Abstract
Background.
Genetic variants at the SCN5A/SCN10A locus are strongly associated with electrocardiographic PR and QRS intervals. While SCN5A is the canonical cardiac sodium channel gene, the role of SCN10A in cardiac conduction is less well characterized.
Methods and Results.
We sequenced the SCN10A locus in 3699 European-ancestry individuals, and identified two intronic SNPs in high linkage disequilibrium (LD, r2=0.86) with each other to be the strongest signals for PR (rs10428132, Beta=−4.74, P=1.52×10−14) and QRS intervals (rs6599251, QRS Beta=−0.73; P =1.2×10−4)) respectively. While these variants were not associated with SCN5A or SCN10A expression in human atrial tissue (n=490), they were in high LD (r2≥0.72) with a common SCN10A missense variant, rs6795970 (V1073A). In total, we identified seven missense variants, four of which (I962V, P1045T, V1073A and L1092P) were associated with cardiac conduction. We replicated our findings in over 21,000 individuals of European ancestry. These four missense variants cluster in the cytoplasmic linker of the second and third domains of the SCN10A protein, and together form six common haplotypes. Using cellular electrophysiology, we found that haplotypes associated with shorter PR intervals had a significantly larger percentage of late current compared with wild-type (I962V+V1073A+L1092P, 20.2±3.3%, P=0.03, and I962V+V1073A, 22.4±0.8%, P=0.0004 vs. WT 11.7 ± 1.6%), and the haplotype associated with the longest PR interval had a significantly smaller late current percentage (P1045T, 6.4±1.2%, P=0.03).
Conclusions.
Our findings suggest an association between genetic variation in SCN10A, the late sodium current, and alterations in cardiac conduction.
Journal Subject Terms: Atrial Fibrillation, Electrophysiology, Ion Channels/Membrane Transport, Functional Genomics
Keywords: population genetics, electrocardiography, cardiovascular genomics, conduction, Na+ current, SCN10A, PR interval
Clinical Perspective:
Genetic variants at the SCN5A/SCN10A locus are strongly associated with electrocardiographic PR and QRS intervals. In addition, variants in SCN10A have also been linked to atrial fibrillation. While SCN5A is the canonical cardiac sodium channel gene responsible for the upstroke of the cardiac action potential, the role of SCN10A, is less well characterized in cardiac conduction. In this study, we have characterized the electrophysiologic function of several missense coding variants in SCN10A identified from large populations where the PR interval was known. We found that these missense variants all clustered in the cytoplasmic linker between two domains of the channel. When these missense mutations were generated and characterized via patch clamp, they were found to alter the amount of late sodium current present. Genetic haplotypes coding for channels that were associated with shorter PR intervals had a larger late sodium current, and therefore our results suggest that there is an association between the amount of late sodium current and the PR interval. Clinically, this may be relevant as longer PR intervals are associated with a higher risk of atrial fibrillation and therefore further characterization of the role of SCN10A may provide a novel target for future therapies.
Introduction
The PR and QRS intervals on the surface electrocardiogram reflect atrioventricular conduction and ventricular depolarization, respectively. PR interval prolongation is associated with increased risk of atrial fibrillation (AF), the need for pacemaker implantation, and death.1 Similarly, QRS interval prolongation is associated with increased risk for heart failure and death.2, 3 Genome wide association studies (GWAS) have identified a locus on chromosome 3 associated with the PR and QRS intervals, with the most significant signal mapping to an intronic region of SCN10A.4–7 SCN10A encodes for the voltage-gated sodium channel, Nav1.8, and is flanked by SCN5A and SCN11A, both of which encode for voltage-gated sodium channels, Nav1.5 (cardiac) and Nav1.9 (neuronal), respectively.8, 9
Until recently, Nav1.8 channels were thought to be restricted to the dorsal root ganglia and peripheral sensory neurons.10, 11 However, we have previously shown that SCN10A is expressed in the myocardium, and preferentially in the specialized Purkinje fibers of the cardiac conduction system.6 Further, blocking the Nav1.8 channel in mice with a selective antagonist, A-803467, prolongs the PR and QRS intervals, reduces the late sodium current (INa,L) in ventricular myocytes, and slows action potential firing in intracardiac neurons, suggesting a direct role for SCN10A in cardiac electrophysiology.6, 12, 13 In GWAS of PR and QRS intervals, two common SCN10A coding variants have been identified, both significantly associated with cardiac conduction.4–7 Recent work has also found that rs6801957, the most significant intronic single nucleotide polymorphism (SNP) in SCN10A associated with the QRS interval, alters a transcription factor binding site for Tbx5 in mice that regulates the expression of SCN5A.14–16
To further define the relationship of variation in SCN10A with cardiac conduction, we sequenced the SCN10A gene in the Cohorts for Heart and Aging Research in Genomic Epidemiology Targeted Sequencing Study (CHARGE-TSS) to identify rare and common variants in SCN10A. We examined the association of these variants with atrioventricular conduction. We then replicated our findings among European and African-ancestry subjects sequenced as part of the Exome Sequencing Project (ESP) and by direct genotyping in a larger sample of CHARGE participants. Next, we examined whether coding variants are functionally related to cardiac conduction using cellular electrophysiology. Finally, we sought to determine whether the most significant SCN10A signal is associated with SCN5A and SCN10A transcript expression in humans.
