Abstract
G protein-coupled receptors (GPCRs) are the largest family of signaling proteins targeted by more clinically used drugs than any other protein family. GPCR signaling via G proteins is quenched (desensitized) by the phosphorylation of the active receptor by specific GPCR kinases (GRKs) followed by tight binding of arrestins to active phosphorylated receptors. Thus, arrestins engage two types of receptor elements: those that contain GRK-added phosphates and those that change conformation upon activation. GRKs attach phosphates to serines and threonines in the GPCR C-terminus or any one the cytoplasmic loops. In addition to these phosphates, arrestins engage the cavity that appears between trans-membrane helices upon receptor activation and several other non-phosphorylated elements. The residues that bind GPCRs are localized on the concave side of both arrestin domains. Arrestins undergo a global conformational change upon receptor binding (become activated). Arrestins serve as important hubs of cellular signaling, emanating from activated GPCRs and receptor-independent.
Keywords: arrestin, GPCR, phosphates, receptor specificity, signaling, protein engineering
Introduction
The functions of the first member of the arrestin family, arrestin-1a, were discovered in the visual system. Arrestin-1 (under the name of 48 kDa protein) was found to specifically bind light-activated rhodopsin along with the visual G protein transducin and rhodopsin kinase (current systematic name GRK1 (Gurevich et al., 2012)) (Kuhn, 1978). Its binding was found (also by Kuhn’s group) to be greatly enhanced by rhodopsin phosphorylation (Kuhn et al., 1984) and then shown to inhibit (arrest) light-induced rhodopsin signaling (Wilden et al., 1986). This function was later satisfactorily explained by direct competition between arrestin-1 and visual G protein transducin (Krupnick et al., 1997; Wilden, 1995), which arrestin wins when rhodopsin has three or more attached phosphates (Mendez et al., 2000; Vishnivetskiy et al., 2007). The first non-visual arrestin (under the name of β-arrestin, later retroactively renamed β-arrestin1; systematic name arrestin-2) was discovered later (Lohse et al., 1990). It preferentially outcompetes cognate G protein, Gs, at the β2-adrenergic receptor (β2AR) (Lohse et al., 1992) phosphorylated by an earlier discovered β2AR kinase (βARK; systematic name GRK2 (Gurevich et al., 2012)) (Benovic et al., 1989). This evidence suggested that two-step desensitization, phosphorylation of active G protein-coupled receptor (GPCR) by a specific kinase, followed by the high-affinity binding of an arrestin protein, is the common mechanism in the GPCR family (Carman and Benovic, 1998). Even though later arrestins were shown to interact with numerous non-receptor partners (Xiao et al., 2007), their binding to active phosphorylated GPCRs, which precludes further receptor interactions with cognate G proteins, remains their key biological function. Therefore, here we focus on the molecular mechanisms of arrestin binding to GPCRs, discussing both receptor and arrestin elements involved, as well as the resulting conformational rearrangements in the arrestin molecule and their functional implications.
The mechanics of GPCR activation and receptor elements engaging arrestins
The original idea that GPCRs exist in two states, active and inactive, which was the basis of the extended ternary complex model of GPCR signaling (Samama et al., 1993), turned out to be an over-simplification. Interestingly, early structures and biophysical studies of rhodopsin (Altenbach et al., 2008; Farrens et al., 1996; Palczewski et al., 2000; Scheerer et al., 2008) and β2AR (Rasmussen et al., 2007; Rosenbaum et al., 2011) appeared to support this two-state idea: the most notable conformational rearrangement accompanying GPCR activation was the outward movement of transmembrane helices V and VI, opening a cavity on the cytoplasmic side for the signaling proteins to dock to (Fig. 1). So, GPCRs were considered as on-off switches, with one active and one inactive conformation (Samama et al., 1993). However, detailed biophysical studies of the prototypical non-visual GPCR, β2AR, showed that in the absence of an agonist it exists as an ensemble of several conformations (Manglik et al., 2015). The agonist perturbs this equilibrium; although the greatest changes were visible in the presence of a G protein. Even upon G protein binding more than one receptor conformation was present (Manglik et al., 2015). Light-activated rhodopsin is also conformationally heterogenous, with the equilibrium significantly shifted by the cognate G protein, but conformational homogeneity could not be achieved even in this situation (Van Eps et al., 2017). The fact that there are likely numerous “active” conformations of any GPCR, supported by the structural studies of receptor-G protein (Carpenter et al., 2016; Koehl et al., 2018; Liang et al., 2017; Rasmussen et al., 2011; Van Eps et al., 2018; Zhang et al., 2017) and receptor-arrestin complexes (Kang et al., 2015; Zhou et al., 2017), is consistent with the idea that by “pushing” the receptor into specific subsets of these conformations activating ligands might have functional bias: preferentially facilitate GPCR interactions with particular G proteins or GRKs/arrestins (Smith et al., 2018; Wisler et al., 2018). However, documented direct competition between G proteins and arrestins (Krupnick et al., 1997; Lohse et al., 1992; Wilden, 1995) suggests that these subsets must significantly overlap.
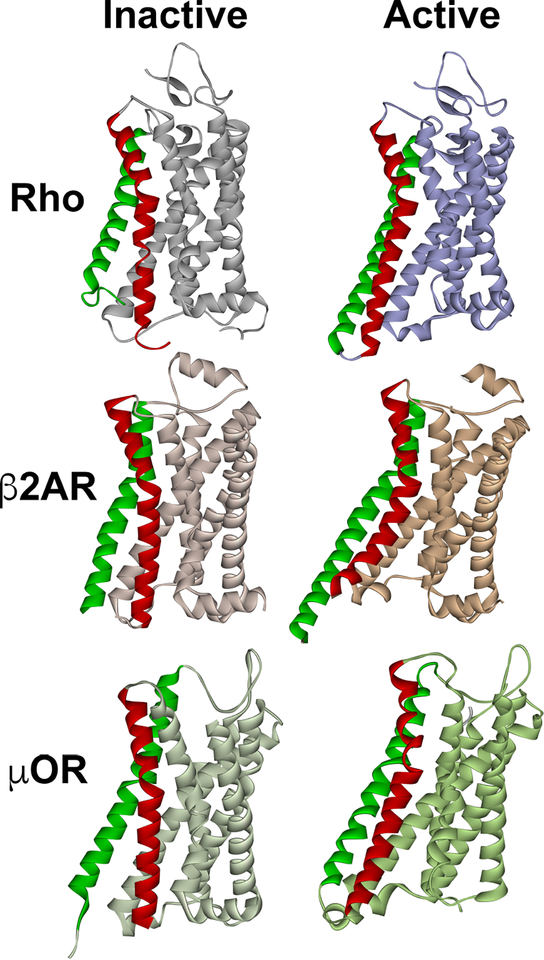
Fig. 1. The structure of inactive and active GPCRs.
The most pronounced activation-induced conformational change in all GPCRs is the outward movement of TM5 (green) and TM6 (red). In case of TM6, activation-induced shift is ∼ 14 A in Gs-coupled β2AR (Rasmussen et al., 2011), but smaller in Gi-coupled receptors, ∼10 A in both rhodopsin (Kang et al., 2018; Kang et al., 2015) and μ-opioid receptor (Koehl et al., 2018). The structures of inactive (dark) rhodopsin (Rho) (PDB ID: 1GZM (Li et al., 2004)), active rhodopsin (from complex with arrestin-1; PDB ID: 4ZWJ (Kang et al., 2015)), inactive β2AR (PDB ID: 2RH1 (Rasmussen et al., 2007)), active β2AR (from complex with Gs; PDB ID: 3SN6 (Rasmussen et al., 2011)), inactive μ-opioid receptor (μOR) (PDB ID: 4DKL (Manglik et al., 2012)), and active μOR (from complex with Gi; PDB ID: 6DDE (Koehl et al., 2018)) are shown. The common feature of active GPCR structures is the cavity between helices that opens on the cytoplasmic side. In case of Gi-coupled GPCRs (Rho and μOR) the outward movement of the helices is smaller, so that this cavity is narrower than in case of Gs-coupled β2AR. The dimensions of this cavity might underlie GPCR selectivity for particular G proteins, whereas non-visual arrestins likely accommodate GPCRs that couple to all types of G proteins.
