Abstract
Particle size distribution and stability are key attributes for the evaluation of the safety and efficacy profile of medical nanoparticles (Med-NPs). Measuring particle average size and particle size distribution is a challenging task which requires the combination of orthogonal high-resolution sizing techniques, especially in complex biological media. Unfortunately, despite its limitations, due to its accessibility, low cost, and easy handling, batch mode dynamic light scattering (DLS) is still very often used as the only approach to measure particle size distribution in the nanomedicine field. In this work the use of asymmetric flow field flow fractionation coupled to multiangle light scattering and dynamic light scattering detectors (AF4-MALS-DLS) was evaluated as an alternative to batch mode DLS to measure the physical properties of lipid-based nanoparticles. A robust standard operating procedure (SOPs) developed by the Nanomedicine Characterization Laboratory (EUNCL) was presented and tested to assess size stability, batch to batch consistency, and the behavior of the lipid-based nanoparticles in plasma. Orthogonal sizing techniques, such as transmission electron microscopy (TEM) and particle tracking analysis (PTA) measurements, were performed to support the results. While batch mode DLS could be applied as a fast and simple method to provide a preliminary insight into the integrity and polydispersity of samples, it was unsuitable to resolve small modifications of the particle size distribution. The introduction of nanoparticle sorting by field-flow fractionation coupled to online DLS and MALS allowed assessment of batch to batch variability and changes in the size of the lipid nanoparticles induced by the interaction with serum proteins, which are critical for quality control and regulatory aspects. In conclusion, if a robust SOP is followed, AF4-MALS-DLS is a powerful method for the preclinical characterization of lipid-based nanoparticles.
Keywords: nanomedicine, particle size distribution, stability, nanoparticle−protein interactions, lipid nanoparticles, asymmetric flow field-flow fractionation, standard operating procedure, European Nanomedicine Characterisation Laboratory
Introduction
The use of nanotechnology in healthcare is expanding and has raised hopes to address unmet medical needs by contributing to better diagnostic and therapeutics tools. However, the number of approved medical nanoparticles (Med-NPs) is still very limited if compared to the tremendous research activity in the field.2,3 One of the major causes of failures of Med-NPs in late clinical phases is the lack of suitable protocols for their characterization at physicochemical and biological levels. Due to their uniqueness, Med-NPs requires the use of complex methodological approaches to characterize their physical–chemical properties. To complicate matters, their properties should be tested not only in their pristine state but also when dispersed and interacting with complex biological media, since the presence of organic molecules and proteins may affect the particle properties, changing their behavior once they are in contact with the human body. The development of reliable suitable standard protocols suitable for the preclinical characterization of Med-NPs is therefore a key prerequisite to assess the quality and safety of emerging Med-NPs, and to support their translation into the clinic.2,4,5
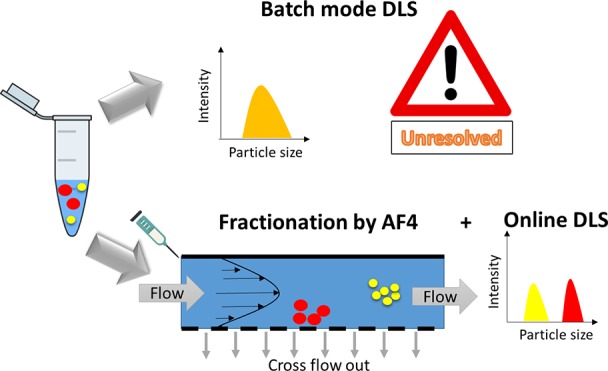
Average particle size, particle size distribution (PSD), and stability are the most monitored parameters in the preclinical characterization of Med-NPs,2,6 due to their impact on body absorption, biodistribution, and excretion of nanomaterials.7−9 Batch mode dynamic light scattering (DLS) is the most widely used sizing technique.2 It measures the autocorrelation function of time-dependent fluctuations in the light scattered by a suspension of nanoparticles, which are associated with the Brownian motion of the particles. The hydrodynamic diameter of the particles is then calculated, via the Stokes–Einstein equation, which is only applicable to spherical particles (for more details we suggest to refer to the recent work by Maguire and co-workers).10 Due to the strong dependence of light scattering intensity on particle size, in batch mode DLS, where all the contributions of the light scattered by a polydispersed sample are measured together, large objects mask the smaller ones. Batch mode DLS can be applied as the first preliminary analysis to check the size range, polydispersity, and integrity of Med-NP formulations. It may also be useful to determine the tendency of a nanoformulation to undergo significant aggregation or degradation when exposed to conditions of high ionic strength and unsuitable pH that may happen in physiological conditions.11 However, the users should be aware of its limitations: in the presence of multiple populations in similar size ranges, the size distribution may appear as a global distribution without fine distinction between population of particles or between single particles and small aggregates.4,12 Moreover, batch mode DLS is not suitable to measure changes of PSD in complex biological media, such as in plasma.4,13 Unfortunately, despite its low-resolution, due to its accessibility, low cost, and easy handling, batch mode DLS is still nowadays very often used as the only approach to measure particle size distribution in the nanomedicine field, especially by industry which often adopts limited characterization steps in order to reduce development costs.2
The coauthors feel that in the nanomedicine community, there is still a strong need to raise awareness of the limitation of batch mode DLS and to develop a reliable approach for the evaluation of particle size distribution in simple and complex biological media. In this context, the USA Nanotechnology Characterization Lab (NCI-NCL, https://ncl.cancer.gov) and the European Nanomedicine Characterisation lab (EUNCL, http://www.euncl.eu) have joined their efforts by developing multiple protocols to measure PSD and foster the use of robust standard operation procedures (SOPs). EUNCL and NCI-NCL also provide a platform for educating the community.
In order to characterize the average size and particle size distribution of Med-NPs, a multistep approach of incremental complexity was developed by EUNCL and NCI-NCL, which consists of a fast prescreening to check sample integrity and stability by batch mode DLS, followed by the integration of multiple orthogonal high resolution sizing measurements to resolve PSD in simple and complex media representative of the clinical administration of the product.
Among the high resolution sizing techniques available on the market, separation techniques coupled to online sizing measurements are powerful to overcome the inherent limitations of batch mode DLS, thanks to the introduction of a fractionation step to separate the particles according to their size, prior to detection. Asymmetric flow field flow fractionation (AF4) is a robust fractionation method based on the generation of a parabolic flow profile in a narrow ribbon-like channel, via the application of a liquid cross-flow. It is a liquid chromatography separation technique, where nonspecific interactions are reduced to a minimum, due to the absence of a stationary phase.14 AF4 is becoming widely used in the field of nanomedicine, as demonstrated by a rapidly increasing number of publications on different types of medical nanoparticles (Med-NPs), including liposomes, lipid nanoparticles (LNPs), polymeric NPs, virus like particles (VLPs), metallic NPs, and metal oxide NPs.4,15−21 Med-NP-sorting by AF4, coupled to online sizing detectors such as dynamic light scattering (DLS or quasi elastic light scattering QELS) and multiangle light scattering (MALS) allows resolution of the PSD of very complex samples and analysis of small changes in particle size, which are critical for quality control and regulatory aspects of Med-NPs.
The EUNCL, in collaboration with the NCI-NCL, has developed a standard operating procedure (SOP) to determine the size distribution of Med-NPs by coupling AF4 with online size measurement (DLS and/or MALS),22 in line with the new ISO standard published in 2018 (ISO/TS 21362:2018).23 This protocol, when applicable to Med-NPs systems, could be considered as an alternative to the DLS alone for regulatory purposes.4 The SOP developed by EUNCL has the potential to characterize multiple Med-NP systems and can be applied with the aim to (i) resolve the PSD of complex mixtures and/or (ii) discriminate between populations of particles with different shapes and/or (iii) discriminate between larger particles and small aggregates, as recently described by Gioria et al.4,12,24,25 In addition, the SOP allows investigation of the behavior of the PSD of Med-NP in the presence of serum proteins, which is crucial to understand their efficacy and safety in vitro and in vivo.22
The aim of this work is to present the AF4-MALS-DLS SOP developed by EUNCL and to show its applicability to lipid-based nanoparticles, called Lipidots26−29 a formulation which is representative of the organic Med-NPs under development in the nanomedicine field. The coauthors strongly support use of orthogonal techniques helps to better assess the particle size distribution of Med-NPs. For this reason, the AF4-MALS-DLS measurements are complemented with transmission electron microscopy (TEM) and particle tracking analysis (PTA). Batch mode DLS measurements are also performed in order to raise awareness of its limitations to measure PSD in simple and complex biological media.
