Abstract
Microglia are the resident immune cells that maintain brain homeostasis and contribute to neurodegenerative disorders. Recent studies of microglia at transcriptomic and epigenetic levels revealed specific molecular pathways that regulate microglia development, maturation and reactive states. The transcriptionfactor PU.1 plays a key role in regulating several microglial functions. Environmental factors such as microbiota, early life stress and maternal immune activation can dysregulate PU.1 and innate immune response. This review discusses the epigenetic regulation of key transcriptional factors in human and murine microglia, highlighting their networks for shaping the microglial function. PU.1 and other microglia-specific transcriptional factors can be further studied to determine their therapeutic applications in neurologic disorders.
Keywords: Alzheimer’s disease, autism spectrum disorders, early life stress, epigenetic, microglia, neurodegenerative disorders, PU.1, transcription factors
The transcriptomic and epigenetic landscape of microglia
Microglia are the resident immune cells that maintain homeostasis in the central nervous system (CNS). Microglia share several markers with monocytes, but are derived from the embryonic yolk sac and have distinct lineage from bone marrow-derived myeloid cells [1]. Microglia support neuronal functions and networks in addition to maintaining brain homeostasis [2]. Under physiological conditions, microglia actively monitor their surroundings, respond to perturbations, clear cell debris, and regulate synaptic transmission [3]. During development, they modulate synapse formation and elimination, revealing microglia’s role in shaping the brain circuitry [4]. Microglial dysfunction contributes to the pathogenesis of neurodegenerative disorders including Alzheimer’s disease (AD) and neurodevelopmental disorders including autism spectrum disorder and schizophrenia [5-7]. In neurodegenerative state, microglia can enhance neuroinflammation leading to more cell deaths [6, 8]. Although not as clear in neurodevelopmental disorder, microglia can have impaired pruning or secrete molecules that contribute to hyperactivity in the CNS [7, 9]. This further emphasizes the need to define the molecular mechanisms that control microglial functions. Investigation on how microglial function is regulated at transcriptome and epigenome (See Glossary level is invaluable in developing therapeutic strategies for neurological disorders.
Advancement in defining microglia-specific markers has enabled scientists to sort, characterize and determine the molecular phenotype of microglia at different states. RNA-sequencing studies have identified microglia-specific genes such as olfactomedin-like protein 3 (Olfml3), which is involved in early development patterning, and sialic acid binding Ig-like lectin H (Siglech), which is involved in innate immune cell differentiation [10]. Microglial markers such as purinergic receptor P2Y G-protein coupled 12 (P2ryl2), transmembrane protein 119 (Tmem119), and Fc receptor-like S, scavenger receptor (Fcrls), are used to define microglia homeostatic state. Whereas, inflammatory marker Clec7a is used to define microglia in neurodegenerative state [11]. Transcriptomic analysis of isolated mouse microglia revealed that microglia express a unique genes sets called the “sensome” that enable them to sense endogenous ligands and interact with their environment [12].
In mice, microglia undergo a step-wise program of maturation that is synchronized with the developmental phases of the brain [13, 14], which consists of three distinct temporal stages: early microglia spanning until embryonic day 14, premicroglia from embryonic day 14 to a few weeks after birth, and adult microglia from a few weeks after birth onward [13]. The transcriptomic profile can be shaped by intrinsic factors, such as age and sex, pathogenic and tissue-specific cues including protein aggregates, stress signals, nutrients, and external cues such as signals from the microbiota colonizing gastrointestinal and urogenital tracts [15]. Under pathological circumstances, cues such as debris or stress signals from surrounding tissue can lead to transcriptomic and morphological changes in microglia, which can switch from homeostatic to reactive states [16, 17]. Here, this review is focused on summarizing the recent discoveries on the molecular mechanisms of how environmental cues can affect microglial function.
