Abstract
Passive bioaerosol samplers can improve environmental and health protection by enhancing the practicality and cost-effectiveness of air sampling. Here, we present the outdoor field testing of a novel, passive bioaerosol sampler, the Rutgers Electrostatic Passive Sampler (REPS), based on the use of polarized, ferroelectric polymer film (poly(vinylidene fluoride)). Four 10-day-long field campaigns were conducted to compare total (culturable + non-culturable) and culturable bioaerosol collection efficiencies of REPS to passive samplers (PTFE settling filters and agar settling plates). These collection efficiencies were calculated relative to performance of an active, reference Button Sampler. Compared to passive PTFE filters, which exclusively rely on gravitational particle deposition, REPS collected a 7-fold higher total microorganism quantity. Relative to the Button Sampler, REPS collected 25% of the total number of bacteria and fungi and 65% of the culturable bacteria. Furthermore, REPS achieved this performance without any air movers, pumps, batteries or external power. Since the Button Samplers operated at 4 L/min, REPS was calibrated to have equivalent sampling rates of 2.6 L/min and 1.0 L/min for culturable bacteria and total microorganisms, respectively. These results suggest that REPS can passively collect airborne microorganisms, including culturable bacteria, with high efficiency over long-term sampling durations. REPS can provide better preservation of bacterial culturability because it has no active airflow, which desiccates microbes in active samplers. Since there are limited options available for long-term, unattended bioaerosol sampling, REPS can complement currently available bioaerosol sampling technologies for numerous environmental health applications, such as exposure assessment for epidemiology and monitoring aeroallergen trends.
Keywords: bioaerosol sampling, passive aerosol sampling, biosampler, field measurements, polarized ferroelectric polymer film, Rutgers Electrostatic Passive Sampler (REPS)
1. Introduction
Bioaerosols are airborne particles of biological origin, including viable and nonviable microorganisms, their fragments, agglomerates and toxins, and particulates shed or produced by living organisms, such as bacteria, mold spores, and pollen grains (Cox and Wathes 1995; Reponen et al. 2011). Exposure to bioaerosols is of concern as they can cause adverse health effects. For example, infectious diseases, such as influenza, can be spread by dispersal of microorganisms through coughing and sneezing; acute toxic effects can result from inhalation of fungal spores or organic dust, while health effects like asthma, allergies, and even cancers can result from chronic inhalation of allergens and mycotoxins (Cox and Wathes 1995; Douwes et al. 2003; Linaker and Smedley 2002). Healthcare facilities, agricultural animal houses and residences with post-flooding moisture problems are particularly prone to having high microbial loads which include microorganisms of concern (Cox and Wathes 1995). Additionally, widespread airborne dispersal of pathogens through bioterrorism has been called the most under-addressed threat related to terrorism (United Nations 2006).
At the same time, environmental bioaerosols constitute a substantial fraction (about 5 to 35% on average) of all airborne particulate matter and can be found throughout the atmosphere (Bauer et al. 2008; Elbert et al. 2007; Fröhlich-Nowoisky et al. 2009; Held et al. 2008; Hock et al. 2008; Jaenicke 2005; Kellogg and Griffin 2006; Smith et al. 2010; Womiloju et al. 2003). For example, about 20% of all flowering plants depend on wind pollination, resulting in the widespread dispersal of aeroallergens (Ackerman 2000). The ubiquitous presence of bioaerosols and their subsequent environmental and health impacts calls for air sampling technologies that can provide data over spatiotemporal scales that are relevant for environmental and health studies.
However, there is a lack of sampling technologies available to meet the needs of the aerobiology community; particularly, there are technology gaps in the monitoring of long-term trends for bioaerosols, especially for culturable samples (Douwes et al. 2003; Peccia et al. 2008). Short sampling times can decrease sample representativeness and increase sample variability (Lee et al. 2006). The development of bioaerosol sampling techniques that allow for longer sampling times is especially needed to improve the representativeness of personal monitoring (Pasanen 2001).
One of the main reasons for this difficulty in conducting long-term bioaerosol sampling is reliance on active, pump-based samplers, which require extensive personnel time and resources (Cartwright et al. 2009; Kilburg-Basnyat et al. 2015a; Kilburg-Basnyat et al. 2015b; Noss et al. 2008). Such samplers require noisy, heavy pumps, and external power (Plog and Quinlan 2001; Reponen et al. 2011), which limit their application. Personal pumps are also available, but these can be cumbersome, provide limited sampling flowrates and the wearing of personal sampling pumps may affect exposure estimates by causing the wearers to modify their behaviors (Cherrie et al. 1994; Wood 1977). Moreover, continuous airflow in active sampling desiccates collected microbes and limits sampling time. Frequent sampler media replacement can be done (Cartwright et al. 2009; Jensen and Schafer 2003; Stewart et al. 1995; Zhen et al. 2013), but this can be costly and time consuming for applications such as epidemiological research, where many cost-effective replicate samples are desired or required (Adams et al. 2015). Recent development towards pump-free, low-profile bioaerosol sampling technologies, such as the simple “plug and play” ionic sampler described by Gordon et al. (2015), highlights the current interest and prospects in the field for simplifying and improving long-term bioaerosol sampling capabilities.
A passive bioaerosol sampler, where no pumps or power sources are required, would significantly reduce the logistical burdens and financial costs of air sampling and allow for long-term sampling in any indoor or outdoor location. For passive sampling to be practical, it has to allow for detection of the biological agents of concern while maintaining simplicity in its use (Tovey et al. 2003). A new type of passive bioaerosol sampler, herein termed the Rutgers Electrostatic Passive Sampler (REPS), has been developed based on the use of ferroelectric polymer film (poly(vinylidene fluoride), or PVDF), which remains permanently polarized for typical environmental applications (Therkorn et al. 2017). This new bioaerosol sampler is designed to substantially improve passive capture of airborne biological particles as well as their analysis due to: 1) the use of polarized PVDF to electrostatically attract charged particles in addition to capture of particles settling under gravity, 2) use of a spiral film shape to increase total collection surface area, and 3) a sampler design that fits into 50 mL conical centrifuge tubes to expedite pre- and post-sampling sampler transport and analyses. Our earlier research showed that in a laboratory setting, this sampler was able to collect particles representing the full-size spectrum of typical bioaerosols (nano to micron-size), and the collected particles were removed for analyses without any losses (Therkorn et al. 2017).
Here, we present the results of the first outdoor field testing of REPS. Outdoor field testing was performed in four 10-day-long campaigns over highly varied weather conditions. There were three specific objectives for this research. The first was to compare long-term bioaerosol collection of REPS versus passive sampler controls for total (culturable and non-culturable) and culturable bacteria and fungi. The second was to perform outdoor field testing across varied weather conditions so that the obtained performance results would be broadly applicable in various sampling environments. The third was to evaluate REPS performance using a conventional, active sampler metric – flowrate; here, the relative collection efficiencies of REPS as well as the passive PTFE settling filters were compared to that of an active reference sampler with a known flowrate. This allowed for the calculation of equivalent sampling flowrates for the tested passive samplers as has been done in past studies investigating passive aerosol sampler performances (Yamamoto et al. 2011). This research is not only the first field application of REPS, but, we believe, it is also the first use of any passive sampling technique to investigate integrated, long-term trends in total (culturable + non-culturable) and culturable bioaerosol.
2. Materials and Methods
2.1. Sampling Site
Fig. 1 presents the setup of the outdoor sampling site. Four 10-day-long outdoor field sampling campaigns were conducted by an organic garden on an agricultural college campus in New Jersey. The rationale for choosing 10-day-long campaigns was three-fold. First, this allowed for investigation of passive sampling as a method for studying long-term bioaerosol trends. Second, 10 days is within the range considered to be ideal for application of passive samplers to assess personal and environmental exposures (USEPA 2016). Third, the campaign length was chosen to ensure there would be sufficient biological particles collected for analyses based on preliminary sampling at the study site. The field campaigns were conducted between September and December of 2015. Samplers were placed on a table about 1 m above the ground inside a protective metal enclosure. The top of the enclosure was covered with a tarp that protected the samplers from large falling debris but did not impede air movement through the enclosure.