Materials and Methods
Materials
Suppocire NB was purchased from Gattefossé (Saint-Priest, France), Lipoid S75 (soybean lecithin at >75% phosphatidylcholine) from Lipoid (Ludwigshafen, Germany), Myrj S40 (polyethylene glycol 40 stearate), and super refined soybean oil from Croda Uniqema (Chocques, France). ICG, IR780 iodide dye, fetal bovine serum (FBS), phosphate buffered saline (PBS) 10× and other chemicals were purchased from Sigma-Aldrich (Saint-Quentin-Fallavier, France). The IR780-lipid dye was synthesized as described in ref (26).26
Production of the Lipidots
The LNPs were synthesized by ultrasonication as previously described.26,30,31 An oil premix with, respectively, 85, 255, and 65 mg of Suppocire NB, oil, and lecithin was prepared. For Lipimage815 and ICG Lipidots, 200 μL of a 10 mg/mL IR780-Oleyl dye solution or 300 μL of a 10 mM ICG solution in ethanol was respectively poured in a 5 mL vial and mixed with the oil premix melted at 50 °C. The mixture was homogenized, and the solvent was then evaporated under argon flux. After homogenization at 50 °C, the continuous aqueous phase, composed of 345 mg of Myrj S40 and the appropriate amount of 154 mM NaCl buffer was introduced. The vial was placed in a 50 °C water bath, and the mixture was sonicated for 5 min using a VCX750 Ultrasonic processor (power output 190 W, 3 mm probe diameter, Sonics). The lipid nanoparticles were dialyzed overnight at room temperature against 1000 times their volume in the appropriate aqueous buffer (12 to 14 000 Da MW cut off membranes, ZelluTrans, Carl Roth, France). Finally, the nanoparticle dispersions were filtered through a 0.22 μm Millipore membrane for sterilization. The total mass concentration, the drug loading, the chemical composition, and zeta potential were measured as described elsewere.26,31,32,32,33 To assess batch to batch consistency, three batches of Naked Lipidots, three batches of Lipimage815, and two batches of ICG-Lipidots were synthesized following the same manufacturing procedure.
Batch Mode DLS
The batch mode DLS measurements were performed using a Malvern Zetasizer Nano-ZS instrument equipped with a 633 nm HeNe laser, as described in the SOP of EUNCL.34 The original particle suspension was diluted 200 times in PBS (phosphate buffered saline 1×, pH 7.4) immediately before the measurement, in order to reach a concentration of 0.5 mg/mL. The final particle concentration was optimized in order to obtain an attenuator factor of 6 or 7, and a value of the intercept of the autocorrelation function >0.9, as described in the SOP. The particles were not filtered prior to the measurements, to avoid material losses in the filter (e.g., aggregates). To study NP interactions with serum proteins, 10 μL of the NP stock solutions was diluted in 200 μL of pure fetal bovine serum (FBS), incubated for 1 h at 37 °C, and then diluted at RT with 1.8 mL of PBS 1× immediately before the measurements. All the measurements were performed at 25 °C. An equilibration step was performed for 5 min before the measurements, as described in the SOP. Quality control measurements with polystyrene particles of 60 nm (Thermo Scientific Nanosphere NIST traceable standard, Series 3000, 3060A) were performed at the beginning of each measurement session to verify the instrument performances.34 Size distribution results were generated by averaging 10 consecutive measurements of 12 times 10 s runs, as indicated in the ISO22412:2008.1 The results of cumulant analysis (z-ave, PdI) and of the intensity based-PSD analysis are reported, as described in the SOP.
Asymmetric Flow Field Flow Fractionation Measurements
A Wyatt eclipse asymmetric field flow fractionation (AF4) system online connected to (i) an UV–vis absorbance detector, (ii) a Wyatt Dawn Helios multiangle light scattering (MALS) with an integrated quasi elastic light scattering (QELS or DLS) detector, and (iii) a Wyatt Optilab rEX differential Refractive Index detector (dRI) was used to carry out AF4-UV–vis-MALS-DLS-dRI measurements, as described in the SOP from EUNCL.22
To measure the particle size distribution, each particle suspension was diluted 200 times in PBS (final concentration of 0.5 mg/mL) and then injected through a 30 μL loop into the AF4 channel (350 μm spacer W type, 10 kDa regenerated cellulose membrane). To study changes in the PSD induced by Med-NP interactions with serum proteins, 10 μL of the Lipidots suspensions were diluted in 200 μL of pure fetal bovine serum (FBS), incubated for 1 h at 37 °C, and then diluted at RT with 1.8 mL of PBS 1× immediately before the injection (final concentration of 0.5 mg/mL in 10% buffered serum).
Phosphate buffered saline (PBS), (pH 7.4) was used as mobile phase. The optimized elution method is described in Figure S1. After a focus step at 1.2 mL/min (cross-flow), a gradient elution imposing a cross-flow decreasing linearly between 1.2 mL/min and 0 mL/min was applied for 40 min (detector flow: 1 mL/min). The absorbance of the eluted fractions was monitored at 230 nm. The hydrodynamic diameter and the geometric diameter (Dg) of the particles were determined from the online DLS and MALS measurements in the elution window comprised between 22 and 34 min, by using the software Astra 6.0 (particle method) and applying the Zimm model or the cumulant model to fit the MALS and the online DLS data, respectively.
The amount of mass recovery was estimated by calculating the area under the UV–vis peak of the samples eluted with and without an applied cross-flow:
The mean of Dg and Dh were calculated by averaging the values obtained across the full width at half-maximum (fwhm) for each resolvable eluting peak, as indicated in the ISO/TS 21362:2018 standard document. Each sample was injected 3 times. The mean value and the standard deviation of the mean over three replicates were determined, in order to evaluate the precision of the method (minimum repeatability condition tested as described in the ISO/TS 21362:201823 and in the SOP developed by EUNCL).
Transmission Electron Microscopy
Negatively stained grids were prepared using the mica-carbon flotation technique by injecting 3 μL of Lipidots dispersion at the mica–carbon interface. The grid was then stained in 2% uranyl acetate for less than a minute. The carbon with the negatively stained LNP was gathered on a copper grid for TEM observation.35 The images were processed by manually scoring the size of more than 400 not overlapping NPs. Particle size distribution by number was obtained by plotting the diameter of an equivalent sphere obtained for the area of each particle, making use of ImageJ software. The mean, the mode, and the X10, X50, and X90 values (see Size Glossary) were estimated from each PSD. Each PSD was fitted by using a LogNormal distribution. The mode of the distribution was calculated by estimating the peak of the LogNormal distribution associated with each sample. In the case of the particles incubated with serum, the negative staining of the proteins introduced some noise and very small aggregates <10 nm in diameter which were excluded from the analysis.
Particle Tracking Analysis
The average hydrodynamic diameter and the PSD of Naked Lipidots and of Lipimage815 were characterized using PTA developed by NanoSight (Malvern-Pannalytical Instruments Ltd.) according to the validated EUNCL SOP (EUNCL PCC-023).36 This equipment utilizes the properties of light scattering and Brownian motion to obtain particle size distributions of samples in liquid suspension. In brief, the samples were diluted in particle free PBS; analysis was conducted directly after dispersion in PBS as reported in ref (36). PTA was carried out by means of a NS500 system equipped with a 405 nm laser, with PTA version 3.0. Six by 60 second videos were recorded at an NP concentration sufficient to obtain a minimum of 200 completed tracks per video for statistical significance. The mean, the mode, and the X10, X50, and X90 values were estimated from each PSD.