GRKs evolved from Ser/Thr-specific AGC kinases (Mushegian et al., 2012) and phosphorylate serine and threonine, but not tyrosine residues in their targets. Activation-dependent phosphorylation of Ser and Thr residues by GRKs in various GPCRs has been demonstrated in vivo, in cultured cells, and in vitro (Azevedo et al., 2015; Fredericks et al., 1996; Hausdorff et al., 1989; Inagaki et al., 2015; Kim et al., 2004; Li et al., 2015; Moro et al., 1993; Pals-Rylaarsdam and Hosey, 1997; Seibold et al., 1998; Seibold et al., 2000; Wilden and Kühn, 1982). Receptor-attached phosphates (Shukla et al., 2013; Zhou et al., 2017), as well as several potentially phosphorylatable (Innamorati et al., 2001; Walther et al., 2010; Wanka et al., 2018) or non-phosphorylated receptor elements were directly implicated in arrestin binding by mutagenesis in rhodopsin and non-visual GPCRs (DeGraff et al., 2002; Raman et al., 2003; Raman et al., 1999; Schmidlin et al., 2003; Wanka et al., 2018), as well as by the crystal structure of the arrestin-1-rhodopsin complex (Kang et al., 2015; Zhou et al., 2017). Interestingly, relevant phosphates could be on the receptor C-terminus (Azevedo et al., 2015; Mendez et al., 2000; Wilden and Kühn, 1982), or any intracellular loop (Celver et al., 2001; Inagaki et al., 2015; Lee et al., 2000; Mukherjee et al., 2002; Pals-Rylaarsdam et al., 1997). This varied localization of the phosphates engaged by arrestins is consistent with the finding that the only thing that really matters is the number of phosphates (Mendez et al., 2000; Vishnivetskiy et al., 2007) and spacing between the phosphates and/or negative charges that need to fit the spacing of the positively charged patches on the arrestin surface (Zhou et al., 2017).
Structural basis of the competition between arrestins and G proteins for GPCRs
Arrestins and G proteins can compete for the GPCR only if they engage some of the same elements of the receptor. Judging by the structures of GPCR complexes with G proteins (Carpenter et al., 2016; Koehl et al., 2018; Liang et al., 2017; Rasmussen et al., 2011; Van Eps et al., 2018; Zhang et al., 2017) and rhodopsin complex with visual arrestin-1 (Kang et al., 2015; Zhou et al., 2017), both types of proteins engage the cavity that opens upon receptor activation on the intracellular side between the helices that move outward (Fig. 1). Interestingly, if the part of the arrestin protein that engages this cavity (see below) is deleted, arrestin no longer competes with the G protein, so that the receptor can interact with both proteins simultaneously (Cahill et al., 2017; Kumari et al., 2016; Thomsen et al., 2016). Thus, the engagement of this receptor cavity appears to be the key prerequisite for the homologous desensitization, i.e., arrestin competition with the G proteins.
The suppression of GPCR coupling to G proteins by direct competition is the first and arguably the most established function of arrestins. All arrestins preferentially bind phosphorylated active GPCRs, and GRKs are the key kinases that specifically phosphorylate active, but not inactive, receptors. Thus, for arrestins to play the key role in homologous GPCR desensitization, and that is hardly in doubt (Carman and Benovic, 1998; Gurevich and Gurevich, 2004), G proteins, GRKs, and arrestins must bind significantly overlapping subsets of GPCR conformations. This certainly limits the potential of proposed biased signaling (Smith et al., 2018; Wisler et al., 2018), as this concept implies that some ligands must induce non-overlapping GPCR conformations preferentially coupling to G proteins or arrestins (and in the latter case by necessity GRKs, although this aspect is often overlooked). At the moment, we have very few structural studies revealing the details of the GPCR-G protein complexes (Carpenter et al., 2016; Koehl et al., 2018; Liang et al., 2017; Rasmussen et al., 2011; Van Eps et al., 2018; Zhang et al., 2017), and only one structure of the receptor-arrestin complex (Kang et al., 2015; Zhou et al., 2017). Thus, we cannot determine which conformations are specific for G protein-coupled GPCRs and which represent arrestin-preferring conformations, as conformational differences among activated GPCRs might reflect the difference inherent to the receptors themselves, rather than the differences that predetermine the binding partner. It should also be noted that the conformations of Gs- and Gi-bound receptors appear to be different (Koehl et al., 2018; Van Eps et al., 2018) (Fig. 1), which might underlie documented bias towards particular G proteins of GPCRs that couple to more than one class of G proteins (e.g., (Violin and Lefkowitz, 2007) and references therein). It is also noteworthy that double electron-electron resonance (DEER) measurements between selected points in rhodopsin and bound arrestin-1 always yield several distances (Kang et al., 2015; Zhou et al., 2017). While the most populated ones invariably match the structure, to the delight of crystallographers, the existence of the others strongly suggests that the arrestin-receptor complex likely has several “flavors”, only one of which was so far captured in the crystal. Thus, we need a lot more structures of GPCRs in complex with their binding partners before any general conclusions can be drawn.
Receptor-binding elements of arrestins
The first attempts to determine which parts of arrestin are involved in receptor binding had been made before any structural information became available. Differential chemical modification and hydrogen-deuterium exchange of free and rhodopsin-bound arrestin-1 identified numerous arrestin-1 residues likely involved in rhodopsin binding (Ohguro et al., 1994). A cluster of positive charges was identified as a phosphate-binding element by site-directed mutagenesis (Gurevich and Benovic, 1995), and large central part of the arrestin molecule was found to determine receptor preference of arrestin-1/2 chimeras (Gurevich et al., 1995). Remarkable similarity of the three-dimensional structure of all arrestin proteins established later (Granzin et al., 1998; Han et al., 2001; Hirsch et al., 1999; Milano et al., 2002; Sutton et al., 2005; Zhan et al., 2011a) explained why all chimeras fold successfully and are functional. All arrestins turned out to be elongated two-domain molecules, with each domain consisting of seven β-strands and having a concave and convex side (Fig. 2A). Several arrestin-1 peptides were shown to compete with full-length arrestin-1 and transducin for rhodopsin (Pulvermuller et al., 2000). Later studies of a new set of the arrestin-1/2 chimeras identified two elements on the concave sides of the two arrestin domains that are largely responsible for their preferential binding to rhodopsin or M2 muscarinic acetylcholine receptor (Vishnivetskiy et al., 2004). Surprisingly, few side chains within these elements were found to be immobilized by receptor binding (Fig. 2B) or play a direct role in receptor selection (Vishnivetskiy et al., 2011) (Fig. 2A). Replacement of the “receptor-discriminator” residues with alanines showed that they play key role in the binding of all arrestins to all GPCRs tested (Vishnivetskiy et al., 2011), suggesting that their replacement has a potential to yield non-visual arrestins with modified receptor specificity. Indeed, manipulation of some of these residues significantly changed receptor specificity of the most promiscuous non-visual subtype, arrestin-3, and generated mutants with 50–60-fold preference for some GPCRs over others, in contrast to parental arrestin-3 that binds all these receptors with comparable affinity (Gimenez et al., 2014; Gimenez et al., 2012). Interestingly, a totally different experimental approach, based on the site-directed spin labeling, also identified the concave sides of both arrestin domains as the site of receptor binding (Hanson et al., 2007; Hanson et al., 2006) (Fig. 2B). In these experiments, surface residues were replaced with cysteines, which were chemically modified with a spin label. Spin labels were fairly mobile, as could be expected of labels placed on the surface of a protein where nothing obstructs their movement, and became immobilized to various degrees within the receptor “footprint” when arrestins were bound by active phosphorylated rhodopsin (Hanson et al., 2007; Hanson et al., 2006) (Fig. 2B). In addition, the effect on rhodopsin binding of the elimination or reversal of arrestin surface charges clearly supported the localization of the receptor footprint to the concave sides of both arrestin domains (Hanson and Gurevich, 2006). An NMR study of free and receptor-bound arrestin-1 also supported this localization of the receptor-binding elements (Zhuang et al., 2013).