Fig. 1.
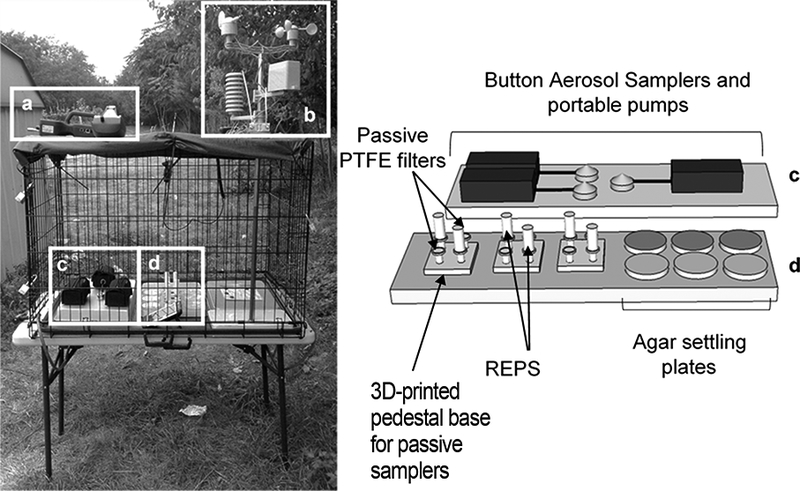
The outdoor sampling site setup. 1a: the real-time aerosol monitoring instruments operated each day for one hour, including the Aerotrak Handheld Optical Particle Counter (OPC, model 9306, TSI Inc., Shoreview, MN), and a DustTrak DRX Aerosol Monitor (DRX, model 8534, TSI Inc.). 1b: portable weather station (WS-2080 Wireless Weather Station, Ambient Weather, Chandler, AZ). 1c: active reference samplers for bioaerosol sampling operated in 24-hour intervals: three Button Aerosol Samplers (SKC Inc., Eighty Four, PA) operated at 4 L/min with PTFE filters (2 μm pore size, SKC Inc.) and AirChek XR5000 Sample Pumps (SKC Inc.). 1d: the passive samplers: six passive settling filters (25 mm PTFE filters with 2 μm pore size (SKC Inc.)), six REPS, and six 90 mm agar settling plates (three for culturable bacteria and three for culturable fungi).
A portable weather station (WS-2080 Wireless Weather Station, Ambient Weather, Chandler, AZ) was also operated at the sampling site to continuously record the temperature, relative humidity, wind speed and peak wind gust in 5-minute intervals (Fig. 1b). There was precipitation for 13% (5/40) of all sampling days; on these days, plastic rain covers were placed over the enclosure to protect the samplers from direct exposure to rain. Openings were cut into the sides of the covers to allow free airflow.
2.2. Passive Samplers and Sample Analysis
The tested passive samplers were: 1) six REPS with polarized PVDF film (Therkorn et al. 2017) (Fig. 2), 2) six passive (i.e., no pumps attached) poly(tetrafluoroethylene) (PTFE) filters (2 μm pore size, 25 mm diameter, SKC Inc., Eighty Four, PA), and 3) six agar settling plates – three for culturable bacteria and three for culturable fungi (90 mm in diameter, Fisher Scientific, Waltham, MA). The passive PTFE filters and REPS were placed onto 3D-printed pedestals 2 cm high to allow for free air movement around the samplers (Fig. 1d). The pedestal onto which each passive sampler was placed was chosen in a randomized manner for each campaign. Each passive sampler was secured to its pedestal base with a small, autoclaved metal clip. The pedestals were cleaned before each new campaign by wiping them with 70% ethanol. Prior to deployment, the passive PTFE filters were autoclaved, and the PVDF films were cleaned and assembled into the 3D-printed film holders as described elsewhere (Therkorn et al. 2017). Briefly, the PVDF films were wiped with 70% ethanol and rinsed with sterile, Milli-Q water (EMD Millipore Corp., Milli-Q Direct 8, Billerica, MA). The 3D-printed film holders were cleaned before each campaign by scrubbing with a bristle brush in 10% Alconox detergent solution (Alconox Inc., White Plains, NY), then sonicating in sterile, Milli-Q water for 8-hours to ensure that all contaminants on the surface and in the interstitial spaces (voids) of the 3D-printed film holders were removed. The passive PTFE filters and REPS were transported to/from the field in individual, sterile 50 mL conical centrifuge tubes (Falcon Tubes, Corning Inc., Tewksbury, MA).
Fig. 2.

Front and top views of the 3D-printed REPS film holder. Film holder diameter, X = 22 mm, and height, Y = 75 mm, are indicated in the figure as dimensions X and Y. As illustrated in the top view, one piece of polarized, ferroelectric poly(vinylidene fluoride) (PVDF) film (130 × 70 mm) is spiraled through the openings. This results in the formation of multiple layers of film with sides of opposite polarization facing each other across air channels of optimized width (2.25 mm) (Therkorn et al. 2017).
The passive PTFE filters and REPS were deployed for the entirety of a given sampling campaign (10 days); after each campaign, particles were immediately eluted from these samplers and the resulting suspensions were subdivided for microscopy and culture analyses. For the purpose of this study, the term elution refers to the removal of particles, including microbes, from the surfaces of the samplers and filters. Particle elution, staining with Acridine Orange to count total bacteria and fungi, and counting procedures were the same as described by Therkorn et al. (2017). Briefly, particles were eluted from samplers and filters using a 2-minute vortex and 10-minute ultrasonic agitation in sterile, Milli-Q water. Then, a 1 mL aliquot of elution suspension was used for Acridine Orange staining of bacteria and fungi, and these microbes were then counted by epifluorescence microscopy. Acridine Orange staining by the CAMNEA method for quantification of bioaerosols is a common method in the bioaerosol field for evaluating total (culturable + non-culturable) bacteria and fungi, and it has been previously used to discern culturable versus non-culturable microbes from the same samples (Palmgren et al. 1986).
In summary, elution volume of each REPS was 40 mL, and it was divided as follows: 1 mL for staining/counting, and for the two REPS with the highest number of stained bacteria + fungi, 600 μL in total was taken from each of these REPS to cultivate on spread plates. Spread plates were made by evenly spreading 100 μL aliquots of the particle elution suspension onto each of the six plates: three for culturable bacteria (tryptic soy agar (TSA, Becton, Dickinson and Co., Sparks, MD) with 200 mg Cycloheximide antifungal agent (Sigma-Aldrich Co., St. Louis, MO) per liter agar) and three for culturable fungi (malt extract agar (MEA, Becton, Dickinson and Co.)). All plates were incubated at room temperature for five days and new colony forming units (CFU) were counted every 24-hours. For each of the passive PTFE filters, the elution volume was 5 mL, and it was divided as follows: 1 mL for staining/counting, and for the two PTFE filters with the highest number of stained bacteria + fungi, 600 μL total was taken from each of these filter’s elution suspensions for spread plating. The REPS sampler’s elution suspensions were 40 mL of sterile Milli-Q water, instead of the 5 mL used for the filters, because 40 mL was the minimum volume needed to fully submerge REPS in the centrifuge tubes (Therkorn et al. 2017). For discussion on the limitations of using such a relatively large elution volume, see Section 5 below: Current REPS Limitations and Future Development.
The third type of passive sampler was the agar settling plates. Agar settling plates were included in this study because these have traditionally been the most common method for passive bioaerosol sampling (Andon 2006; Cox and Wathes 1995). Three TSA and three MEA settling plates were deployed each sampling day between 0800 and 1200 hours for a 4-hour sampling period. Four hours is the maximum recommended amount of time for which settling plates can be used without significantly affecting microbial growth through agar desiccation (Baird et al. 2000; Sandle 2015). After the 4-hour exposure, settling plates were closed, returned to the lab, and incubated at room temperature for five days. New CFU were counted every 24-hours. The total number of CFU for the bacteria and fungi for a campaign were determined as the sum of CFU from individual sampling days from each campaign and then multiplied by a factor of six to scale up the 4-hour exposure periods to estimate continuous 24-hour sampling. This scaling up was done so that the agar settling plate exposure time frame could be compared to the other tested passive samplers that sampled continuously for 10 days. An assumption was made that the 4-hour period is representative of 24-hours, which it most likely is not (Fierer et al. 2008) – an inherent limitation in using settling plates for long-term, continuous bioaerosol sampling. For ~3% of all settling plates (7/240), the number of CFU were too high to count, so these plates were divided into four quadrants, one quadrant was counted, and the CFU multiplied by four for that plate.
