Summary
Western lifestyle is linked to autoimmune and metabolic diseases, driven by changes in diet and gut microbiota composition. Using a Toll-like receptor 7 (TLR7)-dependent mouse models of lupus, we dissect dietary effects on the gut microbiota and find that Lactobacillus reuteri can drive autoimmunity but is ameliorated via dietary resistant starch (RS). Culture of internal organs and 16S rDNA sequencing revealed TLR7-dependent translocation of Lactobacillus reuteri in mice and fecal enrichment of Lactobacillus in a subset of lupus patients. L. reuteri colonization worsened autoimmune manifestations under specific-pathogen-free and gnotobiotic conditions, notably increasing plasmacytoid dendritic cells (pDCs) and interferon signaling. However, RS suppressed the abundance and translocation of L. reuteri via short-chain fatty acids, which inhibited its growth. Additionally, RS decreased pDCs, interferon pathways, organ involvement and mortality. Thus, RS exerts beneficial effects in lupus-prone hosts through suppressing a pathobiont that mediates interferon pathways implicated in the pathogenesis of human autoimmunity.
eTOC
The role of commensals in autoimmunity remains unclear. Zegarra-Ruiz et al. report that Lactobacillus reuteri is enriched in lupus models driven by innate inflammation. L. reuteri engages type I interferon pathways worsening autoimmune manifestations. L. reuteri overgrowth is suppressed by metabolites from a special starch diet, thereby improving lupus outcome.
Introduction
Western lifestyle is linked to various inflammatory conditions such as autoimmune and metabolic diseases, driven by changes in diet and gut microbial composition, as suggested by the hygiene hypothesis (Lambrecht and Hammad, 2017; Thorburn et al., 2014). Diet shapes microbial communities in the gut, influencing immune processes (Carmody et al., 2015; Hooper et al., 2012; Kau et al., 2011). A Western diet is characterized by a low-fiber and high-fat composition, which affects gut microbial communities impacting immune functions and promoting disease (Hooper et al., 2012; Kau et al., 2011; Thorburn et al., 2014). Gut microbiota and their metabolites mediate mucosal and systemic immune responses, and alterations in its composition have been linked to multiple autoimmune disorders (Ruff and Kriegel, 2015; Yurkovetskiy et al., 2015). How diet influences autoimmunity via the microbiota remains unclear despite longstanding associations between dietary effects on immunity and immune diseases (Vieira et al., 2014).
A decrease in consumption of major dietary components (e.g., fiber) has led to changes in the microbial community composition and an increase in the incidence of metabolic and immune-related diseases such as colitis (Desai et al., 2016; Koh et al., 2016). Resistant starch (RS) is a type of fiber that is resistant to digestion in the small intestine. Instead, it is fermented by the microbiota in the lower gastrointestinal tract, and thus represents a useful tool to modulate the gut microbiota in experimental settings. Colonic fermentation leads to multiple effects on the host including production of the short-chain fatty acids (SCFAs) acetate, propionate and butyrate (Fuentes-Zaragoza et al., 2010). These metabolites are key mediators of the gut microbiota on host immunity. SCFAs exert their effects not only in the gut but also in distant organs by dampening immune responses that are uncontrolled in patients on a low-fiber Western diet (Thorburn et al., 2014). High-fiber diets have been shown to ameliorate metabolic and allergic diseases mainly through their microbiota-dependent fermentation to SCFAs that promote tissue barrier integrity, mucus production, IgA secretion, and regulatory T cell (Treg) differentiation, thereby supporting an anti-inflammatory environment (Mariño et al., 2017; Thorburn et al., 2014). However, little is known about the beneficial effects of a high-fiber diet in autoimmune diseases.
Systemic lupus erythematosus (SLE) is a prototypical autoimmune disease characterized by a type I interferon (IFN) signature mediated by plasmacytoid dendritic cells (pDCs) (Crow, 2014). These cells are located at sites of injury, which exacerbate proinflammatory responses. In a feed-forward cycle together with autoantibodies, they promote the formation of immune complexes that deposit in the kidneys promoting lupus nephritis, in vascular endothelium leading to vasculitis and atherosclerosis, or in the brain contributing to neuropsychiatric manifestations (Crow, 2014).
We set out to test the role of diet and the microbiota in TLR7-dependent mouse models of lupus and dissect its mechanisms. Using antibiotics and germ-free (GF) mice, we demonstrate that gut microbiota depletion ameliorates the IFN pathway and autoimmunity in these models. A genetic link between human SLE and type I IFN has been established by multiple studies (Crow, 2014). In addition, we characterized the gut and tissue microbiota and identified specific bacterial taxa that were linked to lupus development and associated with human SLE microbiomes. The autoimmune phenotype was transferable via the gut microbiota and gavage of a single commensal identified by these studies, L. reuteri, drove the pathogenesis in the gnotobiotic setting. Finally, using RS as a dietary approach to modulate autoimmunity via the gut microbiota, we demonstrate that RS prevented IFN pathways, disease manifestations and mortality in these models. L. reuteri abundance was inhibited by RS in vivo and its growth by SCFAs in vitro and vivo, thus providing a mechanistic link between diet, the microbiota, and its effects on an autoimmune host.
Results
The gut microbiota exacerbates lupus-related mortality and pathogenesis
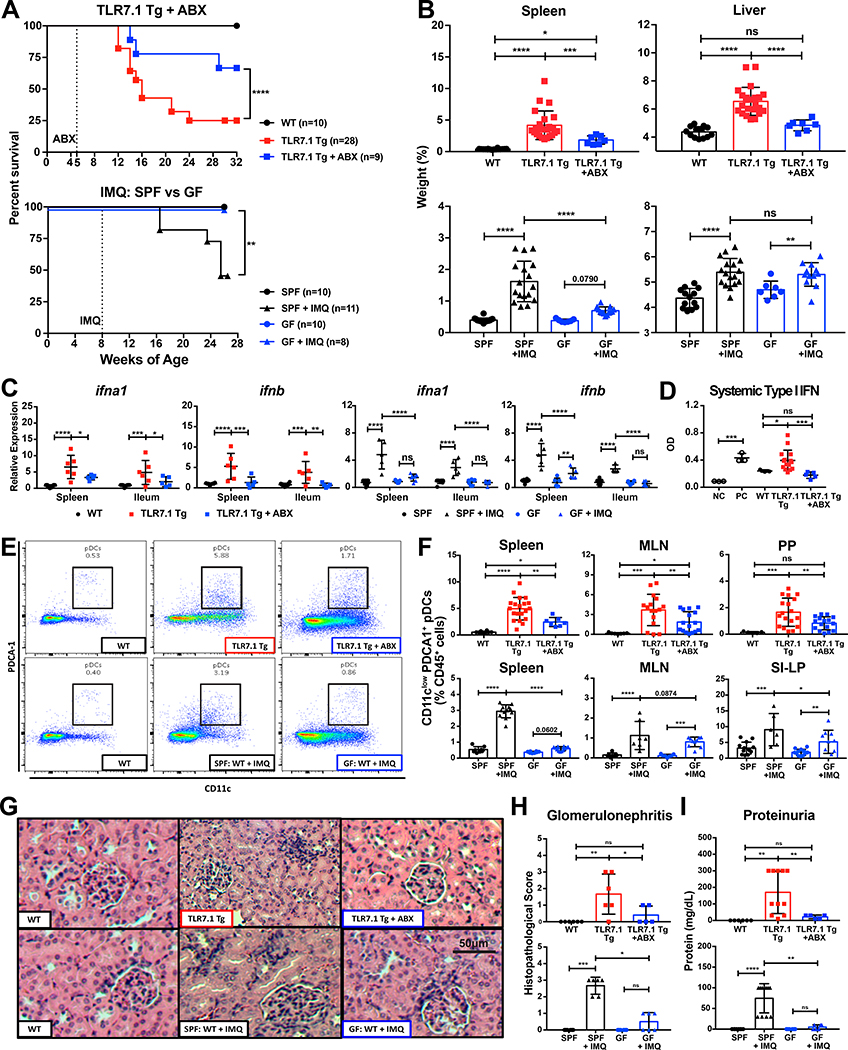
The TLR7/IFN-pathway is central in the pathogenesis of human SLE (Crow, 2014). Thus, we used a TLR7-dependent spontaneous and inducible mouse model to study the role of the microbiota in systemic autoimmunity. Lupus-prone TLR7.1 Tg C57B1/6 mice exhibit an 8- to 16-fold increase in TLR7 expression and spontaneously develop systemic signs of SLE starting at 6 weeks of age (Deane et al., 2007). To induce lupus-like disease in mice not genetically prone to excessive TLR7 signaling, we used topical imiquimod (IMQ), a TLR7 agonist, which was applied to the skin three times a week to 8-week-old wild type (WT) C57B1/6 mice (Yokogawa et al., 2014). In order to assess the role of the gut microbiota, we suppressed its growth in both models using broad-spectrum antibiotics (ABX) or a germ-free (GF) state (Figure S1A). These interventions led to significantly increased survival (Figure 1A), decreased splenomegaly and hepatomegaly (Figure 1B), and type I IFN gene expression in ileum and spleen as well as type I IFN secretion in the blood (Figures 1C-1D). Consistent with systemic IFN levels, pDCs accumulated in spleen, mesenteric lymph nodes (MLN), and Peyer’s patches (PP) in both models and were also suppressed by antibiotics or a GF state (Figures 1E-1F).
Figure 1. The gut microbiota exacerbates lupus-related mortality and pathogenesis.
TLR7.1 Tg lupus-prone mice were treated with broad-spectrum antibiotics (ABX). C57B1/6 (SPF vs. GF) mice were treated with topical imiquimod (IMQ) to induce a lupus-like syndrome. (A) Survival of both lupus-like models. Experimental cohorts of TLR7.1 Tg and C57B1/6 mice were sacrificed for organ weights, immunologic, and histologic analyses. (B) Weights of spleens and livers. (C) Relative mRNA expression of type I IFN in spleen and ileum. (D) Systemic type I IFN responses in serum measured with a reporter cell line. (E-F) Cells from spleen, MLN, PP, and SI-LP were isolated and analyzed by flow cytometry. (E) Representative FACS plots from splenic pDCs. (F) Frequencies of pDCs in spleen, MLN, PP, and SI-LP. (G) Representative H&E staining of kidney tissue. (H) Histopathologic scores for glomerulonephritis. (I) Quantifications of proteinuria. The results are expressed as mean ± SEM (survival cohorts n=8–28 mice per group, experimental cohorts n=3–20 mice per group). The results are representative of at least two independent experiments. *P<0.05 was considered statistically significant; **P <0.01; ***P<0.001; ****P<0.0001; ns=not significant; NC=negative control; PC=positive control. See also Figure S1.
Blood disorders (anemia and leukocytosis) were normalized in antibiotics- treated mice and in the germ-free setting, suggesting that particularly these manifestations are microbiota-dependent (Figure S1B). Antibiotics decreased bone marrow cellular depletion, and restored granulocyte-monocyte and megakaryocyte- erythrocyte progenitor populations (Figures S1C-S1E), which could be linked to lower type I IFN levels (Buechler et al., 2013). Finally, depletion of the gut microbiota decreased renal injury due to immune cell infiltration and glomerulonephritis (Figures 1G-1H and S1F) and improved renal physiology by decreasing proteinuria (Figure 1I).
These data indicate that the gut microbiota is needed for TLR7-dependent systemic autoimmunity. We next defined the gut microbial community structure in both models in order to identify potential pathobionts driving lupus pathogenesis.
