Summary:
In many eukaryotes the centromere is epigenetically specified and not strictly defined by sequence. In contrast, budding yeast has a specific 125bp sequence required for kinetochore function. Despite the difference in centromere specification, budding yeast and multicellular eukaryotic centromeres contain a highly conserved histone H3 variant, CENP-A. The localization of budding yeast CENP-A, Cse4, requires the centromere DNA binding components, which are not conserved in multicellular eukaryotes. Here, we report that Cse4 localizes and functions at a synthetic kinetochore assembly site that lacks centromere sequence. The outer kinetochore Dam1-DASH and inner kinetochore CBF3 complexes are required for Cse4 localization to that site. Furthermore, the natural kinetochore also requires the outer kinetochore proteins for full Cse4 localization. Our results suggest that Cse4 localization at a functional kinetochore does not require the recognition of a specific DNA sequence by the CBF3 complex; rather, its localization depends on stable interactions among kinetochore proteins.
Keywords: kinetochore, centromere, Cse4, CENP-A, synthetic kinetochore
Introduction
The accurate segregation of chromosomes depends on the kinetochore, a large protein complex that links spindle microtubules to chromatin. Kinetochores assemble on centromeres and are regulated to ensure that sister chromatids attach to opposite spindle poles in metaphase and travel along depolymerizing microtubules in anaphase (Biggins, 2013). Many kinetochore proteins are conserved among eukaryotic organisms; however, the underlying centromere sequence is highly variable. Centromeres of multicellular eukaryotes are epigenetically maintained and embedded in highly repetitive AT-rich DNA lacking strict sequence identity (De Rop et al., 2012). In contrast, the budding yeast centromere is a defined ~125-bp sequence consisting of three elements (CDEI, CDEII, and CDEIII), sufficient for centromere specification (Biggins, 2013).
Despite the intrinsic differences between the epigenetically maintained centromeres of multicellular eukaryotes and sequence-specified centromeres of budding yeast, both types of centromeres incorporate a histone H3 variant called CENP-A (De Rop et al., 2012). The incorporation of nucleosomes with CENP-A (Cse4 in budding yeast) is required for proper kinetochore assembly. CENP-A integrates at the centromere with the assistance of a histone chaperone, known as HJURP in vertebrates and Scm3 in budding and fission yeast (De Rop et al., 2012). HJURP/Scm3 has a highly conserved domain that binds CENP-A. In budding yeast, the inner kinetochore CBF3 complex facilitates the deposition of Cse4 at the centromere. A mutation in CDEIII eliminates Cse4 localization, suggesting that the CEN sequence is needed for Cse4 deposition (Ortiz et al., 1999). Since the CBF3 complex binds CDEIII and also binds Scm3, the current model is that CBF3 binds to the centromere and recruits Scm3, which deposits Cse4 in centromere chromatin (Camahort et al., 2007; Lechner and Carbon, 1991).
Here, we address the question of whether recognition of the centromere by the CBF3 complex is essential for Cse4 localization and function. Others and we previously demonstrated that recruiting a microtubule-binding complex to DNA allows the assembly of a functional kinetochore at a non-centromeric locus (Kiermaier et al., 2009; Lacefield et al., 2009). By fusing Ask1, a member of the Dam1-DASH microtubule-binding complex, to the lactose repressor (Ask1-LacI) and by placing tandem repeats of the lactose operator (LacO) on a chromosome, we created a ‘synthetic kinetochore’; Ask1-LacI binds LacO and recruits other kinetochore proteins (Lacefield et al., 2009). The synthetic kinetochore can replace an endogenous kinetochore on a chromosome and segregate sister chromatids in mitosis. Using the synthetic kinetochore, we tested whether Cse4 incorporates into a non-centromeric DNA locus.
We find that Cse4 localizes and functions at the synthetic kinetochore, independent of centromere sequence. The full localization of Cse4 to the synthetic kinetochore requires the Dam1-DASH and CBF3 complexes, suggesting that binding of CBF3 to CDEIII is not required for Cse4 incorporation. Rather, recruitment of CBF3 through kinetochore protein interactions allows Cse4 incorporation. The Dam1-DASH complex is also required for full Cse4 localization at the natural centromere, suggesting that outer microtubule-binding components stabilize the inner kinetochore for Cse4 localization. Our results suggest that the localization and function of Cse4 at the centromere is not strictly specified by centromere sequence but relies on the interactions of kinetochore protein complexes.
Results
Cse4 localizes to a synthetic kinetochore assembly site, independent of centromere sequence.