2.3. Active Reference Samplers and Sample Analysis
Three Button Aerosol Samplers (SKC Inc., Eighty Four, PA) with PTFE filters (2 μm pore size, 25 mm diameter (SKC Inc.)) operating side by side at 4 L/min served as the reference active samplers for field campaigns (Fig. 1c). The main reason for selecting the Button Aerosol Sampler as the active reference sampler was because it is a well-characterized sampler; the Button Sampler has a curved porous inlet that is minimally affected by wind direction, and the sampler has high precision and filter loading uniformity (Hauck et al. 1997; Kalatoor et al. 1995). Its inlet efficiency closely matches the inhalable convention of ACGIH/CEN/ISO (Aizenberg et al. 2000a; Toivola et al. 2004), and has been validated for bioaerosol sampling for viable and non-viable analyses, including bacteria and fungi (Adhikari et al. 2003; Aizenberg et al. 2000b; Lee et al. 2006; Toivola et al. 2004; Wang et al. 2001). Additionally, when used with appropriate filters, the Button Sampler could be operated in 24-hour intervals without excessive backpressure, thus minimizing chances that its battery-operated pump would fail– an important feature since this is a study of long-term bioaerosol trends. Other recently published manuscripts performing long-term field testing of electrostatic dust cloths for quantification of airborne endotoxins also chose the Button Sampler or a similar filter-based sampler as their active reference control (Kilburg-Basnyat et al. 2015b; Samadi et al. 2010).
Other types of active bioaerosol samplers, such as impingers or impactors, were not selected as the active, reference samplers for this study as they would require frequent media replacement and stationary power outlets, making this field testing impractical. Other samplers which have been used for long-term bioaerosol sampling were considered, such as the Burkard Spore Trap which is run in 7-day intervals (Sterling et al. 1999). However, this sampler would not allow for enumeration of total and culturable collection as the spore sampler deposits particles onto a rolling sticky tape. Another consideration is that whatever sampler was chosen to be used as the active reference had to be left mostly unattended for the entire duration of field testing. This is why active, real-time sensors, like the WIBS or UVAPS were also not appropriate for this study.
The number of microorganisms (bacteria + fungi) collected by the three Button Samplers were determined and averaged each day to estimate variability in the collocated, reference active sampler results. Each 24-hour sampling period started with three clean Button Samplers and three fully recharged, calibrated pumps (AirChek XR5000 Sample Pump, SKC Inc.). For cleaning, the Button Samplers were autoclaved, soaked overnight in ethanol, and then wiped dry with Kim Wipes (Kimberly-Clark Professional, Roswell, GA) to remove any residue. Immediately before field deployment, new PTFE filters were loaded into each Button Sampler in a laminar flow hood (NuAire Class II, Type A2, Plymouth, MN) using autoclaved forceps, and the Button Samplers were transported to and from the field in a clean, unused plastic zip bag.
After each 24-hour sampling interval, the Button Samplers were returned to the lab, the filters were removed with autoclaved forceps and placed into separate 50 mL conical centrifuge tubes containing 5 mL of autoclaved Milli-Q water (the same elution volume as used for the passive PTFE filters), and the microorganisms were eluted from the filters. All data presented here for the Button Sampler have been adjusted for the anticipated 20% loss due to incomplete elution of bacteria and fungi from the 2 μm PTFE filters (Therkorn et al. 2017). The 5 mL elution liquid was subdivided into aliquots for microscopy and culture analysis. Total bacteria and fungi were stained and counted as described above; however, 100 μL of elution suspension was used for staining/counting due to the expectation of higher total yield numbers. It is desirable to have about 20 stained microbes per microscope view field for counting (Palmgren et al. 1986). To estimate the culturable fraction of bacteria and fungi for each sampling day, agar spread plates were also made, incubated and counted as described above using 100 μL aliquots of elution liquid from the Button Sampler with the highest number of observed bacteria and fungi determined by microscopy. The culturable fraction was determined as a ratio of the culturable microorganism number determined by culture technique versus the total number of microorganisms determined by microscopy.
In summary, while the passive samplers were left unattended for 10 days of collection, the Button Samplers had their filters removed, particles eluted, and total (staining + microscopy) and culturable analyses performed every 24 hours. Thus, the passive sampler’s results represent continuous 10-day sampling while the Button Sampler’s results represent a sum of ten 24-hour intervals.
2.4. Real-Time Aerosol Monitoring Instruments
The real-time aerosol monitoring instruments were 1) an Aerotrak Handheld Optical Particle Counter (OPC, model 9306, TSI Inc., Shoreview, MN), and 2) a DustTrak DRX Aerosol Monitor (DRX, model 8534, TSI Inc.) (Fig. 1a). The DRX and OPC were run for 60-minutes each sampling day, starting between 0800 and 1200. The OPC monitored particle number concentrations in the 0.3 to 10 μm size range. This data was used to estimate the daily biological fraction of particulate matter by dividing the average total number of stained bacteria and fungi obtained daily from the Button Samplers by the average total number of particles larger than 1 μm, registered by the OPC, across a given sampling campaign. Single and agglomerated bacterial and fungal particles are typically expected to be in the size range of 1 to 10 μm (Gorny et al. 1999; Reponen et al. 2011; Yamamoto et al. 2014). Since the OPC was only run for 60-minutes each sampling day, the daily OPC values were averaged across each campaign to reduce uncertainty in calculated daily biological fractions.
The DRX monitored PM10 and PM2.5 mass concentrations. These data were compared to a nearby New Jersey PM2.5 sampling station (NJDEP Air Quality Monitoring Station at Rutgers University, New Brunswick, NJ), which provides continuous 1-hour averages for PM2.5 using beta attenuation monitoring (BAM-1020 Continuous Particulate Monitor, Met One Instruments Inc., Grants Pass, OR). The NJDEP station is located about 1.10 km from the sampling site and therefore served as a reference to investigate local particulate pollutant levels at the sampling site in relation to the nearby station. The DRX is factory calibrated with the respirable fraction of standard ISO 12103–1, A1 test dust. Thus, assumptions are made about the relationship between the light scattering of the measured particles and their mass concentration. Since the sampling was conducted on an agricultural site, the DRX was calibrated for the site using two gravimetric PM2.5 Personal Modular Impactors (PMI, SKC Inc., Eighty Four, PA). A correction factor of (−)12% was applied to the DRX measurements for PM2.5 (and for PM10, which had similar mass concentration to the PM2.5 across all campaigns). For more details on the DRX calibration, see the Supplemental Material.
2.5. Blanks
For each campaign, one PTFE filter and one REPS were used as field blank controls: they were placed onto a sampling pedestal, then immediately removed and taken to the lab for analysis. Field blanks were also taken for the Button Samplers on three randomly chosen days in each campaign by assembling and transporting an extra Button Sampler to the field, placing it onto the sampling site table, and then immediately returning it in its transport bag. Blanks for agar settling plates, representing 20% of the total number of agar plates prepared, were randomly chosen. Blanks for agar spread plates were prepared from each of the blank samplers. Mean blank values were subtracted from results. Blank values for total stained bacteria and fungi were ≤ 10% of the post-sampling values for all samplers. No contamination was detected for agar settling plates or agar spread plates.
2.6. Theoretical Limits of Detection (LODs)
Assuming detection of one microorganism or one CFU and well-mixed samples, and given the differences in volumes used for elution and analyses, the theoretical LODs for REPS were 40 microbes for microscopy-based analyses and 400 culturable microbes for culture-based analyses. For the passive PTFE filters, the LODs were 5 and 50 microbes for microscope and culture-based analyses, respectively. For the PTFE filters removed from the Button Samplers, the LODs were 50 microbes for both microscope and culture-based analyses. For the agar settling plates, the LOD was one CFU.