Methods
Study Populations and Genetic Assays
Discovery Study Sample: CHARGE Targeted Sequencing Study
The Cohorts for Heart and Aging Research in Genetic Epidemiology (CHARGE) Consortium is a consortium of population- and community-based cohorts designed to facilitate studies of genetic epidemiology.17 CHARGE-TSS included a random sample, stratified by sex, of at least 1000 Atherosclerosis Risk in Communities (ARIC) Study, 500 Cardiovascular Health Study (CHS), and 500 Framingham Heart Study (FHS) participants of European ancestry. In each cohort, investigators selected an additional sample of individuals from the extremes of the phenotype distribution of several cardio-metabolic traits, including PR and QRS intervals. Loci for targeted sequencing were identified by GWAS conducted by the CHARGE consortium for a number of phenotypes. The present analysis focuses on targeted sequencing of regions of the SCN10A gene for PR and QRS intervals. In total 3,699 participants of European ancestry were used in this analysis, including individuals selected on the basis of extreme PR, QRS, or other cardio-metabolic traits and a cohort random sample. All studies performed were approved by the Institutional Review Board of the participating institutions. Details of case and cohort selection, and sequencing can be found in Supplementary Methods.
Replication and Haplotype Study Sample: Exome Sequencing Project
The NHLBI Exome Sequencing Project (ESP), a parallel exome sequencing consortium, served as the replication cohort. In total, our replication sample included 607 ESP participants of European ancestry and 972 African American participants. Haplotypes were derived from all ESP participants, increasing the ESP-Haplotype sample to 4,306 participants of European ancestry and 972 African American participants. Full details of the ESP cohort selection are available in the Supplementary Methods.
Extension Study Sample: CHARGE Exome chip cohorts
To increase the sample size, we examined directly genotyped data from the entire ARIC, CHS and FHS cohorts. Participants with a QRS interval >120ms were excluded from the QRS interval analyses. A total of 20,666 European ancestry participants (CHARGE-Exome sample) were analyzed. Full details of cohort selection are available in Supplementary Methods.
Haplotype Analysis
Genotypes of both European and African descent populations from ESP were phased separately by race using PHASE.18 PHASE estimates of haplotype frequency were reported. Full details are available in the Supplementary Methods.
Gene expression analysis
We performed an expression quantitative trait loci (eQTL) analysis using the most significant 5 SNPs at the SCN10A locus associated with PR or QRS intervals in this study or in prior GWAS efforts of PR or QRS intervals (rs10428132, rs6795970, rs6801957, rs6800541, rs6599250). Please see the Supplemental Methods for details.
Human SCN10A clone, mutagenesis, expression, and characterization
The human SCN10A alpha subunit cDNA built-in a pCMV6-XL5 vector was purchased from OriGene. The SCN10A DII-DIII linker variants/haplotypes (IV, PT, VA, LP, VA+LP, IV+VA and IV+VA+LP) were generated using appropriately designed mutagenic primers. The SCN10A constructs were expressed in a Neuroblastoma 2a (N2a) cell line derived from mouse (ATCC, catalog number CCL-131). N2a cells were used because these neuronal cells expressed functional SCN10A channels after transfection. SCN10A expression was verified in freshly obtained human atrial tissue via immunohistochemistry. Wild type and variant/haplotype Nav1.8 currents were measured using the whole-cell patch clamp recordings. Tetrodotoxin (TTX, 150 nM) was used to block endogenous sodium currents. The data are presented as mean ± standard error and a student’s t-test was used to determine a significant difference (P<0.05). Full details of patch clamping methods and analysis are available in the Supplementary Methods.
Results
Sequence Results in Discovery Samples
We sequenced an approximately 117 kilobase region encompassing the SCN10A gene in a discovery sample of 3699 individuals of European ancestry from the Atherosclerosis Risk in Communities (ARIC) study, the Cardiovascular Health Study (CHS), and the Framingham Heart Study (FHS) (Table S1). We identified 3005 non-coding (intronic/intergenic) and 204 coding variants (Tables S2, S3A). There was evidence of selection pressure against putatively functional variants; nonsynonymous variants tended to have a lower minor allele frequency than synonymous variants (P=0.046), and rare nonsynonymous variants were more likely to be predicted as having adverse effects compared to common nonsynonymous variants, P=0.014–0.000013 (Table S3B).