We investigated whether Cse4 is recruited to a non-centromeric sequence, the synthetic kinetochore assembly site at the LacO array. We performed chromatin immunoprecipitation (ChIP), comparing two strains, one expressing Ask1-LacI (assembling a synthetic kinetochore at LacO) and one that does not express Ask1-LacI (no synthetic kinetochore). Both strains have 256 repeats of LacO integrated at the LEU2 locus, which serves as the site of kinetochore assembly since Ask1-LacI binds LacO and recruits kinetochore proteins (Lacefield et al., 2009). Cse4 is internally tagged with GFP (GFP-Cse4), making a fully functional protein that can be immunoprecipitated with anti-GFP antibody (Chen et al., 2000). Due to the repetitive sequence of the LacO array, we used primers that amplify a unique region at one end of the array to avoid the amplification of different numbers of repeats, which interferes with quantification (Suppl. Fig. S1A). As a positive control, we used primers that amplify CEN15 since GFP-Cse4 localizes at centromeres. As a negative control, we used primers that amplify ADE2, a locus in which Cse4 is not enriched (Fig. 1A). The minimal level of Cse4 at ADE2 measures non-specific binding. As expected, there was an enrichment of Cse4 at CEN15 when compared to the ADE2 locus (Suppl. Fig. S1B).
Figure 1. Cse4 localizes to the site of synthetic kinetochore assembly.
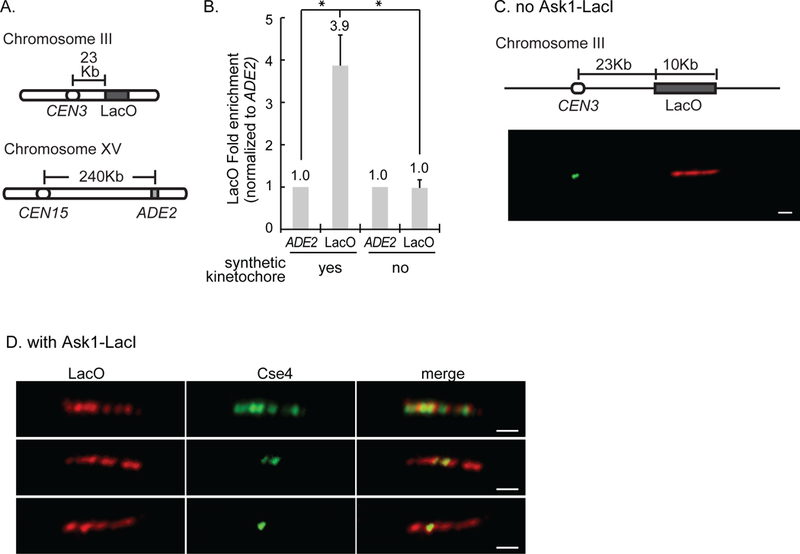
A) The three loci analyzed by ChIP. B) Quantification of ChIP-qPCR averaged from three experiments ± S.D, showing the LacO enrichment over non-specific binding at the ADE2 locus. * represents statistical significance. C, D) Representative images of chromatin fiber immunofluorescence. Strains have the LacO array and express GFP-Cse4 and mCherry-LacI. Cse4 and LacI were detected using anti-GFP and anti-mCherry antibodies. Scale bar 1μm. C) Chromatin fiber from a strain without Ask1-LacI showing Cse4 at CEN3 and LacO ~23Kb away. D) Images of chromatin fibers from a strain with Ask1-LacI with Cse4 along the entire array (top), part of the array (middle), and in one focus (bottom).
ChIP-qPCR showed a 3.9-fold increase of Cse4 at LacO in cells with the synthetic kinetochore compared to cells without the synthetic kinetochore (Fig. 1B). The enrichment of GFP-Cse4 at LacO versus ADE2 is statistically significant (t test, p<.0001). The level of Cse4 at LacO shown is likely an underestimate due to the necessity of having ChIP primers that amplify one end of the LacO array. In the strain without the synthetic kinetochore, the level of LacO in the IP is not statistically different from that of ADE2 (t test, p = 0.8), suggesting that only background levels of Cse4 are present at LacO (Fig. 1B). In summary, the ChIP demonstrates that Cse4 localizes to a non-centromeric sequence, LacO, when the synthetic kinetochore is present.
To further test Cse4 localization to the site of synthetic kinetochore assembly, we performed chromatin fiber immunofluorescence. Chromatin fibers were spread from strains with and without the synthetic kinetochore. Both strains have 256 repeats of LacO ~23kb from CEN3 and express GFP-Cse4 and mCherry-LacI. Immunostaining with anti-GFP and anti-mCherry antibodies detect Cse4 and LacO, respectively. Chromatin fibers from a strain without the synthetic kinetochore showed a stretch of mCherry signal representing LacO and Cse4 positioned at the centromere (Fig. 1C). From a strain with the synthetic kinetochore, 51% of LacO arrays showed Cse4 at the array. We are likely under-estimating the percent of LacO arrays with Cse4 due to loss of some nucleosomes in the preparation of stretched chromatin. Of those LacO arrays with Cse4 signal, Cse4 was along the array, in multiple foci, or in one focus (n= 64) (Fig. 1D). In contrast, in a strain without the synthetic kinetochore, only 6% of LacO arrays showed Cse4 at LacO, mostly in one focus, likely representing immunofluorescence background (n= 59). These results demonstrate that Cse4 localizes to a non-centromeric sequence with an assembled synthetic kinetochore.