2.7. Statistical Analyses
Residual plots evaluated statistical model assumptions, including assumptions for normality and homogeneity of variance. Some of the data sets had unequal residual variances. Therefore, independent samples t-tests for equality of means, assuming two-tailed significance (equal variances not assumed), were conducted to compare the collection efficiencies of REPS versus PTFE filters across campaigns. Collection efficiencies of the passive samplers were determined by comparing the average total bacteria and fungi collected relative to that from the Button Samplers. To compare the performances of REPS and the PTFE settling filters to that of the agar settling plates, Welch’s one-way analysis of variance (ANOVA) followed by Games-Howell post-hoc test was used to compare differences in average total bacteria and fungi CFU/cm2 of deposition surface; projected surface area was used for REPS since it has a bottom diameter of 22 mm which is similar to the 25 mm PTFE filter diameter. Average equivalent sampling flowrates (mL/min) were determined for REPS and the passive PTFE filters by estimating the passive sampler collection efficiencies relative to the Button Aerosol Samplers and multiplying this value by the Button Sampler flowrate (4,000 mL/min). All analyses were performed using SPSS Statistics Premium Edition, v23 (IBM Corporation 2015) with α = 0.05.
3. Results
3.1. Sampling Site Conditions
Mean temperature ranged from 20°C ± 4°C for the first campaign conducted in September to 9°C ± 5°C for the final campaign carried out in December. The number of days on which temperature went below freezing was 0, 1, 3 and 4 for campaigns 1–4, respectively. These freezing temperatures tended to occur overnight. The mean relative humidities, wind speeds, and peak wind gusts were similar across the four campaigns, with combined campaign averages of 67% ± 21%, 0.5 m/s ± 0.7 m/s and 1.1 m/s ± 1.1 m/s, respectively. Throughout each campaign, the daily relative humidity was highly variable and ranged from about 20% to 99%. The total precipitation was different for each campaign with totals of 0, 17, 21 and 39 mm for campaigns 1 to 4, respectively (The Rutgers Gardens Weather Tower 2016). Due to the variability in weather conditions during field testing and the relatively low number of field campaigns (4 total), robust conclusions on the effect of meteorological conditions on passive sampler performance cannot be drawn with the present dataset.
According to the Button Sampler data, the average total bioaerosol concentration for the field campaigns (bacteria + fungi) was 5×104 ± 3×104/m3 and it ranged from 104 to 105/m3. For a summary table of sampling site meteorological conditions, and more discussion and details on conditions for particulate pollution, biological fractions of particulate matter, and culturable fractions of bacteria + fungi, please see the Supplemental Material.
3.2. Sampler Performance Comparisons
For sampler performances across the four campaigns, there was no statistically significant effect of campaign for any given sampler metric. Therefore, to increase power, all statistical results described below are pooled sampler data across all four campaigns. For illustrative purposes, the data in figures 3–5 show the results by campaign.
Fig. 3.
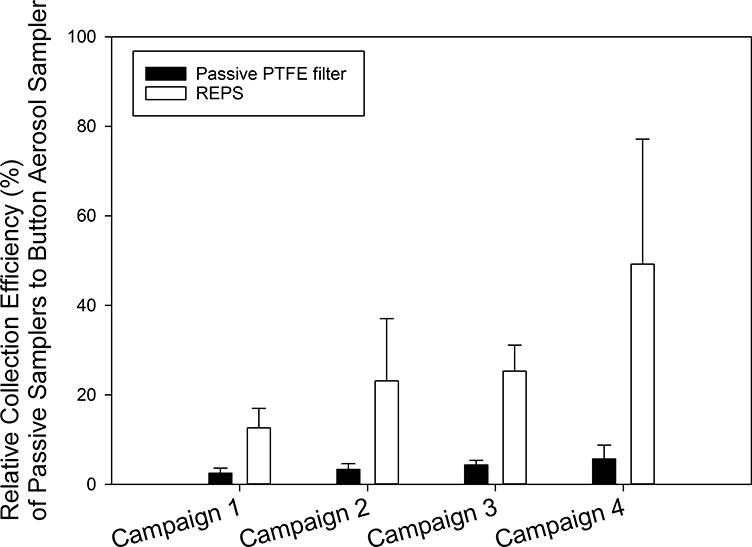
Relative collection efficiency (%, mean ± 1SD) of passive samplers when capturing total bioaerosol particles (culturable + non-culturable bacteria and fungi) as enumerated by Acridine Orange staining and epifluorescence microscopy. The reference Button Aerosol Samplers were operated at 4 L/min. The number of samples presented here by sampler are: Button Sampler (n=30 per campaign), passive PTFE filter (n=6 per campaign), REPS (n=6 per campaign). For the Button Sampler, three were deployed each 24-hours, the average of those three was determined each 24-hours, and then the campaign total was taken as the sum across the 10 days of averaged daily intervals. For the passive samplers, an average was found across the six deployed samplers for each campaign.
Fig. 5.
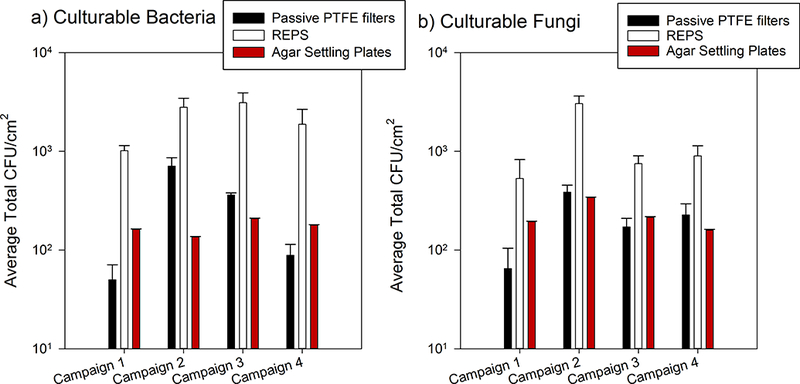
Average total CFU normalized to sampler’s horizontal surface area (CFU/cm2, mean ± 1SD) of culturable bacteria (a) and culturable fungi (b) collected by passive samplers. For REPS, projected surface area was used (380 mm2, bottom of sampler has a 22 mm diameter). The number of samples presented here by sampler are: agar settling plates (n=30 per campaign per microbe type), passive PTFE filter (n=6 per campaign per microbe type), REPS (n=6 per campaign per microbe type). Agar settling plates were deployed daily for four hours in triplicate for bacteria and fungi. Campaign total was determined as the sum of averaged daily enumerated CFU multiplied by six to scale up to continuous 24-hour sampling. For the passive samplers, agar spread plates were made at the end of each campaign for the two out of six REPS and the two out of six passive PTFE filters with the highest number of stained bacteria + fungi as determined by microscopy.
Fig. 3 presents the collection efficiencies (mean ± 1SD) of REPS and passive PTFE filters relative to the Button Aerosol Samplers for total bacteria and fungi across the four campaigns. The mean relative collection efficiency of REPS (29% ± 21%) was statistically significantly greater than that of the passive PTFE filters (4% ± 2%, t(23.45) = −5.72, p < 0.001).
Fig. 4 demonstrates the collection efficiencies of REPS versus the passive PTFE filters for culturable fungi and bacteria relative to the Button Aerosol Samplers for each sampling campaign. REPS had a mean relative collection efficiency for culturable fungi and bacteria of 24% ± 13% and 65% ± 41%, respectively, while the passive PTFE filters had collection efficiencies of 5% ± 2% and 11% ± 17% for the same culturable microbes. The mean culturable collection efficiency of REPS was statistically significantly greater than the mean collection efficiency of the passive PTFE filters for both fungi (t(24.29) = −7.16, p < 0.001) and bacteria (t(30.82) = −5.99, p < 0.001).
Fig. 4.