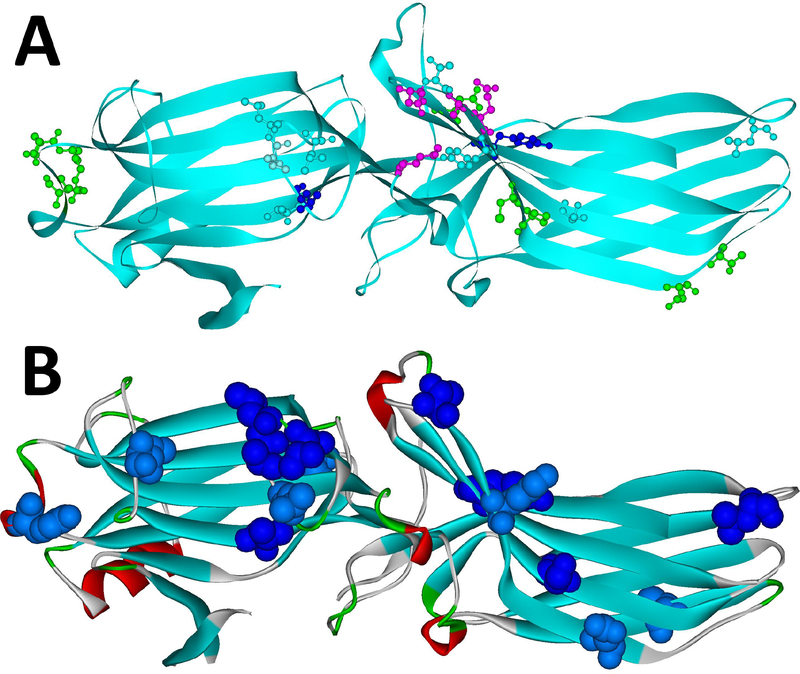
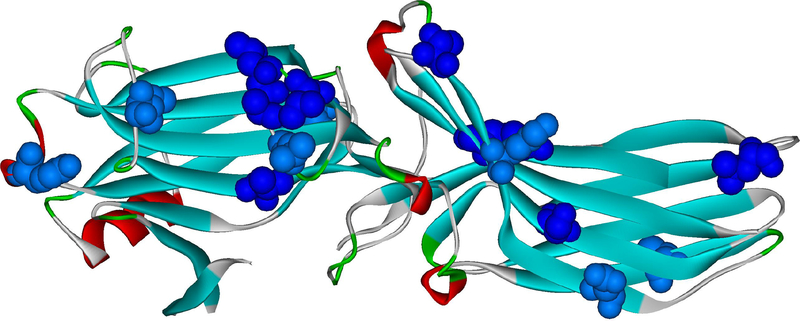
Fig. 2. Arrestin elements implicated in receptor binding.
A. The residues in positions identified as important for the receptor preference of different arrestin proteins (the structure of arrestin-2 is shown; PDB ID: 1G4M (Han et al., 2001); the residues indicated are those of arrestin-2) are shown as ball-and stick models colored as follows: light blue, those identified by reduced mobility of the spin label in receptor-bound arrestins using EPR (Val70, Leu71, Leu73, Val167, Leu191, Ser234, Thr246, Tyr249) (Hanson et al., 2007; Hanson et al., 2006); green, those identified by site-directed mutagenesis and receptor binding in vitro and in cells (Leu48, Glu50, Arg51, Asp240, Cys251, Pro252, Asp259, Thr261) (Gimenez et al., 2014; Gimenez et al., 2012; Prokop et al., 2017; Vishnivetskiy et al., 2011); dark blue, those identified by both of these methods (Leu68, Tyr238); magenta, those identified by direct contact with the receptor in the crystal structure of the arrestin-1 complex with rhodopsin (Lys138, Asn245, Ala247, Gln248) (Kang et al., 2015), including those confirmed by in-cell assays (Lys138, Gln248) (Prokop et al., 2017). Receptor-binding residues identified by several independent methods are localized on the concave sides of the two arrestin domains, with the highest concentration in the central crest of the receptor-binding side of arrestins. B. The residues that change mobility upon receptor binding, according to SDSL-EPR study (Vishnivetskiy et al., 2011) are shown as CPK models (dark blue – strong immobilization; light blue – less pronounced immobilization). The same crystal structure of arrestin-2 (PDB ID: 1G4M) was used. Both panels were generated using Accelrys DS ViewerPro 6.0 (Dassault Systemes, BIOVIA Corp, San Diego, CA).
Interestingly, the crystal structure of the arrestin-1 complex with rhodopsin revealed a much smaller footprint than deduced on the basis of mutagenesis, EPR, and NMR experiments, mostly involving the loops in the central crest of the receptor-binding side of arrestin (Kang et al., 2015; Zhou et al., 2017) (Fig. 3). The contact site included some residues that were not previously identified as contributing to receptor discrimination, including the “finger loop” and the “middle loop” (the latter was previously termed “139-loop” in arrestin-1 (Kim et al., 2012; Vishnivetskiy et al., 2013)). Site-directed mutagenesis of these residues showed that they indeed participate in receptor binding, differentially affecting the interactions with different GPCRs and/or functional states of the same receptor (Chen et al., 2017; Prokop et al., 2017). Many residues, particularly those in the C-domain, that were found to affect receptor preference, did not appear to touch the receptor in the crystallized complex (compare Figs. 2 and 3). This apparent controversy might be explained by the heterogeneity of the complex, only one flavor of which was crystallized (see above).
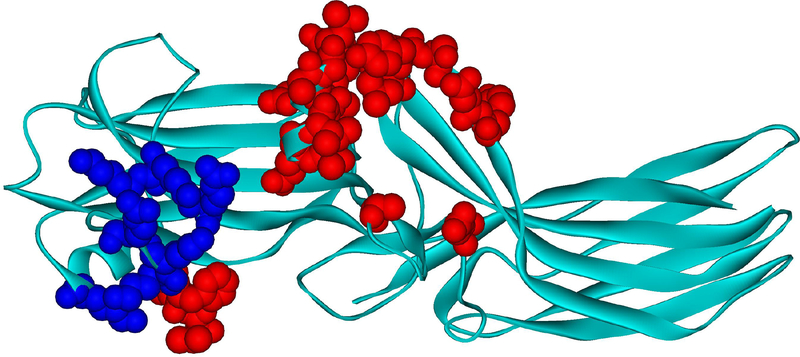
Fig. 3. Arrestin-1 residues contacting rhodopsin in co-crystal.
Mouse arrestin-1 residues that directly interact with rhodopsin are shown as CPK models (based on rhodopsin-bound mouse arrestin-1 structure from complex A, PDB ID: 4ZWJ (Kang et al., 2015)). Those that bind receptor-attached phosphates or negative charges in the rhodopsin C-terminus are colored dark blue (Zhou et al., 2017)(mouse arrestin-1 residues Lys15, Lys16, Arg19, Lys111, Lys167, Lys168, Arg172), those that interact with unphosphorylated parts of the rhodopsin molecule are colored red (Kang et al., 2015; Zhuo et al., 2014) (Val12, Ile13, Phe14, Gln70, Glu71, Ile73, Asp74, Met76, Gly77, Leu78, Arg82, Asp83, Leu84, Lys142, Leu250, Tyr251, Ser252, Asp254, Tyr255, Arg292, Thr320).
Arrestin transition into the “active” state
The four vertebrate arrestin subtypes demonstrate high sequence conservation (Gurevich and Gurevich, 2006; Indrischek et al., 2017) and share virtually identical three-dimensional structure in their basal state (Granzin et al., 1998; Han et al., 2001; Hirsch et al., 1999; Milano et al., 2002; Sutton et al., 2005; Zhan et al., 2011a), although careful comparison revealed subtle differences (Sente et al., 2018). High Arrhenius activation energy of the arrestin-1 binding to rhodopsin discovered as early as in 1989 has suggested that arrestin must undergo a global conformational change in the process of receptor binding (Schleicher et al., 1989). The finding that both phosphorylated rhodopsin and polyanion heparin greatly increase the accessibility of the arrestin-1 C-terminus to protease suggested that its release might be part of this conformational change (Palczewski et al., 1991). Interestingly, Arrhenius activation energy of rhodopsin binding of the short splice variant of arrestin-1 p44 lacking the C-terminus was found to be roughly half of that of the full-length arrestin-1 (Pulvermuller et al., 1997), clearly indicating that the C-terminus release is not the only conformational change in arrestin-1 induced by the receptor binding.