Results and Discussion
Particle Size Distribution by Batch Mode DLS
Lipidots are lipid-based nanoparticles (LNP) composed by a lipid core, made of soybean oil and wax, and a surfactant shell, consisted of phospholipids and PEGylated surfactants.30,33 Since their development in 2008, they have shown a high potential to incorporate lipophilic drugs and dyes for medicinal and imaging applications.26−29 Lipidots loaded with Indocyanine green (ICG-Lipidots) and IR780-Oleyl dye (Lipimage815) have been initially developed for in vivo imaging purposes.28 Lipimage815, being the most promising formulation, has shown efficacy as an in vivo NIR nanotracer for intraoperative fluorescence imaging in the surgical excision of malignant masses, as demonstrated in in vivo studies performed in mice and dogs.37,38 In order to translate its potential efficacy to the clinical utility, an extensive preclinical characterization has been initiated with the development and validation of a large panel of analytical and physicochemical methods in collaboration with EUNCL.32 In this frame, AF4-MALS-DLS was chosen to measure the PSD of Lipidots. Here, we would like to show the potential of AF4-MALS-DLS to (i) assess accurately the PSD of three formulations, e.g. Naked Lipidots, Lipimage815, and ICG-Lipidots, (ii) to monitor small changes associated with batch to batch variability or instability during long-term storage and (iii) to study the Lipidots behavior in the presence of serum proteins.
In our study the three Lipidots formulations were synthesized at the lab scale by ultrasonication, as previously reported.30,33 The composition of the oily and aqueous phase allows varying the size between 30 and 120 nm. In our case, the composition which would allow production of Lipidots of 50 nm (previous analysis performed by batch mode DLS) was selected. Their physical and chemical properties are summarized in Table S1. The physical properties of the Lipidots were not affected by dye loading.26,27,31,39 The neutral charge is induced by the presence of a dense PEG coating of the particle surface, which also ensured a high colloidal stability for the particles.
As preliminary measurement, batch mode DLS was used to check the average size range and the polydispersity of the samples dispersed in phosphate buffered saline (PBS, pH 7.4) at a final particle concentration of 0.5 mg/mL. Sample preparation and measurement were performed following the conditions reported in the EUNCL’s SOP (http://www.euncl.eu/about-us/assay-cascade/PDFs/Prescreening/EUNCL-PCC-001.pdf?m=1468937875&). The instrument performances were checked prior to each measurement session by measuring 60 nm polystyrene standard nanoparticles as certified reference materials (CRM). Temperature equilibration prior to the measurement was set to 5 min in order to ensure thermal stability. Sample concentration was selected to be in the optimal range for the measurements, as described in the Materials and Methods section, and the absence of precipitation was assured by a stable count rate during 10 repeated measurements. Only the intensity-based analysis together with the relative plots were reported, in order to avoid any uncertainty which could come from the transformation to volume or number based particle size distribution by using the Mie theory.
As shown in Figure S2, all the formulations are characterized by the presence of one population, possessing a z-ave of 50 nm and a PdI of 0.14 (cumulant analysis), indicative of a size distribution of moderate width. Even if the ISO standards only requires reporting the mean and the standard deviation of the mean of z-ave and the PdI values obtained by the cumulant analysis,1 the coauthors recommend to always compare the results of the cumulant analysis with the values calculated by the analysis of the intensity-based PSD. Usually, if close values are obtained in the two different data treatments, it is indicative of well-monodispersed samples, whereas significantly different values indicate that the sample particle distribution is polydispersed. In the case of the three Lipidots formulations analyzed, the mean Dh obtained by the intensity based PSD analysis was 15% larger than the average z-ave obtained by cumulant analysis (Table 1), suggesting that the z-ave value may not be representative of the most probable size of the nanoparticles contained in the samples, due to their intrinsic polydispersity.
Table 1. Particle size distribution obtained by multiple techniques: cumulant analysis and distribution analysis by batch mode DLS; mean, mode, minimum and maximum diameters obtained by online MALS (Dg by Zimm model) and online DLS (Dh by cumulant analysis) PSD. Mean, mode and D10, D50 and D90 obtained by number based-PSD measured by TEM and PTA.
| Batch
mode DLS (Dh in nm) |
AF4-MALS
(Dg in nm) |
AF4-DLS
(Dh in nm) |
TEM
(Diameter of an equivalent sphere in nm) |
PTA
(Dh in nm) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cumulant
analysis |
PSD
analysis (Intensity based PSD) |
Mass
based PSD |
Intensity
based PSD |
Number
based PSD |
||||||||||||
| Sample | z-avea (SD) | PdIa (SD) | Pk1-Inta (σ) | % of peak area | Modeb | Meanc(SD) | Dmin, Dmax | Modeb | Meanc (SD) | Dmin, Dmax | Moded | Meand | D10, D50, D90 | Mode | Mean | D10, D50, D90 |
| Naked Lipidots | 54 (1) | 0.142 (0.005) | 62 (26) | 100 | 35 | 43 (3) | 24, 60 | 42 | 48 (2) | 30, 70 | 21 | 29 | 14, 24, 45 | 45 | 56 | 32, 43, 67 |
| Naked Lipidots in serum | 41 (2) | 0.279 (0.003) | 61 (28) | 93 | 37 | 38 (3) | 32, 43 | 40 | 44 (2) | 40, 56 | 25 | 28 | 18, 27, 36 | NAe | NA | NA |
| Lipimage815 | 51 (1) | 0.139 (0.018) | 60 (24) | 100 | 24 | 33 (4) | 10, 52 | 40 | 43 (4) | 27, 67 | 29 | 32 | 21, 29, 45 | 47 | 65 | 36, 46, 104 |
| Lipimage815 in serum | 40 (1) | 0.281 (0.003) | 61 (30) | 93 | 38 | 40 (4) | 36, 45 | 40 | 45 (4) | 38, 56 | 25 | 29 | 17, 25, 41 | NA | NA | NA |
| ICG Lipidots | 52 (1) | 0.138 (0.006) | 61 (25) | 100 | 31 | 37 (4) | 17, 59 | 40 | 48 (4) | 22, 70 | 21 | 29 | 12, 24, 44 | NA | NA | NA |
| ICG Lipidots in serum | 42 (1) | 0.258 (0.03) | 60 (26) | 94 | 44 | 42 (4) | 40, 46 | 50 | 52 (4) | 40, 66 | 20 | 26 | 11, 21, 41 | NA | NA | NA |
Mean and standard deviation of the z-ave and PdI means calculated over 10 replicates; σ = standard deviation of the PSD by intensity, representative of the distribution with:
Value of the diameter calculated at the fractogram maximum (UV–vis detector).
Mean and standard deviation of the mean over 3 replicates. The mean values are values calculated by averaging the diameter over the fwhm of one elution peak as indicated by the ISO/TS 21362:2018.
Mode value estimated by fitting the PSD number by a LogNormal distribution.
NA= not available
It has been previously shown that, even in the cases where the sample shows a nicely monodispersed sample with a PdI < 0.1, and thus seems suitable for batch mode DLS measurements, multiple populations can be hidden in the scattering signal due to the low resolution of the technique.4,12,8 Using a fractionation step, which separates the particles according to their hydrodynamic radius, and a continuous online detection of the eluted fractions by sizing detectors, such as the multiangle light scattering (MALS) and dynamic light scattering (DLS or QELS) detectors could help to assess the particle size distribution with higher resolution.4
Optimization of AF4-MALS-DLS Measurements
As a second step, the Lipidots were analyzed by AF4-MALS-DLS, adapting the SOP developed by the EUNCL infrastructure (http://www.euncl.eu/about-us/assay-cascade/PDFs/PCC/EUNCL-PCC-022.pdf?m=1468937868&).22
The optimization of the AF4 elution conditions can be laborious, and instrument performances should be checked in order to assess the suitability of the conditions for each particular system. AF4-MALS-DLS analysis is not applicable to formulations that possess big aggregates, when a significant portion of the material is lost on the membrane of the AF4 channel or when particles are affected by the application of the cross-flow during focusing and elution. For this reason, initiatives like EUNCL foster the use of robust standard operating procedures, with proper quality controls to ensure the robustness and suitability of the method to characterize each specific nanoparticle system. Method precision depends on the nature of the sample and of the analytical methods utilized and should be evaluated under repeatability conditions as indicated in the ISO 21362:2018 (i.e., at a minimum, multiple replicates should be performed by the same analyst, same instrument, same location, same method, and same conditions, over short time intervals).23 Moreover, the users should be aware of the possible sample loss in the channel, by always checking for mass recovery. Material loss may be more significant for positively charged particles, which may be lost during separation, due to their irreversible interaction with the negatively charged membranes in the AF4 separation channel. Appropriate analysis of the data obtained by light scattering should be verified by checking the normalization of the detector prior to each measurement session.22 Finally it is also strongly recommended to run appropriate standards of similar size of your sample in order to check the performances of the AF4 fractionation system and of the online detectors in the specific working size range of interest.22
Based on EUNCL’s SOP, the AF4 elution conditions had to be adapted in the case of Lipidots. Phosphate buffered saline (PBS, pH 7.4) was chosen as an elution buffer, being representative of physiological conditions. During method optimization, multiple elution methods were tested (isocratic vs gradient elution profiles) by varying the cross-flow and the focus flow in the 0.8–1.5 mL/min range. Finally the elution program was optimized as reported in Figure S1. Before each measurement session, the normalization of the MALS detector was performed by injecting an aliquot of BSA at 5 mg/mL as described in the SOP.22 The performances of online MALS and DLS detectors were checked by injecting 60 nm polystyrene particles dispersed in SDS 0.05%, as described in the SOP.22
In order to check the precision of the optimized elution method, 3 repeated injections of each sample dispersed in PBS were measured (Figure S3). For each replica, the mean particle size was calculated by averaging the size values obtained across the elution peak at the fwhm (Table 1). The estimated precision was lower than 10% for all the samples tested, showing an acceptable repeatability of the elution method. The mass recovery of each formulation was estimated to be >75% (Table S2), thus higher than the accepted threshold according to ISO 21362:2018.
