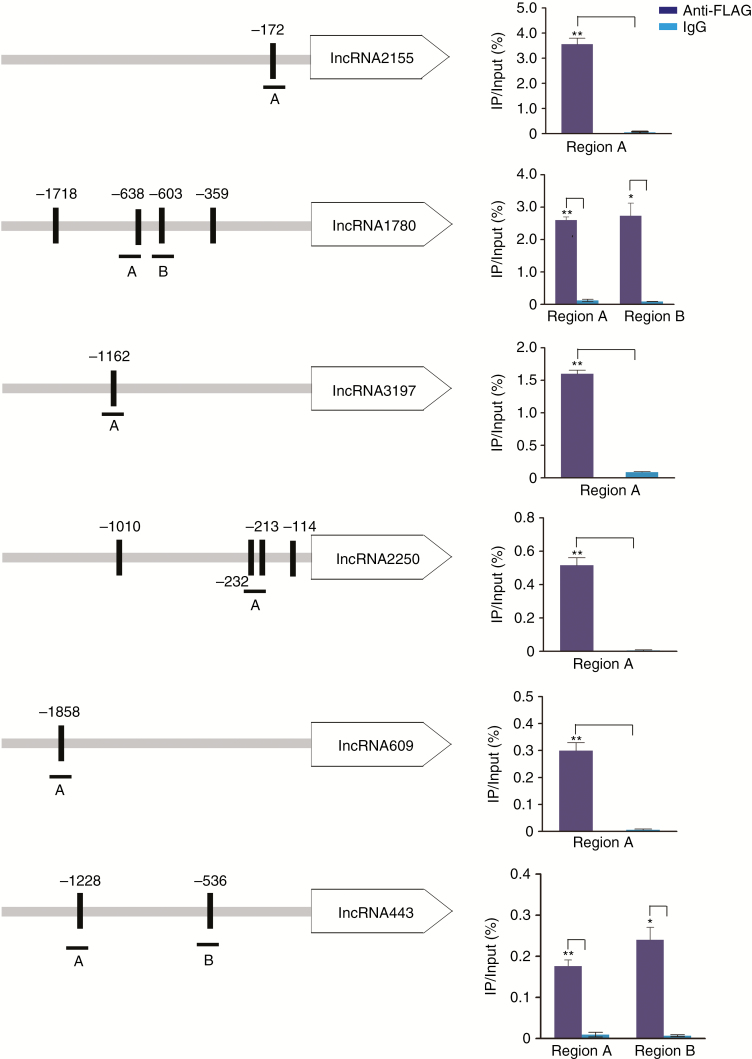
RIN binds to the promoters of six target lncRNAs in vivo
To test the hypothesis that RIN could regulate the six target lncRNAs as shown previously, we performed ChIP in vivo and investigated if the promoter region (2000 bp upstream region starting from the lncRNA start site) contains the C-A/T-rich-G (CArG box) motif, which is known as the MADS box protein target (Ito et al., 2008). The qPCR amplified region was confirmed by the FUZZNUC program with one or more CArG boxes (Table S6). Chromatin from 35S-driven RIN-overexpressing tomato fruit picked 3 d after the breaker stage was immunoprecipitated with anti-FLAG monoclonal antibody and pre-immune mouse IgG was set as the negative control (Fig. S3). The promoter region of E8, a known RIN binding sequence (Ito et al., 2008), was tested with RIN enrichment by ChIP-qPCR as a positive control and Actin was used as negative control, which supported the reliability of the detection and suggested that the ChIP-qPCR data warranted further study (Fig. S4). We detected RIN binding to the following six targets, lncRNA2155, lncRNA1780, lncRNA3197, lncRNA2250, lncRNA609 and lncRNA443 (Fig. 4). Intriguingly, the relative enrichment of these lncRNAs by the anti-FLAG antibody compared with IgG was quite different. The relative enrichment of lncRNA2250, lncRNA609 and lncRNA443 was low, while that of lncRNA3197, lncRNA1780 and lncRNA2155 was much higher. The relative enrichment determined by ChIP qPCR was consistent with the ChIP-seq enrichment score. These data suggested that RIN exhibited differential binding ability to the promoter fragments of these lncRNAs and suggested that the ChIP-seq data were accurate. Taken together, these data clearly demonstrated that RIN binds to the promoter regions of the six target lncRNAs in vivo, which suggested a direct regulation of lncRNAs in tomato fruit by RIN.