Elucidating critical transcriptional regulators of microglial gene expression
The list of known microglia-specific genes is constantly being updated based on improved microglial isolation techniques and labeling methods. Microglial transcriptomes are tightly regulated by multiple key transcription factors such as Sall1, Spi1, Mafb, and Mef2c [5, 15]. Recent genomic and proteomic studies revealed microglia express a unique set of genes distinct from infiltrating monocytes and other immune cells [18, 19]. Barres group recently identified Tmem119++/Sall1+/Gpr56+/Apoe+ as yolk sac-derived microglia, whereas hematopoietic stem cell-derived microglia-like cells express Tmem119+/Ms4a7+/Clec12a+/Apoe+++ suggesting lineage-specific microglia markers that can be useful in determining the origin of microglia [18, 19]. In addition to microglia, new CD38+ resident myeloid cells called CNS border-associated macrophages (BAMs) are embryonically-derived cells that line along the meninges, choroid plexus, and perivascular spaces [20]. Both microglia and BAMs are CD11b+/CD45low, making it difficult to distinguish between them without localization patterns. Based on current FACS and immunohistochemical analysis, the transcription factor SALL1 was identified as a key marker for microglia. High expression of SALL1 in microglia allows distinguishing SALL1+ microglia from SALL1− BAM cells and infiltrating peripheral monocytes [20, 21]. Currently, FACS and immunocytochemistry are used to distinguish microglia, macrophages and BAM based on RNA-seq identified gene markers including differential expression of CD11b, CD45, CD38, CLEC7A, FCRLS, P2RY12, SALL1 and TMEM119.
In addition to FACS and immunohistochemical approaches, assay for transposase accessible chromatin sequencing (ATAC-seq) can evaluate transcriptional networks that control microglial signature at an epigenetic level determining the chromatin accessibility landscape in microglia [5, 22, 23]. Using this approach, Gosselin et al. reported PU.1 transcriptional network as the most abundantly expressed ETS domain transcription factor in both murine and human microglia [22]. PU.1 is an ETS-domain transcription factor encoded by SPI1 (human) or Spi1 (murine) gene. PU.1 binds to purine-rich sequence (PU-box) and activates gene expression during immune cell development [24, 25]. Microglia development is largely controlled by the transcription factors IRF8 and PU.1. The microglia-enriched PU.1 binding regions were also enriched with Smad, Mef2, and Ctcfl family-binding motifs, identifying interactions between the transcription factors [5]. Furthermore, four binding motifs Mafb, Stat3, Usf1, and Smad2 were preferentially associated with PU.1 binding specifically in microglia in mice, suggesting these transcription factors are microglia-specific co-regulators that cooperate with PU.1 for establishing microglia-specific gene enhancer profiles [5].
The role of PU.1 in the maintenance of homeostatic microglia
Spi1 (PU.1) mRNA is continuously expressed from EMPs to adulthood microglia, suggesting the necessity of PU.1 for microglial development and functional maintenance [1]. Chromatin immunoprecipitation (ChIP)-seq analysis of immortalized mouse microglial cell line BV2 cells revealed that about 63% of microglial sensome genes are PU.1 targets, suggesting that PU.1 regulates specific microglial functions [26]. PU.1-deficient mice display multiple immune-developmental abnormalities, including parenchymal microglia [27]. Accordingly, PU.1-deficient mice have reduced expression of Csf1r, Csf2r and Csf3r, suggesting a critical role of PU.1 for the expression of essential cell survival receptors [28].
Alterations in the PU.1 expression level can change microglial morphology and function. PU.1 expression levels are similar in isolated human glial cells from biopsy and postmortem adult human brain tissues [29]. However, silencing of PU.1 by siRNA reduced the number of microglia and their ramification and phagocytosis of amyloid-beta peptide 1-42 (AB42) in mixed glial culture in their study [29]. Silencing of PU.1 in human microglia culture suppressed expression of myeloid adapter protein DAP12/TYROBP, TREM2, LST1, CD11b, MRC1, CEBPA, PTPRC and MHC class II genes HLA-DR, HLA-DP and HLA-DQ [30, 31], suggesting its role in antigen presentation and pro-inflammatory response by PU.1 [30, 31].
Beside PU.1 as a key transcription factor, other transcriptional regulators are involved in the precise tuning of microglial homeostasis, making microglial transcriptome tightly regulated (Table 1). These molecules are necessary to understand how microglial transcriptome can be regulated in various states. In vitro and in vivo studies show that PU.1 act as a master regulator and controls the development and maintenance of homeostatic function in microglia, such as CSF1R-mediated cell survival, phagocytosis, antigen presentation, and ramified morphology, but has no effect on TLR4-mediated inflammatory responses. But further research is required to determine how these different transcription factors interact to maintain microglial identity and determine if there are differences between human and murine microglia.