Cse4 functions at the synthetic kinetochore
If Cse4 functions at the synthetic kinetochore, a Cse4 loss-of-function mutant would disrupt synthetic kinetochore activity. We characterized a temperature-sensitive allele of Cse4, with a Myc tag at the C-terminus (cse4-Myc), to determine if the C-terminal tag creates a conditional loss-of-function allele of Cse4. The temperature sensitivity of cse4-Myc cells can be rescued by GFP-Cse4, with GFP internally integrated into Cse4 (Chen et al., 2000; Wisniewski et al., 2014) (Fig. S2A). At 37°C, cse4-Myc cells arrest with large buds. The arrest depends on spindle checkpoint protein Mad2, suggesting that cse4-Myc cells have defective kinetochore-microtubule attachments and signal the spindle checkpoint (Fig. S2B). By ChIP using anti-Myc antibody, Cse4-Myc localizes to CEN15 at 25°C and 37°C, albeit at somewhat reduced levels at 37°C (Fig. S2C). The Cse4 protein levels are similar in cse4-Myc, GFP-Cse4, and CSE4 cells at 25°C and 37°C (Fig S2D). Therefore, the mutant phenotype is due to altered protein function and not decreased protein levels. We conclude that cse4-Myc is a conditional partial loss-of-function allele of CSE4 that can be used to test the role of Cse4 at the synthetic kinetochore.
To determine if Cse4 functions at the synthetic kinetochore, we first analyzed growth of cse4-Myc cells dependent on the synthetic kinetochore for segregation of a chromosome. As previously described, to make chromosome III segregation dependent on the synthetic kinetochore, we integrated the GAL1,10 promoter upstream of CEN3 (GAL-CEN3) (Lacefield et al., 2009). CEN3 is repressed when cells are grown in galactose medium (CEN3 off, Fig. 2A) (Hill and Bloom, 1987). Our previous experiments showed that cells with the synthetic kinetochore grow in galactose medium, but cells without the synthetic kinetochore grow poorly (Lacefield et al., 2009). We tested whether Cse4-Myc disrupted growth of cells dependent on the synthetic kinetochore for survival. We spotted dilutions of a yeast culture onto glucose and galactose plates and grew them at 25, 30, and 37°C. On glucose plates, the natural kinetochore is on and cse4-Myc cells grow similarly to CSE4 cells at 25 and 30°C but grow worse at 37°C, suggesting that Cse4-Myc does not interfere with the function of the natural kinetochore at 25 and 30°C (Fig. 2B, S2A). On galactose plates, the natural kinetochore is off and Cse4-Myc localizes to LacO when the synthetic kinetochore is present (Fig. S2E). The cse4-Myc cells grow worse than CSE4 cells at 25, 30, and 37°C when segregation of chromosome III depends on the synthetic kinetochore (Fig 2B). The growth-defect of cse4-Myc cells compared to CSE4 cells at 25 and 30°C on galactose plates suggests that functional Cse4 is required for the synthetic kinetochore to segregate a chromosome.
Figure 2. Cse4-Myc disrupts synthetic kinetochore function.
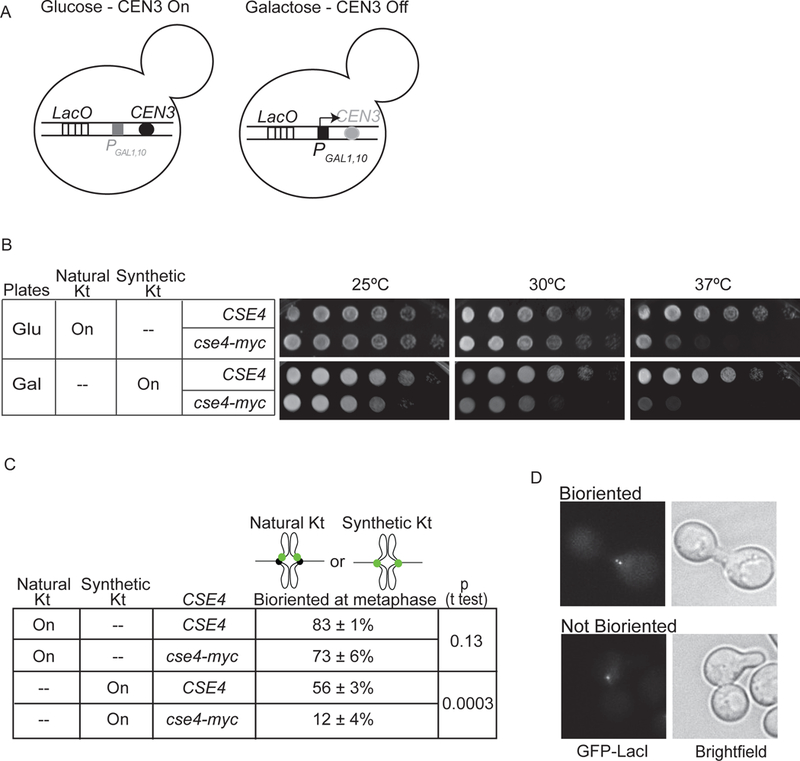
A) Depiction of natural kinetochore function in cells with the GAL1,10 promoter at CEN3 (PGAL1,10CEN3) and grown in glucose versus galactose-containing medium. B) Serial dilutions of cultures spotted on glucose and galactose plates at 25, 30, and 37°C. C) The percent biorientation of natural or synthetic kinetochores in metaphase-arrested cells grown at 37°C (average of 3 experiments ± S.D., 100 cells counted per experiment). D) Images show a cse4-Myc cell with (top) and without (bottom) bioriented synthetic kinetochores.