Increased abundance and translocation of lactobacilli in lupus
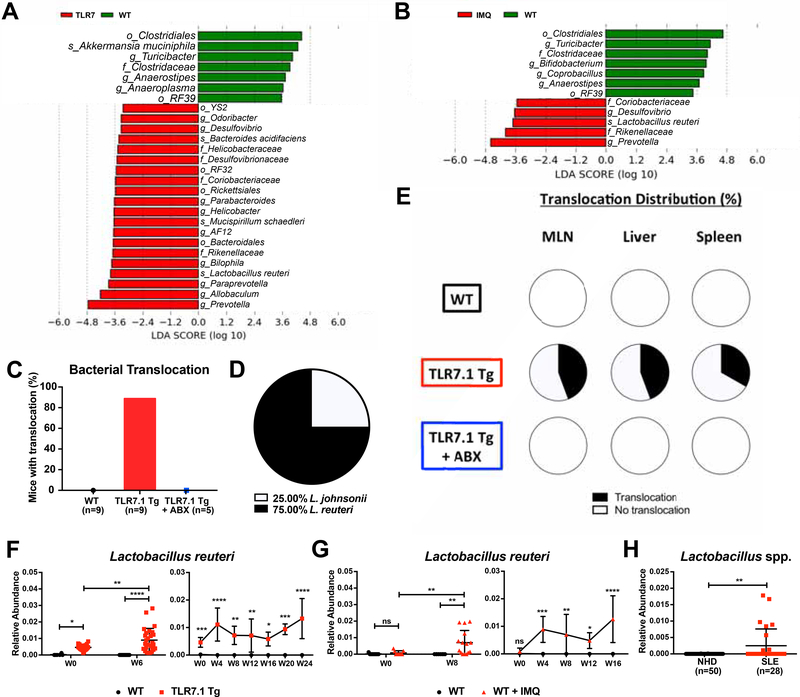
We collected fecal samples longitudinally from mice of both the spontaneous and inducible model, isolated bacterial DNA, and performed high-throughput 16S ribosomal DNA sequencing. 12-week-old TLR7.1 Tg mice carried altered gut microbial communities compared to age- and sex-matched WT specific-pathogen free (SPF) C57B1/6 mice, among them L. reuteri, Desulfovibrio, and Rikenellaceae (Figures 2A, S2A, and Table S1). Interestingly, lupus-prone mice harbored a more diverse and different microbial composition compared to WT mice (Figure S2C). Fecal microbiomes from IMQ-treated WT mice revealed a similar community structure as TLR7.1 Tg mice with regards to diversity and individual taxa, among them L. reuteri, Desulfovibrio, and Rikenellaceae (Figures 2B, S2B, S2D, and Table S2).
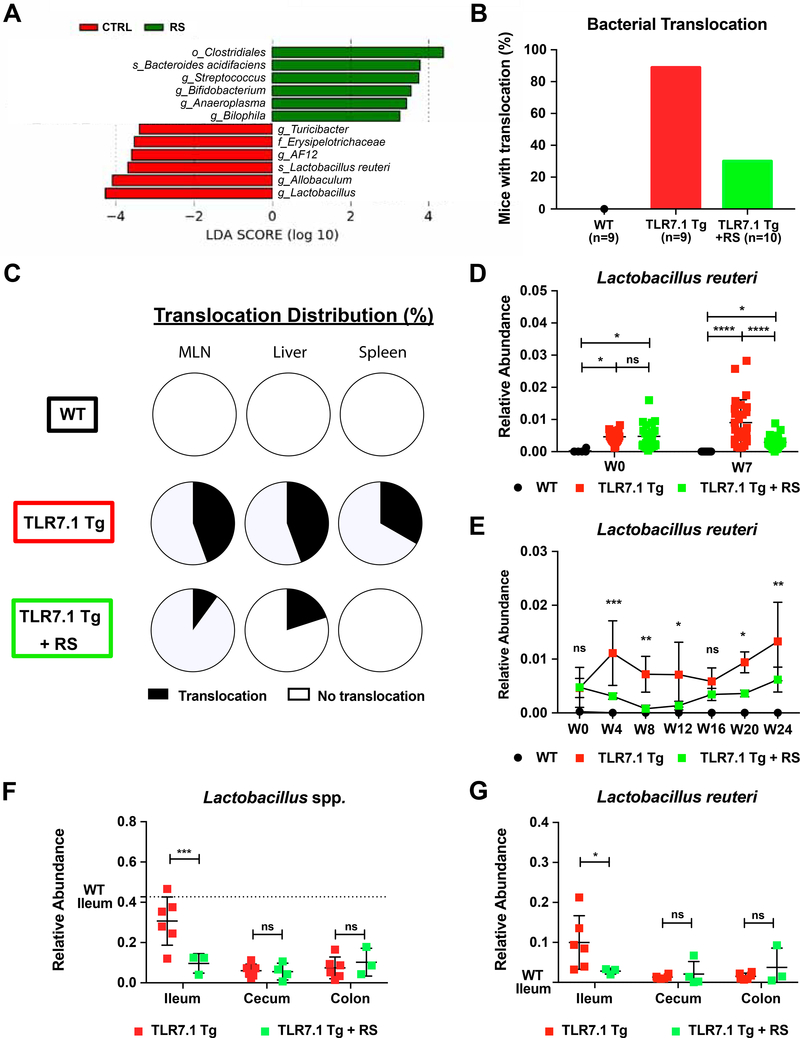
Figure 2. Increased abundance and translocation of lactobacilli in lupus.
Fecal pellets from WT C57B1/6, TLR7.1 Tg, and C57B1/6 mice treated with IMQ were collected; bacterial DNA was isolated and sequenced. Linear Discriminant Analysis Effect Size (LDA) scores of altered taxa in (A) TLR7.1 Tg compared to WT mice and (B) C57B1/6 mice treated with or without IMQ. Shown are significantly altered taxa (increased in red, decreased in green). (C) Total percentage of bacterial translocation as determined by anaerobic and aerobic non-selective cultures. (D) Profile of translocating bacterial species. (E) Distribution of Lactobacillus spp. translocation in MLN, liver, and spleen from WT, TLR7.1 Tg, and TLR7.1 Tg mice treated with or without ABX. (F) Relative abundances of fecal L. reuteri in TLR7.1 Tg compared to WT mice and (G) C57B1/6 mice treated with or without IMQ evaluated by 16S rDNA gene sequencing. (H) Relative abundances of fecal Lactobacillus spp. in normal healthy donors (NHD) and systemic lupus erythematosus (SLE) patients. The results are expressed as mean ± SEM (n=3–20 mice and 28–50 human samples per group). Data are representative of at least three independent experiments. *P<0.05 was considered statistically significant; **P<0.01; ***P<0.001; ****P<0.0001; ns=not significant; W0=week of disease onset. See also Figure S2 and Tables S1 and S2.
Given the importance of gut commensal translocation in autoimmunity (Costa et al., 2016; Manfredo Vieira et al., 2018), we next assessed the integrity of the intestinal epithelium using fluorescein isothiocyanate (FITC)-dextran. Both models displayed signs of a functionally impaired gut barrier with FITC-dextran leakage into the systemic circulation (Figure S2E). To determine if not only small molecules but whole bacteria can escape the gut in these lupus models, we sterilely cultured mesenteric lymph nodes (MLN), liver, and spleen at 15–18 weeks of age aerobically and anaerobically in different media. Commensal bacteria grew consistently in tissues from TLR7.1 Tg mice, which were suppressed by ABX (Figure 2C). Using different non-selective culture media, we found that only Lactobacillus spp. translocated with L. reuteri being the dominant species (Figures 2D-2E).
We then proceeded to determine if L. reuteri was longitudinally enriched in fecal microbiomes of mice from both lupus models as already supported by the 16S rDNA sequencing data collected at study end points (Figures 2A-2B, and S1A). L. reuteri increased over time in the feces of mice from both lupus models as their disease progressed (Figures 2F-2G). In addition, 16S rDNA sequencing of fecal samples showed that Lactobacillus spp. were increased in a longitudinal cohort of SLE patients compared to healthy controls (Figure 2H), suggesting a potential role for these taxa in SLE pathogenesis. The results from the murine studies confirmed that mice in both the spontaneous and the inducible lupus model exhibit similar gut microbial community changes, gut leakiness and Lactobacillus spp. enrichment and translocation to internal organs. We next investigated if the enriched gut bacterial communities play a role in systemic autoimmunity.
Bacterial communities increased in TLR7-dependent lupus models are transferable and exacerbate lupus-related pathogenesis
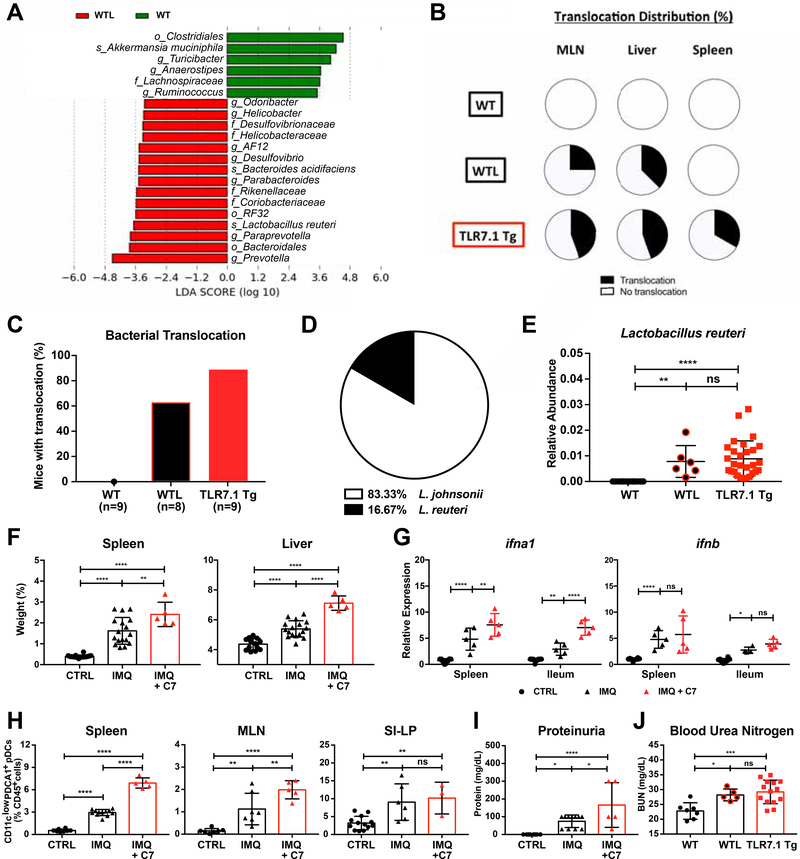
To test if the bacterial taxa increased in the gut microbiota of both lupus models are transferable to other, non-autoimmune-prone mice, we cohoused 5-week-old TLR7.1 Tg mice with their WT SPF C57B1/6 littermates for 7 weeks (Figure S3A). We then performed 16S rDNA sequencing from fecal pellets of cohoused WT littermates (WTL) and age- and sex-matched WT SPF C57B1/6 mice. Cohousing transferred the majority of the bacterial taxa that were enriched in lupus prone mice, among them L. reuteri, Desulfovibrio, and Rikenellaceae (Figures 3A, S3B, and Table S3). Cohousing also induced a more diverse microbial community composition with an increase in observed species (Figure S3C). FITC-dextran gavage revealed that cohousing increased gut leakiness and IgG serum levels, while decreasing fecal IgA levels (Figures S3D-S3F). Importantly, TLR7 expression was required for this phenomenon because TLR7 KO mice were not susceptible to increased gut leakiness after cohousing (Figure S3G). Accordingly, we found through organ cultures that cohoused WT C57B1/6 mice displayed bacterial translocation similar to TLR7.1 Tg C57B1/6 mice (Figure 3C).