Table 1.
Key transcriptional factors for microglial development
| Key Transcription Factors |
Expression of Transcription Factor |
Signaling Molecules | Role in Microglial Function | Effect on Microglial Population and Transcriptome |
Ref |
|---|---|---|---|---|---|
| Interferon regulatory factor 8 (IRF8) | Highly expressed in the hematopoietic precursors | Heterodimeric partner of PU.1 and is a molecular target downstream of PU.1 | Regulates early maturation of microglial precursor cells; regulates survival of microglia in early stage | IRF8-deficiency showed reduced number of mature precursor cells that are CD45+, CX3CR1HIGH, F4/80HIGH | [13, 18, 20, 77] |
| Myocyte enhancer factor 2a MEF2a | Specifically expressed in adult microglia | TGFB | Maintaining matured microglial phenotype and shapes epigenomic landscape | MEF2A motif analysis showed increased fraction of promoters associated with the regulator regions of adult microglia genes | [13, 34, 78] |
| Myocyte enhancer factor 2c MEF2C | Reduced in aged mice due to upregulation of type-1 interferon (IFN-α/β) expression | ↑IFN1 > ↓MEF2C; TGFB | Aged microglia phenotype with increased secretion of pro-inflammatory cytokines; regulating neuronal synaptic plasticity | Injection of TNFa, a proinflammatory cytokine associated with aging, in MEF2C-deficient mice led to IBA1 intensity. Following LPS stimulation in vtiro, isolated from MEF2C-deficient microglia secreted more proinflammatory cytokines and reduced social preference behavior |
[34, 79] |
| PU.1 | Continuously expressed from EMPs to adulthood microglia | Downstream of RUNX1 during early development; TGFB | Maintains homeostatic genes and suppresses pro-inflammatory pathways | 63% of microglial sensome genes are PU.1 targets: Lst1, Hla-Dra, Cd11b, Mrc1, Cebpa, Trem2, Ptprc, Tyrobp | [22] |
| Runt-related transcription factor 1 (RUNX1) | Highly expressed in the hematopoietic precursors | TGFB | Involved in proliferation and differentiation into ramified morphology | RUNX1-binding motif is enriched at the enhancer landscape of adult mouse and human microglia, suggesting a role in the maintenance of adult microglial phenotype | [22, 80-82] |
| Spalt like transcription factor 1 (SALL1) | Expressed during embryogenesis, upregulated in adult phase | TGFB | Maintains homeostatic genes and suppresses pro-inflammatory pathways | Increased homeostatic genes including P2ry12, Serpinf1 or Slc2a5; reduced macrophage-related genes such as Msr1, Cd69, Igf1 and Spic And reduced pro-inflammatory genes: Axl, Nox2 and Irf7 | [21, 24, 57, 83] |
| V-maf musculoapon eurotic fibrosarcoma oncogene homolog B (MAFB) | Specifically expressed in adult microglia, increased expression during aging | ↑GM-CSF > ↓MAFB; TGFB | Role in adulthood microglia maturation and differentiation; suggested antagonistic relationship with the interferon pathway; “off-state factor” for regulating microglia response under stressed or pathological conditions, which regulates microglial gene signature, self-renewal and morphology | Promotes anti-inflammatory phenotypes and regulates expression of immune and viral genes; increased Ctsh and Pmepa1 in the later adult stage indicating that Mafb suppresses antiviral response pathways; MAFB-deficient microglia show increased self-renewal and reduced P2ry12 and Ccl2 expression in the presence of GM-CSF treatment | [13, 84-88] |
Epigenetic regulation of transcription factors in microglia
Immune memory in microglia and macrophages are shaped by epigenetically mediated alterations in the enhancer repertoire of targeted genes [11, 32]. Microglial gene enhancers form complexes with lineage-determining transcription factors including PU.1 and SALL1, and become co-activators to regulate the transcription of targeted genes [33]. ATAC-seq of isolated microglia across different developmental stages showed that accessibility of promoter regions by transcriptional complexes was largely conserved over time [13, 34]. Acetylation of histone H4 on the promoter and intron-1 region of the PU.1 locus are tightly regulated for allowing the interaction between the locus and RNA polymerase II. Inhibition of histone deacetylase (HDAC) increases acetylation of H4 and disrupts the interaction between the locus and RNA polymerase II, leading to the suppression of PU.1 transcription in murine microglia [35, 36]. Valproic acid treatment, which selectively inhibits the catalytic activity of class I HDACs and induces proteasomal degradation of HDAC2, of primary adult human microglia reduces phagocytic capacity and expression of PU.1 and CD45, suggesting the its regulation of microglial phagocytosis via epigenetic mechanism [36-38]. Furthermore, Rustenhoven et al. identified another HDAC class I/II/IV inhibitor vorinostat as an effective attenuator of PU.1 in primary human microglia-enriched culture [30, 39]. However, vorinostat treatment showed reduction in DAP12 expression, whereas expression of CD45 and HLA-DR, HLA-DP or HLA-D1 remained unchanged [30]. This discrepancy could be due to the difference in the coverage of HDAC classes or off-target effect of the drugs. Studies with HDAC inhibitors show that microglial transcriptome can be epigenetically regulated, possibly through PU.1 suppression. More precise epigenetic studies are necessary by genetic targeting of specific HDACs in microglia.