To determine if Cse4-Myc directly alters synthetic kinetochore function, we analyzed the synthetic kinetochore biorientation at metaphase. We monitored GAL-CEN3 LacO cells that express both Ask1-LacI and GFP-LacI to mark a synthetic kinetochore with GFP. We previously found that the synthetic kinetochore biorients at metaphase, as shown by the separation of two sister GFP foci ~0.5 μm apart (Lacefield et al., 2009). To determine if Cse4-Myc disrupts biorientation, we allowed CSE4 and cse4-Myc cells to reach metaphase arrest (by depletion of Cdc20) in galactose to turn off CEN3. We counted the percent of cells with separated GFP foci at 37°C (Fig. 2C,D). With Cse4, 56 ± 3% of cells bioriented synthetic kinetochores. In contrast, only 12 ± 4% of cse4-Myc cells bioriented synthetic kinetochores. We monitored biorientation of a natural kinetochore in cells with a LacO array at CEN15 and expressing GFP-LacI and found that 83 ± 1% of CSE4 cells and 73 ± 6% of cse4-Myc cells had bioriented natural kinetochores at 37°C (Fig. 2C). Our data suggest that Cse4-Myc more severely disrupts synthetic kinetochore function than natural kinetochore function. We conclude that Cse4 is required for synthetic kinetochore function.
The CBF3 and Dam1-DASH complexes are important for Cse4 localization at the synthetic kinetochore.
The current model for targeting Cse4 to the centromere depends on the interaction of the inner kinetochore CBF3 complex with the CDEIII DNA element. With CBF3 bound to CDEIII, the histone chaperone Scm3 binds to CBF3 component Ndc10 and mediates the deposition of Cse4 at the centromere (Camahort et al., 2007) (Fig. 3A). Despite the absence of CDEIII at the synthetic kinetochore assembly site, we tested the role of the CBF3 complex in Cse4 localization. We first analyzed Ndc10 localization to the synthetic kinetochore. We tagged Ndc10 with GFP and performed ChIP-qPCR with antibody against GFP. There was a 9-fold enrichment of Ndc10 at LacO in cells with the synthetic kinetochore compared to cells without the synthetic kinetochore (Fig. 3B). This result shows that Ndc10 does not bind LacO independently, but localizes to LacO through the assembled synthetic kinetochore.
Figure 3). The CBF3 complex and the Dam1-DASH complex are required for Cse4 localization at LacO.