Association of Common Variants with PR and QRS Intervals
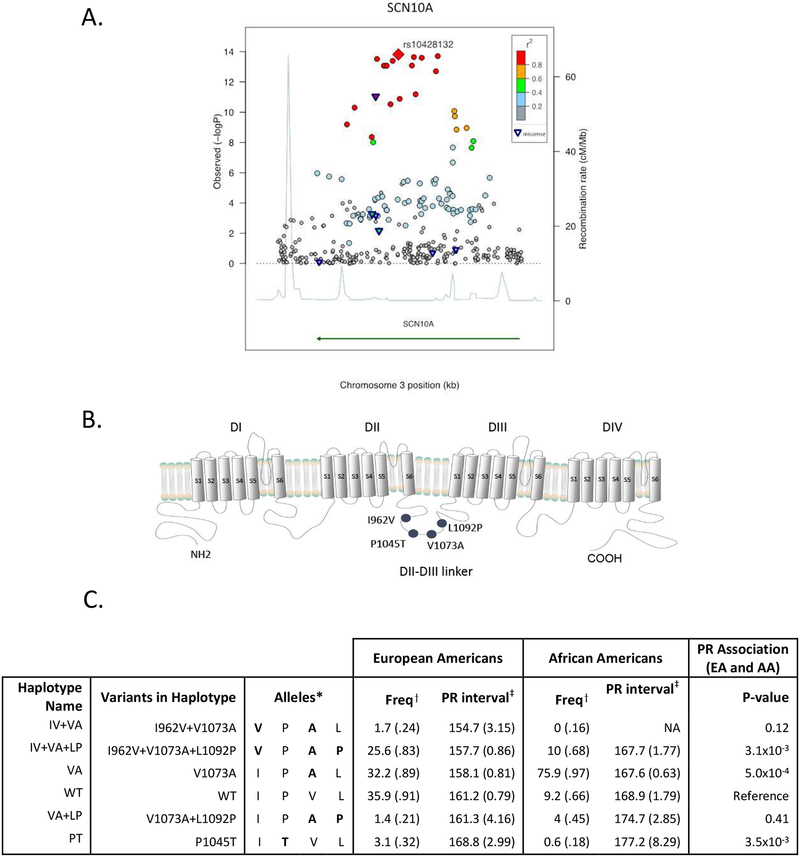
We examined the association of cardiac conduction with common and rare SCN10A variants. The SNP with the most significant association with the PR (rs10428132, Beta=−4.74; P =1.5×10−14; n=3699, Figure 1A, Table S4) and QRS (rs6599251, Beta=−0.73; P=1.2×10−4; n=3699, Table S4) intervals were both intronic and in high linkage disequilibrium (LD) with each other (r2=0.86). These variants are also in high LD (r2≥0.72) with the most significant SNPs identified by prior GWAS of the PR or QRS intervals, and importantly, with the most significant nonsynonymous SCN10A coding SNP rs6795970, V1073A (Table 1, Tables S4–S5).4–7 The most significant SCN10A SNPs associated with PR (rs10428132) and QRS (rs6599251) were not in LD with common variants in two neighboring loci, SCN5A and SCN11A, which also encode for voltage-gated sodium channels (Figure S1).
Figure 1. Association of genetic variation in SCN10A with the PR interval.
A) Regional association plot of SCN10A with PR interval in the CHARGE-TSS discovery sample. Each shape represents a single variant. The rhombus marks the index variant. Triangles denote nonsynonymous coding variants. Color-coding denotes the correlation between each variant and the index SNP. B) Schematic of the Nav1.8 channel α-subunit, which is comprised of four repeating domains (DI-DIV), each consisting of six transmembrane spanning segments (S1–S6). The four nonsynonymous variants significantly associated with PR interval are clustered within a cytoplasmic linker between the second and third domains (DII-DIII linker). C) Six haplotypes were observed in the ESP European American (EA) and African American (AA) populations. The table lists the SNPs where the haplotypes differ from WT, the specific alleles of the four common coding variants carried on each haplotype, haplotype frequency (%), mean PR interval and standard error among EA and AA. PR association results combine both ethnicities; each haplotype is compared to WT. Two rare haplotypes, IV+VA and VA+LP, were identified but had limited power to compare differences in PR interval with WT (frequency among EA 2% and 1% respectively). Neither I962V nor L1092P variant occurred in isolation. *Specific alleles listed in order from 5’ end of gene for the four variants: I962V, P1045T, V1073A, L1092P; †% Frequency (SE); ‡Mean, msec (SE)
Table 1.
Common SCN10A Coding Variant Associations with the PR interval
| CHARGE Targeted Sequencing (EA, n=3699) | Exome Sequencing Project (EA, n=607) | Exome Sequencing Project (AA, n=972) | CHARGE Exome Chip Project (EA, n=20666) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Amino acid substitution | A1/A2 | Freq* (%) | β (SE) [msec] | P value | β (SE) [msec] | P value | Freq† (%) | β (SE) [msec] | P value | Freq† (%) | β (SE) [msec] | P value |
| PR interval | |||||||||||||
| rs57326399 | I962V | T/C | 25.2 | −2.22 (0.94) | 6.7×10−3 | −2.83 (1.54) | 0.07 | 10.2 | 0.01 (1.83) | 0.99 | 25.8 | −1.93 (0.25) | 9.75×10−15 |
| rs73062575 | P1045T | G/T | 2.4 | 5.36 (2.03) | 6.8×10−4 | 12.69 (3.70) | 6.5×10−4 | 0.6 | 10.79 (6.98) | 0.12 | 2.7 | 3.4 (0.68) | 4.87×10−7 |
| rs6795970 | V1073A | A/G | 58.3 | −3.94 (0.67) | 8.9×10−12 | −5.13 (1.43) | 3.6×10−4 | 90.2 | −2.14 (1.89) | 0.26 | 60.2 | −3.31 (0.22) | 2.73×10−49 |
| rs12632942 | L1092P | A/G | 23.6 | −2.12 (0.74) | 5.2×10−4 | −2.75 (1.56) | 0.08 | 13.9 | 2.06 (1.59) | 0.19 | 26.0 | −1.69 (0.25) | 1.02×10−11 |
| rs74717885 | I206M | T/C | 1.3 | −6.74 (3.03) | 0.12 | −1.19 (5.48) | 0.83 | 0.2 | NA | 1.3 | −1.87 (2.86) | 0.51 | |
| rs7630989 | S509P | A/G | 2.7 | −4.75 (2.00) | 0.20 | −2.03 (4.46) | 0.65 | 20.9 | −1.6 (1.31) | 0.22 | 3.1 | −2.23 (1.92) | 0.25 |
| rs77804526 | V1697I | C/T | 1.0 | −5.60 (3.44) | 0.80 | 1.40 (5.95) | 0.82 | 0.2 | NA | 1.3 | −1.51 (3.0) | 0.61 | |
Frequency in CHARGE-TSS population,
Frequency in ESP African American population
Abbreviations: EA, European American; AA, African American; Freq, coded allele frequency; A1 non-coded allele, A2 coded allele
Expression QTL (eQTL) analyses for SCN5A and SCN10A and Nav1.8 channel protein expression
Examining transcripts from human atria (n=369 samples), ventricle (n=313 samples) and other tissues, we found no evidence that SCN5A transcript expression is associated with the PR and QRS index SNPs identified in the current study (rs10428132 or rs6599251), nor with the most significant SCN10A SNPs from prior studies of PR or QRS, including rs6801957 (Figure S2, Supplemental Methods, Results, and Table S6).14, 15 While we found expression of Nav1.8 channel protein in freshly obtained human atrial tissues using immunohistochemistry (Figures S3–S4), expression of SCN10A transcript was low in atrial and ventricular tissue, therefore hampering our ability to identify a SNP SCN10A eQTL association. Nonetheless, in an additional 121 samples from human left atrial appendage, we found no association by quantitative PCR between SCN5A and SCN10A transcript expression and the most significant SCN10A SNP (rs6800541) identified by prior GWAS of the PR interval (Figure S5, Table S7).5, 19