Figure 3. Bacterial communities increased in TLR7-dependent lupus models are transferable and exacerbate lupus-related pathogenesis.
Fecal pellets from WT mice and WT littermates (WTL) cohoused with TLR7.1 Tg were collected; bacterial DNA was isolated and sequenced. (A) LDA scores of altered taxa in WTL compared to WT. Shown are significantly altered taxa (increased in red, decreased in green). (B) Distribution of Lactobacillus spp. translocation in MLN, liver and spleen from WT, WTL, and TLR7.1 Tg mice. (C) Total percentage of bacterial translocation as determined by culture. (D) Profile of translocating bacterial species in WTL mice. (E) Relative abundances of fecal L. reuteri (LR) in WT, WTL and TLR7.1 Tg mice evaluated by 16S rDNA gene sequencing. (F-J) SPF C57B1/6 mice were treated with IMQ and gavaged with cecal microbial content from 12-week-old TLR7.1 Tg mice (C7). Mice were sacrificed for organ weights, immunologic, and histologic analyses. (F) Weights of spleen and liver. (G) Relative mRNA expression of type I IFN in spleen and ileum. Cells from spleen, MLN, and SI-LP were isolated and analyzed by flow cytometry. (H) Frequency of pDCs in spleen, MLN, and SI-LP. (I) Quantification of proteinuria. Q) Blood urea nitrogen levels in WTL. The results are expressed as mean ± SEM (n=5–20 mice per group). The results are representative of at least two independent experiments. *P<0.05 was considered statistically significant; **P<0.01; ***P<0.001; ****p<0.0001; ns=not significant. See also Figure S3 and Table S3.
In parallel to the TLR7-dependent lupus models, Lactobacillus spp. were the only species found to translocate in cohoused WT C57B1/6 mice, but unlike the lupus models it was predominantly L. johnsonii as opposed to L. reuteri that translocated (Figure 3D) and translocation was limited to MLN and liver (Figure 3B). 16S rDNA sequencing showed that L. reuteri was enriched in fecal samples of cohoused mice supporting that this species was transferable (Figure 3E).
To test if the gut microbiota (including this species) of lupus-prone TLR7.1 Tg mice were able to exacerbate systemic autoimmunity, we performed cecal transfer studies followed by IMQ treatment. We anaerobically collected all cecal content from 12-week-old TLR7.1 Tg mice and performed cecal microbiota transfer to WT SPF C57B1/6 mice after weaning and before IMQ treatment (Figure S3A). C57B1/6 mice that received microbiota derived from TLR7.1 Tg mice developed more severe splenomegaly and hepatomegaly (Figure 3F), enhanced type I IFN gene expression in spleen and ileum (Figure 3G), and more pDCs accumulating in spleen and MLN (Figure 3H). Cecal microbiota transfer further increased gut leakiness (Figure S3H) and worsened renal disease as reflected by increased proteinuria, resembling cohousing effects on blood urea nitrogen levels (Figures 3I-3J).
These results show that the microbiota of lupus-prone mice is transferable and able to exacerbate systemic autoimmunity. Since L. reuteri was consistently increased in both the spontaneous and inducible lupus model and translocated to tissues outside the gut, we next studied the individual effects of Lactobacillus spp. in lupus pathogenesis using SPF and GF mice.
L. reuteri exacerbates lupus-related pathogenesis
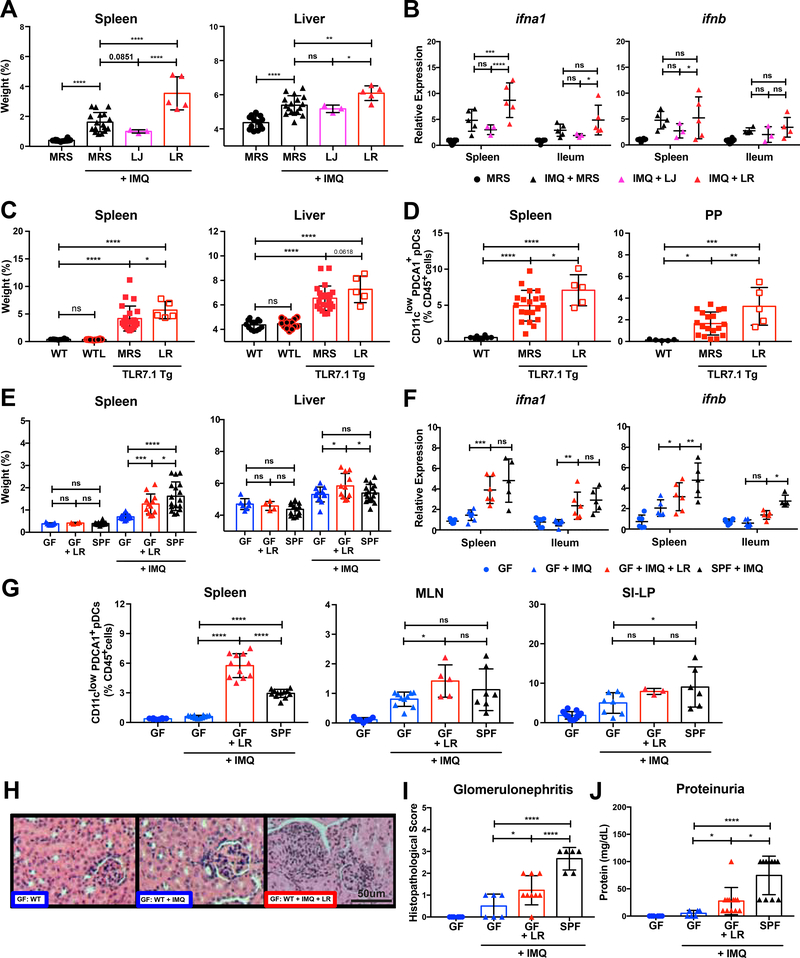
5-week-old SPF C57B1/6 (B6) mice were gavaged daily with either L. reuteri (LR) or L. johnsonii (LJ) primary tissue isolates from TLR7.1 Tg mice and concomitantly treated with topical IMQ (Figure S4A). IMQ-treated WT B6 mice receiving L. reuteri developed increased splenomegaly and hepatomegaly (Figure 4A), and an increased type I IFN gene expression in the spleen and ileum (Figure 4B) compared to media alone or gavage with L.johnsonii. Consistent with the in vivo data, L. reuteri, but not L. johnsonii supernatant induced also IFN-related gene signatures in vitro in IMQ-stimulated pDCs (Figure S4B). Instead, L. johnsonii induced Tregs in vivo whereas L. reuteri did not (Figure S4C). Also, only L. reuteri worsened IMQ-induced anemia and gut permeability in WT B6 mice under SPF conditions (Figures S4D-S4E). In TLR7 Tg mice, L. reuteri gavage increased splenomegaly and pDCs accumulation in spleen and PP (Figures 4C-4D, and S4A); similarly, leukocyte recruitment to the kidney was exacerbated (Figure S4F).
Figure 4. L. reuteri exacerbates lupus-related pathogenesis.
(A-B) SPF C57B1/6 mice were treated with IMQ and were gavaged with De Man, Rogosa and Sharpe broth (MRS), L. reuteri (LR), or L. johnsonii (LJ) primary tissue isolates harvested from TLR7.1 Tg mice. Mice were sacrificed for organ weights and immunologic analyses. (A) Weights of spleen and liver. (B) Relative mRNA expression of type I IFN in spleen and ileum. (C-D) TLR7.1 Tg mice were gavaged with MRS or LR. Mice were sacrificed for organ weights and immunologic analyses. (C) Weights of spleen and liver. Cells from spleen and PP were isolated and analyzed by flow cytometry. (D) Frequencies of pDCs in spleen and PP. (E-I) GF C57B1/6 mice were treated with IMQ and monocolonized with LR. Mice were sacrificed for organ weights, immunologic, and histologic analyses. (E) Weights of spleen and liver. (F) Relative mRNA expression of type I IFN in spleen and ileum. Cells from spleen, MLN, and SI-LP were isolated and analyzed by flow cytometry. (G) Frequencies of pDCs in spleen, MLN, and SI-LP. (H) Representative H&E staining of kidney tissue. (I) Histopathologic scoring for glomerulonephritis. Q) Quantification of proteinuria. The results are expressed as mean ± SEM (n=3–16 mice per group). The results are representative of at least two independent experiments. *P<0.05 was considered statistically significant; **P<0.01; ***P<0.001; ****p<0.0001; ns=not significant. See also Figure S4.
Since L. reuteri gavage into SPF B6 and TLR7.1 Tg B6 mice was sufficient to exacerbate lupus-related symptoms, we investigated if L. reuteri alone could drive systemic autoimmunity in the gnotobiotic setting. We monocolonized 6-week-old GF B6 mice with L. reuteri, treated them with IMQ, and sacrificed them at 16 weeks of age (Figure S4A). Monocolonization with L. reuteri also led to splenomegaly, hepatomegaly (Figure 4E), as well as splenic and ileal type I IFN gene expression (Figure 4F). Hepatosplenomegaly was not seen after monocolonization with an unrelated gut bacterium, B. thetaiotaomicron (BT) (Figure S4G) that mainly triggers cross-reactive adaptive as opposed to innate signals related to autoimmunity (Greiling et al., 2018). L. reuteri-monocolonized mice accumulated more pDCs in spleen and MLN compared to B. thetaiotaomicron (Figures 4G and S4H) and suffered from worsened signs of lupus nephritis (Figures 4H-4J).
Collectively, these data support that L. reuteri alone was sufficient to exacerbate systemic autoimmunity under both SPF and GF conditions. Given that selective members of the gut microbiota in both the spontaneous and inducible lupus model were sufficient to exacerbate disease, we studied next if dietary modulation of pathobionts instead of broad depletion of the entire gut microbiota could ameliorate lupus-like disease and gut commensal translocation similarly to broad-spectrum antibiotics.
Resistant starch modulates the gut microbiome and decreases L. reuteri translocation
We used a diet enriched with RS, a type of fiber that resists digestion by the host and is fermented by commensals in the distal gut, as a tool to modulate bacterial communities in the lupus-prone models. We first isolated DNA from fecal pellets of TLR7.1 Tg mice fed with RS for 7 weeks and performed 16S rDNA sequencing (Figure S5A). L. reuteri was decreased by RS compared to age- and sex-matched TLR7.1 Tg mice fed with a standard diet controlled for the same ingredients except for RS (Figures 5A, S5B, and Table S4). Interestingly, neither microbial diversity nor microbial composition among groups were significantly affected by RS (Figure S5C). However, RS decreased bacterial translocation in MLN, liver, and spleen ofTLR7.1 Tg mice similarly to oral antibiotics (Figures 5B and 5C).
Figure 5. Resistant starch modulates the gut microbiome and decreases L. reuteri translocation.
Fecal pellets and gut segments from WT C57B1/6, TLR7.1 Tg, and TLR7.1 Tg mice fed with resistant starch (RS) were collected; bacterial DNA was isolated and sequenced. (A) LDA scores of altered taxa in TLR7.1 Tg mice fed with RS compared to control (CTRL) diet. Shown are significantly altered taxa (increased in green, decreased in red). (B) Total percentage of bacterial translocation as determined by culture. (C) Distribution of Lactobacillus spp. translocation in MLN, liver, and spleen from WT, TLR7.1 Tg, and TLR7.1 Tg mice fed with RS. Relative abundances of fecal L. reuteri (LR) in TLR7.1 Tg mice fed with RS compared to CTRL diet evaluated by 16S rDNA gene sequencing in (D) experimental or (E) survival cohorts. (F) Relative abundances of Lactobacillus spp. and (G) LR in gut tissues evaluated by 16S rDNA gene sequencing. The results are expressed as mean ± SEM (n=3–20 mice per group). Data are representative of at least three independent experiments. *P<0.05 was considered statistically significant; **P <0.01; ***P<0.001; ****P<0.0001; ns=not significant; W0=week RS begins. See also Figure S5 and Tables S4-S5.