Relative collection efficiency (%, mean ± 1SD) of passive samplers when capturing culturable bacteria (a) and culturable fungi (b) as enumerated by CFU. The reference Button Aerosol Samplers were operated at 4 L/min. The number of samples presented here by sampler are: Button Sampler (n=30 per campaign per microbe type), passive PTFE filter (n=6 per campaign per microbe type), REPS (n=6 per campaign per microbe type). Agar spread plates were made after each 24-hour Button Sampler deployment from one of the three deployed Button Samplers; the selected unit had the highest number of bacteria and fungi, as determined by microscopy, among the three Button Samplers. Campaign total was determined as the sum of average CFU across daily spread plates made in triplicate for bacteria and fungi. For the passive samplers, agar spread plates were made at the end of each campaign for the two out of six REPS and the two out of six passive PTFE filters with the highest number of stained bacteria + fungi as determined by microscopy. Campaign total was determined as the average total CFU across the spread plates made in triplicate for bacteria and fungi.
Fig. 5 shows the number of culturable microorganisms (mean ± 1SD) across the three passive sampling methods – REPS, passive PTFE filters and agar settling plates – normalized to each sampler’s horizontal surface area: average total CFU/cm2. The mean values for average total CFU/cm2 for REPS, passive PTFE filters, and settling plates for culturable bacteria were 2204 ± 1720, 303 ± 425, and 17 ± 8, respectively; for fungi, these values were 1308 ± 1099, 213 ± 140, and 22 ± 18, respectively. Compared to deposition by gravitational settling alone, as used by the passive PTFE filters and agar settling plates, REPS provided statistically significantly greater collection of culturable bacteria (Welch’s F(2, 30.67) = 24.29, p < 0.001) and fungi (Welch’s F(2, 30.77) = 37.80, p < 0.001). Compared to the agar settling plates and passive PTFE filters, REPS provided a one and two order of magnitude higher collection of culturable microorganisms per surface area, respectively. Post-hoc Games-Howell multiple comparisons of means showed that the mean collection efficiencies of all three passive samplers were statistically significantly different for both culturable bacteria and fungi (p < 0.001 for all comparisons, except p = 0.009 for passive PTFE filters versus agar settling plates for culturable bacteria collection).
Fig. 6 demonstrates equivalent sampling flowrates (mean ± 1SD) for REPS and passive PTFE filters across all sampling campaigns. Both samplers had a much better sampling performance for culturable bacteria; REPS provided a mean equivalent sampling flowrate of 2594 ± 714 mL/min while the passive PTFE filters had a mean equivalent sampling flowrate of 443 ± 128 mL/min. For each passive sampler, the mean equivalent sampling flowrate for collection of total bacteria + fungi was similar to that for culturable fungi collection. For REPS, the mean equivalent sampling flowrate for total bacteria + fungi was 1157 ± 673 mL/min, while for culturable fungi the mean equivalent sampling flowrate was 959 ± 394 mL/min; for passive PTFE filters, the mean equivalent sampling flowrate for total bacteria + fungi was 159 ± 74 mL/min, while for culturable fungi the mean equivalent sampling flowrate was 191 ± 71 mL/min.
Fig. 6.

Average equivalent sampling flowrates (mL/min, mean ± 1SD) for passive samplers calibrated against the Button Aerosol Sampler operated at 4,000 mL/min. It should be noted that, as with all calibrations of this kind, the presented equivalent sampling flowrates are likely location and sampling condition specific. The number of samples presented here by each sampler for the total collection of bacteria + fungi (culturable + non-culturable) as enumerated by Acridine Orange staining and counting by microscopy are: Button Sampler (n=120 across campaigns 1–4), passive PTFE filter (n=24 across campaigns 1–4), REPS (n=24 across campaigns 1–4). The number of samples presented here by sampler for the culturable collection of bacteria and fungi as enumerated by CFU are: Button Sampler (n=120 across campaigns 1–4 per microbe type), passive PTFE filter (n=24 across campaigns 1–4 per microbe type), REPS (n=24 across campaigns 1–4 per microbe type).
4. Discussion
4.1. Comparison of REPS with Other Passive Aerosol Samplers
Compared to the performances of the passive PTFE filters and agar settling plates, which exclusively rely on gravitational settling of particles, REPS provided substantially and significantly greater passive collection of total bacteria and fungi. This greater capture of microorganisms by REPS was observed in all campaigns despite highly variable meteorological conditions. The average total number of stained bacteria and fungi collected by REPS and the passive PTFE filters across campaigns were 5×105 and 9×104, respectively. As the LODs for these two samplers were 40 and 5 microbes, respectively, it can be seen that these levels of collection were more than sufficient to meet the minimum. Similarly, the average total CFU collected by REPS and the passive PTFE filters across campaigns were 7×103 and 1×103, respectively, as compared to their respective LODs of 400 and 50 CFU. The average total number of CFU on the agar settling plates across campaigns were ~200 which was not only above the LOD of 1 CFU, it was within the recommended range for the number of CFU/plate for satisfactory counting (30–300 CFU/plate) (Breed and Dotterrer 1916). Since the REPS samplers and the passive PTFE filters have similar projected diameters (22 and 25 mm, respectively), the enhanced biological particle collection of REPS reflects its optimized electrostatic particle capture and increased collection surface area (Therkorn et al. 2017).
Previous studies have also investigated equivalent sampling flowrates of other types of passive aerosol samplers. The Personal Aeroallergen Sampler (PAAS) has been compared to a two-stage cyclone sampler and found to have an equivalent sampling flowrate about two orders of magnitude less than that of REPS: 32 ± 6 mL/min to 66 ± 44 mL/min when sampling fungal species > 5 μm in aerodynamic diameter (Yamamoto et al. 2011). The equivalent sampling flowrate of an electret sampler (Brown et al. 1994; Brown et al. 1995; Brown et al. 1996) has been calibrated to be 22.5 mL/min when testing its performance in the laboratory versus the United Kingdom’s Conventional Methods for the Determination of Hazardous Substances (MDHS) guidance 39/4 membrane filter pump sampling method using asbestos fibers as the test dust (Burdett and Revell 1999).
Electret samplers and electrostatic dust cloths are similar in principle to REPS as they also rely on electrostatic capture of particles. Electrets are not permanently polarized but use charge injection to create the electrostatic attraction; however, this charge steadily dissipates throughout use (Burdett and Bard 2007; Kilburg-Basnyat et al. 2016; Therkorn et al. 2017). In addition, this high surface charge can damage microorganisms (Mainelis et al. 2002). On the other hand, REPS particle capture relies on the electric fields present at the film’s surfaces produced by its (internal) bulk polarization and, therefore, has no charge to damage microorganisms. The electric field at the REPS film surfaces does not deliver any charge and, to the best of our knowledge, does not damage any captured microorganisms.
The published electret calibration experiments were only conducted for up to 180 minutes (Burdett and Bard 2007). Thus, the difference in estimated equivalent sampling flowrates between the electret and REPS is most likely not a result of the electret losing substantial charge as would be expected during longer sampling campaigns. The diameter of the electret (25 mm) investigated in the above study is similar to the projected diameter of REPS (22 mm), but the total collection surface area of REPS is much greater: ~30,765 mm2 including film and 3D-printed film holder versus 491 mm2 of the electret. As suggested by Burdett and Revell (1999), the total collection of the electret sampler can be increased by increasing the total surface area of the electret, but this was not practical, given their chosen methods for preparation and analysis of the corona-poled poly(propylene) disc. Here, the design of REPS offers an alternative and improvement to using a one-dimensional flat sampling disc. By coiling PVDF film into a 3D-printed film holder, the total sampling collection surface area is increased, but the entire sampler still fits into a 50 mL conical centrifuge tube for practical sampler transport.
4.2. Comparison of REPS with Active Aerosol Samplers
The average total number of stained bacteria and fungi collected by the Button Samplers each day was 3×105 which is well above the Button Sampler’s LOD of 50 microbes for microscopy-based analysis. The LOD for the culture-based analyses for the Button Sampler’s was also 50 CFU, and the average total number of CFU collected by the Button Sampler’s across the campaigns was also well above this minimum (2×104 CFU, average for bacteria and fungi).