Missense SNPs
While the most significant intronic PR and QRS SNPs were not associated with expression of the two cardiac sodium channels, SCN5A and SCN10A, they were in high LD (r2=0.83 and 0.72, respectively) with a common SCN10A missense variant, rs6795970 (V1073A). We therefore examined common SCN10A missense variants for association with cardiac conduction. We identified seven common missense SNPs in SCN10A among individuals of European descent, and four – I962V (IV), P1045T (PT), V1073A (VA) and L1092P (LP) – were associated with PR interval duration (Table 1, Table S2). The four SNPs associated with PR interval duration clustered within a 130 amino acid residue region of the cytoplasmic linker that connects the second and third domains (DII-DIII linker) of the Nav1.8 channel (Figure 1B). The V1073A and P1045T variants were associated with the largest effects on PR interval and were nominally associated with QRS duration (Table 1, Table S4).
We sought to replicate and extend our significant findings from the CHARGE-TSS in additional samples using two approaches. First, the coding regions of SCN10A were sequenced among those of European and African descent as part of ESP. Both the V1073A and P1045T variants were associated with PR interval in 607 European-descent individuals who were part of our ESP-replication sample. The other two coding variants, I962V and L1092P, while not reaching statistical significance in the smaller sample size of ESP, showed an equal or larger magnitude of effect on PR interval. We did not find significant associations among African Americans (n=972); however, power to identify a significant association was more limited due to lower minor allele frequencies and smaller sample sizes (Table 1).
In our second approach, we directly genotyped the seven SCN10A common coding variants in 20,666 individuals from the CHARGE Exome Chip Project (FHS, ARIC and CHS) using the Illumina HumanExome v1.0 array (Table 1, Table S1). We found that all four common coding variants (I962V, P1045T, V1073A and L1092P) were significantly associated with both PR (P-values ranging from 10−7 to 10−49, Table 1) and QRS (p-values ranging from 10−4 to 10−14, Table S4) intervals.
To determine whether the four common coding SCN10A SNPs associated with the PR interval were independent from SNPs in neighboring sodium channel genes, SCN5A and SCN11A, we performed a conditional analysis of the PR interval using the ESP cohort and common SNPs in SCN5A and SCN11A. We found that both V1073A and P1045T remained significantly associated with PR interval duration and that the effect size was preserved among the four common coding SNPs (Table S8).
Haplotype Analysis
Three of the four SNPs associated with the PR interval were in moderate to high LD, Table S5, and six common haplotypes were present among individuals of European or African ancestry (Figure 1C). Haplotype analyses were performed using 5278 individuals of European and African ancestry sequenced in ESP. The most frequent haplotype among individuals of European ancestry (frequency of 36%) was designated the wild type (WT) SCN10A haplotype and was associated with a mean PR interval of 161 milliseconds (msec) among European Americans and 169 msec among African Americans. The VA haplotype and IV+VA+LP haplotype both had mean PR intervals that were shorter compared to WT in both ethnic groups (Figure 1C). By contrast, the PT haplotype had a significantly longer mean PR interval compared with WT (Figure 1C). There was suggestive evidence that haplotype structure influenced PR interval given that the VA+LP haplotype had a longer mean PR interval compared with the VA only haplotype (P=0.02). Similar to the PR findings, the PT haplotype tended to have the longest QRS interval; however no significant haplotype-QRS associations were detected, due in part to the smaller effect sizes with QRS (Table S9).
Biophysical Effects of SNPs and Haplotypes on Channel Function
Given the common location of the four nonsynonymous variants in the DII-DIII linker of SCN10A (Figure 1B), we sought to determine the biophysical effects of the haplotype combinations on channel function using cellular electrophysiology. SCN10A wild type and haplotype whole-cell currents were acquired by heterologous expression in N2A cells using patch clamp electrophysiology (See Supplemental Methods). Analyses were performed for all six common haplotype combinations observed, as well as for the two variants, I962V and L1092P, which were not seen in isolation among European Americans and African Americans (Table 2, Figure S6).
Table 2.