Consistent with these findings, RNA expression studies showed that RS improved markers of gut epithelial integrity and antimicrobial defense including Muc2 and Reg3g (Figures S5D-S5F). Functionally, gut leakiness in both the spontaneous and inducible TLR7-dependent lupus model was prevented by RS (Figure S5G). L. reuteri was persistently decreased in fecal pellets of RS-fed mice throughout the study (Figures 5D-5E, and Table S5). To determine L. reuteri colonization in different gut segments, we performed 16S rDNA sequencing of the ileum, cecum and colon. L. reuteri was abundant in the ileum of TLR7.1 Tg mice and profoundly suppressed in its niche by RS (Figures 5F-5G). These results demonstrated that RS suppressed the gut pathobionts that were enriched in both lupus models. Furthermore, RS tightened the gut epithelial barrier and decreased lupus-related gut leakiness and L. reuteri translocation to distal organs. Since growth of L. reuteri, a bacterium linked to the pathogenesis of lupus-like disease, was suppressed by RS, we investigated if feeding RS was able to control systemic autoimmunity.
Resistant starch suppresses lupus-related mortality and pathogenesis
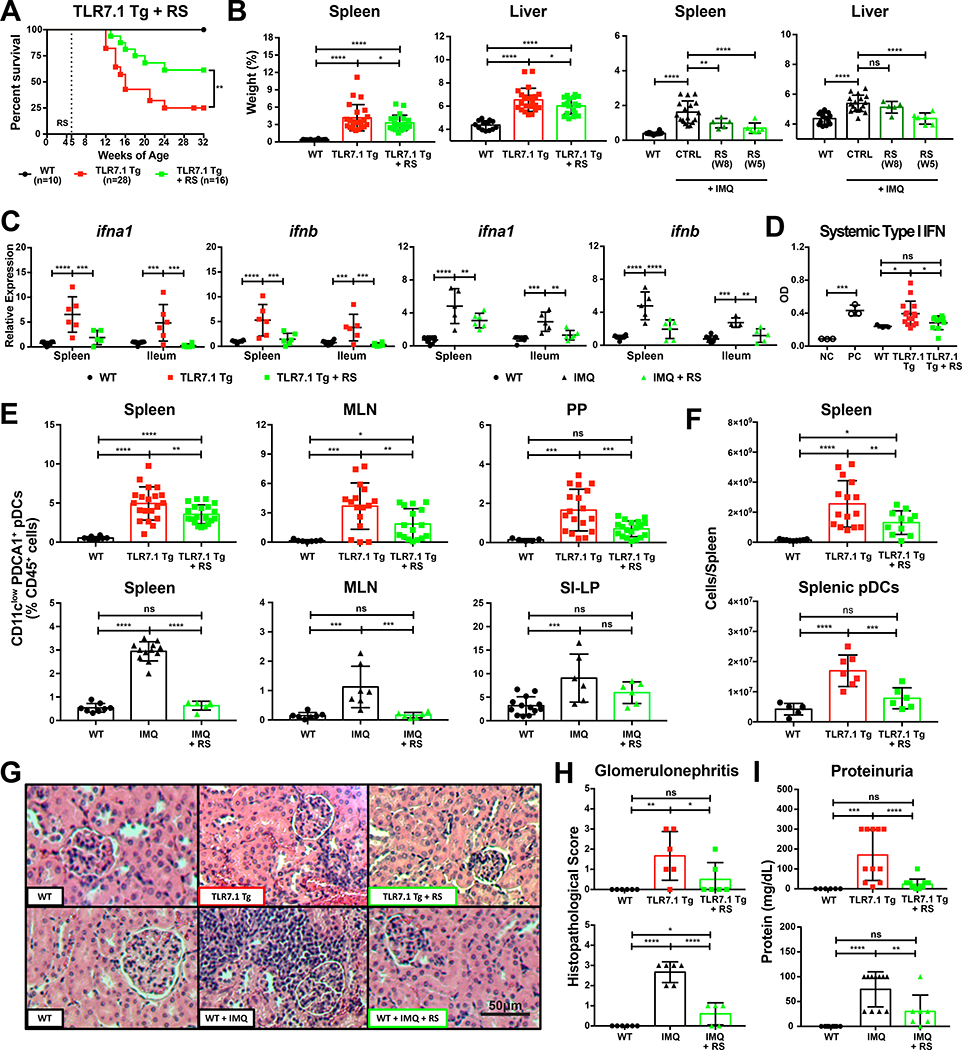
We continuously fed RS to 5-week-old TLR7.1 Tg mice and followed them until 32 weeks of age. We found a significant survival advantage compared to control diet-fed TLR7.1 Tg mice (Figure 6A). 5-week-old WT SPF B6 mice treated with IMQ were also fed with RS and sacrificed at 12 and 16 weeks of age, respectively, to study the impact of RS on the inducible model (Figure S5A). RS led to less severe organomegaly and type I IFN gene expression in both models (Figures 6B-6C) and to normalization of systemic type I IFN levels in TLR7.1 Tg mice (Figure 6D). In addition, relative and absolute pDCs frequencies of RS-fed mice normalized (Figures 6E-6F), as did Thl7, neutrophils, and CD44+ T cells, but not Tregs frequencies (Table S6). The dependence of the lupus phenotype on TLR7/IFN-related signals was reflected by reversal of hepatosplenomegaly, anemia and gut barrier leakiness when IMQ was added to RS-fed TLR7.1 Tg mice (Figure S6A).
Figure 6. Resistant starch suppresses lupus-related mortality and pathogenesis.
TLR7.1 Tg and C57B1/6 mice treated with IMQ were fed RS. (A) Survival analysis of TLR7.1 Tg mice fed with RS compared to CTRL diet. Experimental cohorts of TLR7.1 Tg and C57B1/6 mice were sacrificed for organ weights, immunologic, and histologic analyses. (B) Weights of spleen and liver. (C) Relative mRNA expression of type I IFN in spleen and ileum. (D) Systemic type I IFN responses in serum measured with a reporter cell line. (E-F) Cells from spleen, MLN, PP, and SI-LP were isolated and analyzed by flow cytometry. (E) Frequency of pDCs in spleen, MLN, PP, and SI-LP. (F) Total spleen and pDCs numbers. (G) Representative H&E staining of kidney tissue. (H) Histopathologic scoring for glomerulonephritis. (I) Quantification of proteinuria. The results are expressed as mean ± SEM (Survival cohorts n=8–28 mice per group, Experimental cohorts n=3–20 mice per group). The results are representative of at least two independent experiments. *P <0.05 was considered statistically significant; **P<0.01; ***P<0.001; ****P<0.0001; ns=not significant; NC=negative control; PC=positive control. See also Figure S6 and Table S6.
RS feeding also decreased the relative expression levels of proinflammatory molecules in the spleen and gut (Figures S6B-S6C). This suggests a systemic anti-inflammatory effect of RS. Reduced inflammation could explain a restoration in bone marrow physiology reflected by normalization of blood disorders (anemia and leukocytosis), bone marrow cellular repletion, and restored granulocyte-monocyte and megakaryocyte-erythrocyte progenitors populations (Figures S6D-S6F) (Soehnlein et al., 2017; Trompette et al., 2018; Ueda et al., 2005). Importantly, RS also significantly reduced renal immune infiltration and glomerulonephritis as well as proteinuria levels in TLR7.1 Tg mice (Figures 6G-6I). Furthermore, RS reduced the infiltration of multiple immune cell populations including total leukocyte counts, pDCs, myeloid cells and CD8+ T cells (Figure S6G). Interestingly, dietary modulation of the gut microbiota did not affect the increase in autoantibody levels or total IgG (Figures S6H-S6I).
These data support that a dietary intervention before onset of lupus was sufficient to prevent the development of innate inflammation and lupus-like disease. Since L. reuteri exacerbates systemic autoimmunity whereas RS decreased L. reuteri abundance and lupus pathogenesis, we next investigated if RS fermentation could control L. reuteri growth, thereby limiting L. reuteri-dependent progression of lupus.
Resistant starch-induced SCFAs inhibit L. reuteri growth in vitro and L. reuteri- mediated autoimmunity in vivo
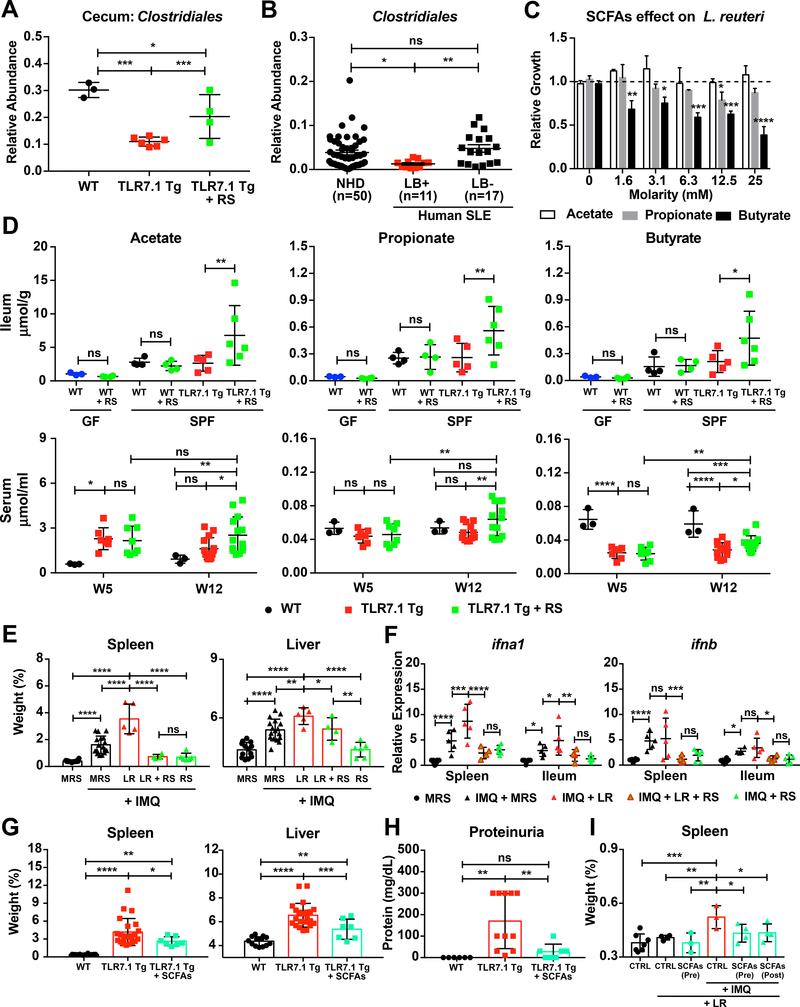
The main metabolites of fermentation of RS by the gut microbiota are the SCFAs acetate, propionate, and butyrate (Thorburn et al., 2014). We thus measured the levels of SCFAs after RS feeding and the abundance of their bacterial producers with the hypothesis that RS suppresses pathobiont growth via these metabolites in both lupus models. 16S rDNA sequencing of fecal pellets from RS-fed TLR7.1 Tg mice revealed an increase in Clostridiales among other bacterial taxa capable of fermenting RS into SCFAs (Figures 5A and S5B).