Fig. 4.
RIN directly binds to the promoter regions of six lncRNAs as revealed by chromatin immunoprecipitation. ChIP qPCR shows the binding of RIN to the promoter regions of six lncRNAs. The promoter structures of the target genes are presented. Boxes represent CArG box elements and numbers indicate the position of these motifs. Fragments with upper-case letters A or B represent the regions used for ChIP qRT-PCR. Values are the percentage of DNA fragments that co-immunoprecipitated with anti-FLAG antibody (blue bars) or non-specific antibodies (pre-immune mouse IgG; cyan bars) relative to the input DNAs. *P < 0.05, **P < 0.01, difference from IgG, by Student’s t-test.
RIN binds to the promoters of six target lncRNAs in vitro
We further carried out EMSAs to detect if RIN could bind to the promoters of six target lncRNAs in vitro. Recombinant RIN protein with GST was purified (Fig. S5). Previous studies have shown that RIN can bind the CArG box of DNA sequences (Fujisawa et al., 2013). The probes were designed to contain one or more CArG-box motifs in the promoter region of lncRNAs (Table S1). The results indicated that RIN can bind to the six target lncRNAs (Fig. 5). To determine if the binding was specific, unlabelled CArG-box probes (cold probes) and mutant probes were added. The binding bands weakened when the amount of cold probes was increased, suggesting that the unlabelled CArG-box probes can competitively bind with RIN. By contrast, the binding of GST-RIN to the CArG-box was not decreased when the mutant probes were added, highlighting the binding specifcity of RIN to the target lncRNAs in vitro. (Fig. 5). In addition, the fact that RIN bound strongly to the GArG boxes of lncRNA2155, lncRNA1780 and lncRNA3197 (Fig. 5) strengthened the result of ChIP qPCR. These data collectively indicated that RIN bound to six lncRNA targets both in vivo and in vitro, and that the binding ability might be positively related to the ChIP-seq enrichment score.
Fig. 5.
Electrophoretic mobility shift assay of RIN binding to the promoter regions of six target lncRNAs. Binding of RIN to the promoters of six target lncRNAs containing a RIN binding motif (GArG-box element). The sequences of the wild-type probes containing the GArG-box were labelled with biotin. Competition for RIN binding was performed with 100× and 1000× cold probes containing the wild-type CArG-box (CNNNNNNNNG) or mutated CArG-box (ANNNNNNNNT), respectively. The symbols – and + represent absence or presence, respectively, and +++ indicates increasing amounts.
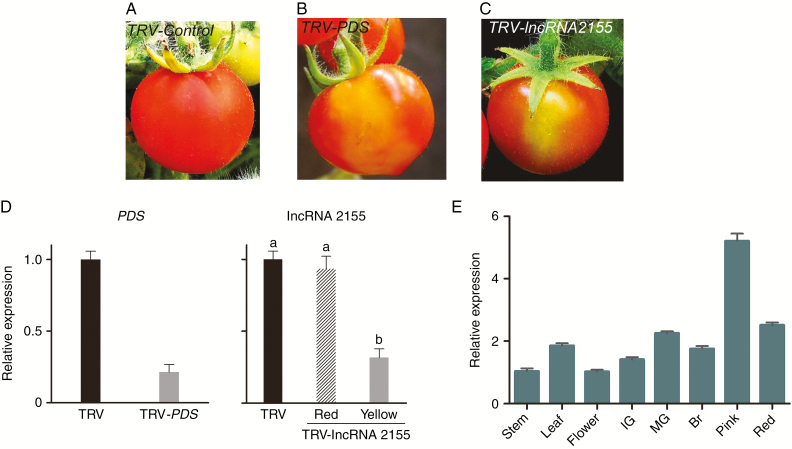
Silencing of a novel ripening-related RIN target lncRNA