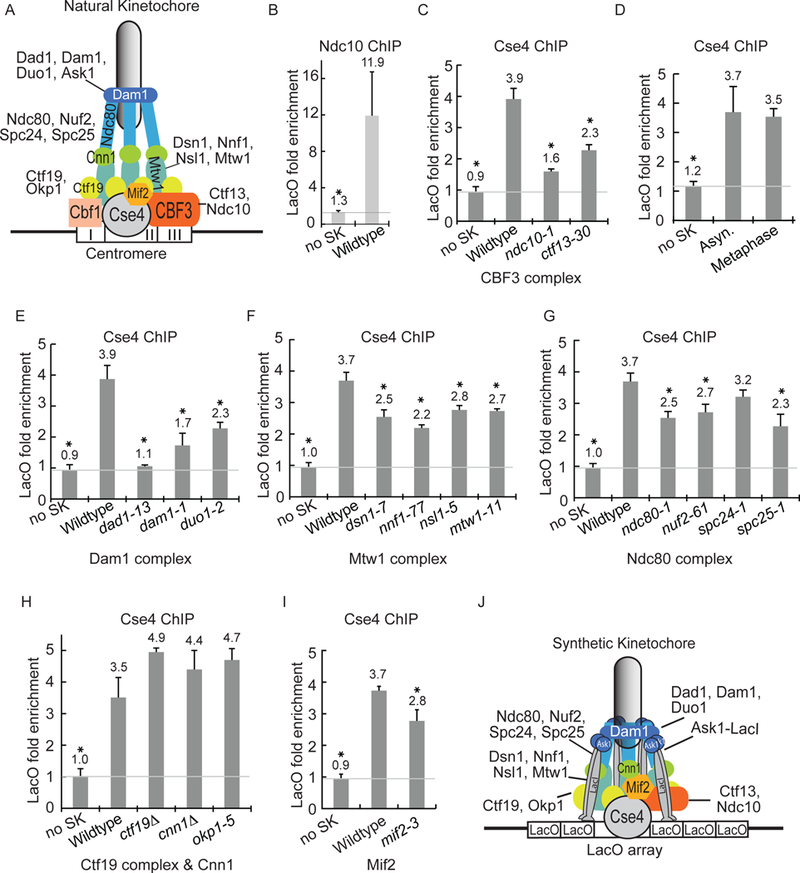
A) Model of the natural kinetochore. B-I) The quantification of ChIP-qPCR averaged from at least three experiments ± S.D. * represents statistical significance (t test, p< .05). B) The ChIP of Ndc10-GFP with anti-GFP antibody. C) The ChIP of GFP-Cse4 with anti-GFP antibody, comparing asynchronous cells with kinetochore mutations at 37°C. D) The ChIP of GFP-Cse4 with anti-GFP antibody, comparing asynchronous to metaphase-enriched cells. E-I) The ChIP of GFP-Cse4 with anti-GFP antibody, comparing asynchronous cells with kinetochore mutations at 37°C. J) Model of the synthetic kinetochore.
We analyzed the localization of Cse4 to LacO in ndc10–1 and ctf13–30 temperature-sensitive mutants. If the kinetochore protein has a role in Cse4 localization at LacO, Cse4 localization should be reduced in the mutant strains. At 37°C, Cse4 localization at LacO is reduced 77% in ndc10–1 cells and 53% in ctf13–30 cells compared to wildtype cells (Fig. 3C). These results suggest that the CBF3 complex, which normally binds CDEIII, is important for Cse4 localization at a synthetic kinetochore assembly site, even in the absence of the CDEIII element.
We next tested the role of other kinetochore components in Cse4 localization to the synthetic kinetochore. Since some kinetochore mutations activate the spindle checkpoint and cause an enrichment of metaphase cells, we compared the level of Cse4 localization in a wildtype asynchronously growing strain to that of a MET25 promoted CDC20 strain, which arrests in metaphase with methionine addition due to the depletion of Cdc20. The results showed that asynchronous and metaphase-enriched cultures had similar levels of GFP-Cse4 at LacO (Figure 3D).
Since Ask1 is a component of the Dam1-DASH outer kinetochore complex, we tested whether recruitment of other Dam1-DASH components was important for Cse4 localization to LacO. In the dad1–13 strain, Cse4 levels at LacO were decreased 93% compared to the wildtype strain (Fig. 3E). In the dam1–1 and the duo1–2 strains, Cse4 levels at LacO were decreased 73% and 53% relative to that of a wildtype strain, respectively. The difference in the reduction of Cse4 in the dad1–13, dam1–1, and duo1–2 mutants likely reflects the severity of the temperature-sensitive mutations. To ensure that the decreased Cse4 localization was not due to differences in cell cycle stage, we repeated the ChIP in cells enriched in metaphase (due to the depletion of Cdc20). The metaphase-enriched cells mutant for Dam1-DASH components also show a significantly decreased level of Cse4 at LacO when compared to wildtype cells (Fig. S3A).
Since dad1–13 mutant cells have a defect in chromosome-microtubule attachments (Janke et al., 2002), we tested whether the loss of microtubule attachments causes decreased Cse4 localization at the synthetic kinetochore. We added microtubule depolymerizing drugs to dad1–13 and wildtype cells (Fig. S3B). The level of Cse4 at LacO was not reduced in wildtype cells with nocodazole, suggesting that disruption of microtubules does not decrease Cse4 levels at LacO (Fig. S3A,B). With nocodazole, dad1–13 cells have an 88% reduction of Cse4 levels at LacO compared to wildtype cells. These results suggest that the Dam1-DASH complex is required for Cse4 localization at LacO, likely due to its role in recruiting other kinetochore proteins to the synthetic kinetochore.