Particle Size Distribution by AF4-MALS-DLS
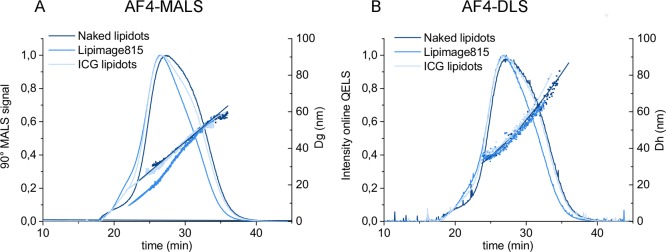
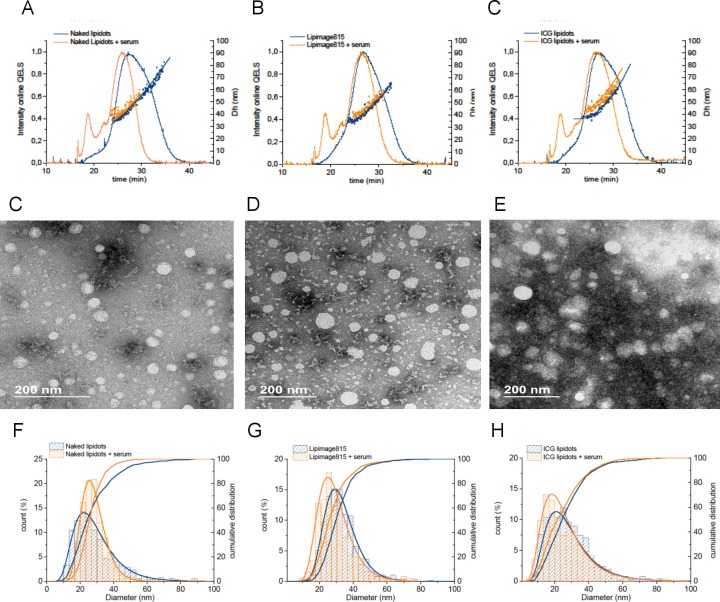
The AF4-MALS-DLS fractograms of the Naked Lipidots, Lipimage815 and ICG-Lipidots are reported in Figure 1, showing an upward trend of Dg and Dh over the elution peak associated with each fraction eluted between the 22nd and 34th minutes. All the samples tested were characterized by one main population, with a geometric diameter (Dg) in the 10–60 nm range and a hydrodynamic diameter (Dh) in the 25–70 nm range, in agreement with batch mode DLS (Figure 1, Table 2). A small shoulder was detected at the beginning of the elution peak of the Lipidots, which may be associated with a small population of NPs possessing similar sizes but different surface properties (e.g., a PEG and lecithin to lipid ratio). However, in the 0–22 min elution window, the scattering signal was too low to allow analysis of the PSD.
Figure 1.
Elution behaviors of the Lipidots formulation detected by AF4-MALS and AF4-DLS. (A) Intensity of the 90° LS detector and calculated geometric diameter (Dg); (B) Intensity of DLS online detector and calculated hydrodynamic diameter (Dh) of the Naked Lipidots (dark blue), Lipimage 815 (blue) and ICG lipidot (light blue).
The mode and the mean values of Dh and Dg, measured across the full width at half-maximum (fwhm) of the peak are reported in Tables 1 and S2. However, since the data indicates a clear upward trend of Dg and Dh over the elution peak, care should be taken in making quantitative comparison between the samples by only considering the calculated Dg and Dh mean, and the standard deviation of the means, since those values may not represent the true mean for the peak. Such a trend indicates that the population of nano-objects is polydispersed with larger particles exiting the channel after smaller ones, and their relative fractions might not be properly weighted.23 Therefore, if the sample is polydispersed, we suggest to refer to the elution profile, analyzing the Dg and Dh trend across the peak in order to have qualitative indications of the physical properties of the Med-NP analyzed.
Interestingly, AF4-MALS-DLS analysis provides indirect information on particle shape (e.g., spherical vs elongated particles) by calculating the shape factor, which is the ratio of the geometric diameter (Dg) determined by MALS and the hydrodynamic diameter (Dh) measured by DLS.4,12,16,19 The calculated Dg to Dh ratio of the all Lipidots formulations studied was in the 0.7–0.9 range, suggesting that they all possess a compact homogeneous core characterized by a spherical shape (theoretical compact sphere ratio= 0.778).4,12,22
Particle Size Distribution by PTA
Particle tracking analysis (PTA) was chosen as a complementary technique to measure the particle size distribution of Naked Lipidots and of Lipimage815. The calculated mode and mean are reported in Table 1, while the number-based PSD is shown in Figure S4. PTA results are in line with the results obtained by AF4-MALS-DLS. According to PTA measurements, the two samples were characterized by one main population possessing a mode of 45 nm. No significant differences were detected in the mode and in the D50 of the naked vs the dye-loaded samples.
PTA was not chosen as the main orthogonal technique to support AF4-MALS-DLS findings, since PTA limitation is the incapability to measure PSD of small organic particles in the presence of serum proteins (<30 nm). In fact in the buffer of choice there must be an inhomogeneity in the refractive indices of a particle and the medium within which it exists in order for light scattering to occur, but unfortunately this condition is not satisfied if small organic particles are dispersed in biological complex media containing proteins of comparable sizes. Without such an inhomogeneity, no measurement can be made by the PTA. Therefore, PTA can be a valid alternative to the AF4 technique to measure the PSD of lipid based NPs in simple media (e.g., PBS), batch to batch consistency, and stability, but, in the present setup, it cannot be used to measure their PSD in biological complex media, due to their small size.10
Particle Size Distribution by TEM
In order to support the AF4-MALS-DLS findings in simple and complex media, we measured the particle size distribution with transmission electron microscopy (TEM).