Six lncRNAs were identified as direct targets of RIN, some of which might be involved in tomato fruit ripening. To test this prediction, a VIGS method was used to silence these six target lncRNAs. VIGS of the PDS gene was used as a positive control. Among the target lncRNAs, we found that silencing of lncRNA2155 resulted in obvious phenotypes in tomato fruit. Compared with TRV-control fruit (Fig. 6A), a clear phenotype, orange and partly yellow fruit skin, could be detected in TRV-PDS and TRV-lncRNA2155 fruits (Fig. 6B, C). mRNA levels were measured by qRT-PCR. The results showed that expression of PDS and lncRNA2155 were reduced by approximately 80 % and 65 %, respectively, in the yellow part of tomato fruits compared with the TRV-control fruits (Fig. 6D). The results suggested that transient silencing of lncRNA2155 could affect fruit ripening. In addition, lncRNA2155 was expressed in all major tissues during tomato growth and development (Fig. 6E), although the abundance of lncRNA2155 mRNA was relatively higher in fruits than in roots, stems and leaves. In particular, the mRNA level of lncRNA2155 was highest at the pink stage of ripening (Fig. 6E). Moreover, it has recently been reported that several lncRNAs could function as protein-coding genes. lncRNA2155 does not seem to encode a protein and RIN was used as a positive control (Data Fig. S6).
Fig. 6.
Compared with TRV control tomato fruit (A), virus-induced gene slicing (VIGS) of lncRNA2155 tomato fruits infiltrated with TRV-PDS (B) and TRV-lncRNA2155 (C) revealed partial ripening. (D) qRT-PCR analysis of PDS transcript in TRV control and TRV-PDS tomato fruits (yellow sections). (E) lncRNA2155 expression in major organs analysed by qRT-PCR. Actin was the internal control. Error bars indicate ±s.d. of three biological replicates. Different lowercase letters indicate values with significant differences.
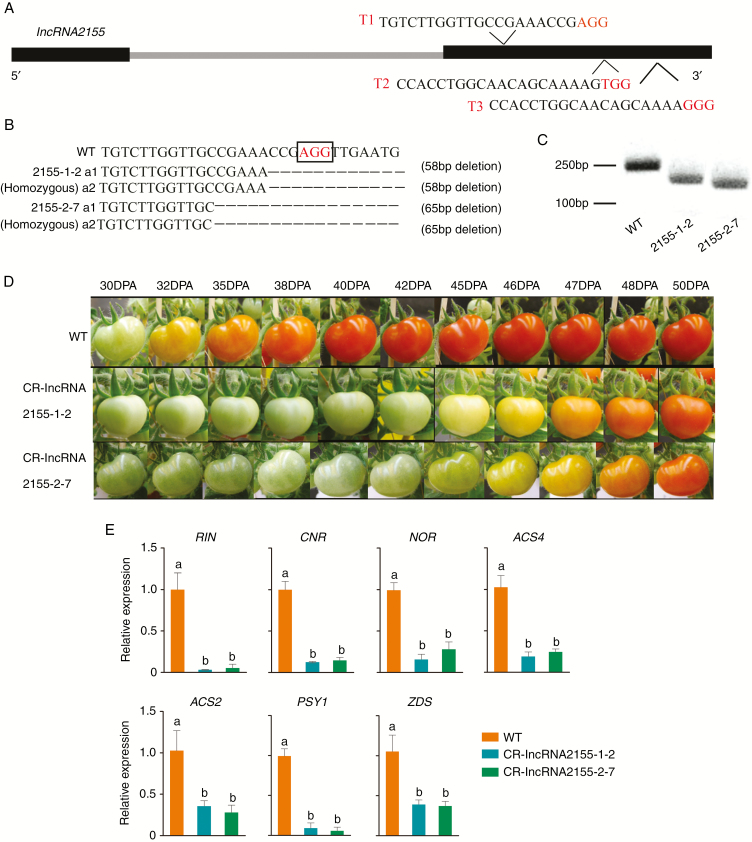
Generation of stable loss-of-function lncRNA2155 mutants using the CRISPR/Cas9 gene-editing system
To further investigate the function of lncRNA2155 in tomato fruit ripening, we generated transgenic mutants using CRISPR/Cas9 gene-editing technology. Three target sites were designed for lncRNA2155 (Fig. 7A; Table S2), and two CRISPR/Cas9 CR-lncRNA2155 mutant lines were detected from first-generation transgenic plants (T0), including chimeric and heterozygous mutants. CR-lncRNA2155-1 was a chimeric mutant with a 58-bp deletion on one allele and CR-lncRNA2155-2 was a heterozygous mutant with a 8-bp deletion on one allele (Fig. S7). Compared with WT fruits, the ripening period of the two mutants was much longer. In addition, we selected two putative off-targets to verify the specific knockout of lncRNA2155 in modified T0 plants and no mutation was found in any putative off-target site (Table S2).