Several groups have identified epigenetic regulators that can contribute to microglial function (Table 2). Methyl-CpG binding protein 2 (MECP2) regulates microglial responsiveness to environmental stimuli. MECP2-deficiency, as seen in Rett Syndrome, can lead to microglial activation and dysfunction [17]. Global loss of Mecp2 leads to reduced numbers of microglia, perivascular meningeal macrophages, intestinal macrophages and LY6Clo circulating monocytes [17]. MECP2 acts as a microglia-specific transcriptional repressor of SNAT1, a major glutamate transporter. MECP2-deficient microglia have impaired glutamate synthesis and mitochondria function [40]. Further examination with ChIP-seq analysis revealed that Mecp2-deletion increased histone acetylation at enhancer regions of Fkbp5, a canonical glucocorticoid target gene, and recruitment of nuclear receptor co-repressor2 (NCOR2) and HDAC3 complex [17]. These results show dysregulation of genes involved in glucocorticoid signaling, hypoxia response and inflammatory responses, suggesting Mecp2 is critical for maintaining immune cell function including microglia [17].
Table 2.
Epigenetic modification in microglia
| Stimulation | Epigenetic Modification | Microglia Phenotype and Effect on Transcription Factor | Refs |
|---|---|---|---|
| Aβ accumulation and 1X or 4XLPS injection | H3K4me1 increased H3K4me1 levels in microglia from 1xLPS versus 4xLPS wildtype animals showed enrichment for the ‘thyroid hormone signaling pathway’, including a putative enhancer for hypoxia inducible factor-1α (HIF-1α). 4xLPS-treated APP animals showed increased H3K4me1 levels in putative enhancers related to phagocytic function. | Activation of HIF-1a and mTOR pathways leading to DAM or MGnD phenotype with enhanced phagocytosis | [11] |
| Aβ accumulation | Similar levels of H3K4me2 in AD microglia and DAM microglia | Suggested to be involved in priming of DAM phenotype since similar level of active H3K4me2 in AD microglia and DAM microglia | [41] |
| MECP2 deletion | Increased histone H4 acetylation at cis-regulatory regions of Fkbp5, a canonical glucocorticoid target gene, and recruitment of nuclear receptor co-repressor2 and histone deacetylase 3 (Hdac3) complex | Dysregulation of genes involved in glucocorticoid signaling, hypoxia response and inflammatory responses | [17] |
| Dying neurons | Clearance-specific genes display negligible amounts of H3K27me3 in cbMg, with higher cell death rate, as compared to in stMg. Dying cells in vitro increases the expression of H3K27me3-specific demethylases, which leads to reduced H3K27me3 activity. | Suppression of clearance genes and progressive induction of phagocytic genes | [42] |
Histone H3 at lysine 4 (H3K4) is used to package DNA in eukaryotic cells including human cells, and modifications to the histone alter the accessibility of genes for transcription. Recent ChIP-seq results have revealed active H3K4me2 regions shared in both MGnD and wild-type microglia, suggesting that the disease-associated regions primed in MGnD can be primed even in homeostatic microglia [41]. Peripheral immune stimulation and cerebral beta-amyloidosis can lead to epigenetic modification in microglia. Cerebral beta-amyloidosis activates HIF-1a and mTOR pathways, which leads to transcriptional and functional alterations associated with increased inflammatory and immune response genes in MGnD [11]. In addition, APP mice with four-time LPS treatment showed increased H3K4me1 levels in putative enhancers related to phagocytic function [11]. These data suggest the differential effect of immune training versus tolerance due to multiple peripheral immune stimulation, which is reflected in the epigenetic landscape of MGnD in AD mouse models. Further research can reveal how other stimuli may lead to chronic modulation of microglia response and contribute to the progression of the neurodegenerative disorder.