Electrophysiology parameters of the six haplotypes and I962V (IV) and L1092P (LP) variants
| Current density | Voltage dependent activation | Voltage dependent inactivation | Late current | Time course of recovery from inactivation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pA/pF | V1/2 (mV) | k (mV) | n | V1/2 (mV) | k (mV) | n | INa, L (%) | INa,L (pA/pF) | n | τ, f (ms) | τ, s (ms) | n | |
| PT | −24.6 ± 5.1 | 11.8 ± 1.8* | 12.5 ± 1.1 | 7 | −68.5 ± 1.3 | 10.4 ± 1.1 | 6 | 6.4 ± 1.2* | −3.3 ± 0.8 | 5 | 17.3 ± 2.2 | 348.6 ± 79.3* | 5 |
| VA+LP | −60.1 ± 12.5 | −2.3 ± 1.3* | 8.0 ± 1.1 | 7 | −70.1 ± 2.6 | 10.4 ± 2.3 | 6 | 12.0 ± 1.8 | −7.2 ± 1.5 | 5 | 10.3 ± 2.7 | 226.5 ± 75.1 | 3 |
| WT | −25.6 ± 6.7 | 3.0 ± 1.0 | 10.5 ± 0.8 | 7 | −68.9 ± 1.4 | 12.2 ± 1.3 | 6 | 11.7 ± 1.6 | −5.5 ± 1.0 | 7 | 10.7 ± 2.7 | 124.5 ± 34.7 | 6 |
| VA | −53.4 ± 8.7* | −6.3 ± 1.4* | 8.5 ± 1.3 | 6 | −70.5 ± 1.1 | 9.3 ± 1.0 | 5 | 13.5 ± 3.2 | −7.8 ± 1.8 | 6 | 20.2 ± 5.8 | 326.5 ± 115.6 | 6 |
| IV+VA+LP | −44.2 ± 12.1 | −3.5 ± 1.0* | 10.2 ± 0.9 | 6 | −74.0 ± 3.6 | 13.4 ± 2.7 | 6 | 20.2 ± 3.3* | −8.7 ± 1.8+ | 5 | 11.7 ± 4.5 | 245.0 ± 100.0 | 4 |
| IV+VA | −26.1 ± 6.7 | 1.1 ± 1.1 | 10.2 ± 0.9 | 8 | −67.6 ± 2.3 | 10.9 ± 2.2 | 6 | 22.4 ± 0.8* | −9.7 ± 2.2+ | 5 | 7.5 ± 1.6 | 167.9 ± 36.2 | 5 |
| IV | −33.6 ± 11.3 | 2.8 ± 1.1 | 10.1 ± 0.9 | 7 | −70.2 ± 1.7 | 10.1 ± 1.5 | 6 | 15.4 ± 1.7 | −6.3 ± 2.4 | 3 | 4.2 ± 1.6 | 84.6 ± 16.3 | 3 |
| LP | −17.8 ± 3.5 | 8.3 ± 2.1 | 12.4 ± 1.4 | 7 | −70.7 ± 2.4 | 12.1 ± 2.1 | 4 | 12.9 ± 3.1 | −5.3 ± 1.4 | 6 | 11.5 ± 2.4 | 196.2 ± 53.8 | 6 |
Current density at +10 mV, INaL (%) is percentage of INaL and INaL (pA/pF) at +20 mV. τf and τs are the fast and slow time constants for recovery from inactivation, respectively. Data displayed as mean ± standard error, n=number of cells.
statistically different from WT (P>0.05).
significant difference compared to PT haplotype (P<0.05).
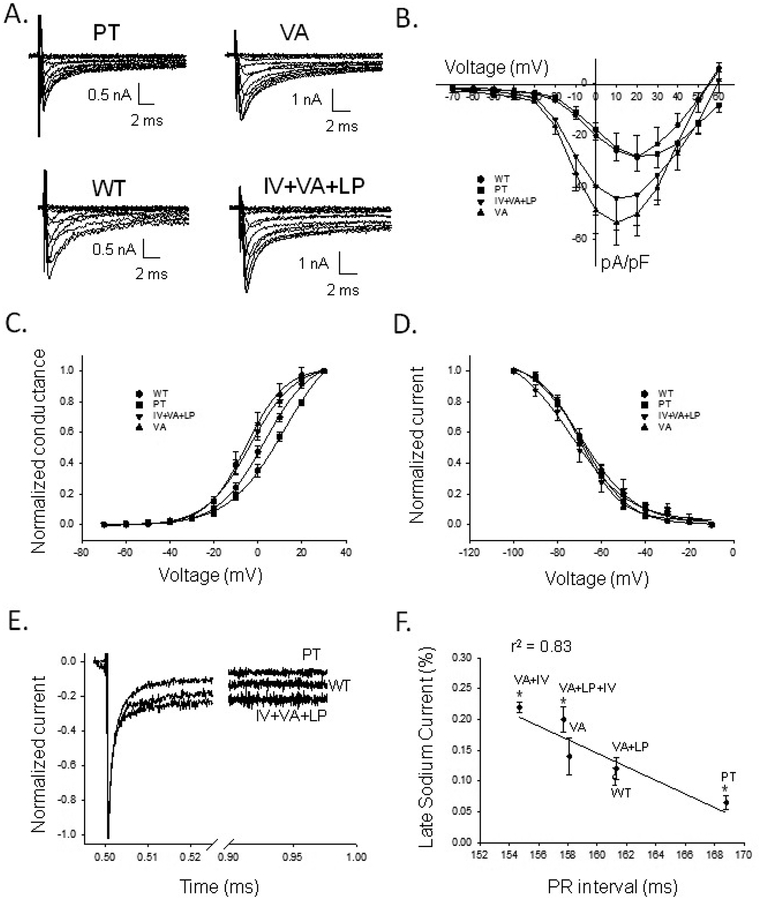
Representative current traces of the most frequently observed haplotypes are illustrated in Figure 2A. Plots of current density versus voltage for the four most common haplotypes are shown in Figure 2B. The VA haplotype had a significantly larger current density than the WT at +10 mV, while the other haplotypes were not significantly different from WT (Table 2). Plots of conductance-voltage (G-V) relationships are shown in Figure 2C. We observed a subtle relationship in voltage-dependent opening (V1/2) and PR interval among the haplotypes (Figures S6–S7, Table 2). The midpoint of channel opening (V1/2) of the G-V relationship for PT was significantly right-shifted (+11.8 ± 1.8 mV; P=0.001; n=7), and both VA (−6.3 ± 1.5 mV; P=0.0002; n=6) and IV+VA+LP (−3.5 ± 1.0 mV; P=0.0008; n=7) were significantly left-shifted compared to WT (+3.0 ± 1.0 mV; n=7). There was no difference in the slope for voltage-dependent opening (Table 2). Furthermore, there was no relation between haplotype differences in the PR interval and voltage-dependent inactivation (Figure 2D, Table 2), the recovery from inactivation (Figure S8, Table 2), or open-state inactivation (Figure S9).