We also sequenced the microbiota of cecal tissue from RS- and control diet-fed TLR7.1 Tg mice and compared them to age- and sex-matched WT SPF C57B1/6 mice. Cecal abundance of bacteria from the order Clostridiales was reduced in lupus- prone mice and rescued by RS (Figure 7A). Remarkably, Clostridiales from Lactobacillus-positive stool samples of SLE patients were also decreased compared to Lactobacillus-negative samples (Figure 7B).
Figure 7. Resistant starch-induced SCFAs inhibit L. reuteri growth in vitro and L. reuteri-mediated autoimmunity in vivo.
Fecal pellets and gut segments from TLR7.1 Tg mice fed with RS were collected; bacterial DNA was isolated and sequenced. (A) Relative abundances of Clostridiales in cecal tissue evaluated by 16S rDNA gene sequencing. (B) Relative abundances of fecal Clostridiales in NHD and SLE patients separated by the presence or absence of Lactobacillus spp. (LB) determined by 16S rDNA gene sequencing. (C) Relative growth of L. reuteri (LR) cultured anaerobically with increasing doses of SCFAs acetate, propionate, and butyrate compared to MRS media alone. (D) SCFA levels measured in ileum and serum. (E-F) SPF C57B1/6 mice were treated with IMQ, fed RS, and gavaged daily with LR. Mice were sacrificed for organ weights and immunologic analyses. (E) Weights of spleen and liver. (F) Relative mRNA expression of type I IFN in spleen and ileum. (G-I) TLR7.1 Tg mice were administered SCFAs in their drinking water. Mice were sacrificed for organ weights and immunologic analyses. (G) Weights of spleen and liver. (H) Quantification of proteinuria. GF C57B1/6 mice were given SCFAs in their drinking water before (Pre) or after (Post) LR monocolonization and treated with IMQ. Mice were sacrificed for organ weights analyses. (I) Weights of spleen. The results are expressed as mean ± SEM (n=3–20 mice and 11–50 human samples per group). The results are representative of at least two independent experiments. *P<0.05 was considered statistically significant; **P<0.01; ***P<0.001; ****P<0.0001; ns=not significant. See also Figure S7 and Table S7.
In vitro, L. reuteri growth was inhibited by SCFAs, in particular butyrate, in a dose-dependent manner (Figures 7C and S7B). Analysis of SCFA levels in fecal pellets, gut segments, and peripheral blood revealed a selective increase of acetate, propionate, and butyrate in serum and ileum of RS-fed TLR7.1 Tg mice (Figure 7D, Table S7). To test if RS prevents LR-dependent exacerbation of lupus, we fed RS to 5-week-old SPF C57B1/6 mice treated with IMQ and gavaged daily with L. reuteri starting at 6 weeks of age until 16 weeks of age (Figure S7A). RS significantly inhibited splenomegaly and type I IFNs (Figures 7E-7F). Accumulation of pDCs, anemia, lupus nephritis, and gut leakiness were also reduced in RS-fed mice (Figures S7C-S7G).
Next, we supplied SCFAs to the drinking water of SPF TLR7.1 Tg mice as well as L. reuteri-monocolonized mice treated with IMQ (Figure S7A). LR-induced effects in both models were mitigated by SCFA administration in vivo. In TLR7.1 Tg mice, hepatosplenomegaly, pDCs numbers in spleen, MLN, and PP, and proteinuria were profoundly suppressed to levels in WT mice (Figures 7G-7H and S7H). In L. reuteri-monocolonized B6 mice, SCFAs administered before or after colonization inhibited spleen enlargement (Figure 7I). Thus, a dietary intervention was sufficient to prevent the development of TLR7-dependent systemic autoimmunity exacerbated by L. reuteri.
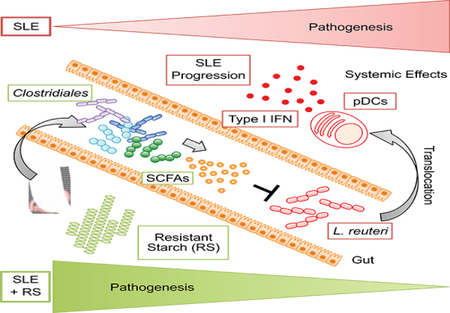
Overall, we demonstrated that RS fermentation to SCFAs and its effects on gut microbial communities improved outcomes in TLR7-dependent lupus models. RS decreased the abundance of L. reuteri, gut leakiness, type I IFN and proinflammatory responses, pDC infiltrations, and organ pathology, thereby preventing the development of systemic autoimmunity and decreasing mortality.
Discussion
In this study, we show the importance of diet-sensitive gut microbiota in systemic autoimmunity using two different TLR7-dependent lupus models. We were able to identify key bacterial taxa that were increased in both models and were transferable to WT mice by cohousing. Cohousing led to increased susceptibility to lupus-like manifestations. Profiling fecal microbiomes from TLR7.1 Tg mice and their co-housed WT littermates revealed an enrichment of mainly three bacterial taxa: the genera Lactobacillus and Desulfovibrio, and the family Rikenellaceae. WT mice colonized with microbiota containing these pathobionts developed increased gut leakiness with translocation of pathobionts.
Interestingly, the gut leakiness was TLR7-dependent based on studies with TLR7 KO mice. Little is known about the physiologic function of TLR7 in the gut, in particular on the gut epithelium. Endogenous retroviruses surge in TLR7 KO mice, a phenomenon dependent on the gut microbiota (Young et al., 2013). If, however, regulation of the barrier integrity is implicated in this process is unclear. Translocating bacteria have recently been shown in the (NZW x BXSB)F1 model to induce endogenous retroviral protein components, suggesting this possibility but a link to TLR7 remains to be determined (Manfredo Vieira et al., 2018). In this model, E. gallinarum drove autoimmunity whereas lactobacilli were absent from the microbiomes of these mice (Manfredo Vieira et al., 2018). The genetic and microbial differences likely reflect different SLE phenotypes described in humans (Banchereau et al., 2016).
In the TLR7.1 Tg model, we found that culture of internal organs revealed two Lactobacillus species, L. reuteri and L. johnsonii, which translocated outside the gut. Importantly, enrichment with L. reuteri, but not L. johnsonii promoted systemic autoimmunity and innate pathways. L. reuteri was also able to exacerbate lupus pathogenesis in the monocolonized setting compared to monocolonization with another gut bacterium, B. thetaiotaomicron. A RS-rich diet prevented L. reuteri outgrowth, pDC/IFN pathways, and lupus-related mortality. Interestingly, RS decreased L. reuteri abundance through generation of SCFAs that inhibit L. reuteri growth in vitro and vivo. In summary, we were able to link a diet-sensitive gut pathobiont to lupus pathogenesis, in particular to the TLR7-pDC-IFN axis that is central to human SLE (Crow, 2014) as well as autoinflammatory syndromes called interferonopathies (Rodero and Crow, 2016). The phenotypic heterogeneities of interferonopathies might also be influenced by the gut microbiota.
Supportive of a role for lactobacilli in the pathogenesis of lupus, taxa in this genus were found to be enriched in female NZB/W FI mice, another model of systemic lupus. In this study, Lactobacillus spp. were associated with more severe disease whereas they were reduced as disease is controlled with dexamethasone (Luo et al., 2017). The microbiota of MRL/lpr mice, on the other hand, were described to be depleted of Lactobacillaceae and a mix of Lactobacillus strains improved outcomes in this lupus model (Mu et al., 2017b, 2017a; Zhang et al., 2014). These studies did not include gnotobiotic experiments or define which strain mediates the observed effects, making a causal relationship difficult to establish. In addition, the MRL/lpr model likely represents a different subset of human SLE (Banchereau et al., 2016). While sensitive to postnatal antibiotic depletion of the microbiota, removal all microbiota from birth via germ-free rederivation did not affect lupus phenotypes based on previous studies (Maldonado et al., 1999). Interestingly, L. reuteri has been shown to have anti-inflammatory and Treg-inducing properties and is thus promoted as a probiotic in inflammatory bowel disease (Livingston et al., 2010; Lorea Baroja et al., 2007). Individual species in the microbiome are known to have diametrical effects depending on the disease studied, pointing to the importance of the interactions with the host (Ley, 2016). Consistent with this notion, we did not find alterations in Treg levels after colonization with L. reuteri in the TLR7-dependent lupus models (Table S6). Instead, we mechanistically connected L. reuteri in vivo with a pathogenic innate pathway involved in systemic autoimmunity, suggesting host- and context-dependent effects of gut commensal species in chronic disease states. The pDC/IFN-promoting properties of L. reuteri in the context of a lupus-prone host suggest a paradigm in which a bacterium that is normally considered a probiotic may become harmful under certain genetic or environmental conditions.
Because diet can shape gut microbial communities and also influence host immune processes (Kau et al., 2011), we used a dietary intervention to suppress lupus-related pathobionts. Fiber-rich diets such as RS are beneficial for pathogen resistance and metabolic diseases that share inflammatory features with SLE (Desai et al., 2016; De Filippo et al., 2010; Ganguly, 2018; Sonnenburg and Sonnenburg, 2014). We were able to show that RS suppressed the abundance of L. reuteri in its ileal niche and prolonged survival of lupus-prone mice. Besides inhibiting the abundance of lupus-related pathobionts, RS also increased bacterial taxa capable of fermenting fiber into SCFAs; loss of their abundance have been linked to disease (De Filippo et al., 2010; Koh et al., 2016; Macia et al., 2015; Walker et al., 2011). Interestingly, bacteria from the order Clostridiales, which are known to promote mucus thickening (Wlodarska et al., 2015), were reduced in Lactobacillus-colonized SLE patients and lupus-prone mice. RS feeding of mice increased this order not only in fecal but also in cecal microbiomes. All three major SCFAs - acetate, butyrate, and propionate - were significantly elevated in the ileum of RS-fed mice, supporting local effects in the niche of L. reuteri.
One of the best described host effects of SCFAs are induction of Tregs (Arpaia et al., 2013; Smith et al., 2013; Thorburn et al., 2014). Treg frequencies, however, were not altered by RS feeding in the gut or secondary lymphoid organs of lupus-prone mice (Table S6). Instead, we found that L. reuteri growth was inhibited in vitro by SCFAs in a dose-dependent manner, suggesting the local effects of SCFAs in the ileum suppresses pathobiont growth in vivo, thereby mediating indirectly its protective effects on the host. A direct effect on the host tissue is also possible, which is not mutually exclusive. SCFAs are known to improve gut barrier integrity, thereby restraining bacteria or bacterial components to traffic from the gut to the liver (Kelly et al., 2015; Liou et al., 2013). RS decreased L. reuteri-induced gut leakiness in the IMQ-inducible lupus model, possibly via the barrier-tightening effect of SCFAs. Supporting these results, RS increased tight junctional components of the ileal epithelial barrier, a process previously shown to be mediated by SCFAs (Wang et al., 2012). RS also suppressed gut leakiness functionally that was induced by TLR7 signals and L. reuteri in vivo.