Fig. 7.
Loss of lncRNA2155 function greatly delayed tomato fruit ripening. (A) Three targets in the lncRNA2155 genome sequence. (B) Editing type analysis of CR-lncRNA2155-1-2 and CR-lncRNA2155-2-7. (C) DNA large deletions are validated in CR-lncRNA2155 by PCR. (D) Compared with WT, the CR-lncRNA2155 mutants show delayed fruit ripening. (E) qRT-PCR revealed different expression of ripening-related genes in CR-lncRNA2155 and WT. Error bars indicate ±s.d. of three biological replicates, each measured in triplicate. Different lower-case letters indicate statistically significant differences based on an ANOVA followed by Duncan’s test (P < 0.05).
To further confirm that lncRNA2155 can promote tomato fruit ripening, we identified homogenous mutants with a 58-bp deletion and 65-bp deletion at both alleles of the lncRNA2155 locus in the second generation (T1) of CR-lncRNA2155-1 and CR-lncRNA2155-2 plants (Fig.7B, C). Fruit ripening time was much slower in the CR-lncRNA2155 mutants than in the WT (Fig. 7D). A comparison of WT and lncRNA2155 mutant fruits at different stages of ripening revealed that fruit colour change in the mutant was postponed for approx. 14 d compared to the WT fruits. To elucidate how lncRNA2155 regulated fruit ripening, we performed qRT-PCR of several key genes associated with fruit ripening from WT and CR-lncRNA2155 fruits at 35 dpa (three biological replicates for each sample) (Fig. 7E). The down-regulated genes included nor, Cnr and RIN that encoded TFs, and ACS2, ACS4, phytoene synthase (PSY1) and ζ-carotene desaturase (ZDS) associated with ethylene and lycopene biosynthesis. Overall, the silencing of lncRNA2155 clearly resulted in a delayed maturity phenotype. These data demonstrated that lncRNA2155 delays fruit ripening and might play an essential role in the regulation of tomato fruit ripening.
DISCUSSION
In this study, we identified 627 lncRNAs as significantly enriched in RIN in a genome-wide study through ChIP-seq analysis, 187 of which were differentially expressed between wild-type and the rin mutant (Fig. 1A; Table S4). RIN was further shown to bind with the promoter regions of several target lncRNAs in vivo and in vitro. Using CRISPR/Cas9-mediated lncRNA2155 deletion in wild-type tomato fruits, we found that lncRNA2155 inactivation delayed fruit ripening by modulating the expression of ripening-related genes. Our data revealed the transcriptional regulation of lncRNAs in plants and demonstrated a unique role for lncRNA in fruit ripening, providing a new biological resource and a novel means to regulate fruit ripening.