Greatly reduced receptor binding of arrestin-1 (Vishnivetskiy et al., 2002) and arrestin-2 and -3 (Hanson et al., 2007) mutants with deletions in the inter-domain hinge suggested that receptor binding might require the movement of the two domains relative to each other. Two hypotheses regarding the nature of this movement were proposed: it was hypothesized to be clam-shell like, so that arrestin grabs the cytoplasmic tip of the receptor like a pincer (Gurevich and Gurevich, 2004), or the twisting of the two domains relative to each other (Modzelewska et al., 2006). DEER measurements of the distances within arrestin-1 (Kim et al., 2012), as well as non-visual arrestin-2 and -3 (Zhuo et al., 2014) in free and receptor-bound form did not detect a clam-shell-like movement of the domains upon receptor binding. However, distance measurements in all three arrestins revealed other significant conformational changes, particularly the movement of the “finger loop”, “139-loop” (Kim et al., 2012; Vishnivetskiy et al., 2013) (termed “middle loop” in arrestin-2 (Shukla et al., 2013)), and the loops at the tips of both arrestin-1 domains (Kim et al., 2012) (Fig. 4). Similar conformational changes were detected upon receptor binding in non-visual arrestin-2 and -3 (Zhuo et al., 2014). There was one striking difference, though: the released C-terminus in receptor-bound arrestin-1 appeared to just flop around without moving to any preferred position relative to the rest of the molecule, which yielded broad distributions of the distances between the C-terminus and selected sites in the rest of the molecule without any noticeable peaks that would reflect preferred distances (Hanson et al., 2006; Kim et al., 2012; Vishnivetskiy et al., 2010). In contrast, in both non-visual subtypes similar distance measurements showed a preference for a particular position upon release, resulting in a distinct distance (Zhuo et al., 2014). This might be related to the fact that the released C-terminus of non-visual arrestins binds clathrin (Goodman et al., 1996) and clathrin adapotor AP2 (Laporte et al., 1999), whereas the C-terminus of arrestin-1 does not bind clathrin (Goodman et al., 1996) and binds AP2 only with fairly low affinity (Moaven et al., 2013).
Fig. 4. Elements of arrestin-1 that move upon rhodopsin binding.
Bovine arrestin-1 structure in it basal conformation (PDB ID: 1CF1 (Hirsch et al., 1999)) is shown, with the elements that move upon rhodopsin binding (Kim et al., 2012) colored dark red. These include the finger loop (residues 67–78 (Hanson et al., 2006)), the 139-loop (Kim et al., 2012) (a.k.a. the middle loop in arrestin-2 (Shukla et al., 2013)), as well as loops at the distal tips of the N-domain (residues 155–168) and C-domain (residues 336–344).
Crystal structures of naturally pre-activated short splice variant of arrestin-1 (Kim et al., 2013) and C-terminally truncated arrestin-2 in complex with the phosphorylated receptor peptide (Shukla et al., 2013) suggested that one of the key changes in “active” arrestins is approximately 20° twist of the two domains relative to each other. Although in these cases the receptor was absent, subsequent structure of the arrestin-1 complex with rhodopsin (Kang et al., 2015; Zhou et al., 2017) also revealed similar domain twist and confirmed loop movements detected by the EPR studies (Kim et al., 2012; Zhuo et al., 2014). This structure also supported earlier idea that arrestin finger loop assumes α-helical conformation upon receptor binding (Szczepek et al., 2014) (Fig. 3), with the helix inserting itself into the cavity between GPCR α-helices created by helix movements upon activation (Fig. 1). The structure of the arrestin-3 trimer in the presence of an abundant cytoplasmic small molecule inositol hexakisphosphate (IP6) revealed that all three protomers in this trimer are in the active receptor-bound-like conformation, with characteristic twist of the two domains (Chen et al., 2017). The tips of the finger loops of the three molecules formed α-helices, which stabilized each other via hydrophobic interactions, similar (but not identical) to the hydrophobic interactions with rhodopsin of the α-helical tip of the finger loop of bound arrestin-1 (Chen et al., 2017; Kang et al., 2015; Zhou et al., 2017). It is also worth noting that while the mutations that preclude the formation of the α-helix in the finger loop invariably reduce arrestin-3 binding to GPCRs, the effects of the replacement of the hydrophobic leucine with charged residues are receptor subtype-specific (Chen et al., 2017). This structure solved a long-standing mystery in the field. It was shown by different labs that arrestin-3 facilitates the activation of JNK3 (Breitman et al., 2012; Miller et al., 2001; Seo et al., 2011; Song et al., 2009), as well as ubiquitous JNK1/2 (Kook et al., 2014) independently of GPCR stimulation. However, it was unclear how arrestin can become active without GPCR binding. The structure of the arrestin-3 trimer with IP6, where there were two IP6 molecules clearly resolved in each of the three interfaces between protomers, identified one possible non-receptor activator. Interestingly, the phosphates of the IP6 molecule engage the same positive charges in the arrestin-3 (Chen et al., 2017) that are engaged by the rhodopsin-attached phosphates in arrestin-1 (Zhou et al., 2017) and the phosphates on the receptor peptide in arrestin-2 (Shukla et al., 2013) in respective complexes. However, this structure does not fully explain the molecular mechanism of arrestin-3-dependent JNK3 activation: purified arrestin-3 was shown to scaffold MKK4-JNK3 and MKK7-JNK3 modules in the absence of IP6 (Zhan et al., 2011b; Zhan et al., 2013), and a short 25-residue N-terminal peptide of arrestin-3, that was found to increase JNK phosphorylation by both MKKs and facilitate JNK3 activation in cells (Zhan et al., 2016), does not contain residues implicated in the IP6 binding or trimerization.
Thus, it appears well established that in the active state of arrestins the two domains twist relative to each other by about 20°, several loops move, and the tip of the finger loop assumes α-helical conformation. The comparison of all available structures of arrestins in their active and basal state also identified several elements on the non-receptor-binding side of the molecule that show significantly different conformations (Chen et al., 2017).
Arrestins and cellular signaling: where do non-receptor partners bind?
More than a hundred non-receptor partners of each non-visual arrestin subtype were identified (Xiao et al., 2007). However, the elements of arrestin engaging these proteins were determined in very few cases. Only the interaction sites of clathrin (Goodman et al., 1996), clathrin adaptor AP2 (Laporte et al., 1999), microtubules (Hanson et al., 2007), calmodulin (Wu et al., 2006), and critical residues engaging cAMP phosphodiesterase (Baillie et al., 2007), MEK1 (Meng et al., 2009), and cRaf1 (Coffa et al., 2011b) were identified. It appears reasonable to hypothesize that any protein that binds the arrestin-receptor complex must engage the parts that are not shielded by the receptor, i.e., interact with the non-receptor-binding side of the molecule (Gurevich and Gurevich, 2003). This notion is supported by the demonstration that all three JNK3-binding elements of arrestin-3 are localized on that side (Zhan et al., 2014), suggesting that receptor-associated arrestin-3 can also bind the kinases of this pathway, which explains the reported JNK3 phosphorylation in response to GPCR activation (McDonald et al., 2000). As numerous non-receptor partners were found to preferentially bind either active or basal arrestins (Ahmed et al., 2011; Coffa et al., 2011a; Song et al., 2006), it stands to reason that the elements that these partners engage have different conformations in these two states (Fig. 5). Thus, arrestin activation by GPCRs (or any other activators) must induce conformational changes that promote or suppress the binding of the partners with the preference for a particular arrestin conformation.
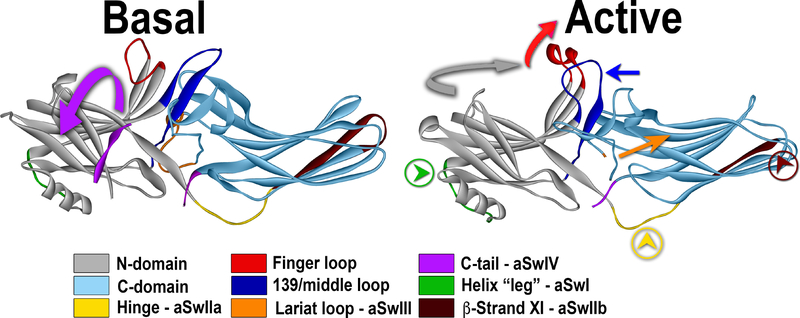
Fig. 5. Activation of arrestin-3 and switch regions.
Conformational changes upon arrestin-3 activation are shown: the release of the arrestin C-terminus (magenta; magenta arrow shows the direction of the movement; aSwIV) anchored to the body of the molecule via the three-element interaction in the basal state (PDB ID: 3P2D (Zhan et al., 2011a)), the movement of the finger loop and helix formation in its tip (red; red arrow shows the direction of the movement), the twist of the two domains relative to each other (N-domain, gray; C-domain, teal). Movements of several loops, likely creating effector docking sites, are also shown: middle loop (dark blue; dark blue arrow indicates the direction of movement); the “leg” of the α-helix I (green, with green arrowhead in a circle; aSwI), inter-domain hinge (yellow with yellow arrowhead in a circle; aSwIIa), register-shifted β-strand XI in the C-domain (dark brown; aSwIIb), and the lariat loop (orange with orange arrow; in the active structure (PDB ID: 5TV1 (Chen et al., 2017)) it is not resolved; aSwIII). All arrestin subtypes have switches II, III, and IV, whereas only arrestin-1 has polyproline motifs in the switch I. Note that only the interactions of clathrin and AP2 with the aSwIV were experimentally confirmed so far (Kim and Benovic, 2002).