In summary, the current study highlights the relevance of diet-microbiota-host interactions in the development of autoimmunity and defines a link between pathobiont outgrowth and disease manifestations. Mechanistically, the pathobiont instigates the pDC-IFN axis in lupus-prone animals with excessive TLR7 signaling that results in impaired gut barrier integrity. The rapid rise of chronic immune diseases, which is paralleled by major changes in modern diets, might be related to mechanisms such as those we uncovered here. A lack of dietary fiber might allow for outgrowth of pathobionts that promote immune pathways in genetically prone individuals. These processes ultimately lead to onset of systemic inflammatory conditions. Dietary or other targeted approaches towards the gut microbiota would restore homeostasis by restraining disease-promoting pathobionts (Lemon et al., 2012). Dietary interventions such as resistant starches could bypass major side effects related to broad-spectrum antibiotics (e.g., antibiotic resistance or systemic toxicities). The appropriate diet would be expected to enrich beneficial commensals or metabolites that naturally control pathobiont overgrowth. In the future, asymptomatic but autoimmune-prone individuals with heightened pDC and type I IFN pathways might benefit from personalized diets tailored to their microbiota composition (Bourn and James, 2016; Kau et al., 2011; Robertson and James, 2014). Because the type I IFN pathway is involved in various chronic inflammatory and metabolic diseases, identifying strains in the microbiota that fuel this pathway might have implications beyond autoimmunity (Ganguly, 2018).
STARS Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Martin Kriegel (martin.kriegel@yale.edu).
Experimental Model and Subject Details
Animals.
Five-week-old female TLR7.1 Tg, TLR7 KO, and C57B1/6 mice were used in this study except if noted otherwise in the figures. All mice were healthy before initiation of the study, were not subjected to previous procedures, and were naive to drugs. TLR7.1 Tg and TLR7 KO mice were kindly provided by Silvia Bolland (NIH) (Deane et al., 2007) and Akiko Iwasaki (Yale) (Lund et al., 2004). All strains were maintained on a C57B1/6 background. C57B1/6 mice were originally obtained from the Jackson Laboratory and bred at Yale. All mice were maintained in an SPF environment. Mice were cohoused for 2 weeks after weaning and bedding was swapped twice among experimental cages to equilibrate the microbiomes from different cages and parental origin. Mice were provided with water and a standard laboratory diet ad libitum (2018 Harlan Teklad, South Easton, MA, USA) except if noted otherwise. They were supplied with hardwood chips as bedding and were housed in a temperature-controlled, air-conditioned room on a 12-hr light-dark cycle. GF C57B1/6 mice were kept under sterile conditions in CBC (Class Biologically Clean, Madison, WI, USA) Quad flexible film isolator (Softwall), exposed to a 12-hr light/dark cycle and provided standard, autoclaved tap water and mouse chow (2018S Harlan Teklad, South Easton, MA, USA) ad libitum. All experiments with mice were performed using protocols approved by the Yale University Institutional Animal Care and Use Committee.
Human subjects.
All human subject studies were approved by the Yale Human Investigations Committee and in accordance with the Declaration of Helsinki. Only female subjects were studied because adult SLE is a highly female-biased autoimmune disease with a female-to-male ratio of approximately 9:1. Stool samples were collected from SLE patients (n = 12 females, 18 years of age and older) and sex- and age-matched (±5 years) healthy controls (n = 22) from up to three study visits (baseline, week 4, and week 8). The SLE patient cohort, longitudinal study design, inclusion and exclusion criteria as well as sampling protocol and 16S rDNA sequencing were previously described (Greiling et al., 2018) (ClinicalTrials.gov identifier: NCT02394964). In brief, all patients were diagnosed by a healthcare provider and thoroughly evaluated at each study visit at the Yale Center for Clinical Investigation. Number of visits, sex, age, race, ethnicity, weight, height, body mass index, autoantibody profile, complete blood counts, kidney function, inflammatory markers, complement levels, constitutional symptoms, cutaneous, mucosal, musculoskeletal, vascular, and neurologic manifestations, immunomodulatory drugs, and adjunctive medications, as well as a detailed dietary history have been captured and were previously described (Greiling et al., 2018). Exclusion criteria were ongoing chronic infection, antibiotic or probiotic use in the last 90 days, topical antibiotic or antimicrobial use in the last 7 days, bathing or tooth brushing in the last 8 hours, major gastrointestinal surgery in the last 5 years, gastrointestinal bleeding history, inflammatory bowel disease, bulimia or anorexia nervosa, morbid obesity, uncontrolled diabetes mellitus, malignancy in the past year, and known excessive alcohol use.
Method Details
Diets.
Custom diets for SPF and GF mice were based on the 2018 and 2018S diets (Harlan Teklad, South Easton, MA, USA), respectively. The RS diet (TD.150492 for SPF and TD.160604 for GF mice) is enriched for Hi-Maize 260 (40 g/kg). Mice were cohoused for 2 weeks after weaning and bedding was swapped twice among experimental cages to equilibrate the microbiomes from different cages and parental origin. Afterwards, mice were fed with RS for 7 or 11 weeks according to the experimental setup or up to 32 weeks of age or time of death for survival studies.
Induction of lupus-like disease.
SPF C57B1/6 mice received a topical treatment of 1.25 mg of 5% imiquimod cream (HealthWarehouse) on the skin of the left ear three times a week from 8 to 16 weeks of age, which induces lupus-like disease as previously described (Yokogawa et al., 2014). The model under SPF conditions was similarly applied to the GF setting as previously described (Greiling et al., 2018). Mice under both conditions were followed until 24 weeks of age for survival analysis.
Antibiotic treatment.
TLR7.1 Tg mice of the treatment and control groups were cohoused for 2 weeks after weaning and bedding was swapped twice among experimental cages to equilibrate the microbiomes from different cages and parental origin. Mice at 5 weeks of age were administered broad-spectrum antibiotic consisting of metronidazole (1 g/1; Fisher Scientific), neomycin (1 g/1; Fisher Scientific), ampicillin (1 g/1; Sigma), and vancomycin (0.5 g/1; Acros Organics) in drinking water for 7 weeks or up to 32 weeks of age for survival studies. Sweetener (Equal, 4 g/1) was added to both the antibiotics and control water due to the metallic taste of metronidazole.
Short-chain fatty acids treatment.
TLR7.1 Tg mice of the treatment and control groups were cohoused for 2 weeks after weaning and bedding was swapped twice among experimental cages to equilibrate the microbiomes from different cages and parental origin. Mice at 5 weeks of age were administered a mixture of short-chain fatty acids consisting of sodium acetate (68 mM; Sigma), sodium propionate (26 mM; Sigma), and sodium butyrate (40 mM; Sigma) in the drinking water for 7 weeks. Sweetener (Equal, 4 g/1) was added to both the SCFAs and control water due to the bitter taste of butyrate. GF B6 mice at 5 or 6 weeks of age received the same mixture of SCFAs in their drinking water until they reached 10 weeks of age.
Blood cell counts.
Blood was collected in a 1.5-ml tube without anticoagulant through tail vein bleeding after 5 minutes of light warming-induced vasodilation. For red blood cell counting, 2 μl of blood was transferred to 500 ul of PBS and diluted 1:250. For white blood cell counting, 2 μl of blood was transferred to 100 μl of ACK Lysing Buffer (Gibco) to remove red cells before counting. Cells were counted manually using a hemocytometer.
Histopathology.
Tissues were dissected at autopsy and fixed in 10% neutral formalin, and histological sections were stained with hematoxylin and eosin (H&E). The severity of glomerulonephritis was scored in a blinded fashion on a scale of 0–4 based on the intensity and extent of histopathological change as previously described (Klopfleisch, 2013). Grade of 0 was given to kidneys without glomerular lesions; grade 1 consisted of minimal thickening of the mesangium; grade 2 contained noticeable increases in both mesangial and glomerular cellularity; grade 3 was characterized by the preceding conditions with superimposed inflammatory exudates and capsular adhesions; and grade 4 consisted of obliterated glomerular architecture in greater than 70% of glomeruli. Twenty glomeruli within one area were graded according to this classification and used to calculate the mean glomerular histopathological score for each mouse, and those scores were used to calculate mean scores for each experimental group.
Enzyme-linked immunosorbent assays.
IgG and IgG2a serum autoantibodies against dsDNA and RNA, as well as total IgA and IgG antibodies were determined using standardized enzyme-linked immunosorbent assays (ELISAs). For anti-dsDNA ELISAs, DNA-BIND™ (Corning) plates were coated with 100 μg/ml salmon sperm DNA (Fisher Scientific) and blocked in 0.1M PBS, 3.0% BSA, and 5.0% FBS. Serum was diluted 1:1000 and incubated for 1 h at 37°C. Wells were washed and incubated for 1 h at 37°C with rabbit anti-mouse IgG or IgG2a (Fisher Scientific) conjugated with HRP and developed with TMB substrate solution. 2M sulfuric acid (Sigma) solution was used to stop development. For anti-RNA ELISAs, DNA-BIND™ (Corning) plates were coated with 5 mg/ml yeast RNA (Ambion) and blocked in 0.1M PBS, 0.05% Tween-20, 5.0% FBS. Serum was diluted 1:1000 and incubated for 1 h at 37°C. Wells were washed and incubated for 1 h at 37°C with rabbit anti-mouse IgG or IgG2a (Fisher Scientific) conjugated with HRP and developed with TMB substrate solution. 2M sulfuric acid (Sigma) solution was used to stop development. For total anti-IgG and anti-IgA ELISAs, Ready-Set-Go! Elisa kits (eBioscience) were used. Serum was diluted 1:5000 and incubated for 1 h at 37°C. 2M sulfuric acid (Sigma) solution was used to stop development. Concentrations of all antibodies were determined by reading the absorbance at 450 nm/570 nm reference.
Proteinuria and blood urea nitrogen.
Urine from TLR7.1 Tg mice and WT C57B1/6 mice was tested semiquantitatively for proteinuria with albumin test strips (Multistix1OSG Siemens) at the end of the experimental or survival studies. The test strips were placed in the urine and evaluated after 60 seconds for negative, trace, 30 mg/dl (+), 100 mg/dl (++), 300 mg/dl (+++) and 2000 mg/dl or more (++++) proteinuria. For quantitative determination of blood urea nitrogen (BUN), 50 μl of serum was analyzed at the George M. O’Brien Kidney Physiology Core at Yale using an Excel Chemistry Analyzer (Standbio Laboratory).
In vivo gut permeability assay with Fluorescein isothiocyanate-dextran.
Mice were weighed and fasted for 4 hours prior to oral Fluorescein isothiocyanate (FITC)-dextran (4 KDa; Sigma) administration. A total concentration of 250 mg/kg body weight was administered through gavage. 3 hours after administration, blood samples were collected through the tail vein and 50 μl of serum was placed in a fluorescence plate reader to determine the concentration by fluorescence excitation at 495 nm/519 nm reference.
Assessment of bacterial translocation.
TLR7.1 Tg mice and WT C57B1/6 mice at 15–18 weeks of age underwent sterile laparotomy and mesenteric lymph nodes (MLN), liver, and spleen were aseptically collected in BBL Mycoflask Thioglycollate (Fluid) Prepared Media (BD Diagnostic Systems) and kept in aerobic and anaerobic conditions at 37°C. After 72 hours of incubation, 20 μl of thioglycollate broth was streaked on non-selective Gifu (Sigma-Aldrich), De Man, Rogosa and Sharpe (MRS) (Sigma-Aldrich), or Mega media (Goodman et al., 2011) agar plates and incubated for an additional 24 hours at 37°C. Single colonies were picked and Sanger sequencing of full-length 16S rDNA was performed to identify bacterial species comparing the 16S rDNA to the RDP database vll.5.