Figure 2. Electrophysiological characteristics of Nav1.8 channel haplotypes.
A) Four haplotype Nav1.8 whole-cell sodium currents recorded from N2a cells in the presence of 150 nM tetrodotoxin. Currents were elicited from a holding potential of −100 mV with depolarizing steps between −70 and +60mV in 10mV increments. B) Current density (pA/pF) versus voltage for WT, PT, VA, and IV+VA+LP constructs. C) Conductance versus voltage for WT, PT, VA, and IV+VA+LP, fit with a Boltzmann function (see Supplemental Methods). V1/2 for PT was shifted rightward, and VA and IV+VA+LP were both shifted leftward compared to WT. Slope factors (k) were not significantly different. D) Voltage-dependent inactivation as normalized current versus voltage. The V1/2 and k for the haplotype variants were not significantly different from WT. E) Normalized INa,L of PT, WT, and IV+VA+LP, measured from a holding current of −100mV with pulse to +20mV for 475ms. PT shows a smaller and VA+LP+IV a greater INa,L compared to WT, respectively. F) Percent INa,L versus PR interval. The percent INa,L decreases moving from haplotypes with short to long PR intervals. The data were significantly correlated by the fitted line (r2=0.83, P=0.01). *: statistically different from WT (P>0.05).
A distinct feature of the Nav1.8 channel compared to other sodium channels is the persistence of a significant INa,L. A visual inspection of the recordings (Figure 2A) suggested a difference in the INa,L between haplotypes. To assess this difference systematically, we measured INa,L at the end of a test pulse (+20 mV at 475 msec) when the inactivated current reached steady-state which was then normalized to the peak current. We observed marked differences in INa,L between haplotypes ranging from 6.4 to 22.4%. Compared to WT INa,L (11.7±1.6%, n=7), smaller INa,L for the PT (6.4±1.2%; P=0.03; n=5) and larger INa,L for the IV+VA+LP (20.2±3.3%, P=0.03, n=5) haplotypes were observed (Figure 2E, Table 2). We next sought to determine if there was a relation between haplotype differences in the PR interval and INa,L. When the haplotypes were arranged from the shortest to the longest PR intervals, we observed an inverse linear relation between percent INa,L versus the PR interval, Figure 2F (percent INa,L, r2=0.83, P=0.001).
Secondary Analyses
To comprehensively examine the associations at this locus with cardiac conduction, we secondarily examined association of rare SCN10A variants in aggregate (locus-wide or limited to coding variants) with either the PR or QRS intervals in approximately 3699 individuals from CHARGE-TSS and 1579 individuals from ESP. While no significant association was identified (p>0.05 for all associations tested), power was limited due to a low cumulative allele frequency.
Discussion
By sequencing the SCN10A locus, we identified a cluster of common coding variants in the DIIDIII linker that influence cardiac depolarization and conduction. We have identified an inverse linear relationship between the percent INa,L and the PR interval duration. Haplotypes that reduce the Nav1.8 INa,L were associated with a longer PR interval and conversely, haplotypes that increased the Nav1.8 INa,L were associated with a shorter PR interval compared with WT.
The functional role of SCN10A/Nav1.8 channel on cardiac conduction has only recently begun to be elucidated.20 Increasing experimental data suggests that the SCN10A/Nav1.8 channel may have a direct role in cardiac conduction. First, the specific SCN10A blocker, A-803467, lengthened the PR and QRS intervals in mice,6 and reduced the SCN10A/Nav1.8 INa,L in isolated ventricular myocytes from mouse and rabbit.12 Second, the SCN10A/Nav1.8 channel has been identified in the specialized cardiac conduction fibers and atrial myocytes, as well as in cholinergic, and to a lesser extent, sympathetic intracardiac neurons (Table S10). Third, SCN10A missense variants are associated with PR and QRS intervals. Taken together, these data provide evidence that the SCN10A/Nav1.8 channel participates in cardiac conduction.