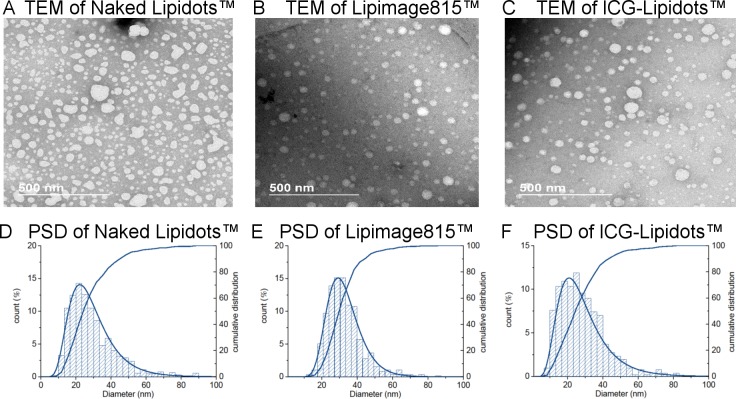
Electron microscopy (EM) is one of the methods always suggested by the regulators,40 since it permits the direct visualization of the particles, gaining direct information about particle size and shape. Moreover, it allows obtaining a number-based particle size distribution, allowing direct calculation of the median diameter (X50). Caution should be taken to make sure that the sample preparation does not induce an artifact, especially for organic particles such as the Lipidots. Differently from other organic particles (e.g., liposomes), the cryo-TEM analysis is not applicable to the Lipidots, because the sample freezing by cryo-vitrification induces the destabilization of the particles (data not shown). To enhance the contrast of images and to preserve the particle integrity during the drying phase, Lipidots were stained by uranyl acetate (negative staining) and then imaged by transmission electron microscopy (TEM), as reported by Arnould et al.41,42 More than 400 nonoverlapping particles were analyzed for each sample. The number-based size distributions obtained are shown in Figure 2, while the calculated mode, mean, and X10, X50, X90 values are reported in Table 1. All the Lipidots samples were characterized by one population of spherical particles in agreement with the AF4-MALS-DLS observations. The mode and mean diameter values calculated by TEM are significantly smaller than the values obtained by AF4-MALS-DLS and PTA analyses. Unfortunately a direct quantitative comparison between the three different techniques is not possible due to the difference in the nature of size parameters measured. PTA and AF4-DLS measure the Dh, which takes into account the hydrated surfactant shell of the particles, while TEM measures the 2D projected area of the particles after staining, which is usually smaller.
Figure 2.
TEM images and particle size distribution of the Lipidots formulations. TEM images of (A) Naked Lipidots, (B) Lipimage815, and (C) ICG Lipidots. Calculated differential and cumulative particle size distribution (%) of the (D) the Naked Lipidots, (E) the Lipimage 815, and (F) and the ICG Lipidots.
Monitoring of Size Changes in Lipidot Formulations: Stability Studies and Batch to Batch Variability
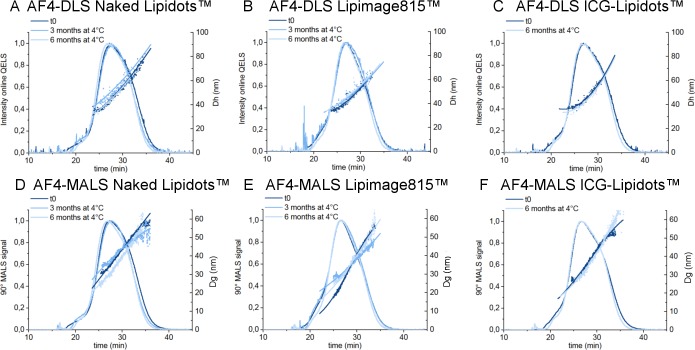
A medical nanoparticle formulation should be stable for multiple months in order to be stored before being used in the clinic. Monitoring changes in size and the leakage of the encapsulated active principle ingredient is critical, especially if the stock solution is stored in the liquid state, as in the case of the Lipidots. Aggregation or destabilization may occur during storage of the product and should be checked by robust methods, which should be able to detect small changes in particle size. In this study, the particle size distribution of the Lipidots was monitored for 6 months by batch mode DLS and AF4-MALS-DLS. Due to light and thermal sensitivity of dyes incorporated,31 all Lipidots solutions were stored in amber vials at 4 °C under argon atmosphere. All data related to colloidal stability of nanoparticles are shown in Figure S2B and in Figure 3, while all the values are summarized in Table S2. No significant changes were detected in elution profiles (e.g., in the retention time, Dh profile vs time) and in the mean particle diameters calculated by online DLS, suggesting that the size of the Lipidots solutions is stable for >6 months, confirming our previous findings by batch mode DLS.31,32 The study on longer time points is ongoing and will be carried out for >1 year.
Figure 3.
Stability over time. Elution profile and diameter by AF4 + online MALS and DLS detectors. On the top: AF4-DLS elution profile and hydrodynamic diameter of (a) the Naked Lipidots, (b) the Lipimage 815, and (c) and the ICG Lipidots. On the bottom: AF4-MALS elution profile and geometric diameter (Dg) of (d) the Naked Lipidots, (e) the Lipimage 815, and (f) and the ICG Lipidots. Dark blue: profile at t0, blue: profile after storage for 3 months at 4 °C, light blue: profile after storage for 6 months at 4 °C.
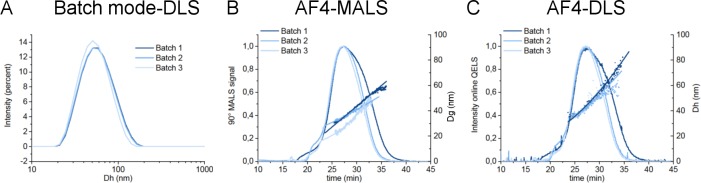
One of the key parameters for the successful translation of nanoparticles in medicinal products is the ability to produce multiple reproducible batches, having the same properties, including the particle size. Batch to batch variability in the manufacturing process has to be monitored from the early stages of development at the lab scale, since significant variation of the PSD between batches can result in different biological effects, as shown by Schädlich et al.43 In this work, multiple batches of each formulation were prepared with the same manufacturing protocol and analyzed by batch mode DLS and AF4-MALS-DLS to evaluate their PSD. No differences were detected in the elution profile of the batches analyzed and in the mean size values (see Table S2), except for one batch of Naked Lipidots which was characterized by a significantly smaller mean size (Dh and Dg), as reported in Figure 4 and in Table S2. Interestingly, batch mode DLS analysis failed to detect such small but significant difference of 5–10 nm. Thus, we can conclude that due to its low resolution, batch mode DLS is often not suitable to detect small changes of size in different batches, while on the contrary, AF4-MALS-DLS allows resolving small differences in PSD between batches. Therefore, AF4 can be considered a promising alternative technique to be adopted for routine quality control of the Med-NPs production. When an AF4 elution method has already being developed and tested for a specific system, batch to batch reproducibility can be easily performed by repetitive 1–2 h measurements, and also AF4-MALS-DLS can be considered an easy handling measurement, similarly to other chromatography techniques used in the field (RP-HPLC, size exclusion chromatography).
Figure 4.
Batch to batch variability of three lots of Naked Lipidots. Particle size distribution by (a) batch mode DLS, (b) AF4-MALS-DLS, and (c) AF4-MALS-DLS of three batches of Naked Lipidots. The batch 3 (light blue) presents a PSD shifted to smaller sizes if compared by the batch 1 (dark blue) and 2 (blue).
Changes of PSD in the Presence of Plasma Proteins
One of the potential routes of administration of Med-NPs, including the Lipidots, is intravenous injection. By this route of administration, nanoparticles are subject, rapidly, to interactions with biological elements included in blood, which affect their behavior in the body and their pharmacokinetics.3 The investigation of interactions of nanoparticle systems incubated in cell culture media and/or with plasma proteins, and modification of their properties due to NP–protein interaction, including particle size and shape, is crucial for regulatory approval since it can modulate cytotoxicity, immune response, and biodistribution of the NPs,44 as clearly mentioned in the last draft regulatory guidance of the FDA.40
Here, we have tested the suitability of AF4-MALS-DLS to detect small changes in Lipidots size after incubation in the presence of serum proteins and then compared the results with the evidence obtained by batch mode DLS and TEM imaging of the sample via negative staining. In order to simulate physiological relevant conditions, the Lipidots were incubated with pure fetal bovine serum (FBS) for an hour at 37 °C and then diluted in phosphate buffered saline immediately before the analysis. In this study only one incubation time point was selected, since the aim was to check the applicability of the SOP and not to perform an extensive study to investigate the behavior of Lipidots in complex biological media. However, to assess the behavior of Med-NPs in the presence of plasma, the coauthors advise to perform kinetic studies considering multiple time points, which should be chosen on a case by case basis and be representative of the specific Med-NP system studied.