We tested proteins in two highly conserved microtubule-binding kinetochore complexes, the Mis12/Mtw1 and Ndc80 complexes, for their role in Cse4 localization at the synthetic kinetochore assembly site. Cse4 localization at LacO was reduced between 33 to 56% in temperature-sensitive mutants of the Mis12/Mtw1 complex (dsn1–7, nnf1–77, nsl1–5, and mtw1–11) compared to wildtype (Fig 3F). Cse4 localization at LacO was reduced between 19 to 51% in temperature-sensitive mutants of the Ndc80 complex (ndc80–1, nuf2–61, spc24–1, and spc25–1) compared to wildtype (Fig 3G). Although the reduction of Cse4 localization in mutants of the Mis12/Mtw1 and Ndc80 complexes is not as severe as that of the Dam1-DASH complex, the results suggest that the Mtw1 and Ndc80 complexes contribute to Cse4 localization.
Next, we tested the inner kinetochore Ctf19 complex proteins, Cnn1, and Mif2 for their role in Cse4 localization at LacO. Deletions of non-essential proteins Ctf19 or Cnn1 or a temperature-sensitive mutation in Okp1, did not reduce Cse4 enrichment at LacO when the synthetic kinetochore is present (Fig. 3H). In mif2–3 cells, Cse4 localization was reduced 37% (Fig. 3I). Therefore, Mif2 may contribute but Cnn1 and the Ctf19 complex are not important for Cse4 localization to the synthetic kinetochore.
We further tested the localization of GFP-Cse4 in Dam1-DASH and CBF3 complex mutants by chromatin fiber immunofluorescence. We prepared chromatin fibers from wildtype, dad1–13, and ndc10–1 cells (Fig. S3C). In wildtype cells with the synthetic kinetochore, 51% have Cse4 at the LacO array (n=64). In contrast, in ndc10–1 and dad1–13 cells, only 27% and 26% of chromatin fibers have Cse4 at the LacO array, respectively, suggesting that fewer cells have Cse4 at LacO (n=72 for ndc10–1, n=113 for dad1–13).
In summary, our results suggest that the Dam1-DASH and CBF3 complexes have an important role in Cse4 incorporation to the chromatin underlying the synthetic kinetochore. The Mis12/Mtw1 and Ndc80 complexes, and Mif2 also contribute to Cse4 localization. Since CBF3 is not known to directly interact with Dam1-DASH, we propose that the Mis12/Mtw1 and Ndc80 complexes have overlapping roles linking the two complexes to recruit Cse4 to the synthetic kinetochore. A model of the synthetic kinetochore is depicted in Figure 3J.
Dam1-DASH complex components contribute to Cse4 localization to the natural centromere
To test if kinetochore proteins involved in Cse4 localization at the synthetic kinetochore also contribute to Cse4 localization at the natural centromere, we analyzed centromere localization of Cse4 in kinetochore temperature-sensitive mutants. We verified that Cse4 localization was substantially reduced in an ndc10–1 strain at the restrictive temperature (Ortiz et al., 1999) (Fig 4A).
Figure 4). The CBF3 and Dam1-DASH complexes are required for Cse4 localization at the natural centromere.
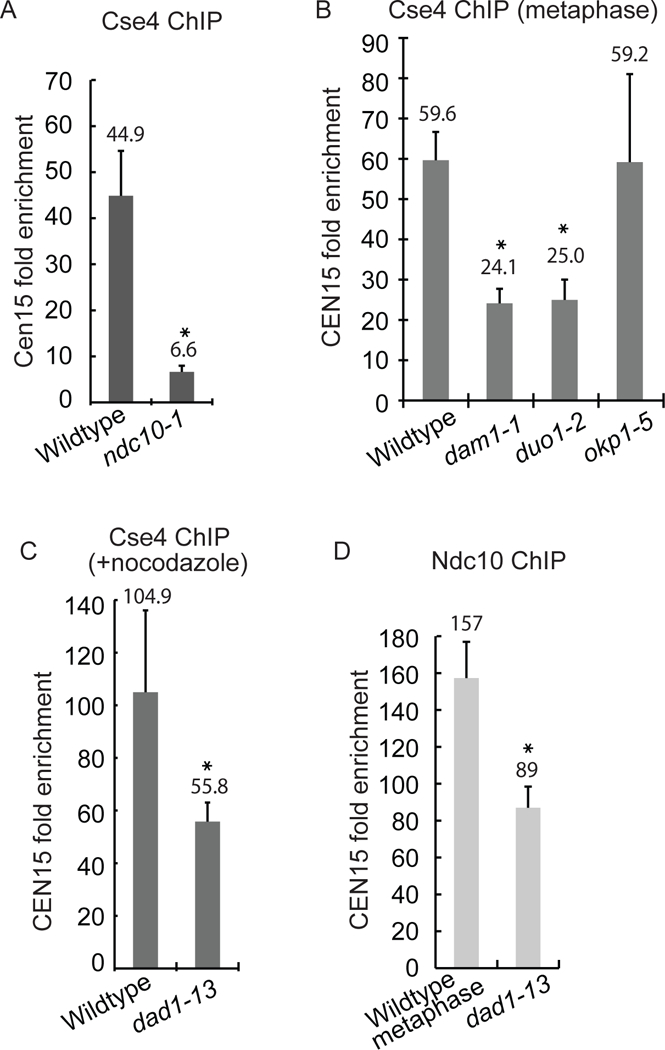
A-C) The quantification of ChIP-qPCR amplifying CEN15 DNA (average of three experiments ± S.D) Anti-GFP antibody was used to IP GFP-Cse4 in cells with kinetochore mutations at 37°C. * represents statistical significance (t test, p< .05). B) Cells were arrested in metaphase. C) Nocodazole and benomyl were added to the cells. D) The quantification of ChIP-qPCR using anti-GFP antibody to IP Ndc10-GFP.