As an important member of non-coding RNA, lncRNA can regulate gene expression at transcriptional, post-transcriptional and epigenetic levels and plays an important functional role in animals and plants (Feng et al., 2006). Although lncRNA has received increasing attention in recent years, research in plants is far behind that in humans and animals (Zhu and Wang, 2012). In recent years, the function of lncRNA in tomato has received greater attention (Jun et al., 2015). Several lncRNAs act as competing endogenous target mimics (eTMs) for tomato miRNAs which participated in tomato yellow leaf curl virus infection (Wang et al., 2015). lncRNA314 is predicted to be involved in tomato fruit ripening because it is highly expressed in fruits at the breaker ripening stage and is suppressed in different ripening mutants (Wang et al., 2016). lncRNA16397-GRXs is demonstrated to play a role in the response of tomato to Phytophthora infestans infection, which provided candidates for breeding to enhance biotic stress resistance in tomato (Cui et al., 2017). Studies also report that plant lncRNAs exert regulatory functions similar to those in humans and animals (Zhang and Chen, 2013). Consistent with this, recent study has showed that protein 53 (p53) can bind to the promoters of some large intervening non-coding RNAs (lincRNAs) and all of these lincRNAs are significantly induced in p53 cells (Guttman et al., 2009). In addition, approximately half resided in the cluster associated with p53-mediated DNA damage response, confirming the validity of the functional inference and suggesting that lincRNAs which are directly bound and regulated by TFs could show functions associated with TFs (Guttman et al., 2009).
Our results showed that six selected target lncRNAs were significantly differentially expressed in the rin mutant compared with AC by qRT-PCR analysis and further validated to bind with RIN in vivo and in vitro by ChIP-qPCR and EMSA. For our selected six lncRNAs, it is noticeable that only four (lncRNA2155, lncRNA1780, lncRNA2250, lncRNA609) are in the overlap group of RIN target lncRNAs by ChIP-seq and RNA-seq of AC and AC RIN RNAi fruits, and the other two (lncRNA443, lncRNA3197) could not be detected by RNA-seq of AC 35S::RIN RNAi and AC fruits (Fig. S1). One of the reasons for this may be that the stage of fruits used for the two RNA-seqs are BR and BR+5, respectively, and the lncRNAs showed decreased expression levels at the BR+5 stage relative to the levels at the BR stage. The result suggested that fruit stage plays an important role for RNA-seq analysis
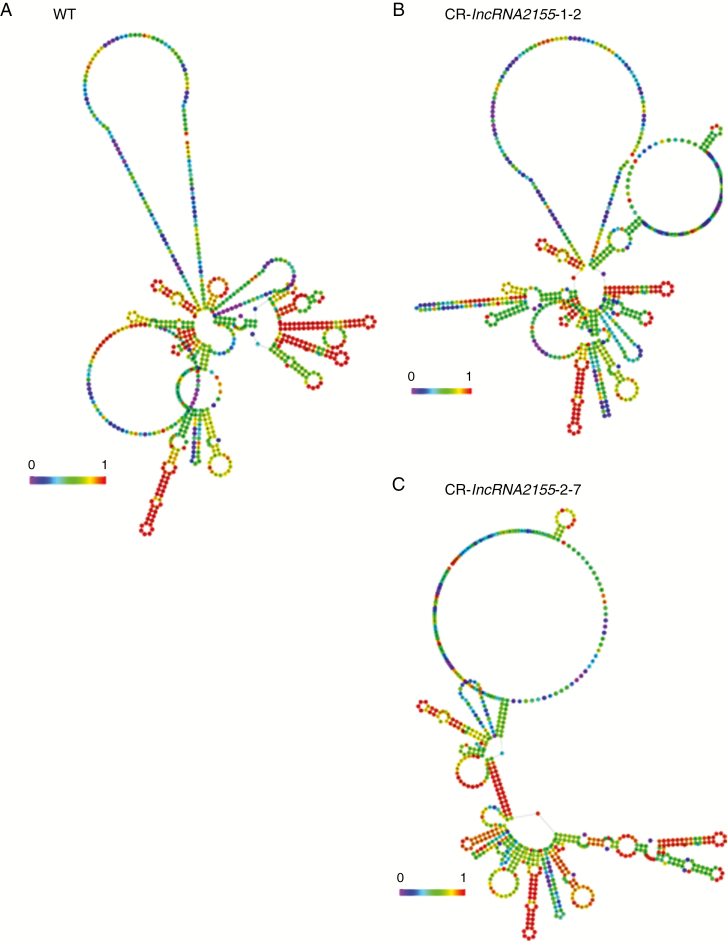
In recent decades, the transcription factor RIN has been recognized as a key component in tomato fruit ripening (Ng and Yanofsky, 2001) and affects many physiological and biochemical processes and metabolic pathways (Vrebalov et al., 2002). RIN regulates the expression of numerous genes participating in all primary ripening pathways, such as ethylene biosynthesis, ethylene perception, cell-wall metabolism, carotenoid accumulation, lycopene accumulation, chlorophyll degradation and other ripening-related TFs (Martel et al., 2011). RIN target genes have been identified as ripening-related characters such as phenotypes and in ethylene production (Fujisawa et al., 2011). In total, 1046 RIN binding sites were detected by ChIP-chip in which ChIPed DNA samples are hybridized with the microarray which have probes designed from the 2-kb upstream regions of all tomato predicted coding genes (Fujisawa et al., 2011, 2013). Although there has been increasingly interest in downstream targets regulated by RIN, few studies have used non-coding RNAs, especially lncRNAs. A total of 32 243 peaks were obtained by ChIP-seq analysis, including promoter regions of both coding and non-coding genes in our study. Six target lncRNAs that were shown to be regulated by RIN directly were analysed for potential function in fruit ripening by VIGS assay. Of these targets, transient silencing of lncRNA2155 in tomato fruit appeared to promote a different skin colour. The yellow did not change to orange, indicating that lncRNA2155 might be involved in the regulation of tomato fruit ripening (Fig. 6). The CRISPR/Cas9 system was used to eliminate lncRNA2155 and investigate its specific role in fruit ripening. The results clearly demonstrated that loss function of lncRNA2155 can generally extend fruit shelf life. Two independent homozygous transgenic mutants (CR-lncRNA2155-1 and CR-lncRNA2155-2) were detected in the T1 generation. Moreover, the two lncRNA2155 mutants with 58-bp and 65-bp deletions exhibited delayed fruit ripening of more than 10 d compared to the time of the colour change in WT fruits. The mutant fruits remained orange at 50 dpa (Fig. 7D). In addition, the mRNA abundance of some key fruit ripening-related genes was up-regulated or down-regulated significantly in lncRNA2155 mutant fruits in comparison to the WT, implying that RIN target lncRNAs might influence fruit ripening by regulating those ripening-related genes directly or indirectly (Fig. 7E). In addition, the secondary structure of CR-lncRNA2155-1-2 and CR-lncRNA2155-2-7 was significantly changed compared with WT (Fig. 8). Because the secondary structure of lncRNA has an important role in function (Li et al., 2016), the dramatic changes in structure suggested that they can affect their functions. In tomato fruit, it has been established that RIN is involved in regulating fruit ripening with a series of TFs and target genes (Giovannoni et al., 2017). In the present study, we suggest a new network model of tomato fruit ripening (Fig. 9). In this model, RIN changes initial ethylene production by regulating ACS4, which is responsible for the ethylene-independent transcriptional regulatory pathway (Ito et al., 2017), and affects fruit ripening by regulating the expression of many coding and non-coding targets, especially lncRNAs. Given the diverse and complex functions of these regulatory networks, further investigations of how target lncRNAs influence fruit ripening would improve our understanding of the mechanism of regulatory networks of fruit ripening by lncRNAs in plants.
Fig. 8.
Secondary structure of WT (A), CRlncRNA2155-1-2 (B) and CRlncRNA2155-2-7 (C).