The PSD by intensity obtained by batch mode DLS on the protein–NP mixtures are summarized in Table 2 and in Figure S2. Not surprisingly, the cumulant analysis was not suitable to analyze such a complex mixture; thus, the z-ave and PdI values became meaningless. The PSD by intensity calculated by batch mode DLS did not change significantly, while a peak associated with serum proteins appeared at 5–10 nm (Table 1 and Figure S2A). Thus, according to the batch DLS analysis (particle size distribution by intensity), no significant changes of the physical properties were induced when Lipidots were incubated with serum proteins. Interestingly, the AF4-DLS-MALS and the TEM analyses showed a slightly different picture from the batch mode DLS (Figure 5). The PSD detected by both TEM and AF4-DLS-MALS became significantly narrower in the presence of serum proteins, especially in the case of the Naked Lipidots. The width of the particle size distribution is reduced (Dmin and Dmax reported in Table 1), while the average size values do not significantly change.
Figure 5.
Effect of NP-protein interactions by TEM and AF4-MALS-DLS. AF4-DLS elution profiles: intensity of the scattered light of the online DLS detector and the calculated hydrodynamic diameter are reported for (a) the Naked Lipidots, (b) the Lipimage 815, and (c) and the ICG Lipidots. TEM images of the (d) the Naked Lipidots, (e) the Lipimage 815, and (f) and the ICG Lipidots after incubation with serum. Calculated differential and cumulative particle size distribution (%) of the (g) the Naked Lipidots, (h) the Lipimage 815, and (i) and the ICG Lipidots. Blue curve: NPs eluted alone; Orange curve: NPs + serum.
These results were unexpected, since an absorption of plasma proteins, such as lipoproteins, on the LNPs surface was expected to rather induce a small increase of PSD, instead of reducing the polydispersity of the samples. The reason why incubation with serum proteins significantly affected the PSD of Naked Lipidots is under investigation and is beyond the scope of this methodological paper. However, we can speculate that the size changes associated with the NP–protein interactions may be induced by the partial release of PEG and lecithin surfactant from the lipid shell. A small variation in the composition of the PEG/Lecithin superficial layer of the different formulations (or batches) could explain the different behavior of the Naked vs dye-loaded Lipidots detected by AF4-DLS-MALS analysis and TEM observations.
Importantly, from our evidence, we can conclude that it is not possible to rely on batch mode DLS measurements to detect small changes of the PSD of Med-NPs when they are dispersed in biologically relevant medium containing serum proteins, since the mixture of particles and proteins is too polydispersed for the low resolution batch mode DLS.45 In this context, AF4-MALS-DLS supported by TEM could provide accurate size information for polydispersed samples in physiological media. By fractionating the free protein and the NPs in the complex mixture before sizing the NPs, AF4-DLS-MALS allows us to successfully obtain accurate size information on small particle size modifications after protein binding in a complex protein–NP mixture.
Conclusions and Future Perspectives
One of our objectives was to raise consciousness of the pitfalls of batch mode DLS when applied to measure the PSD of medical nanoparticles, and to support the use of alternative approaches. In this context, we have shown the potential of AF4-MALS-DLS to measure the PSD of lipid-based nanoparticles of moderate polydispersity. The results were compared with those collected from batch mode DLS analysis. The capability of the two techniques to measure small difference in mean size, PSD, and PdI of the Lipidots (e.g., batch to batch consistency and stability) and to analyze changes in the PSD in complex biological media was assessed. The AF4-MALS-DLS technique was able to successfully measure the PSD of the Lipidots with high resolution, being able to detect batch to batch variability induced by the manufacturing procedure, as well as to find out changes in the PSD induced by plasma proteins in complex biological media, avoiding the misinterpretation of the results which may result from batch mode DLS analysis. Thus, we can conclude that AF4-MALS-DLS possesses the potential to become a future standard for synthesis optimization, quality control, and to monitor the stability of organic nanoparticle systems, including the lipid-based nanoparticle system investigated in this work. An ISO standard is now available (Application of field flow fractionation for characterization of nanomaterial contents, ISO/DTS 21362)5, as an evidence of the interests of regulatory and standardization bodies for this technique.
However, the users should be aware that AF4 fractionation requires trained experts and is less easy to handle than batch mode DLS: the ideal elution conditions for each Med-NPs are different, and method optimization can be moderately laborious and time-consuming. Due to the complexity of the AF4-MALS-DLS set up and to the method optimization required, EUNCL fosters the use of robust standard operating procedures, with proper quality controls to ensure the robustness and suitability of the method to characterize each specific nanoparticle system. SOPs like the one applied in this work are publicly available online (http://www.euncl.eu/) in order to support to the nanomedicine stakeholders, and the organization also offers their direct expertise to users. Importantly, we strongly encouraged users to support the obtained AF4-MALS-DLS results with additional complementary high resolution sizing techniques, including the direct visualization of the nanoparticles by electron microscopy.
Acknowledgments
This research was in part funded by the project EUNCL 2015, a European Union’s Horizon 2020 Research and Innovation Program under grant agreement No 654190. The authors thank Luigi Calzolai, Dora Mehn, and Jeffrey D. Clogston for contributing to the development of the standard operating procedures applied in this work. The authors also thank Sylvie Motellier for her support and fruitful discussion. LETI/DTBS is part of the Arcane Labex program, funded by the French National Research Agency (ARCANE project n° ANR-12-LABX-003) and supported by the French National Research Agency in the framework of the “Investissements d’avenir” program Glyco@Alps (ANR-15-IDEX-02). This work used the CEA LITEN nanosafety platform to perform the batch mode DLS and AF4-DLS-MALS measurement. The AF4-DLS-MALS instrument is part of the NanoID platform that was supported by the French Investissement d’Avenir ANR-10-EQPX-39. This work used the platforms of the Grenoble Instruct-ERIC Centre (ISBG; UMS 3518 CNRS-CEA-UGA-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB) for the preparation of the sample for EM observations. The IBS electron microscope facility is supported by the Rhône-Alpes Region, the Fonds FEDER, the Fondation Recherche Médicale (FRM), and the GIS-IBISA. The authors thank Guy Schoehn, Sylvie Motellier, and Jean-François Damlencourt for access to the platforms.
Glossary
List of Abbreviations
- AF4
asymmetric flow field flow fractionation
- DLS
dynamic light scattering
- EUNCL
European Nanomedicine Characterisation laboratory
- FBS
fetal bovine serum
- ICG
Indocyanine green
- LNPs
lipid-based nanoparticles
- MALS
multi angle light scattering
- Med-NPs
medical nanoparticles
- NCI-NCL
Nanotechnology Characterization Lab
- NPs
nanoparticles
- PBS
phosphate buffered saline
- PSD
particle size distribution
- PTA
particle tracking analysis
- QELS
Quasi-elastic light scattering
- SOP
standard operating procedure
- TEM
transmission electron microscopy
- fwhm
full width at half-maximum
Size Glossary
Diameter of an equivalent sphere measured by TEM
Equivalent spherical diameter corresponding to the diameter of a circle with the same area than the 2D projected area of the visualized particles by electron microscopy.
Hydrodynamic diameter (Dh) measured by DLS (batch and online) and PTA
Diameter of a sphere that possesses the same diffusion coefficient (derived by the Stokes–Einstein equation) as the particles in the applied conditions. It is indicative of the apparent size of the hydrated/solvated particles.
Polydispersity index (PdI) measured by batch DLS
dimensionless measure of the broadness of the size distribution calculated from a simple 2 parameter fit of the correlation data (cumulants analysis), as defined in the ISO13321:1996 E and ISO22412:2008.1
Geometric diameter (Dg) measured by MALS
the geometric diameter is defined as the root mean square distance of the object’s parts from its center of mass.
Mode
the mode of a particle size distribution is defined as the size value x at which the size distribution takes its maximum value.
Median value (X50), X10, and X90
the median value X50 is defined as the value where half of the population resides above this point and half resides below this point. Similarly X10 and X90 are defined as the values where 10% and 90% of the population respectively reside above these points.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.molpharmaceut.8b01033.