It is well known that the Button Aerosol Sampler is expected to desiccate the captured bacteria because its collection airflow continuously passes through the filters onto which the microorganisms are deposited. Desiccation and sampling stress on bacteria have been reported in previous bioaerosol studies (Jensen et al. 1992; Li 1999; Stewart et al. 1995; Zhen et al. 2013), and bacteria may generally be more sensitive to sampling stress than fungi (Chen and Li 2005; Wang et al. 2001). Here, we present novel data on the long-term field collection and analyses of bioaerosol samples using a new passive bioaerosol sampler; our data suggest that REPS, as well as the other tested passive bioaerosol samplers and the Button Sampler, do have the potential to allow for studying long-term bioaerosol trends, including potential investigation of sample viability and culturability. However, the data show an enhanced performance for REPS for culturable bacteria collection as compared to both the passive samplers to which it is compared and its relative performance compared to the Button Sampler. This is an important finding that may allow for future elucidation in trends in microbial activity or infectivity, and REPS may fulfill a current technology gap for long-term study of bioaerosol trends.
By calibrating the collection efficiency of REPS against the active Button Aerosol Samplers with a known flowrate of 4,000 mL/min, an equivalent sampling flowrate is calculated for REPS to allow its comparison to conventional, active bioaerosol samplers. With an equivalent sampling flowrate of ~1,000 mL/min for total bacteria and fungi and ~2,600 mL/min for culturable bacteria, this study has shown that REPS may be used for bioaerosol sampling campaigns up to 10 days in length to passively collect microorganisms with overall performance comparable to that of active samplers. For example, the Button Aerosol Sampler has been previously used to investigate concentrations of and exposure to outdoor aeroallergens, bacteria, fungal spores, and endotoxin levels in various environments (Adhikari et al. 2003; Adhikari et al. 2004; Aizenberg et al. 2000b; Kilburg-Basnyat et al. 2015b; Lee et al. 2006; Toivola et al. 2002).
Other active aerosol samplers with which REPS’ performance in this study may be compared include the IOM Inhalable Sampler and the Personal Environmental Monitor (PEM) sampler (both SKC Inc.). These samplers are designed to sample the inhalable fraction of particulate matter and PM10 or PM2.5, respectively, and operate with flowrates of 2,000 mL/min. The IOM Sampler has been used for sampling airborne fungi in occupational settings (Green et al. 2005) and near waste composting facilities (Taha et al. 2006). The PEM sampler has been used to investigate the ambient and indoor biological content of particulate matter (Menetrez et al. 2009) and microbial species diversity (Hoisington et al. 2014). Two critical and important differences between REPS and active aerosol samplers are that REPS does not need an air pump or power supply: highly beneficial and desirable features that reduce two of the major logistical burdens of active sampling.
5. Current REPS Limitations and Future Development
5.1. Future Collocated Sampler Testing
The present manuscript does not address several important factors for REPS’s field performance which must be addressed in future studies; these include collocated, correlated passive and active sampler performances. This could not be addressed in the present study due to the low overall number of total experimental repeats (four campaigns). Experimental follow-up will be conducted by a larger number of shorter duration collocated sampler field trials. During this follow-up study, the coefficient of variation (CV) across collocated samplers of the same type must also be investigated. Future research will investigate by laboratory tests the best platform for assemblage of multiple REPS to minimize CV%. Then, collocated REPS performance will be investigated through outdoor field sampling campaigns with a more precise analysis method (e.g., qPCR). Future study is also needed to better understand how meteorological conditions impact REPS performance. Such experiments could be first performed in laboratory conditions in controlled temperature, wind and humidity environments, and then carried out in the field.
5.2. Long-Term Sampling Effects on Sample Viability and Culturability
Since the Button Samplers were replaced daily around 8:00AM, perhaps the culturable microbes we counted only represent a short time period before sampler collection. Future study is needed to better understand how passive and active samplers alike influence bioaerosol sample viability and culturability over sampling campaigns of various length. Through these field sampling campaigns, it is also necessary to investigate how desiccation, in addition to loading of different types of particulate pollution onto the Button Sampler’s filter, effects its elution efficiency and sample culturability after 24-hour sampling periods.
5.3. REPS’ Sampling Time Requirements
In each sampling campaign, REPS was continuously operated for 10 days without any attendance. The 10-day campaign length was chosen because it was within the range considered to be ideal for application of passive samplers to assess long-term personal and environmental exposures (USEPA 2016) and was estimated to be sufficient for collecting enough biological particles. It is recommended to collect a total sample volume of at least 0.1 m3 of air when conducting active sampling; at this volume, concentrations of about 100 CFU/m3 are detectable (Burge et al. 1989; Cox and Wathes 1995). At its equivalent sampling flowrate calibrated in this study (average of 1 to 2.6 L/min), REPS would have a minimum required sampling time of about 38 to 100 minutes to collect the minimum recommended sample volume in typical environments. Furthermore, this recommended volume is for culturable microorganisms, and REPS may be better at maintaining microbial culturability than filter-based active samplers. Future studies will investigate REPS usability at shorter sampling times, beginning at 30 minutes and going up to an 8-hour simulated work shift. Use of REPS for longer sampling periods (several weeks and up to one month) will also be investigated to gauge its applicability to bioaerosol monitoring and trend observation over longer durations.
5.4. Elution Volume for Removing Particles from REPS
One of the limiting factors for reducing REPS’ sampling durations is the relatively large elution volume needed to remove particles from REPS surfaces (40 mL). Different sample concentration methods are being investigated, such as the use of new types of technologies including a sample concentrator and a liquid evaporator. These newer technologies hold great promise, but also have drawbacks such as particle loss and long sample preparation times, respectively. Methods to filter the liquid sample have been considered, but unless microscopy will be used to analyze the sample, this would require another elution of filtered particles into liquid for other analyses. Centrifugation for pelleting samples is also being explored for a concentration technique. Alternatively, different methods for removing particles from REPS using a smaller liquid volume are being considered, such as different types of mechanical agitation (i.e., shaking). Another option is to investigate how to modify the sampler’s design to allow for better elution liquid access to all surfaces of the sampler during vortex mixing.
5.5. Potential for REPS Surface Saturation
Increasing REPS’ sampling time may cause concern for saturation of the sampler film surfaces. The average total number of microorganisms (bacteria + fungi) collected by each REPS per sampling campaign was ~6×105. Assuming the typical biological fraction of particulate matter is 0.1 (Fig. S2) (Bauer et al. 2008; Jaenicke 2005), this would mean that there could be a total of approximately 6×106 total particles on each REPS, including non-biological particles. However, even assuming that all of these particles were 10 μm in diameter, which is unlikely, their total projected surface area would be ~6×108 μm2, which would be less than 1% of the entire REPS film surface area of ~1010 μm2. Even this projected surface area of ~6×108 μm2 of captured particles is a substantial overestimation, because most particles were <10 μm, (see Fig. S3 in the Supplemental Material for the normalized particle number size concentrations averaged across campaigns). These data suggest that REPS could be used for much longer sampling campaigns without concern about saturation of its collection surfaces. Nevertheless, it is still a factor that requires investigation through high concentration particle loading testing.
6. Conclusions
In conclusion, we describe successful testing of a novel, passive bioaerosol sampler in highly varied meteorological conditions with broad applicability to different sampling environments. The sampling concept embodied by REPS can significantly improve the field of bioaerosol sampling by allowing for easy, versatile sampling over broad spatiotemporal scales and less burdensome personal sampling as no pumps or power supplies are needed. Our data suggest that REPS, as well as the other tested passive bioaerosol samplers and the Button Sampler, do have the potential to allow for studying long-term bioaerosol trends, including potential investigation of sample viability/culturability. However, the data show an enhanced performance for REPS for culturable bacteria collection as compared to both the passive samplers to which it is compared and its relative performance compared to the Button Sampler.
In this study, REPS had an equivalent sampling flowrate of ~2,600 mL/min for capture of culturable bacteria which is comparable to and even greater than that of some active samplers. While there are certain limitations for using REPS as discussed above in section 5, and more research is needed to understand the predictability of its performance, it has a high collection efficiency through passive sampling means. This high collection efficiency, in combination with future research on reducing the necessary elution liquid volume, will allow REPS to quickly reach the detection limits needed for microbial analyses. Also, the ability of REPS to better preserve bacterial culturability is a major benefit as viability is a key determinant for bioaerosol infectivity (Cox and Wathes 1995) and most of the active sampling technologies struggle to maintain bioaerosol viability. This is an important finding that may allow for future elucidation of trends in microbial activity or infectivity. REPS may fulfill a current technology gap for long-term study of bioaerosol trends given the available active bioaerosol samplers. Potential applications of interest for the aerosol science community include epidemiological research with widespread area or personal monitoring, studies of aeroallergen/pollen patterns, investigations of sources of Legionella across a cityscape, citizen science programs or microbiome studies.