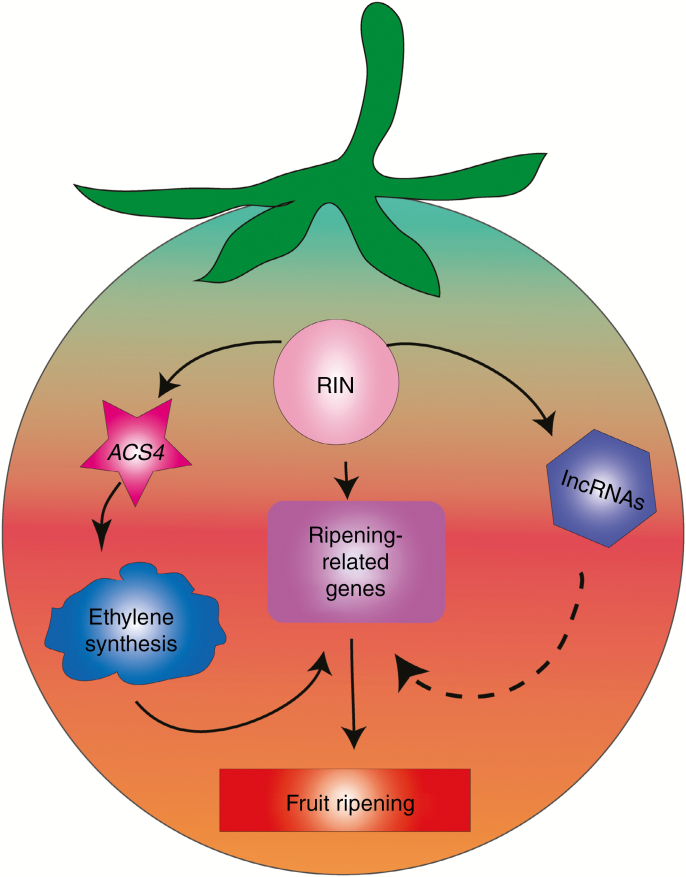
Fig. 9.
Schematic representation of the proposed model for a regulatory mechanism of tomato fruit ripening. Full-line arrows indicate RIN regulates fruit ripening by ethylene via ACS4 and ripening-related target genes. Dotted-line arrow indicates that the regulatory mechanism of lncRNAs during fruit ripening is unclear.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: Primers used in this study. Table S2: Characteristics of CR-lncRNA2155. Table S3: List of lncRNAs with RIN enrichment in promoter regions by ChIP-seq data. Table S4: List of potential direct lncRNA targets with RIN. Table S5: List of lncRNA targets expressed differentially in AC RNA RNAi and AC. Table S6: Predicted RIN binding motifs within the 2000-bp upstream region of lncRNAs analysed in this study. Fig. S1: Venn diagram of potential target lncRNAs by RIN compared with ChIP-seq and RNA-seq. Fig. S2: Visual verification of the ChIP-seq peaks and RNA-seq expression of RNA. Fig. S3: Western blot showing the specificity of the anti-FLAG monoclonal antibody used for ChIP assay. Fig. S4: Preparation and validation of ChIPed DNA samples recovered with the anti-FLAG antibodies. Fig. S5: Gel electrophoresis and western blot demonstrating antibody for EMSA. Fig S6: Protein-coding potential in the lncRNA2155 transcript. Fig. S7: CR-lncRNA2155 mutants were generated using the CRISPR/Cas9 system in the T0 generation of transgenetic tomato plants.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (grant nos. 91540118, 31622050 31672208 and 31471921) to H.Z. We thank Yaoguang Liu (South China Agriculture University) for providing the binary vector pYLCRISPR/Cas9 system, and Prof. Guozheng Qin, Dr Yuying Wang, Dr Weihao Wang (Institute of Botany, CAS) and Zhongqi Fan, Xiaoli Tan (South China Agricultural University) for material and experimental assistance. We also thank Shan Li, Tian Wang and Xindi Li and for technical assistance.