Table S1: Particle composition, surface charge, and dye loading characteristics. Table S2: Particle size distribution obtained by multiple techniques in all the conditions tested. Figure S1: Elution methods used in the AF4 analysis. Figure S2: Stability by batch mode DLS. Figure S3: AF4 reproducibility of sequential injections. Figure S4: PSD of Naked Lipidots and Lipimage815 measured by PTA. Figure S5: Elution profile of free serum proteins (FBS) and of Naked Lipidot ± FBS (PDF)
Author Contributions
FC wrote the paper and performed the DLS and AF4-MALS-DLS experiments and associated data analysis. ER synthesized the nanoparticles analyzed in this study. MB, WLL, and AA performed the TEM experiments and the associated data analysis. APM performed PTA measurements and the associated data analysis. ACC and FC conceived the project. IT contributed to discussion on experiments and presentation. All the authors participated in the development of the scientific work, contributed to discussion on experiments and presentation, and approved the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- ISO22412:2018 Particle Size Analysis — Dynamic Light Scattering (DLS). [Google Scholar]
- D’Mello S. R.; Cruz C. N.; Chen M.-L.; Kapoor M.; Lee S. L.; Tyner K. M. The Evolving Landscape of Drug Products Containing Nanomaterials in the United States. Nat. Nanotechnol. 2017, 12 (6), 523–529. 10.1038/nnano.2017.67. [DOI] [PubMed] [Google Scholar]
- Coty J.-B.; Vauthier C. Characterization of Nanomedicines: A Reflection on a Field under Construction Needed for Clinical Translation Success. J. Controlled Release 2018, 275, 254–268. 10.1016/j.jconrel.2018.02.013. [DOI] [PubMed] [Google Scholar]
- Gioria S.; Caputo F.; Urbán P.; Maguire C. M.; Bremer-Hoffmann S.; Prina-Mello A.; Calzolai L.; Mehn D. Are Existing Standard Methods Suitable for the Evaluation of Nanomedicines: Some Case Studies. Nanomedicine 2018, 13 (5), 539–554. 10.2217/nnm-2017-0338. [DOI] [PubMed] [Google Scholar]
- Halamoda-Kenzaoui B.; Holzwarth U.; Roebben G.; Bogni A.; Bremer-Hoffmann S.. Mapping of the Available Standards against the Regulatory Needs for Nanomedicines. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2019, e1531. 10.1002/wnan.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner K. M.; Zou P.; Yang X.; Zhang H.; Cruz C. N.; Lee S. L. Product Quality for Nanomaterials: Current U.S. Experience and Perspective: Product Quality for Nanomaterials. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015, 7 (5), 640–654. 10.1002/wnan.1338. [DOI] [PubMed] [Google Scholar]
- De Jong W. H.; Hagens W. I.; Krystek P.; Burger M. C.; Sips A. J. A. M.; Geertsma R. E. Particle Size-Dependent Organ Distribution of Gold Nanoparticles after Intravenous Administration. Biomaterials 2008, 29 (12), 1912–1919. 10.1016/j.biomaterials.2007.12.037. [DOI] [PubMed] [Google Scholar]
- Alexis F.; Pridgen E.; Molnar L. K.; Farokhzad O. C. Factors Affecting the Clearance and Biodistribution of Polymeric Nanoparticles. Mol. Pharmaceutics 2008, 5 (4), 505–515. 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitragotri S.; Lahann J. Physical Approaches to Biomaterial Design. Nat. Mater. 2009, 8 (1), 15–23. 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire C. M.; Rösslein M.; Wick P.; Prina-Mello A. Characterisation of Particles in Solution – a Perspective on Light Scattering and Comparative Technologies. Sci. Technol. Adv. Mater. 2018, 19 (1), 732–745. 10.1080/14686996.2018.1517587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzolai L.et al. EUNCLSOP-PCC21. Measuring the NP Stability against Aggregation as a Function of Time and Buffers with Batch-Mode DLS. http://www.euncl.eu/about-us/assay-cascade/PDFs/PCC/EUNCL-PCC-021.pdf?m=1468937870. Last visited in December 2018.
- Mehn D.; Caputo F.; Rösslein M.; Calzolai L.; Saint-Antonin F.; Courant T.; Wick P.; Gilliland D. Larger or More? Nanoparticle Characterisation Methods for Recognition of Dimers. RSC Adv. 2017, 7 (44), 27747–27754. 10.1039/C7RA02432K. [DOI] [Google Scholar]
- Gollwitzer C.; Bartczak D.; Goenaga-Infante H.; Kestens V.; Krumrey M.; Minelli C.; Pálmai M.; Ramaye Y.; Roebben G.; Sikora A.; et al. A Comparison of Techniques for Size Measurement of Nanoparticles in Cell Culture Medium. Anal. Methods 2016, 8 (26), 5272–5282. 10.1039/C6AY00419A. [DOI] [Google Scholar]
- Podzimek S.Light Scattering, Size Exclusion Chromatography and Asymmetric Flow Field Flow Fractionation: Powerful Tools for the Characterization of Polymers, Proteins and Nanoparticles; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. 10.1002/9780470877975. [DOI] [Google Scholar]
- Varenne F.; Makky A.; Gaucher-Delmas M.; Violleau F.; Vauthier C. Multimodal Dispersion of Nanoparticles: A Comprehensive Evaluation of Size Distribution with 9 Size Measurement Methods. Pharm. Res. 2016, 33 (5), 1220–1234. 10.1007/s11095-016-1867-7. [DOI] [PubMed] [Google Scholar]
- Wagner M.; Holzschuh S.; Traeger A.; Fahr A.; Schubert U. S. Asymmetric Flow Field-Flow Fractionation in the Field of Nanomedicine. Anal. Chem. 2014, 86 (11), 5201–5210. 10.1021/ac501664t. [DOI] [PubMed] [Google Scholar]
- Yohannes G.; Jussila M.; Hartonen K.; Riekkola M.-L. Asymmetrical Flow Field-Flow Fractionation Technique for Separation and Characterization of Biopolymers and Bioparticles. J. Chromatogr. A 2011, 1218 (27), 4104–4116. 10.1016/j.chroma.2010.12.110. [DOI] [PubMed] [Google Scholar]
- Zattoni A.; Rambaldi D. C.; Reschiglian P.; Melucci M.; Krol S.; Garcia A. M. C.; Sanz-Medel A.; Roessner D.; Johann C. Asymmetrical Flow Field-Flow Fractionation with Multi-Angle Light Scattering Detection for the Analysis of Structured Nanoparticles. J. Chromatogr. A 2009, 1216 (52), 9106–9112. 10.1016/j.chroma.2009.06.037. [DOI] [PubMed] [Google Scholar]
- Contado C. Field Flow Fractionation Techniques to Explore the “Nano-World.. Anal. Bioanal. Chem. 2017, 409 (10), 2501–2518. 10.1007/s00216-017-0180-6. [DOI] [PubMed] [Google Scholar]
- Till U.; Gibot L.; Mingotaud C.; Vicendo P.; Rols M.-P.; Gaucher M.; Violleau F.; Mingotaud A.-F. Self-Assembled Polymeric Vectors Mixtures: Characterization of the Polymorphism and Existence of Synergistic Effects in Photodynamic Therapy. Nanotechnology 2016, 27 (31), 315102. 10.1088/0957-4484/27/31/315102. [DOI] [PubMed] [Google Scholar]
- Till U.; Gaucher-Delmas M.; Saint-Aguet P.; Hamon G.; Marty J.-D.; Chassenieux C.; Payré B.; Goudounèche D.; Mingotaud A.-F.; Violleau F. Asymmetrical Flow Field-Flow Fractionation with Multi-Angle Light Scattering and Quasi-Elastic Light Scattering for Characterization of Polymersomes: Comparison with Classical Techniques. Anal. Bioanal. Chem. 2014, 406 (30), 7841–7853. 10.1007/s00216-014-7891-8. [DOI] [PubMed] [Google Scholar]
- Mehn D.et al. EUNCLPCC-022. Measurement of Particle Size Distribution of Protein Binding, of Mean Molecular Weight of Polymeric NP Components, Study of Batch to Batch Reproducibility, and Study of Release of Free Coating from NP Surface by FFF- MALS. http://www.euncl.eu/about-us/assay-cascade/PDFs/PCC/EUNCL-PCC-022.pdf?m=1468937868&. Last visited in December 2018.
- ISO TS 21362:2018(E). Nanotechnologies-Analysis of nano-Objects using asymmetrical-flow and centrifugal field flow fractionation.