While there are different models explaining how the changes on the receptor-binding side induce rearrangements on the opposite side of the arrestin molecule (Chen et al., 2018; Latorraca et al., 2018; Scheerer and Sommer, 2017), the elements that change conformation upon activation, which were termed “arrestin switch regions” (aSw) (Chen et al., 2017) (Fig. 5), appear to be the most likely docking sites of partners that differentially interact with arrestins in their basal state and in a different conformation(s) upon GPCR binding or upon activation by other mechanisms, such as IP6 binding (Chen et al., 2017).
High-resolution structure of active arrestin-3 (Chen et al., 2017) and its comparison with other structures of active and inactive arrestins revealed several aSw regions. Arrestin switch region I (aSwI) has unique sequence in arrestin-3 (residues 89–97). It is one of the “legs” attaching the only α-helix in arrestin to the N-domain (Fig. 5). This nine-residue element in arrestin-3 contains seven prolines that create two PPXP motifs, which are consensus recognition sites for SH3 domains. In IP6-activated arrestin-3, the backbone of aSwI shifted by as much as 5.8 Å from its basal position (Chen et al., 2018; Chen et al., 2017). It remains to be elucidated whether this region is involved in binding c-Src, other Src family kinases, or any other arrestin partner with an SH3 domain, but it appears to be a legitimate suspect. Switch region II (aSwII) is shared by all four arrestin subtypes. It consists of two clearly distinct parts, the inter-domain hinge (aSwIIa) that connects the N- and C-domains and the entire β-strand XI (aSwIIb) that extends from the hinge region (Fig. 5). The aSwIIa contains an unconventional poly-proline motif (PGPQP in arrestin-2 and -3, PQP in arrestin-1) that might bind SH3 domains. ASwIIa has nearly identical conformation in all solved active arrestin structures (Chen et al., 2017; Kang et al., 2015; Shukla et al., 2013; Zhou et al., 2017), whereas its conformation in basal arrestins varies (Granzin et al., 1998; Han et al., 2001; Hirsch et al., 1999; Milano et al., 2002; Sutton et al., 2005; Zhan et al., 2011a). The most prominent feature of aSwIIb is a register-shifted β-strand XI (Fig. 5), although this β-strand likely has inherent flexibility: similar register-shift was observed in several structures of basal arrestin-2 (Han et al., 2001; Kang et al., 2009; Milano et al., 2006; Milano et al., 2002). This register shift appears to affect the binding of at least some non-receptor partners. A disulfide bond that traps β-strand XI in the register-shifted “active” position enhanced the binding of JNK3 (Chen et al., 2017). ASwIII is an extension of the lariat loop supplying two negative charges to the polar core (Hirsch et al., 1999) (Fig. 5). It becomes more dynamic upon activation of non-visual arrestin-2 and -3, as interpretable electron density corresponding to this element is lost in the active structures (Chen et al., 2017; Shukla et al., 2013). The C-terminus, which contains the binding sites of the two components of the internalization machinery, clathrin (Goodman et al., 1996) and AP2 (Laporte et al., 1999), is the aSwIV. Arrestin binding to GPCRs was shown to induce the release of the C-terminus (Hanson et al., 2006; Palczewski et al., 1991; Vishnivetskiy et al., 2010; Zhuang et al., 2013; Zhuo et al., 2014), and activated non-visual arrestins show enhanced affinity for both clathrin and AP2 (Kim and Benovic, 2002). Interestingly, in arrestin mutants that engage only receptor-attached phosphates, but do not engage the inter-helical cavity of GPCRs, the C-terminus is likely also released, as these mutants facilitate GPCR internalization (Cahill et al., 2017; Kumari et al., 2016). This is consistent with structural data suggesting that phosphorylated GPCR elements, as well as IP6, displace the C-terminus of arrestins from its basal conformation (Chen et al., 2017; Shukla et al., 2013).
It should be noted that these switches are unlikely to participate in the binding of proteins that interact with basal and active arrestins with similar affinity, which might include several upstream MAP kinases (Coffa et al., 2011a; Song et al., 2009). However, any partner that has interaction sites in both domains must be sensitive to arrestin activation, as activation-associated domain twist would change the position of these sites relative to each other (Chen et al., 2018). If the partners bind receptor-associated arrestins, their docking sites have to localize to the convex non-receptor-binding side of the two arrestin domains.
Thus, the structural studies of arrestins activated by GPCR binding or other means have suggested likely binding sites for partners that discriminate between different states of arrestins, as well as those that bind similarly to the basal and active state. However, the actual elements mediating interactions with most partners still remain to be identified. Site identification is needed not only to satisfy our scientific curiosity, but also to design signaling-biased arrestins with certain functions enhanced or disabled, which have clear potential to become valuable tools for research and therapy (Gurevich and Gurevich, 2015).
Acknowledgements
Supported in part by NIH RO1 grants EY011500, GM077561 and GM109955 (the latter two RO1s were replaced by R35 GM122491) (VVG), NS065868 and DA030103 (EVG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We use systematic names of arrestin proteins, where the number after the dash indicates the order of cloning: arrestin-1 (historic names S-antigen, 48 kDa protein, visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin).
References
- Ahmed MR, Zhan X, Song X, Kook S, Gurevich VV, and Gurevich EV (2011). Ubiquitin ligase parkin promotes Mdm2-arrestin interaction but inhibits arrestin ubiquitination. Biochemistry, 3749–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, and Hubbell WL (2008). High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci U S A 105, 7439–7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo AW, Doan T, Moaven H, Sokal I, Baameur F, Vishnivetskiy SA, Homan KT, Tesmer JJ, Gurevich VV, Chen J, et al. (2015). C-terminal threonines and serines play distinct roles in the desensitization of rhodopsin, a G protein-coupled receptor. Elife 4, doi: 10.7554/eLife.05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GS, Adams DR, Bhari N, Houslay TM, Vadrevu S, Meng D, Li X, Dunlop A, Milligan G, Bolger GB, et al. (2007). Mapping binding sites for the PDE4D5 cAMP-specific phosphodiesterase to the N- and C-domains of beta-arrestin using spot-immobilized peptide arrays. Biochem J 404, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benovic JL, DeBlasi A, Stone WC, Caron MG, and Lefkowitz RJ (1989). Beta-adrenergic receptor kinase: primary structure delineates a multigene family. Science 246, 235–240. [DOI] [PubMed] [Google Scholar]
- Breitman M, Kook S, Gimenez LE, Lizama BN, Palazzo MC, Gurevich EV, and Gurevich VV (2012). Silent scaffolds: inhibition of c-Jun N-terminal kinase 3 activity in the cell by a dominant-negative arrestin-3 mutant. J Biol Chem 287, 19653–19664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill TJ 3rd, Thomsen AR, Tarrasch JT, Plouffe B, Nguyen AH, Yang F, Huang LY, Kahsai AW, Bassoni DL, Gavino BJ, et al. (2017). Distinct conformations of GPCR-β-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc Natl Acad Sci U S A 114, 2562–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman CV, and Benovic JL (1998). G-protein-coupled receptors: turn-ons and turn-offs. Curr Opin Neurobiol 8, 335–344. [DOI] [PubMed] [Google Scholar]
- Carpenter B, Nehmé R, Warne T, Leslie AG, and Tate CG (2016). Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature 536, 104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celver J, Lowe J, Kovoor A, Gurevich VV, and Chavkin C (2001). Threonine 180 is requred for G protein-coupled receptor kinase 3 and b-arrestin mediated desensitization of the mopioid receptor in Xenopus oocytes. J Biol Chem 276, 4894–4900. [DOI] [PubMed] [Google Scholar]