LITERATURE CITED
- Bai Y, Dai X, Harrison AP, Chen M. 2015. RNA regulatory networks in animals and plants: a long noncoding RNA perspective. Briefings in Functional Genomics 14: 91–101. [DOI] [PubMed] [Google Scholar]
- Barry CS, Giovannoni JJ. 2006. Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proceedings of the National Academy of Sciences of the United States of America 103: 7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Karlova R, Ballester AR et al. . 2012. The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24: 4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R et al. . 2010. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant Journal: for Cell and Molecular Biology 64: 936–947. [DOI] [PubMed] [Google Scholar]
- Cui J, Luan YS, Jiang N, Bao H, Meng J. 2017. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant Journal 89: 577–589. [DOI] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. 2006. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes and Development 20: 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB. 2005. Virus-induced gene silencing in tomato fruit. Plant Journal 43: 299–308. [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Ito Y. 2011. Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biology 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y. 2013. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25: 371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Ju Z, Cao D et al. . 2015. MicroRNA profiling analysis throughout tomato fruit development and ripening reveals potential regulatory role of RIN on microRNAs accumulation. Plant Biotechnology Journal 13: 370–382. [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. 2004. Genetic regulation of fruit development and ripening. Plant Cell 16: S170–S180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. 2007. Fruit ripening mutants yield insights into ripening control. Current Opinion in Plant Biology 10: 283–289. [DOI] [PubMed] [Google Scholar]
- Giovannoni J, Nguyen C, Ampofo B, Zhong SL, Fei ZJ. 2017. The epigenome and transcriptional dynamics of fruit ripening. Annual Review of Plant Biology 68: 61–84. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M et al. . 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YC, Kuang JF, Chen JY et al. . 2016. Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiology 171: 1070–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. 2009. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant Journal: for Cell and Molecular Biology 60: 1081. [DOI] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N et al. . 2008. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant Journal 55: 212–223. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nishizawa-Yokoi A, Endo M et al. . 2017. Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nature Plants 3: 866–874. [DOI] [PubMed] [Google Scholar]
- Jin J, Liu J, Wang H, Wong L, Chua N-H. 2013. PLncDB: plant long non-coding RNA database. Bioinformatics 29: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun L, Huan W, Nam-Hai C. 2015. Long noncoding RNA transcriptome of plants. Plant Biotechnology Journal 13: 319–328. [DOI] [PubMed] [Google Scholar]
- Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ. 1994. The never ripe mutation blocks ethylene perception in tomato. Plant Cell 6: 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Joung JG, Mcquinn R et al. . 2012. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant Journal 70: 191. [DOI] [PubMed] [Google Scholar]
- Li R, Zhu H, Luo Y. 2016. Understanding the functions of long non-coding RNAs through their higher-order structures. International Journal of Molecular Sciences 17: E702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xu H, Ju Z et al. . 2018. The RIN-MC fusion of MADS-box transcription factors has transcriptional activity and modulates expression of many ripening genes. Plant Physiology 176: 891–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhou L, Wang Z, Chen J, Qu G. 2016. Oligogalacturonic acids promote tomato fruit ripening through the regulation of 1-aminocyclopropane-1-carboxylic acid synthesis at the transcriptional and post-translational levels. BMC Plant Biology 16: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M et al. . 2006. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nature Genetics 38: 948–952. [DOI] [PubMed] [Google Scholar]
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ. 2011. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiology 157: 1568–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF. 2001. Function and evolution of the plant MADS-box gene family. Nature Reviews. Genetics 2: 186–195. [DOI] [PubMed] [Google Scholar]
- Ng TJ, Tigchelaar EC. 1977. Action of the non-ripening (nor) mutant on fruit ripening of tomato. Journal of the American Society of Horticultural Science 102: 504–509. [Google Scholar]
- Osorio S, Alba R, Damasceno CMB et al. . 2011. Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiology 157: 405–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Gene Consortium 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwerven F, Neuert G, Hendrick N et al. . 2012. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 150: 1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V et al. . 2002. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science 296: 343–346. [DOI] [PubMed] [Google Scholar]
- Wang J, Yu W, Yang Y et al. . 2015. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Scientific Reports 5: 2045–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ai G, Zhang CL et al. . 2016. Expression and diversification analysis reveals transposable elements play important roles in the origin of Lycopersicon-specific lncRNAs in tomato. New Phytologist 209: 1442–1455. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fan X, Lin F et al. . 2014. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proceedings of the National Academy of Sciences of the United States of America 111: 10359–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Chen YQ. 2013. Long noncoding RNAs: new regulators in plant development. Biochemical and Biophysical Research Communications 436: 111–114. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA et al. . 2008. Model-based analysis of ChIP-seq (MACS). Genome Biology 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Fei Z, Chen Y-R et al. . 2013. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nature Biotechnology 31: 154–159. [DOI] [PubMed] [Google Scholar]
- Zhu B, Yang Y, Li R et al. . 2015. RNA sequencing and functional analysis implicate the regulatory role of long non-coding RNAs in tomato fruit ripening. Journal of Experimental Botany 66: 4483–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Wang MB. 2012. Molecular functions of long non-coding RNAs in plants. Genes 3: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI. 2012. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335: 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.