Our results suggest a correlation between the amount of INa,L and the PR interval measured amongst our study cohorts. The PR interval is modulated by many factors, and the correlation observed in this study is not necessarily causative. SCN10A influence on atrioventricular conduction may be secondary to neuronal modulation, a direct effect on cardiomyocyte depolarization, or both.20 Supportive of the cardiomyocyte hypothesis, we found qualitative evidence of Nav1.8 expression in human atrial cardiomyocytes using immunohistochemistry, similar to prior findings with a different antibody.21 While we acknowledge that there is no quantitative data on SCN10A expression in human cardiac conduction tissue, we have documented SCN10A expression in the murine Purkinje system previously.6 Alternatively, SCN10A has also been identified in intracardiac neurons and in cardiac ganglionated plexi.13 Recently, injection of A-803467 into cardiac ganglionated plexi was shown to be protective from AF, underscoring the potential for neuronal SCN10A expression to influence the electrophysiology of the myocardium.22 Future studies of SCN10A expression in the human conduction system and cardiac innervation may further elucidate the role of SCN10A in atrioventricular conduction.
Our findings highlight the role of the DII-DIII linker in Nav1.8 channel inactivation. Previous studies indicated that point mutations in the DII-DIII linker of cardiac (SCN5A) and brain (SCN1A) voltage-gated sodium channels both increased the INa,L in patients with type III long QT syndrome and epilepsy, respectively.23, 24 Crystal structures of sodium channels showed that the S6 transmembrane spanning region forms the inner lining of the pore.25 Since the DII-DIII linker connects the S6 of domain II with S1 of domain III, we hypothesize that this region of the channel has an important role in the process of prolonged inactivation. In support of that conclusion, several variants in this linker are associated with Brugada Syndrome,26 including V1073A, which was independently shown to cause a higher INa,L density with an A in position 1073 compared to V, similar to our findings.27 In that study, the higher current density allele (V1073A) was protective from Brugada Syndrome, signifying that haplotypes at this position may have multiple effects depending on relative expression in atrial, ventricular, and conduction system myocytes.
In this study, we did not find an association between the intronic SCN10A SNP and expression levels of SCN5A in human atrial or ventricular tissue. However, the relation between the SCN5A/SCN10A genomic region and cardiac conduction is highly complex with at least six independent association signals for QRS interval that span both genes.6 The intronic SCN10A SNP associated with the QRS interval, rs6801957, modulates the function of a Tbx5 and Tbx3 enhancer.16 A recent study demonstrated that while no SCN10A transcript could be identified in ventricular cardiomyocytes in either wild-type or SCN10A −/− mice, Nav 1.8 contributes to late sodium current at slow rates, perhaps through modulation of SCN5A.28 In our study, we did not characterize Nav1.8 or Nav1.5 directly in the human ventricular myocardium or Purkinje system, however these will be important future investigations to define the contributions and potential interactions of the Nav1.8 and Nav1.5 channels on ventricular conduction in humans.
Our study was subject to a number of limitations. First, although we examined individuals of both European and African descent, our results may not be generalizable to other ethnicities. Second, despite performing sequencing in 3699 and 1579 individuals in the discovery and replication samples respectively, our power to detect modest associations, particularly with rare variants, was limited. Third, we did not examine whether these genetic variants were related to expression of SCN10A in the conduction system. Fourth, our genetic association and heterologous electrophysiology data regarding SCN10A are correlative and do not imply causality. Finally, in vitro cellular electrophysiology studies may not fully recapitulate the cellular milieu of the cardiomyocyte or the cardiac conduction system.
In conclusion, our sequencing and functional data provide new insights for the physiological role of SCN10A/Nav1.8 channel as a contributing factor to human cardiac conduction. As the SCN10A/Nav1.8 channel has only recently been identified as a novel modifier on cardiac conduction and electrophysiology, further studies are necessary to define the relative contributions of the Nav1.8 and Nav1.5 channels on cardiac conduction.
Supplementary Material
Acknowledgments:
The authors acknowledge support from the National Heart, Lung, and Blood Institute (NHLBI) and the contributions of the research institutions, study investigators, field staff and study participants in creating this resource for biomedical research. We thank the Lifeline of Ohio Organ Procurement Organization and the Division of Cardiac Surgery at The OSU Wexner Medical Center for providing the explanted human cardiac tissue for immunohistochemistry.
Sources of Funding: The current work was funded by NIH grants: 1R01HL088456, 1R01HL111089, 1R01HL116747, 2K24HL105780, R01HL128914, 1RO1HL092577, HL088577, HL10599, HL115580, HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN2682011000010C, HHSN2682011000011C, and HHSN2682011000012C, R01HL087641, R01HL59367, R01HL086694, HL103010, HL102924, HL102925, HL102926, 5RC2HL102419, N01-HC-25195, N02-HL-6–4278, HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01DK089256, R01HL120393, R01AG023629, K23HL114724; by AHA grants: 13EIA14220013, 12FTF11350014; by the Max Schaldach Fellowship in Cardiac Pacing and Electrophysiology and the MGH Fund for Medical Discovery; by the Laughlin Family, and by the Fondation Leducq (14CVD01).
Footnotes
Disclosures: Dr. Ellinor is the PI on a grant from Bayer HealthCare to the Broad Institute focused on the genetics and therapeutics of AF.