Interestingly, Cse4 levels at the centromere are reduced in Dam1-DASH complex mutants. Cells were metaphase-arrested (by Cdc20 depletion) to ensure the comparison of cells at the same cell-cycle stage. Cse4 enrichment at CEN15 was reduced 60% and 59% in dam1–1 and duo1–2 mutants compared to wildtype cells, respectively (Fig. 4B). Since the dad1–13 mutant causes severe microtubule attachment errors (Janke et al., 2002), we depolymerized microtubules with nocodazole in wildtype and dad1–13 cells to compare strains without microtubule attachments. The Cse4 levels at CEN15 were reduced 47% in dad1–13 cells compared to wildtype cells (Fig. 4C). ChIP against Ndc10-GFP shows a 44% reduction of Ndc10 at CEN15 in the dad1–13 strain (Fig. 4D). Since the loss of Dam1-DASH components moderately disrupts centromere clustering (Anderson et al., 2009; Richmond et al., 2013) (Fig. S4), we were concerned that disruption of clustering could decrease Cse4 levels in ChIP assays. However, in okp1–5 mutant cells, centromere clustering is severely disrupted, but the levels of Cse4 at the centromere are not reduced in the ChIP assay (Fig. 4B and Suppl. Fig. S4A,B). The results suggest that the decreased level of Cse4 at the centromere in the Dam1-DASH mutants is not simply a consequence of the loss of centromere clustering. We conclude that kinetochore complexes important for Ndc10 and Cse4 localization to the synthetic kinetochore also contribute to Cse4 localization at the natural kinetochore.
Discussion:
Since a 125bp sequence specifies the centromere in budding yeast, the widely accepted view was that this sequence is required for CENP-A deposition (Biggins, 2013). In other eukaryotes, such as S. pombe, D. melanogaster, and mammals, the centromere is inherited epigenetically through maintenance of CENP-A-containing chromatin (De Rop et al., 2012). Interestingly, HJURP, the histone chaperone required for CENP-A deposition, is also conserved from yeast to humans. Therefore, we tested whether CENP-A integration requires a specific DNA sequence in budding yeast. We demonstrate that CENP-A homolog Cse4 is stably recruited and functions at a non-centromeric sequence, the synthetic kinetochore assembly site. Localization of Cse4 to the synthetic kinetochore assembly site requires the CBF3 complex. Since CBF3 normally binds the centromere to facilitate Cse4 deposition (Camahort et al., 2007), the results suggest that centromere binding of the CBF3 complex is not strictly required for Cse4 localization; instead, the stable interaction of CBF3 with kinetochore proteins allows Cse4 recruitment to DNA.
The role of the CBF3 complex in Cse4 localization seems contradictory to our previous finding that CBF3 components are not required for synthetic kinetochore biorientation (Lacefield et al., 2009). However, the results are consistent when considering the incomplete depletion of Cse4 from LacO in the CBF3 mutants. Although Cse4 levels are reduced by 77% in the ndc10–1 mutant, the remaining Cse4 is likely sufficient for synthetic kinetochore biorientation.
The localization of Cse4 through kinetochore protein interactions is comparable to CENP-A recruitment to epigenetic centromeres in other eukaryotic organisms. In S. pombe and vertebrate cells, the Mis18 complex is required for CENP-A incorporation in the centromere (Barnhart et al., 2011; Fujita et al., 2007; Hayashi et al., 2004; Pidoux et al., 2009). Targeting the kinetochore proteins CENP-C and CENP-I allows the integration of CENP-A to a non-centromeric locus and recruits a fully functioning heritable kinetochore (Hori et al., 2013). Furthermore, Cse4 incorporates into centromeric histones and rescues CENP-A depletion phenotypes in human cells, suggesting that Cse4 can utilize the CENP-A localization pathways of human cells (Wieland et al., 2004). These studies, combined with our results, suggest that although centromere sequence and DNA binding components of the kinetochore have rapidly changed throughout eukaryotic evolution, many features of the histone recruitment pathway remain largely the same.