Supplementary Material
8. Acknowledgement
This work was supported by the United States Environmental Protection Agency’s STAR (Science to Achieve Results) Graduate Fellowship for J. Therkorn [FP-91760601-0], and the Air and Waste Management Association’s Milton Feldstein Memorial Scholarship for J. Therkorn for Air Quality Research. This work was also supported by the NIH-NIEHS funded Center for Environmental Exposure and Disease, P30 ES005022.
7. References
- Ackerman JD (2000). Abiotic pollen and pollination: Ecological, functional, and evolutionary perspectives. Plant Syst. Evol 222:167–185. [Google Scholar]
- Adams RI, Tian Y, Taylor JW, Bruns TD, Hyvärinen A, Täubel M (2015). Passive dust collectors for assessing airborne microbial material. Microbiome 3:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari A, Martuzevicius D, Reponen T, Grinshpun SA, Cho SH, Sivasubramani SK, Zhong W, Levin L, Kelley A. L., St. Clair HG, LeMasters G (2003). Performance of the Button personal inhalable sampler for the measurement of outdoor aeroallergens. Atmos. Environ 37:4723–4733. [Google Scholar]
- Adhikari A, Reponen T, Lee SA, Grinshpun SA (2004). Assessment of human exposure to airborne fungi in agricultural confinements: Personal inhalable sampling versus stationary sampling. Ann. Agric. Environ. Med 11:269–277. [PubMed] [Google Scholar]
- Aizenberg V, Grinshpun SA, Willeke K, Smith J, Baron PA (2000a). Performance characteristics of the Button Personal Inhalable Aerosol Sampler. Am. Ind. Hyg. Assoc. J 61:398–404. [DOI] [PubMed] [Google Scholar]
- Aizenberg V, Reponen T, Grinshpun SA, Willeke K (2000b). Performance of Air-O-Cell, Burkard, and Button Samplers for total enumeration of airborne spores. Am. Ind. Hyg. Assoc. J 61:855–864. [DOI] [PubMed] [Google Scholar]
- Andon BM (2006). Active air vs. passive air (settle plate) monitoring in routine environmental monitoring programs. PDA J. Pharm. Sci. Technol 60:350–355. [PubMed] [Google Scholar]
- Baird R, Hodges N, Denyer S (2000). Handbook of Microbiological Quality Control: Pharmaceuticals and Medical Devices. CRC Press, Boca Raton, FL. [Google Scholar]
- Bauer H, Schueller E, Weinke G, Berger A, Hitzenberger R, Marr IL, Puxbaum H (2008). Significant contributions of fungal spores to the organic carbon and to the aerosol mass balance of the urban atmospheric aerosol. Atmos. Environ 42:5542–5549. [Google Scholar]
- Breed RS and Dotterrer WD (1916). The number of colonies allowable on satisfactory agar plates. J. Bacteriol 1:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown Wake, D., Thorpe A, Hemingway MA, Roff MW (1994). Theory and measurement of the capture of charged dust particles by electrets. J. Aerosol Sci 25:149–163. [Google Scholar]
- Brown Hemingway, M. A., Wake D, Thompson J (1995). Field Trials of an electret-based passive dust sampler in metal-processing industries. Ann. Occup. Hyg 39:603–622. [Google Scholar]
- Brown Hemingway, M., Wake D, Thorpe A (1996). Electret-based passive dust sampler: sampling of organic dusts. Analyst 121:1241–1246. [DOI] [PubMed] [Google Scholar]
- Burdett G and Revell G (1999). Strategy, development and laboratory calibration of a personal passive sampler for monitoring the asbestos exposure of maintenance workers, in Advances in Environmental Measurement Methods for Asbestos, ASTM International. [Google Scholar]
- Burdett G and Bard D (2007). Exposure of UK industrial plumbers to asbestos, Part I: Monitoring of exposure using personal passive samplers. Ann. Occup. Hyg 51:121–130. [DOI] [PubMed] [Google Scholar]
- Burge HA, Feeley JC, Kreiss K, Milton D, Morey P, Otten JA, Peterson K, Tulis JJ, Tyndall R (1989). Guidelines for the Assessment of Bioaerosols in the Indoor Environment. American Conference of Governmental Industrial Hygienists (ACGIH), Cincinnati, OH. [Google Scholar]
- Cartwright C, Horrocks S, Kirton J, Crook B (2009). Review of methods to measure bioaerosols from composting sites (Science report: SC040021/SR3). Environment Agency, United Kingdom. [Google Scholar]
- Chen P-S and Li C-S (2005). Sampling performance for bioaerosols by flow cytometry with fluorochrome. Aerosol Sci. Technol 39:231–237. [Google Scholar]
- Cherrie JW, Lynch G, Bord BS, Heathfield P, Cowie H, Robertson A (1994). Does the wearing of sampling pumps affect exposure? Ann. Occup. Hyg 38:827–838. [DOI] [PubMed] [Google Scholar]
- Cox CS and Wathes CM (1995). Bioaerosols Handbook. Lewis Publishers, United States of America. [Google Scholar]
- Douwes J, Thorne P, Pearce N, Heederik D (2003). Bioaerosol health effects and exposure assessment: progress and prospects. Ann. Occup. Hyg 47:187–200. [DOI] [PubMed] [Google Scholar]
- Elbert W, Taylor PE, Andreae MO, Pöschl U (2007). Contribution of fungi to primary biogenic aerosols in the atmosphere: wet and dry discharged spores, carbohydrates, and inorganic ions. Atmos. Chem. Phys 7:4569–4588. [Google Scholar]
- Fierer N, Liu Z, Rodríguez-Hernández M, Knight R, Henn M, Hernandez MT (2008). Short-term temporal variability in airborne bacterial and fungal populations. Appl. Environ. Microbiol 74:200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich-Nowoisky J, Pickersgill DA, Després VR, Pöschl U (2009). High diversity of fungi in air particulate matter. Proc. Natl. Acad. Sci. USA 106:12814–12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J, Gandhi P, Shekhawat G, Frazier A, Hampton-Marcell J, Gilbert JA (2015). A simple novel device for air sampling by electrokinetic capture. Microbiome 3:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny RL, Dutkiewicz J, Krysinska-Traczyk E (1999). Size distribution of bacterial and fungal bioaerosols in indoor air. Ann. Agric. Environ. Med 6:105–113. [PubMed] [Google Scholar]
- Green BJ, Sercombe JK, Tovey ER (2005). Fungal fragments and undocumented conidia function as new aeroallergen sources. J. Allergy Clin. Immunol 115:1043–1048. [DOI] [PubMed] [Google Scholar]
- Hauck BC, Grinshpun SA, Reponen A, Reponen T, Willeke K, Bornschein RL (1997). Field testing of new aerosol sampling method with a porous curved surface as inlet. Am. Ind. Hyg. Assoc. J 58:713–719. [DOI] [PubMed] [Google Scholar]
- Held A, Zerrath A, McKeon U, Fehrenbach T, Niessner R, Plass-Dülmer C, Kaminski U, Berresheim H, Pöschl U (2008). Aerosol size distributions measured in urban, rural and high-alpine air with an electrical low pressure impactor (ELPI). Atmos. Environ 42:8502–8512. [Google Scholar]
- Hock N, Schneider J, Borrmann S, Römpp A, Moortgat G, Franze T, Schauer C, Pöschl U, Plass-Dülmer C, Berresheim H (2008). Rural continental aerosol properties and processes observed during the Hohenpeissenberg Aerosol Characterization Experiment (HAZE2002). Atmos. Chem. Phys 8:603–623. [Google Scholar]
- Hoisington AJ, Maestre JP, King MD, Siegel JA, Kinney KA (2014). Impact of sampler selection on the characterization of the indoor microbiome via high-throughput sequencing. Build. Environ 80:274–282. [Google Scholar]
- IBM Corporation (2015). IBM SPSS Statistics for Windows, IBM Corp., Armonk, NY. [Google Scholar]
- Jaenicke R (2005). Abundance of cellular material and proteins in the atmosphere. Science 308:73. [DOI] [PubMed] [Google Scholar]
- Jensen PA, Todd WF, Davis GN, Scarpino PV (1992). Evaluation of eight bioaerosol samplers challenged with aerosols of free bacteria. Am. Ind. Hyg. Assoc. J 53:660–667. [DOI] [PubMed] [Google Scholar]
- Jensen PA and Schafer MP (2003). Sampling and characterization of bioaerosols, in NOSH Manual of Analytical Methods (NMAM).