- Ingallina C.; Rinaldi F.; Bogni A.; Ponti J.; Passeri D.; Reggente M.; Rossi M.; Kinsner-Ovaskainen A.; Mehn D.; Rossi F.; et al. Niosomal Approach to Brain Delivery: Development, Characterization and in Vitro Toxicological Studies. Int. J. Pharm. 2016, 511 (2), 969–982. 10.1016/j.ijpharm.2016.08.002. [DOI] [PubMed] [Google Scholar]
- Mehn D.; Iavicoli P.; Cabaleiro N.; Borgos S. E.; Caputo F.; Geiss O.; Calzolai L.; Rossi F.; Gilliland D. Analytical Ultracentrifugation for Analysis of Doxorubicin Loaded Liposomes. Int. J. Pharm. 2017, 523 (1), 320–326. 10.1016/j.ijpharm.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquart A.; Kéramidas M.; Vollaire J.; Boisgard R.; Pottier G.; Rustique E.; Mittler F.; Navarro F. P.; Boutet J.; Coll J.-L.; et al. LipImageTM 815: Novel Dye-Loaded Lipid Nanoparticles for Long-Term and Sensitive in Vivo near-Infrared Fluorescence Imaging. J. Biomed. Opt. 2013, 18 (10), 101311. 10.1117/1.JBO.18.10.101311. [DOI] [PubMed] [Google Scholar]
- Navarro F. P.; Berger M.; Guillermet S.; Josserand V.; Guyon L.; Neumann E.; Vinet F.; Texier I. Lipid Nanoparticle Vectorization of IndoCyanine Green Improves Fluorescence Imaging for Tumor Diagnosis and Lymph Node Resection. J. Biomed. Nanotechnol. 2012, 8 (5), 730–741. 10.1166/jbn.2012.1430. [DOI] [PubMed] [Google Scholar]
- Mérian J.; Boisgard R.; Bayle P.-A.; Bardet M.; Tavitian B.; Texier I. Comparative Biodistribution in Mice of Cyanine Dyes Loaded in Lipid Nanoparticles. Eur. J. Pharm. Biopharm. 2015, 93, 1–10. 10.1016/j.ejpb.2015.03.019. [DOI] [PubMed] [Google Scholar]
- Delmas T.; Fraichard A.; Bayle P.-A.; Texier I.; Bardet M.; Baudry J.; Bibette J.; Couffin A.-C. Encapsulation and Release Behavior from Lipid Nanoparticles: Model Study with Nile Red Fluorophore. J. Colloid Sci. Biotechnol. 2012, 1 (1), 16–25. 10.1166/jcsb.2012.1010. [DOI] [Google Scholar]
- Delmas T.; Couffin A.-C.; Bayle P. A.; Crécy F. de; Neumann E.; Vinet F.; Bardet M.; Bibette J.; Texier I. Preparation and Characterization of Highly Stable Lipid Nanoparticles with Amorphous Core of Tuneable Viscosity. J. Colloid Interface Sci. 2011, 360 (2), 471–481. 10.1016/j.jcis.2011.04.080. [DOI] [PubMed] [Google Scholar]
- Gravier J.; Navarro F. P.; Delmas T.; Mittler F.; Couffin A.-C.; Vinet F.; Texier I. Lipidots: Competitive Organic Alternative to Quantum Dots for in Vivo Fluorescence Imaging. J. Biomed. Opt. 2011, 16 (9), 096013. 10.1117/1.3625405. [DOI] [PubMed] [Google Scholar]
- Varache M.; Escudé M.; Laffont C.; Rustique E.; Couffin A.-C. Development and Validation of an HPLC-Fluorescence Method for the Quantification of IR780-Oleyl Dye in Lipid Nanoparticles. Int. J. Pharm. 2017, 532 (2), 779–789. 10.1016/j.ijpharm.2017.06.019. [DOI] [PubMed] [Google Scholar]
- Delmas T.; Piraux H.; Couffin A.-C.; Texier I.; Vinet F.; Poulin P.; Cates M. E.; Bibette J. How To Prepare and Stabilize Very Small Nanoemulsions. Langmuir 2011, 27 (5), 1683–1692. 10.1021/la104221q. [DOI] [PubMed] [Google Scholar]
- Calzolai L.et al. EUNCLPCC001. Measuring the Size of Nanoparticles Using Batch Mode Dynamic Light Scattering. http://www.euncl.eu/about-us/assay-cascade/PDFs/Prescreening/EUNCL-PCC-001.pdf?m=1468937875&. last visited in December 2018.
- Arnaud C.-A.; Effantin G.; Vivès C.; Engilberge S.; Bacia M.; Boulanger P.; Girard E.; Schoehn G.; Breyton C.. Bacteriophage T5 Tail Tube Structure Suggests a Trigger Mechanism for Siphoviridae DNA Ejection. Nat. Commun. 2017, 8 ( (1), ), 10.1038/s41467-017-02049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire C.EUNCLPCC023. Measuring the Size and Concentration of Nanoparticles Using Particle Tracking Analysis (PTA). http://www.euncl.eu/about-us/assay-cascade/PDFs/PCC/EUNCL_PCC_023.pdf?m=1526712237&. Last visited in December 2018.
- Cabon Q.; Sayag D.; Texier I.; Navarro F.; Boisgard R.; Virieux-Watrelot D.; Ponce F.; Carozzo C. Evaluation of Intraoperative Fluorescence Imaging–Guided Surgery in Cancer-Bearing Dogs: A Prospective Proof-of-Concept Phase II Study in 9 Cases. Transl. Res. 2016, 170, 73–88. 10.1016/j.trsl.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Sayag D.; Cabon Q.; Texier I.; Navarro F. P.; Boisgard R.; Virieux-Watrelot D.; Carozzo C.; Ponce F. Phase-0/Phase-I Study of Dye-Loaded Lipid Nanoparticles for near-Infrared Fluorescence Imaging in Healthy Dogs. Eur. J. Pharm. Biopharm. 2016, 100, 85–93. 10.1016/j.ejpb.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Bunjes H.; Unruh T. Characterization of Lipid Nanoparticles by Differential Scanning Calorimetry, X-Ray and Neutron Scattering☆. Adv. Drug Delivery Rev. 2007, 59 (6), 379–402. 10.1016/j.addr.2007.04.013. [DOI] [PubMed] [Google Scholar]
- FDA/CDER Drug Products, Including Biological Products, That Contain Nanomaterials - Guidance for Industry. 2017. https://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM588857.pdf. Last visited in December 2018. [Google Scholar]
- Arnould A.; Bacia M.; Caputo F.; Couffin A.-C.; Gallet B.; Matei C.; Soulas R.; Damlencourt J.-F.. Comparison of Sample Preparations for TEM Observations of Lipid Nanoparticles (Lipidots). In European Microscopy Congress 2016: Proceedings; European Microscopy Society, Ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2016; pp 61–62. 10.1002/9783527808465.EMC2016.5223. [DOI] [Google Scholar]
- Tezgel Ö.; Szarpak-Jankowska A.; Arnould A.; Auzély-Velty R.; Texier I. Chitosan-Lipid Nanoparticles (CS-LNPs): Application to SiRNA Delivery. J. Colloid Interface Sci. 2018, 510, 45–56. 10.1016/j.jcis.2017.09.045. [DOI] [PubMed] [Google Scholar]
- Schädlich A.; Rose C.; Kuntsche J.; Caysa H.; Mueller T.; Göpferich A.; Mäder K. How Stealthy Are PEG-PLA Nanoparticles? An NIR In Vivo Study Combined with Detailed Size Measurements. Pharm. Res. 2011, 28 (8), 1995–2007. 10.1007/s11095-011-0426-5. [DOI] [PubMed] [Google Scholar]
- Corbo C.; Molinaro R.; Parodi A.; Toledano Furman N. E.; Salvatore F.; Tasciotti E. The Impact of Nanoparticle Protein Corona on Cytotoxicity, Immunotoxicity and Target Drug Delivery. Nanomedicine 2016, 11 (1), 81–100. 10.2217/nnm.15.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondow N.; Brydson R.; Wang P.; Holton M. D.; Brown M. R.; Rees P.; Summers H. D.; Brown A.. Quantitative Characterization of Nanoparticle Agglomeration within Biological Media. J. Nanopart. Res. 2012, 14 ( (7), ), 10.1007/s11051-012-0977-3. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.