Interestingly, the inner CBF3 complex and outer Dam1-DASH complex contribute to Cse4 localization at both the synthetic and natural kinetochores. Therefore, a fully assembled kinetochore contributes to stable Cse4 localization at the natural centromere. Conversely, past studies showed that depletion of Cse4 reduces the localization of most kinetochore proteins at the centromere (Camahort et al., 2007; Collins et al., 2005). Combined, the results suggest that the fully assembled kinetochore and Cse4-containing chromatin reciprocally stabilize each other. We propose that the formation of the assembled kinetochore and the interaction between kinetochore proteins facilitate the stable localization of Cse4 at the centromere.
Experimental Procedures:
Strains.
Strains are listed in Table S1. Synthetic kinetochore strains were previously described (Lacefield et al., 2009). The PCSE4GFP-CSE4 construct, with GFP integrated at residue 83, was integrated at URA3 and the endogenous CSE4 was deleted (Chen et al., 2000). The C-terminally tagged CSE4-Myc was made through PCR integration of the 13Myc epitope (Longtine et al., 1998).
ChIP and qPCR.
ChIP was performed as described, with growth conditions listed in the Supplemental Material (Strahl-Bolsinger et al., 1997). Oligo sequences for qPCR are listed in Table S2. The qPCR was performed as described (Haring et al., 2007). The results shown are the average of at least three ChIP experiments for each genotype ± S.D. The percent reduction of CSE4 enrichment in the mutants was calculated relative to the recruitment to the synthetic kinetochore in comparison to the no synthetic kinetochore control. Statistical significance was determined using the t test.
Chromatin fiber immunofluorescence.
Chromatin fibers were prepared from protoplasts as described (Tsuchiya and Taga, 2001).
Other experimental procedures are listed in the Supplemental Material.
Supplementary Material
Acknowledgements:
We are grateful to Sue Biggins and John Kilmartin for strains and reagents. We thank Brian Calvi, Andrew Murray, Frank Solomon, Claire Walczak, and Mimi Zolan for their critical reading of the manuscript.
References:
- Anderson M, Haase J, Yeh E, and Bloom K (2009). Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell 20, 4131–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, and Foltz DR (2011). HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol 194, 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S (2013). The composition, functions, and regulation of the budding yeast kinetochore. Genetics 194, 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, and Gerton JL (2007). Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell 26, 853–865. [DOI] [PubMed] [Google Scholar]
- Chen Y, Baker RE, Keith KC, Harris K, Stoler S, and Fitzgerald-Hayes M (2000). The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol 20, 7037–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KA, Castillo AR, Tatsutani SY, and Biggins S (2005). De novo kinetochore assembly requires the centromeric histone H3 variant. Mol Biol Cell 16, 5649–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rop V, Padeganeh A, and Maddox PS (2012). CENP-A: the key player behind centromere identity, propagation, and kinetochore assembly. Chromosoma 121, 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, and Yanagida M (2007). Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell 12, 17–30. [DOI] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, and Stam M (2007). Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant methods 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, and Yanagida M (2004). Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118, 715–729. [DOI] [PubMed] [Google Scholar]
- Hill A, and Bloom K (1987). Genetic manipulation of centromere function. Mol Cell Biol 7, 2397–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang WH, Takeuchi K, and Fukagawa T (2013). The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol 200, 45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Tanaka TU, Lechner J, and Schiebel E (2002). Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. Embo J 21, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiermaier E, Woehrer S, Peng Y, Mechtler K, and Westermann S (2009). A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast. Nat Cell Biol 11, 1109–1115. [DOI] [PubMed] [Google Scholar]
- Lacefield S, Lau DT, and Murray AW (2009). Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast. Nat Cell Biol 11, 1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, and Carbon J (1991). A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64, 717–725. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, and Pringle JR (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Ortiz J, Stemmann O, Rank S, and Lechner J (1999). A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev 13, 1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. (2009). Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell 33, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond D, Rizkallah R, Liang F, Hurt MM, and Wang Y (2013). Slk19 clusters kinetochores and facilitates chromosome bipolar attachment. Mol Biol Cell 24, 566–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, and Grunstein M (1997). SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev 11, 83–93. [DOI] [PubMed] [Google Scholar]
- Tsuchiya D, and Taga M (2001). Application of fibre-FISH (fluorescence in situ hybridization) to filamentous fungi: visualization of the rRNA gene cluster of the ascomycete Cochliobolus heterostrophus. Microbiology 147, 1183–1187. [DOI] [PubMed] [Google Scholar]
- Wieland G, Orthaus S, Ohndorf S, Diekmann S, and Hemmerich P (2004). Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol Cell Biol 24, 6620–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J, Hajj B, Chen J, Mizuguchi G, Xiao H, Wei D, Dahan M, and Wu C (2014). Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres. eLife 3, e02203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