L. reuteri and B. thetaiotaomicron monocolonization experiments.
GF C57B1/6 mice were colonized at 6 weeks of age by a single oral gavage with 0.2 ml of thawed 1 × 109 CFU/ml of L. reuteri (LR), isolated from the spleen of a TLR7.1 Tg mouse that showed signs of autoimmune disease. The LR strain was grown overnight at 37°C in MRS broth (Sigma-Aldrich). Mice were assessed for bacterial translocation and burden as described above. Regarding B. thetaiotaomicron (BT) monocolonization, GF C57B1/6 mice were colonized at 6 weeks of age by a single oral gavage with 0.2 ml of thawed 1 × 109 CFU/ml of BT in 20% glycerol. Monocolonization was confirmed by DNA extraction from the fecal pellet, PCR amplification of the full- length 16S rDNA region followed by Sanger sequencing.
Gavage with L. reuteri and L. johnsonii.
At 5 weeks of age SPF C57B1/6 mice were colonized with L. reuteri (LR) via daily oral gavage following the same conditions as for the monocolonization experiments, or with 0.2 ml of thawed 1 × 109 CFU/ml of L. johnsonii (LJ), isolated from the MLN of a TLR7.1 Tg mouse that showed signs of autoimmune disease. LJ was grown at 37°C in MRS broth (Sigma-Aldrich). Mice were assessed for bacterial translocation and burden as described above. For Figure S4C, 7 weeks old SPF WT mice were treated with broad-spectrum antibiotics for 3 weeks and gavaged daily with 1 × 109 CFU/ml of LR or LJ for 3 weeks. For Figures 4C-4D, 5-week-old TLR7.1 Tg mice were colonized with LR under the same conditions as for the monocolonization experiments.
Cecal transfer.
Total cecal content from three 12-week-old TLR7.1 Tg mice that showed signs of autoimmune disease was collected anaerobically and diluted 1:10 in 80% glycerol/PBS. Filtered aliquots were stored at −80°C. At the day of inoculation, cecal contents stocks were mixed anaerobically and diluted 1:5 with PBS. SPF C57B1/6 mice received 0.2 ml of thawed cecal contents at 21 days of age, at 23 days of age, and at 8 weeks of age.
Measurement of metabolites.
Blood, gut tissues and fecal pellets were stored at −80°C. Short-chain fatty acids (SCFAs) acetate, propionate and butyrate were extracted by acidifying samples with 33% hydrochloric acid, performing two cycles of diethyl ether isolation and evaporation with nitrogen gas. SCFAs were measured by gas chromatography mass spectrometry in the Mouse Metabolic Phenotyping Center atYale. Concentrations were calculated from deuterated internal standards.
L. reuteri in vitro growth curves in the presence of SCFAs.
L. reuteri (LR) was grown at 37°C for 24 hours in 10 ml of MRS broth (Sigma-Aldrich) with increasing concentrations of acetate, propionate, and butyrate. Optical densities were measured from the original tubes. Relative growth was assessed by correcting ODs to LR grown in media alone.
Real-Time quantitative PCR analysis.
RNA was isolated from tissue sections stored in Trizol (ThermoFischer Scientific) at −80°C. RNA was then quantified and purified with a DNA-free™ DNA Removal Kit (ThermoFischer Scientific). cDNA was generated using the High Capacity cDNA Reverse Transcription Kit (ThermoFischer Scientific). qPCR was performed using TaqMan primers and probes with TaqMan PCR Master Mix (Applied Biosystems) for genes involved in intestinal epithelial integrity and using SYBR green for all other genes. Reactions were run on an RT-PCR system (QuantiStudio 6 Flex; Applied Biosystems). Gene expression is displayed as fold-increase and normalized to gapdh for Taqman and to actb for SYBR green.
Reporter cell line assay.
Bioactive murine type I IFNs were detected from serum samples using B16-Blue™ IFN-α/β cells (Invivogen), a murine B16 melanoma cell line of C57B1/6 origin after stable transfection with a SEAP reporter gene under the control of the IFN-α/β-inducible ISG54 promoter enhanced by a multimeric ISRE. Serum samples were added to the cells for 24 hours using culture media as a negative control and 5’ppp-dsRNA/LyoVec™ (Invivogen) as a positive control. QUANTI-Blue™ (Invivogen) was used to detect and quantify secreted embryonic alkaline phosphatase after 24 hours of incubation. The optical density of QUANTI- Blue™ was determined by reading the absorbance at 620 nm.
Lactobacilli-plasmacytoid dendritic cell co-culture assay.
SPF C57B1/6 mice were euthanized and spleens were collected, minced into small pieces, and digested in 5 ml of RPMI containing 5% fetal bovine serum, collagenase D (2 mg/ml, Roche) and DNAse I (0.1 mg/ml, Roche) for 30 minutes at 37°C. Samples were filtered through a 100-μm strainer and washed with 20 ml of PBS (50 ml Falcon, room temperature). Finally, the cells were resuspended in 1 ml of culture media. Plasmacytoid dendritic cells (pDCs) were obtained by MACS using positive selection with PDCA-1+ labeled magnetic beads (Miltenyi Biotec, RRID: AB_2747367). 5 × 105 pDCs were plated in triplicates and co-cultured for 18 hours with heat-killed L. reuteri / L. johnsonii (1:10 ratio) or their bacterial supernatant in the presence of IMQ (1 μg/ml) or culture media alone. RNA was isolated and processed as described above for qPCR analysis.
Leukocyte isolation from lamina propria.
Mice were euthanized and small intestine (SI) were collected and placed in ice-cold complete HBSS (HBSS containing 10% fetal bovine serum and 1% penicillin-streptomycin, cHBSS). The lumen of the intestine was flushed with ice-cold cHBSS using a 10-ml syringe and 21G needle. Peyer’s patches were removed and intestines were cut longitudinally in a petri dish containing PBS. Intestine was transferred to a 50 ml Falcon (Fisher) containing cold complete PBS (PBS containing 2% fetal bovine serum and 1% penicillin-streptomycin, cPBS) and mixed on a vortexer for 5 seconds. Tissue was washed three times with 20 ml of cold cPBS. Next, SI was transferred to a 50-ml Falcon containing 20 ml of cPBS at room temperature. 200 μl of 0.5 M EDTA was added to the tube. Tubes were placed horizontally in a shaker (200 rpm, 25 min, 37°C). After incubation, tubes were shaken with a vortexer (Fisher Scientific) and media was discarded. The solution was replaced with 20 ml of cold HBSS and vortexed for 5 seconds. The procedure was repeated two more times using 20 ml of cold HBSS. After the last washing step, the tissue was placed in a 5-cm Petri dish at room temperature and 7 ml of HBSS containing collagenase D (2 mg/ml, Roche) and DNAse I (0.1 mg/ml, Roche) was added. Next, SI was minced into small pieces and incubated in a shaker (50 rpm) for 30 minutes at 37°C. Samples were filtered through a 100-μm strainer and washed with 20 ml of HBSS (50 ml Falcon, room temperature). Next, 10 ml of HBSS were added to the strainer and the suspension transferred to a new tube through a 40-μm strainer and centrifuged 450 x g for 5 minutes at 4°C, then washed one more time with 20 ml of cold HBSS by centrifuging the mixture at 450 x g for 5 minutes at 4°C. Finally, the cells were resuspended in 1 ml of culture media.
Flow cytometry and gating strategy.
Cells isolated from SI-LP, PP, MLN, spleen, blood, kidneys, or bone marrow were surface-stained in FACS buffer (PBS, 0.5% bovine serum albumin, 0.2 mg of sodium azide) for 30 minutes in the dark at room temperature with fluorescently conjugated antibodies specific to CD3 (RRID: AB_312670), CD4 (RRID: AB_312713), CD8 (RRID: AB_469777), CD11b (RRID: AB_893233), CD11c (RRID: AB_2563655), Ly6G (RRID: AB_1877163), I-A/I-E (RRID: AB_493528), CD44 (RRID: AB_469715), CD19 (RRID: AB_314235), B220 (RRID: AB_312991), GR-1 (RRID: AB_313371), NK1.1 (RRID: AB_313393), Ter119 (RRID: AB_313707), CD16/32 (RRID: AB_312806), CD34 (RRID: AB_1727471), Sca-1 (RRID: AB_469669), ckit (RRID: AB_2632809), CD45 (RRID: AB_312973), and PDCA-1 (RRID: AB_1953285). For Tregs/Thl7 identification, after staining for surface antigens, cells were fixed and permeabilized using a Fixation/Permeabilization Solution Kit (BD Biosciences) overnight at 4°C and stained with antibodies specific to FOXP3 (RRID: AB_469793) and IL-17A (RRID: AB_315463) in the presence of a Protein Transport Inhibitor (Containing Monensin) (BD Biosciences) for 1 hour. Cell viability was assessed with a Fixable Viability Dye eFluor™ 780 (eBioscience). Granulocyte-monocyte, common myeloid, and megakaryocyte-erythrocyte progenitors were identified as CD16/32high CD34+ (GMP), CD16/32low CD34+ (CMP), and CD16/32- CD34- (MEP) cells after live/dead staining, doublets exclusion, lineage (CD19, B220, GR-1, NK1.1, Terll9) exclusion, and Sca-1- c-Kit+ selection. pDCs were identified as CD45+ CDllb- CDllclow PDCA-1+ cells after live/dead staining and doublets exclusion. Stained samples were run on an LSR II flow cytometer (Becton Dickinson). All flow cytometry data were analyzed by Flowjo version 10 (Tree Star).
16S rDNA high-throughput sequencing.
Mouse fecal samples and gut segments were collected sterilely and stored at −80°C. DNA isolation from microbiota samples was performed using the MagAttract PowerSoil DNA EP Kit (Qiagen). The V4 region of the 16S rDNA was PCR amplified, normalized, pooled, and sequenced using the Illumina MiSeq with 2 × 250 bp paired-end reads as described (Kozich et al., 2013; Manfredo Vieira et al., 2018). Analysis of 16S rDNA sequencing reads was performed as described with the following minor modifications: Quantitative Insights Into Microbial Ecology (QIIME) (Caporaso et al., 2010) analysis was performed with version 1.8 and a quality score cutoff of 30, assigning taxonomy using the Greengenes gg_13_5_otus reference database (Cullen et al., 2015). Filtered operational taxonomic units (OTUs) were rarefied to a depth of 4,000 sequences per sample and OTUs representing less than 0.01% of total abundance were excluded from further analysis. Analysis of human 16S rDNA sequencing data from SLE patients was performed similarly as above, with the exception that OTUs were rarefied to a depth of 10,000 sequences per sample as previously described (Greiling et al., 2018).
Quantification and Statistical Analysis
Statistical Analysis.
Statistical analysis was carried out with GraphPad Prism (7.0a). All data are presented as means ± SEM and “n” represents the number of mice used for each experiment or the number of human stool samples per group. The exact value of n in each experiment is indicated in corresponding figure legend. An unpaired Student’s t-test was used to evaluate the difference between two groups. For more than two groups, one-way ANOVA was used. For more than two groups under different conditions two-way ANOVA was used. A probability value of p<0.05 was considered significant. For 16S rDNA analysis, differences in the relative abundance of individual taxa between defined groups were tested for significance using the “group significance” algorithm, implemented within QIIME. Tests were performed using the non-parametric Kruskal-Wallis one-way analysis of variance, generating a Benjamini-Hochberg false-discovery rate (FDR) corrected p-value.
Data and Software availability
Data generated by 16S rDNA high-throughput sequencing of fecal and tissue samples have been deposited in the European Nucleotide Archive under the ID code: PRJEB28382. 16S rDNA sequencing of human fecal samples obtained from our human SLE patient cohort at Yale is deposited in the European Nucleotide Archive under the ID code: PRJEB24742.
Supplementary Material
Highlights.