References:
- 1.Cheng S, Keyes MJ, Larson MG, McCabe EL, Newton-Cheh C, Levy D, et al. Long-term outcomes in individuals with prolonged pr interval or first-degree atrioventricular block. JAMA. 2009;301:2571–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhingra R, Pencina MJ, Wang TJ, Nam BH, Benjamin EJ, Levy D, et al. Electrocardiographic qrs duration and the risk of congestive heart failure: The framingham heart study. Hypertension. 2006;47:861–867. [DOI] [PubMed] [Google Scholar]
- 3.Desai AD, Yaw TS, Yamazaki T, Kaykha A, Chun S, Froelicher VF. Prognostic significance of quantitative qrs duration. Am J Med. 2006;119:600–606. [DOI] [PubMed] [Google Scholar]
- 4.Chambers JC, Zhao J, Terracciano CM, Bezzina CR, Zhang W, Kaba R, et al. Genetic variation in scn10a influences cardiac conduction. Nat Genet. 2010;42:149–152. [DOI] [PubMed] [Google Scholar]
- 5.Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, et al. Genome-wide association study of pr interval. Nat Genet. 2010;42:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sotoodehnia N, Isaacs A, de Bakker PI, Dorr M, Newton-Cheh C, Nolte IM, et al. Common variants in 22 loci are associated with qrs duration and cardiac ventricular conduction. Nat Genet. 2010;42:1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, et al. Several common variants modulate heart rate, pr interval and qrs duration. Nat Genet. 2010;42:117–122. [DOI] [PubMed] [Google Scholar]
- 8.Souslova VA, Fox M, Wood JN, Akopian AN. Cloning and characterization of a mouse sensory neuron tetrodotoxin-resistant voltage-gated sodium channel gene, scn10a. Genomics. 1997;41:201–209 [DOI] [PubMed] [Google Scholar]
- 9.Rabert DK, Koch BD, Ilnicka M, Obernolte RA, Naylor SL, Herman RC, et al. A tetrodotoxin-resistant voltage-gated sodium channel from human dorsal root ganglia, hpn3/scn10a. Pain. 1998;78:107–114. [DOI] [PubMed] [Google Scholar]
- 10.Akopian AN, Sivilotti L, Wood JN. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature. 1996;379:257–262. [DOI] [PubMed] [Google Scholar]
- 11.Abrahamsen B, Zhao J, Asante CO, Cendan CM, Marsh S, Martinez-Barbera JP, et al. The cell and molecular basis of mechanical, cold, and inflammatory pain. Science. 2008;321:702–705. [DOI] [PubMed] [Google Scholar]
- 12.Yang T, Atack TC, Stroud DM, Zhang W, Hall L, Roden DM. Blocking scn10a channels in heart reduces late sodium current and is antiarrhythmic. Circ Res. 2012;111:322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verkerk AO, Remme CA, Schumacher CA, Scicluna BP, Wolswinkel R, de Jonge B, et al. Functional nav1.8 channels in intracardiac neurons: The link between scn10a and cardiac electrophysiology. Circ Res. 2012;111:333–343. [DOI] [PubMed] [Google Scholar]
- 14.van den Boogaard M, Wong LY, Tessadori F, Bakker ML, Dreizehnter LK, Wakker V, et al. Genetic variation in t-box binding element functionally affects scn5a/scn10a enhancer. J Clin Invest. 2012;122:2519–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnolds DE, Liu F, Fahrenbach JP, Kim GH, Schillinger KJ, Smemo S, et al. Tbx5 drives scn5a expression to regulate cardiac conduction system function. J Clin Invest. 2012;122:2509–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van den Boogaard M, Smemo S, Burnicka-Turek O, Arnolds DE, van de Werken HJ, Klous P, et al. A common genetic variant within scn10a modulates cardiac scn5a expression. J Clin Invest. 2014;124:1844–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for heart and aging research in genomic epidemiology (charge) consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denny JC, Ritchie MD, Crawford DC, Schildcrout JS, Ramirez AH, Pulley JM, et al. Identification of genomic predictors of atrioventricular conduction: Using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.London B Whither art thou, scn10a, and what art thou doing? Circ Res. 2012;111:268–270. [DOI] [PubMed] [Google Scholar]
- 21.Facer P, Punjabi PP, Abrari A, Kaba RA, Severs NJ, Chambers J, et al. Localisation of scn10a gene product na(v)1.8 and novel pain-related ion channels in human heart. Int Heart J. 2011;52:146–152. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Yu L, Shi S, Jiang H, Huang C, Desai M, et al. Neuronal nav1.8 channels as a novel therapeutic target of acute atrial fibrillation prevention. J Am Heart Assoc. 2016;5:e004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz PJ, Priori SG, Dumaine R, Napolitano C, Antzelevitch C, Stramba-Badiale M, et al. A molecular link between the sudden infant death syndrome and the long-qt syndrome. N Engl J Med. 2000;343:262–267. [DOI] [PubMed] [Google Scholar]
- 24.Lossin C, Wang DW, Rhodes TH, Vanoye CG, George AL Jr. Molecular basis of an inherited epilepsy. Neuron. 2002;34:877–884. [DOI] [PubMed] [Google Scholar]
- 25.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486:135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu D, Barajas-Martinez H, Pfeiffer R, Dezi F, Pfeiffer J, Buch T, et al. Mutations in scn10a are responsible for a large fraction of cases of brugada syndrome. J Am Coll Cardiol. 2014;64:66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behr ER, Savio-Galimberti E, Barc J, Holst AG, Petropoulou E, Prins BP, et al. Role of common and rare variants in scn10a: Results from the brugada syndrome qrs locus gene discovery collaborative study. Cardiovasc Res. 2015;106:520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stroud DM, Yang T, Bersell K, Kryshtal DO, Nagao S, Shaffer C, et al. Contrasting nav1.8 activity in scn10a−/− ventricular myocytes and the intact heart. J Am Heart Assoc. 2016;5:e002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.