- Chen Q, Iverson TM, and Gurevich VV (2018). Structural Basis of Arrestin-Dependent Signal Transduction. Trends Biochem Sci 43, 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Perry NA, Vishnivetskiy SA, Berndt S, Gilbert NC, Zhuo Y, Singh PK, Tholen J, Ohi MD, Gurevich EV, et al. (2017). Structural basis of arrestin-3 activation and signaling. Nat Commun 8, 1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffa S, Breitman M, Hanson SM, Callaway K, Kook S, Dalby KN, and Gurevich VV (2011a). The Effect of Arrestin Conformation on the Recruitment of c-Raf1, MEK1, and ERK1/2 Activation. PLoS One 6, e28723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffa S, Breitman M, Spiller BW, and Gurevich VV (2011b). A single mutation in arrestin-2 prevents ERK1/2 activation by reducing c-Raf1 binding. Biochemistry 50, 6951–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGraff JL, Gurevich VV, and Benovic JL (2002). The third intracellular loop of alpha2-adrenergic receptors determines subtype specificity of arrestin interaction. J Biol Chem 277, 43247–43252. [DOI] [PubMed] [Google Scholar]
- Farrens DL, Altenbach C, Yang K, Hubbell WL, and Khorana HG (1996). Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science 274, 768–770. [DOI] [PubMed] [Google Scholar]
- Fredericks ZL, Pitcher JA, and Lefkowitz RJ (1996). Identification of the G protein-coupled receptor kinase phosphorylation sites in the human beta2-adrenergic receptor. J Biol Chem 271, 13796–13803. [DOI] [PubMed] [Google Scholar]
- Gimenez LE, Babilon S, Wanka L, Beck-Sickinger AG, and Gurevich VV (2014). Mutations in arrestin-3 differentially affect binding to neuropeptide Y receptor subtypes. Cellular signalling 26, 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez LE, Vishnivetskiy SA, Baameur F, and Gurevich VV (2012). Manipulation of very few receptor discriminator residues greatly enhances receptor specificity of non-visual arrestins. J Biol Chem 287, 29495–29505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman OB Jr., Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, and Benovic JL (1996). Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature 383, 447–450. [DOI] [PubMed] [Google Scholar]
- Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, and Buldt G (1998). X-ray crystal structure of arrestin from bovine rod outer segments. Nature 391, 918–921. [DOI] [PubMed] [Google Scholar]
- Gurevich EV, and Gurevich VV (2006). Arrestins are ubiquitous regulators of cellular signaling pathways. Genome Biol 7, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich EV, Tesmer JJ, Mushegian A, and Gurevich VV (2012). G protein-coupled receptor kinases: more than just kinases and not only for GPCRs. Pharmacol Ther 133, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, and Benovic JL (1995). Visual arrestin binding to rhodopsin: diverse functional roles of positively charged residues within the phosphorylation-recignition region of arrestin. J. Biol. Chem 270, 6010–6016. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Dion SB, Onorato JJ, Ptasienski J, Kim CM, Sterne-Marr R, Hosey MM, and Benovic JL (1995). Arrestin interaction with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, b2-adrenergic, and m2 muscarinic cholinergic receptors. J Biol Chem 270, 720–731. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, and Gurevich EV (2003). The new face of active receptor bound arrestin attracts new partners. Structure 11, 1037–1042. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, and Gurevich EV (2004). The molecular acrobatics of arrestin activation. Trends Pharmacol Sci 25, 105–111. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, and Gurevich EV (2015). Analyzing the roles of multi-functional proteins in cells: The case of arrestins and GRKs. Crit Rev Biochem Mol Biol 50, 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, and Schubert C (2001). Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane translocation. Structure 9, 869–880. [DOI] [PubMed] [Google Scholar]
- Hanson SM, Cleghorn WM, Francis DJ, Vishnivetskiy SA, Raman D, Song X, Nair KS, Slepak VZ, Klug CS, and Gurevich VV (2007). Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J Mol Biol, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Francis DJ, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, and Gurevich VV (2006). Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc Natl Acad Sci U S A 103, 4900–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, and Gurevich VV (2006). The differential engagement of arrestin surface charges by the various functional forms of the receptor. J Biol Chem 281, 3458–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausdorff WP, Bouvier M, O’Dowd BF, Irons GP, Caron MG, and Lefkowitz RJ (1989). Phosphorylation sites on two domains of the beta 2-adrenergic receptor are involved in distinct pathways of receptor desensitization. J Biol Chem 264, 12657–12665. [PubMed] [Google Scholar]
- Hirsch JA, Schubert C, Gurevich VV, and Sigler PB (1999). The 2.8 A crystal structure of visual arrestin: a model for arrestin’s regulation. Cell 97, 257–269. [DOI] [PubMed] [Google Scholar]
- Inagaki S, Ghirlando R, Vishnivetskiy SA, Homan KT, White JF, Tesmer JJ, Gurevich VV, and Grisshammer R (2015). G Protein-Coupled Receptor Kinase 2 (GRK2) and 5 (GRK5) Exhibit Selective Phosphorylation of the Neurotensin Receptor in Vitro. Biochemistry 54, 4320–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrischek H, Prohaska SJ, Gurevich VV, Gurevich EV, and Stadler PF (2017). Uncovering missing pieces: duplication and deletion history of arrestins in deuterostomes. BMC Evol Biol 17, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innamorati G, Le Gouill C, Balamotis M, and Birnbaumer M (2001). The long and the short cycle. Alternative intracellular routes for trafficking of G-protein-coupled receptors. J Biol Chem 276, 13096–13103. [DOI] [PubMed] [Google Scholar]
- Kang DS, Kern RC, Puthenveedu MA, von Zastrow M, Williams JC, and Benovic JL (2009). Structure of an arrestin2-clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking. J Biol Chem 284, 29860–29872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Kuybeda O, de Waal PW, Mukherjee S, Van Eps N, Dutka P, Zhou XE, Bartesaghi A, Erramilli S, Morizumi T, et al. (2018). Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Nature 558, 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, et al. (2015). Crystal structure of rhodopsin bound to arrestin determined by femtosecond X-ray laser. Nature 523, 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Vishnivetskiy SA, Van Eps N, Alexander NS, Cleghorn WM, Zhan X, Hanson SM, Morizumi T, Ernst OP, Meiler J, et al. (2012). Conformation of receptor-bound visual arrestin. Proc Nat Acad Sci USA 109, 18407–18412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim OJ, Gardner BR, Williams DB, Marinec PS, Cabrera DM, Peters JD, Mak CC, Kim KM, and Sibley DR (2004). The role of phosphorylation in D1 dopamine receptor desensitization: evidence for a novel mechanism of arrestin association. J Biol Chem 279, 7999–8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Hofmann KP, Ernst OP, Scheerer P, Choe HW, and Sommer ME (2013). Crystal structure of pre-activated arrestin p44. Nature 497, 142–146. [DOI] [PubMed] [Google Scholar]
- Kim YM, and Benovic JL (2002). Differential roles of arrestin-2 interaction with clathrin and adaptor protein 2 in G protein-coupled receptor trafficking. J Biol Chem 277, 30760–30768. [DOI] [PubMed] [Google Scholar]
- Koehl A, Hu H, Maeda S, Zhang Y, Qu Q, Paggi JM, Latorraca NR, Hilger D, Dawson R, Matile H, et al. (2018). Structure of the μ-opioid receptor-Gi protein complex. Nature 558, 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kook S, Zhan X, Kaoud TS, Dalby KN, Gurevich VV, and Gurevich EV (2014). Arrestin-3 binds JNK1 and JNK2 and facilitates the activation of these ubiquitous JNK isoforms in cells via scaffolding. J Biol Chem 288, 37332–37242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupnick JG, Gurevich VV, and Benovic JL (1997). Mechanism of quenching of phototransduction. Binding competition between arrestin and transducin for phosphorhodopsin. J Biol Chem 272, 18125–18131. [DOI] [PubMed] [Google Scholar]
- Kuhn H (1978). Light-regulated binding of rhodopsin kinase and other proteins to cattle photoreceptor membranes. Biochemistry 17, 4389–4395. [DOI] [PubMed] [Google Scholar]
- Kuhn H, Hall SW, and Wilden U (1984). Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett. 176, 473–478. [DOI] [PubMed] [Google Scholar]