- Kalatoor S, Grinshpun SA, Willeke K, Baron P (1995). New aerosol sampler with low wind sensitivity and good filter collection uniformity. Atmos. Environ 29:1105–1112. [Google Scholar]
- Kellogg CA and Griffin DW (2006). Aerobiology and the global transport of desert dust. Trends Ecol. Evol 21:638–644. [DOI] [PubMed] [Google Scholar]
- Kilburg-Basnyat B, Metwali N, Thorne PS (2015a). Effect of deployment time on endotoxin and allergen exposure assessment using electrostatic dust collectors. Ann. Occup. Hyg 59:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburg-Basnyat B, Peters TM, Perry SS, Thorne PS (2015b). Electrostatic dust collectors compared to inhalable samplers for measuring endotoxin concentrations in farm homes. Indoor Air 26:724–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburg-Basnyat B, Metwali N, Thorne PS (2016). Performance of electrostatic dust collectors (EDCs) for endotoxin assessment in homes: Effect of mailing, placement, heating, and electrostatic charge. J. Occup. Environ. Hyg 13:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Grinshpun SA, Martuzevicius D, Adhikari A, Crawford CM, Luo J, Reponen T (2006). Relationship between indoor and outdoor bioaerosols collected with a button inhalable aerosol sampler in urban homes. Indoor Air 16:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-S (1999). Evaluation of microbial samplers for bacterial microorganisms. Aerosol Sci. Technol 30:100–108. [Google Scholar]
- Linaker C and Smedley J (2002). Respiratory illness in agricultural workers. Occup. Med 52:451–459. [DOI] [PubMed] [Google Scholar]
- Mainelis G, Górny RL, Reponen T, Trunov M, Grinshpun SA, Baron P, Yadav J, Willeke K (2002). Effect of electrical charges and fields on injury and viability of airborne bacteria. Biotechnol. Bioeng 79:229–241. [DOI] [PubMed] [Google Scholar]
- Menetrez MY, Foarde KK, Esch RK, Schwartz TD, Dean TR, Hays MD, Cho SH, Betancourt DA, Moore SA (2009). An evaluation of indoor and outdoor biological particulate matter. Atmos. Environ 43:5476–5483. [Google Scholar]
- Noss I, Wouters IM, Visser M, Heederik DJJ, Thorne PS, Brunekreef B, Doekes G (2008). Evaluation of a low-cost electrostatic dust fall collector for indoor air endotoxin exposure assessment. Appl. Environ. Microbiol 74:5621–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren U, Strom G, Blomquist G, Malmberg P (1986). Collection of airborne micro-organisms on Nuclepore filters, estimation and analysis-CAMNEA method. J. Appl. Bacteriol 61:401–406. [DOI] [PubMed] [Google Scholar]
- Pasanen AL (2001). A review: fungal exposure assessment in indoor environments. Indoor Air 11:87–98. [DOI] [PubMed] [Google Scholar]
- Peccia J, Milton DK, Reponen T, Hill J (2008). A role for environmental engineering and science in preventing bioaerosol-related disease. Environ. Sci. Technol 42:4631–4637. [DOI] [PubMed] [Google Scholar]
- Plog BA and Quinlan PJ (2001). Fundamentals of Industrial Hygiene. National Safety Council, USA. [Google Scholar]
- Reponen T, Willeke K, Grinshpun S, Nevalainen A (2011). Biological Particle Sampling, in Aerosol Measurement: Principles, Techniques and Applications, Kulkarni P, Baron P, Willeke K, eds., John Wiley & Sons, Inc., Hoboken, NJ, 549–570. [Google Scholar]
- Samadi S, Heederik DJJ, Krop EJM, Jamshidifard A-R, Willemse T, Wouters IM (2010). Allergen and endotoxin exposure in a companion animal hospital. Occup. Environ. Med 67:486–492. [DOI] [PubMed] [Google Scholar]
- Sandle T (2015). Settle plate exposure under unidirectional airflow and the effect of weight loss upon microbial growth. EJPPS 20:45–50. [Google Scholar]
- Smith DJ, Griffin DW, Schuerger AC (2010). Stratospheric microbiology at 20 km over the Pacific Ocean. Aerobiologia 26:35–46. [Google Scholar]
- Sterling M, Rogers C, Levetin E (1999). An evaluation of two methods used for microscopic analysis of airborne fungal spore concentrations from the Burkard Spore Trap. Aerobiologia 15:9–18. [Google Scholar]
- Stewart SL, Grinshpun SA, Willeke K, Terzieva S, Ulevicius V, Donnelly J (1995). Effect of impact stress on microbial recovery on an agar surface. Appl. Environ. Microbiol 61:1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha MPM, Drew GH, Longhurst PJ, Smith R, Pollard SJT (2006). Bioaerosol releases from compost facilities: Evaluating passive and active source terms at a green waste facility for improved risk assessments. Atmos. Environ 40:1159–1169. [Google Scholar]
- The Rutgers Gardens Weather Tower; (2016). New Brunswick, NJ: http://synoptic.envsci.rutgers.edu/site/ [Google Scholar]
- Therkorn J, Thomas N, Calderon L, Scheinbeim J, Mainelis G (2017). Design and development of a passive bioaerosol sampler using polarized ferroelectric polymer film. J. Aerosol Sci 105:128–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivola M, Alm S, Reponen T, Kolari S, Nevalainen A (2002). Personal exposures and microenvironmental concentrations of particles and bioaerosols. J. Environ. Monit 4:166–174. [DOI] [PubMed] [Google Scholar]
- Toivola M, Nevalainen A, Alm S (2004). Personal exposures to particles and microbes in relation to microenvironmental concentrations. Indoor Air 14:351–359. [DOI] [PubMed] [Google Scholar]
- Tovey ER, Mitakakis TZ, Sercombe JK, Vanlaar CH, Marks GB (2003). Four methods of sampling for dust mite allergen: differences in ‘dust’. Allergy 58:790–794. [DOI] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency (USEPA) (2016). Air Monitoring Methods - Passive Monitoring. http://www3.epa.gov/ttn/amtic/passive.html
- United Nations (2006). Uniting against terrorism: recommendations for a global counter-terrorism strategy, report of the Secretary-General, United Nations, Geneva. [Google Scholar]
- Wang Z, Reponen T Grinshpun AS, Górny LR, Willeke K (2001). Effect of sampling time and air humidity on the bioefficiency of filter samplers for bioaerosol collection. J. Aerosol Sci 32:661–674. [Google Scholar]
- Womiloju TO, Miller JD, Mayer PM, Brook JR (2003). Methods to determine the biological composition of particulate matter collected from outdoor air. Atmos. Environ 37:4335–4344. [Google Scholar]
- Wood JD (1977). A review of personal sampling pumps. Ann. Occup. Hyg 20:3–17. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Schmechel D, Chen BT, Lindsley WG, Peccia J (2011). Comparison of quantitative airborne fungi measurements by active and passive sampling methods. J. Aerosol Sci 42:499–507. [Google Scholar]
- Yamamoto N, Nazaroff WW, Peccia J (2014). Assessing the aerodynamic diameters of taxon-specific fungal bioaerosols by quantitative PCR and next-generation DNA sequencing. J. Aerosol Sci 78:1–10. [Google Scholar]
- Zhen H, Han T, Fennell DE, Mainelis G (2013). Release of free DNA by membrane-impaired bacterial aerosols due to aerosolization and air sampling. Appl. Environ. Microbiol 79:7780–7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