L. reuteri colonizes lupus-prone hosts and translocates to MLN, liver, and spleen
L. reuteri exacerbates TLR7-dependent lupus in conventional and germ-free mice
Resistant starch ameliorates pDCs, type I IFN pathways, and lupus-related mortality
Starch diet-derived short-chain fatty acids suppress L. reuteri in vitro and vivo
Acknowledgements
We would like to thank the O’Brien Kidney Center, the Mouse Metabolic Phenotyping Center, and the Pathology Tissue Microarray Facility at Yale for technical assistance. We thank the Palm lab for the use of an anaerobic chamber, Vivian Lim from the Pereira lab for assistance with bone marrow precursor studies, Minerva Ringland for assistance with in vivo studies, and Fatima Saldana (Baylor College of Medicine) for providing expertise in image processing. We also would like to thank Silvia Bolland (NIH) for providing TLR7.1 Tg mice and Akiko Iwasaki (Yale) for providing TLR7 KO mice. This work was supported by grants from the National Institutes of Health (NIH) (K08AI095318, R01AI118855, T32AI07019, T32DK007017–39), the O’Brien Center at Yale (NIH P30DK079310), the Arthritis National Research Foundation, the Arthritis Foundation, the Lupus Research Institute, the HHMI Medical Research Fellows Program, and the Lupus Foundation of America. The human microbiome study was performed with assistance from the Yale Center for Clinical Investigation, which is supported by the Clinical and Translational Science Awards grant number UL1 RR024139 from the National Center for Research Resources, the National Center for Advancing Translational Science, and the NIH Roadmap for Medical Research.
Footnotes
Declaration of interests
M.A.K. received salary, consulting fees, honoraria and research funds from Roche, Bristol-Meyers Squibb, AbbVie, and Cell Applications.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, DeRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau R, Hong S, Cantarel B, Baldwin N, Edens M, Cepika A, Acs P, Turner J, Vinod P, Kahn S, et al. (2016). Personalized Immunomonitoring Uncovers Molecular Networks That Stratify Lupus Patients. Cell 165, 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourn R, and James JA (2016). Pre-Clinical Lupus. Curr. Opin. Rheumatol 8, 444–454. [Google Scholar]
- Buechler MB, Teal TH, Elkon KB, and Hamerman JA (2013). Type I IFN drives emergency myelopoiesis and peripheral myeloid expansion during chronic Toll-like receptor 7 signaling. J. Immunol 190, 886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. (2010). QIIME allows analysis ofhigh- throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nat. Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Gerber GK, Luevano JM, Gatti DM, Somes L, Svenson KL, and Turnbaugh PJ (2015). Diet Dominates Host Genotype in Shaping the Murine Gut Microbiota. Cell Host Microbe 17, 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FRC, Franfozo MCS, Oliveira G.G. De, Ignacio A, Castoldi A, Zamboni DS, Ramos SG, Camara NO, Zoete M.R. De, Palm NW, et al. (2016). Gut microbiota translocation to the pancreatic lymph nodes triggers NOD2 activation and contributes to T1D onset. J. Exp. Med 213,1223–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow MK (2014). Type I Interferon in the Pathogenesis ofLupus. J. Immunol 192, 5459–5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundeil EA, Trent MS, Degnan PH, Booth CJ, Yu H, and Goodman AL (2015). Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science (80-,). 347,170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane JA, Pisitkun P, Barrett RS, Feigenbaum L, Town T, Ward JMM, Flavell RA, and Bolland S (2007). Control of Toll-like Receptor 7 Expression Is Essential to RestrictAutoimmunity and Dendritic Cell Proliferation. Immunity 27, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. (2016). A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 167,1339–1353.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Baptiste J, and Lionetti P (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. PNAS 107,14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Zaragoza E, Riquelme-Navarrete M, Sánchez-Zapata E, and JA P-Á (2010). Resistant starch as functional ingredient: A review. Food. Res. Int 43, 931–942. [Google Scholar]
- Ganguly D (2018). Do Type I Interferons Link SystemicAutoimmunities and Metabolic Syndrome in a Pathogenetic Continuum? Trends Immunol. 39, 28–43. [DOI] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, and Gordon JI (2011). Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. PNAS 108, 6252–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiling TM, Dehner C, Chen X, Hughes K, Iñiguez AJ, Boccitto M, Ruiz DZ, Renfroe SC, Vieira SM, Ruff WE, et al. (2018). Commensal orthologs ofthe human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transi. Med 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, and Macpherson AJ (2012). Interactions Between the Microbiota and the Immune System. Science (80-.). 336,1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, and Gordon JI (2011). Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, et al. (2015). Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 17, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfleisch R (2013). Multiparametric and semiquantitative scoring systems for the evaluation of mouse model histopathology-a systematic review. BMC Vet Res 9,123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A, De Vadder F, Kovatcheva-Datchary P, and Bäckhed F (2016). From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 165,1332–1345. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, and Schloss PD (2013). Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq. Appl. Environ. Microbiol 79, 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht BN, and Hammad H (2017). The immunology of the allergy epidemic and the hygiene hypothesis. Nat. Immunol 18,1076–1083. [DOI] [PubMed] [Google Scholar]
- Lemon KP, Armitage GC, Reiman DA, and Fischbach MA (2012). Microbiota-Targeted Therapies : An Ecological Perspective. Sci. Transi. Med 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE (2016). Gut microbiota in 2015: Prevotella in the gut: Choose carefully. Nat. Rev. Gastroenterol. Hepatol 13, 69. [DOI] [PubMed] [Google Scholar]
- Liou AP, Paziuk M, Luevano J-M, Machineni S, Turnbaugh PJ, and Kaplan LM (2013). Conserved Shifts in the Gut Microbiota Due to Gastric Bypass Reduce Host Weight and Adiposity. Sci. Transi. Med 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston M, Loach D, Wilson M, Tannock GW, and Baird M (2010). Gut commensal Lactobacillus reuteri 100–23 stimulates an immunoregulatory response. Immunol. Cell. Biol 88, 99–102. [DOI] [PubMed] [Google Scholar]
- Lorea Baroja M, Kirjavainen PV, Hekmat S, and Reid G (2007). Anti-inflammatory effects of probiotic yogurt in inflammatory bowel disease patients. Clin. Exp. Immunol 149, 470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, and Flavell RA (2004). Recognition of single-stranded RNA viruses by Toll-like receptor 7. PNAS 101, 5598–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo XM, Edwards MR, Mu Q, Yu Y, Vieson MD, Reilly CM, Ahmed SA, and Bankoie AA (2017). Gut microbiota in human SLE and a mouse model oflupus. Appl. Environ. Microbiol. AEM 02288–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian McKenzie C, Hijikata A, Wong C, et al. (2015). Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation ofthe inflammasome. Nat. Commun 6, 6734. [DOI] [PubMed] [Google Scholar]
- Maldonado MA, Kakkanaiah V, Glen C, Chen F, Reap EA, Farkas WR, Jennette JC, Michael P, Kotzin BL, Cohen PL, et al. (1999). The Role ofEnvironmental Antigens in the Spontaneous Development ofAutoimmunity in MRL-lpr Mice. J. Immunol 162, 6322–6330. [PubMed] [Google Scholar]
- Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, Costa FRC, Tiniakou E, Greiling T, Ruff W, et al. (2018). Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science (80-.). 359,1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño E, Richards JL, McLeod KH, Stanley D, Yap YA, Knight J, McKenzie C, Kranich J, Oliveira AC, Rossello FJ, et al. (2017). Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol 18, 552–562. [DOI] [PubMed] [Google Scholar]
- Mu Q, Zhang H, Liao X, Lin K, Liu H, Edwards MR, Ahmed SA, Yuan R, Li L, Cecere TE, et al. (2017a). Control oflupus nephritis by changes of gut microbiota. Microbiome 5,1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Q, Tavella VJ, Kirby JL, Cecere TE, Chung M, Li S, Ahmed SA, Eden K, Allen IC, Reilly CM, et al. (2017b). Antibiotics ameliorate lupus-like symptoms in mice. Sei. Rep 7,1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JM, and James JA (2014). Preclinical systemic lupus erythematosus. Rheum. Dis. Clin. North. Am 40, 621–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero MP, and Crow YJ (2016). Type I interferon-mediated monogenic autoinflammation: The type I interferonopathies, a conceptual overview. J. Exp. Med 213, 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff WE, and Kriegei MA (2015). Autoimmune host - microbiota interactions at barrier sites and beyond. Trends. Mol. Med 21, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-y M, Glickman JN, and Garrett WS (2013). The Microbial Metabolites, Short-Chain FattyAcids, Regulate Colonic Treg Cell Homeostasis. Science (80-.). 569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soehnlein 0, Steffens S, Hidalgo A, and Weber C (2017). Neutrophils as protagonists and targets in chronic inflammation. Nat. Rev. Immunol 17, 248–261. [DOI] [PubMed] [Google Scholar]
- Sonnenburg ED, and Sonnenburg JL (2014). Starving our Microbial Self: The Deleterious Consequences of a Diet Deficient in Microbiota-Accessible Carbohydrates. Cell Metab. 20, 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn AN, Macia L, and Mackay CR (2014). Diet, Metabolites, and “Western-Lifestyle” Inflammatory Diseases. Immunity 40, 833–842. [DOI] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Pattaroni C, Lopez-Mejia IC, Riva E, Pernot J, Ubags N, Fajas L, Nicod LP, and Marsland BJ (2018). Dietary Fiber Confers Protection against Flu by Shaping Ly6c -Patrolling Monocyte Hematopoiesis and CD8 + T Cell Metabolism. Immunity 48, 992–1005.e8. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Kondo M, and Kelsoe G (2005). Inflammation and the reciprocal production of granulocytes and lymphocytes in bone marrow. J. Exp. Med 201, 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira S, Pagovich O, and Kriegei M (2014). Diet, Microbiota and Autoimmune Diseases. Lupus 23, 518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, et al. (2011). Dominant and diet-responsive groups ofbacteria within the human colonic microbiota. ISME J. 5, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HB, Wang PY, Wang X, Wan YL, and Liu YC (2012). Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein claudin-1 transcription. Dig. Dis. Sei 57, 3126–3135. [DOI] [PubMed] [Google Scholar]
- Wlodarska M, Willing BP, Bravo DM, and Finlay BB (2015). Phytonutrient diet supplementation promotes beneficial Clostridia species and intestinal mucus secretion resulting in protection against enteric infection. Sei. Rep 5,1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa M, Takaishi M, Nakajima K, Kamijima R, Fujimoto C, Kataoka S, Terada Y, and Sano S (2014). Epicutaneous Application of Toll-like Receptor 7 Agonists Leads to Systemic Autoimmunity in Wild-Type Mice ANew Model of Systemic Lupus Erythematosus.Arthritis. Rheumatol. 66, 694–706. [DOI] [PubMed] [Google Scholar]
- Young GR, Eksmond U, Salcedo R, Alexopoulou L, and Jonathan P (2013). Resurrection of endogenous retroviruses in antibody-deficient mice. Nature 491, 774–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy LA, Pickard JM, and Chervonsky AV (2015). Microbiota and autoimmunity: Exploring new avenues. Cell Host Microbe 17, 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liao X, Sparks JB, and Luo M (2014). Dynamics of Gut Microbiota in Autoimmune Lupus. Appl. Environ. Microbiol 80, 7551–7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated by 16S rDNA high-throughput sequencing of fecal and tissue samples have been deposited in the European Nucleotide Archive under the ID code: PRJEB28382. 16S rDNA sequencing of human fecal samples obtained from our human SLE patient cohort at Yale is deposited in the European Nucleotide Archive under the ID code: PRJEB24742.