- Kumari P, Srivastava A, Banerjee R, Ghosh E, Gupta P, Ranjan R, Chen X, Gupta B, Gupta C, Jaiman D, et al. (2016). Functional competence of a partially engaged GPCR-β-arrestin complex. Nat Commun 7, 13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte SA, Oakley RH, Zhang J, Holt JA, Ferguson s.S.G., Caron MG, and Barak LS (1999). The 2-adrenergic receptor/arrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc Nat Acad Sci USA 96, 3712–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorraca NR, Wang JK, Bauer B, Townshend RJL, Hollingsworth SA, Olivieri JE, Xu HE, Sommer ME, and Dror RO (2018). Molecular mechanism of GPCR-mediated arrestin activation. Nature 557, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KB, Ptasienski JA, Pals-Rylaarsdam R, Gurevich VV, and Hosey MM (2000). Arrestin binding to the M2 muscarinic acetylcholine receptor is precluded by an inhibitory element in the third intracellular loop of the receptor. J Biol Chem 275, 9284–9289. [DOI] [PubMed] [Google Scholar]
- Li J, Edwards PC, Burghammer M, Villa C, and Schertler GF (2004). Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol 343, 1409–1438. [DOI] [PubMed] [Google Scholar]
- Li L, Homan KT, Vishnivetskiy SA, Manglik A, Tesmer JJ, Gurevich VV, and Gurevich EV (2015). G Protein-coupled Receptor Kinases of the GRK4 Protein Subfamily Phosphorylate Inactive G Protein-coupled Receptors (GPCRs). J Biol Chem 290, 10775–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YL, Khoshouei M, Radjainia M, Zhang Y, Glukhova A, Tarrasch J, Thal DM, Furness SGB, Christopoulos G, Coudrat T, et al. (2017). Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature 546, 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MJ, Andexinger S, Pitcher J, Trukawinski S, Codina J, Faure JP, Caron MG, and Lefkowitz RJ (1992). Receptor-specific desensitization with purified proteins. Kinase dependence and receptor specificity of beta-arrestin and arrestin in the beta 2-adrenergic receptor and rhodopsin systems. J Biol Chem 267, 8558–8564. [PubMed] [Google Scholar]
- Lohse MJ, Benovic JL, Codina J, Caron MG, and Lefkowitz RJ (1990). beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science 248, 1547–1550. [DOI] [PubMed] [Google Scholar]
- Manglik A, Kim TH, Masureel M, Altenbach C, Yang Z, Hilger D, Lerch MT, Kobilka TS, Thian FS, Hubbell WL, et al. (2015). Structural Insights into the Dynamic Process of β2-Adrenergic Receptor Signaling. Cell 161, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, and Granier S (2012). Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature 485, 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald PH, Chow CW, Miller WE, Laporte SA, Field ME, Lin FT, Davis RJ, and Lefkowitz RJ (2000). Beta-arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science 290, 1574–1577. [DOI] [PubMed] [Google Scholar]
- Mendez A, Burns ME, Roca A, Lem J, Wu LW, Simon MI, Baylor DA, and Chen J (2000). Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron 28, 153–164. [DOI] [PubMed] [Google Scholar]
- Meng D, Lynch MJ, Huston E, Beyermann M, Eichhorst J, Adams DR, Klusmann E, Houslay MD, and Baillie GS (2009). MEK1 binds directly to betaarrestin1, influencing both its phosphorylation by ERK and the timing of its isoprenaline-stimulated internalization. J Biol Chem 284, 11425–11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano SK, Kim YM, Stefano FP, Benovic JL, and Brenner C (2006). Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding. J Biol Chem 281, 9812–9823. [DOI] [PubMed] [Google Scholar]
- Milano SK, Pace HC, Kim YM, Brenner C, and Benovic JL (2002). Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry 41, 3321–3328. [DOI] [PubMed] [Google Scholar]
- Miller WE, McDonald PH, Cai SF, Field ME, Davis RJ, and Lefkowitz RJ (2001). Identification of a motif in the carboxyl terminus of beta -arrestin2 responsible for activation of JNK3. J Biol Chem 276, 27770–27777. [DOI] [PubMed] [Google Scholar]
- Moaven H, Koike Y, Jao CC, Gurevich VV, Langen R, and Chen J (2013). Visual arrestin interaction with clathrin adaptor AP-2 regulates photoreceptor survival in the vertebrate retina. Proc Natl Acad Sci U S A 110, 9463–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modzelewska A, Filipek S, Palczewski K, and Park PS (2006). Arrestin interaction with rhodopsin: conceptual models. Cell Biochem Biophys 46, 1–15. [DOI] [PubMed] [Google Scholar]
- Moro O, Lameh J, and Sadee W (1993). Serine- and threonine-rich domain regulates internalization of muscarinic cholinergic receptors. J Biol Chem 268, 6862–6865 [PubMed] [Google Scholar]
- Mukherjee S, Gurevich VV, Preninger A, Hamm HE, Bader M-F, Fazleabas AT, Birnbaumer L, and Hunzicker-Dunn M (2002). Aspartic acid 564 in the third cytoplasmic loop of luteinizing hormone/choriogonadotropin receptor is crucial for phosphorylation-independent interaction with arrestin2. J Biol Chem 277, 17916–17927. [DOI] [PubMed] [Google Scholar]
- Mushegian A, Gurevich VV, and Gurevich EV (2012). The origin and evolution of G protein-coupled receptor kinases. PLoS One 7, e33806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohguro H, Palczewski K, Walsh KA, and Johnson RS (1994). Topographic study of arrestin using differential chemical modifications and hydrogen/deuterium exchange. Protein Sci. 3, 2428–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, LeTrong I, Teller DC, Okada T, Stenkamp RE, et al. (2000). Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289, 739–745. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Pulvermuller A, Buczylko J, and Hofmann KP (1991). Phosphorylated rhodopsin and heparin induce similar conformational changes in arrestin. J Biol Chem 266, 18649–18654. [PubMed] [Google Scholar]
- Pals-Rylaarsdam R, Gurevich VV, Lee KB, Ptasienski J, Benovic JL, and Hosey MM (1997). Internalization of the m2 muscarinic acetylcholine receptor: arrestin-independent and -dependent pathways. J Biol Chem 272, 23682–23689. [DOI] [PubMed] [Google Scholar]
- Pals-Rylaarsdam R, and Hosey MM (1997). Two homologous phosphorylation domains differentially contribute to desensitization and internalization of the m2 muscarinic acetylcholine receptor. J Biol Chem 272, 14152–14158. [DOI] [PubMed] [Google Scholar]
- Prokop S, Perry NA, Vishnivetskiy SA, Toth AD, Inoue A, Milligan G, Iverson TM, Hunyady L, and Gurevich VV (2017). Differential manipulation of arrestin-3 binding to basal and agonist-activated G protein-coupled receptors. Cellular signalling 36, 98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvermuller A, Maretzki D, Rudnicka-Nawrot M, Smith WC, Palczewski K, and Hofmann KP (1997). Functional differences in the interaction of arrestin and its splice variant, p44, with rhodopsin. Biochemistry 36, 9253–9260. [DOI] [PubMed] [Google Scholar]
- Pulvermuller A, Schroder K, Fischer T, and Hofmann KP (2000). Interactions of metarhodopsin II. Arrestin peptides compete with arrestin and transducin. J. Biol. Chem 275, 37679–37685. [DOI] [PubMed] [Google Scholar]
- Raman D, Osawa S, Gurevich VV, and Weiss ER (2003). The interaction with the cytoplasmic loops of rhodopsin plays a crucial role in arrestin activation and binding. J Neurochem 84, 1040–1050. [DOI] [PubMed] [Google Scholar]
- Raman D, Osawa S, and Weiss ER (1999). Binding of arrestin to cytoplasmic loop mutants of bovine rhodopsin. Bichemistry 38, 5117–5123. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, et al. (2007). Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature 450, 383–387. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. (2011). Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, et al. (2011). Structure and function of an irreversible agonist-β(2) adrenoceptor complex. Nature 469, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samama P, Cotecchia S, Costa T, and Lefkowitz RJ (1993). A mutation-induced activated state of the beta 2-adrenergic receptor. Extending the ternary complex model. J Biol Chem 268, 4625–4636. [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, and Ernst OP (2008). Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502. [DOI] [PubMed] [Google Scholar]
- Scheerer P, and Sommer ME (2017). Structural mechanism of arrestin activation. Curr Opin Struct Biol 45, 160–169. [DOI] [PubMed] [Google Scholar]
- Schleicher A, Kuhn H, and Hofmann KP (1989). Kinetics, binding constant, and activation energy of the 48-kDa protein-rhodopsin complex by extra-metarhodopsin II. Biochemistry 28, 1770–1775. [DOI] [PubMed] [Google Scholar]
- Schmidlin F, Roosterman D, and Bunnett NW (2003). The third intracellular loop and carboxyl tail of neurokinin 1 and 3 receptors determine interactions with beta-arrestins. Am J Physiol Cell Physiol 285, 945–958. [DOI] [PubMed] [Google Scholar]
- Seibold A, January BG, Friedman J, Hipkin RW, and Clark RB (1998). Desensitization of beta2-adrenergic receptors with mutations of the proposed G protein-coupled receptor kinase phosphorylation sites. J Biol Chem 273, 7637–7642. [DOI] [PubMed] [Google Scholar]
- Seibold A, Williams B, Huang ZF, Friedman J, Moore RH, Knoll BJ, and Clark RB (2000). Localization of the sites mediating desensitization of the beta(2)-adrenergic receptor by the GRK pathway. Mol Pharmacol 58, 1162–1173. [DOI] [PubMed] [Google Scholar]
- Sente A, Peer R, Srivastava A, Baidya M, Lesk AM, Balaji S, Shukla AK, Babu MM, and Flock T (2018). Molecular mechanism of modulating arrestin conformation by GPCR phosphorylation. Nat Struct Mol Biol 25, 538–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J, Tsakem EL, Breitman M, and Gurevich VV (2011). Identification of arrestin-3-specific residues necessary for JNK3 activation. J Biol Chem 286, 27894–27901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla AK, Manglik A, Kruse AC, Xiao K, Reis RI, Tseng WC, Staus DP, Hilger D, Uysal S, Huang LY, et al. (2013). Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature 497, 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Lefkowitz RJ, and Rajagopal S (2018). Biased signalling: from simple switches to allosteric microprocessors. Nat Rev Drug Discov 17, 243–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Coffa S, Fu H, and Gurevich VV (2009). How does arrestin assemble MAPKs into a signaling complex? J Biol Chem 284, 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Raman D, Gurevich EV, Vishnivetskiy SA, and Gurevich VV (2006). Visual and both non-visual arrestins in their “inactive” conformation bind JNK3 and Mdm2 and relocalize them from the nucleus to the cytoplasm. J Biol Chem 281, 21491–21499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, and Gurevich VV (2005). Crystal Structure of Cone Arrestin at 2.3Å: Evolution of Receptor Specificity. J Mol Biol 354, 1069–1080. [DOI] [PubMed] [Google Scholar]
- Szczepek M, Beyriere F, Hofmann KP, Elgeti M, Kazmin R, Rose A, Bartl FJ, von Stetten D, Heck M, Sommer ME, et al. (2014). Crystal structure of a common GPCR-binding interface for G protein and arrestin. Nat Commun 5, 4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ARB, Plouffe B, Cahill III TJ, Shukla AK, Tarrasch JT, Dosey AM, Kahsai AW, Strachan RT, Pani B, Mahoney JP, et al. (2016). GPCR-G Protein-β-Arrestin Super-Complex Mediates Sustained G Protein Signaling. Cell 166, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eps N, Altenbach C, Caro LN, Latorraca NR, Hollingsworth SA, Dror RO, Ernst OP, and Hubbell WL (2018). Gi- and Gs-coupled GPCRs show different modes of G-protein binding. Proc Natl Acad Sci U S A 115, 2383–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eps N, Caro LN, Morizumi T, Kusnetzow AK, Szczepek M, Hofmann KP, Bayburt TH, Sligar SG, Ernst OP, and Hubbell WL (2017). Conformational equilibria of light-activated rhodopsin in nanodiscs. Proc Natl Acad Sci U S A 114, E3268–E3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violin JD, and Lefkowitz RJ (2007). Beta-arrestin-biased ligands at seven-transmembrane receptors. Trends Pharmacol Sci 28, 416–422. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Baameur F, Findley KR, and Gurevich VV (2013). Critical role of the central 139-loop in stability and binding selectivity of arrestin-1. J Biol Chem 288, 11741–11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Francis DJ, Van Eps N, Kim M, Hanson SM, Klug CS, Hubbell WL, and Gurevich VV (2010). The role of arrestin alpha-helix I in receptor binding. J. Mol. Biol 395, 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Gimenez LE, Francis DJ, Hanson SM, Hubbell WL, Klug CS, and Gurevich VV (2011). Few residues within an extensive binding interface drive receptor interaction and determine the specificity of arrestin proteins. J Biol Chem 286, 24288–24299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Hirsch JA, Velez M-G, Gurevich YV, and Gurevich VV (2002). Transition of arrestin in the active receptor-binding state requires an extended interdomain hinge. J Biol Chem 277, 43961–43968. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Hosey MM, Benovic JL, and Gurevich VV (2004). Mapping the arrestin-receptor interface: structural elements responsible for receptor specificity of arrestin proteins. J. Biol. Chem. 279, 1262–1268. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Raman D, Wei J, Kennedy MJ, Hurley JB, and Gurevich VV (2007). Regulation of arrestin binding by rhodopsin phosphorylation level. J Biol Chem 282, 32075–32083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther C, Nagel S, Gimenez LE, Mörl K, Gurevich VV, and Beck-Sickinger AG (2010). Ligand-induced internalization and recycling of the human neuropeptide Y2 receptor is regulated by its carboxyl-terminal tail. J Biol Chem 285, 41578–41590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanka L, Babilon S, Kaiser A, Mörl K, and Beck-Sickinger AG (2018). Different mode of arrestin-3 binding at the human Y1 and Y2 receptor. Cellular signalling 50, 58–71. [DOI] [PubMed] [Google Scholar]
- Wilden U (1995). Duration and amplitude of the light-induced cGMP hydrolysis in vertebrate photoreceptors are regulated by multiple phosphorylation of rhodopsin and by arrestin binding. Biochemistry 34, 1446–1454. [DOI] [PubMed] [Google Scholar]
- Wilden U, Hall SW, and Kühn H (1986). Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proceedings of the National Academy of Sciences 83, 1174–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilden U, and Kühn H (1982). Light-dependent phosphorylation of rhodopsin: number of phosphorylation sites. Biochemistry 21, 3014–3022. [DOI] [PubMed] [Google Scholar]
- Wisler JW, Rockman HA, and Lefkowitz RJ (2018). Biased G Protein-Coupled Receptor Signaling: Changing the Paradigm of Drug Discovery. Circulation 137, 2315–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Hanson SM, Francis DJ, Vishnivetskiy SA, Thibonnier M, Klug CS, Shoham M, and Gurevich VV (2006). Arrestin binding to calmodulin: a direct interaction between two ubiquitous signaling proteins. J Mol Biol 364, 955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, McClatchy DB, Shukla AK, Zhao Y, Chen M, Shenoy SK, Yates JR, and Lefkowitz RJ (2007). Functional specialization of beta-arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci U S A 104, 12011–12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Gimenez LE, Gurevich VV, and Spiller BW (2011a). Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual arrestins. J Mol Biol 406, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Kaoud TS, Dalby KN, and Gurevich VV (2011b). Non-visual arrestins function as simple scaffolds assembling MKK4- JNK3α2 signaling complex. Biochemistry 50, 10520–10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Kaoud TS, Kook S, Dalby KN, and Gurevich VV (2013). JNK3 binding to arrestin-3 differentially affects the recruitment of upstream MAP kinase kinases. J Biol Chem 288, 28535–28547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Perez A, Gimenez LE, Vishnivetskiy SA, and Gurevich VV (2014). Arrestin-3 binds the MAP kinase JNK3alpha2 via multiple sites on both domains. Cellular signalling 26, 766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Stoy H, Kaoud TS, Perry NA, Chen Q, Perez A, Els-Heindl S, Slagis JV, Iverson TM, Beck-Sickinger AG, et al. (2016). Peptide mini-scaffold facilitates JNK3 activation in cells. Sci Rep 6, 21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun B, Feng D, Hu H, Chu M, Qu Q, Tarrasch JT, Li S, Kobilka TS, Kobilka BK, et al. (2017). Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 546, 248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XE, He Y, de Waal PW, Gao X, Kang Y, Van Eps N, Yin Y, Pal K, Goswami D, White TA, et al. (2017). Structural Identification of Phosphorylation Codes for Arrestin Recruitment by G protein-Coupled Receptors. Cell 170, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang T, Chen Q, Cho M-K, Vishnivetskiy SA, Iverson TI, Gurevich VV, and Hubbell WL (2013). Involvement of Distinct Arrestin-1 Elements in Binding to Different Functional Forms of Rhodopsin. Proc Nat Acad Sci USA 110, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo Y, Vishnivetskiy SA, Zhan X, Gurevich VV, and Klug CS (2014). Identification of receptor binding-induced conformational changes in non-visual arrestins. J Biol Chem 289, 20991–21002. [DOI] [PMC free article] [PubMed] [Google Scholar]