Microglia can efficiently clear apoptotic cells and debris, which is regulated by H3K27me3, a histone methylation occurring on the amino (N) terminal tail of the core histone H3. This tri-methylation is associated with the downregulation of nearby genes via the formation of heterochromatic regions. In the adult brain, clearance activity of microglia is suppressed in areas with low frequency of neuronal deaths [42]. This region-dependent phagocytic activity is regulated via suppression of clearance-related genes by epigenetic modification of H3K27me3 based on ChIP-Seq analysis. In contrast to striatum microglia (stMG), cerebellar microglia (cbMg) displayed a phagocytic microglia phenotype with reduced ramifications and cell volume that is characteristic of immature microglia and MGnD, but lack enrichment of pro-inflammatory genes in mice [42]. Single-nuclei RNA-seq and proteomic analysis validated that mouse cbMg expressed higher levels of clearance-associated proteins such as AXL, LC3, APOE, CD74, MHCII, and MRC1. These studies demonstrate the epigenetic regulation of clearance-associated genes for the regulation of microglial phagocytosis in a brain region-specific manner.
In addition to histone modifications, microRNAs (miRNAs) are also involved in controlling the epigenetic landscape in the CNS. miRNAS are small non-coding RNA molecules involved in silencing and post-transcriptional regulation of gene expression via base-pairing with complementary sequences within mRNA molecules. Through miRNA and mRNA microarray assays, gene regulators such as miR-124 and miR-155 have been identified in microglial activation pathways [24, 43]. Several miRNAs including miR-124 and miR-155 modulate the development of microglial cells. miR-124 is suggested to directly inhibit C/EBPa and PU.1 to prevent acquisition of a reactive microglia state [44]. These studies suggest that miRNAs including miR124 and miR155 act as key regulators of microglia quiescence in the CNS.
Microglia in neurodegenerative state
In diseased states, microglia can become dysfunctional and neurotoxic, which are referred as disease-associated microglia (DAM) or microglia neurodegenerative phenotypes (MGnD). DAM or MGnD are found in neurodegenerative disorders including AD, amyotrophic lateral sclerosis (ALS) and multiple sclerosis [41, 45]. In AD brains, microglia surrounding amyloid plaques display activated amoeboid morphologies with enlarged soma and retracted processes, indicating microglial response to protein accumulation or increased inflammatory mediators [41, 46-48]. In mouse models of AD such as amyloid precursor protein (APP)/presenilin-1 (PS1) and 5XFAD transgenic mice, there is a subset of microglia surrounding amyloid plaques with elevated phagocytic and microglial activation markers including Galectin-3 and CLEC7A, and reduced expression of homeostatic markers such as P2RY12. This suggests a switch from homeostatic to reactive microglial state in specific population associated with amyloid plaques [46]. Krasemann et al. found dystrophic neurites stimulate TREM2 –APOE pathway that results in phenotypic switch from homeostatic to MGnD microglia [45]. Customized NanoString-based gene expression analysis, which uses molecular barcodes and microscopic imaging to detect and count unique transcripts in hybridization reactions, revealed APOE and TGFβ as the major upstream regulators of MGnD microglia. Upregulation of APOE suppresses gene expression of microglial homeostatic transcriptional factors including Spi1, Mef2a, Mafb, Smad3, and induces inflammatory response via induction of transcription factors Bhlhe40, Tfec, Atf, and inflammatory miRNA miR-155, which if genetically ablated, can reverse abnormal microglia signature and ameliorate disease in SOD1 mice, a mouse model for ALS [12, 45]. More recently, Litvinchuk et al. also identified microglia-specific transcription factors Irf8, Spi1, Runx1 are significantly upregulated in FACS-isolated microglia in PS19 mice, a model for neurodegenerative tauopathy and Alzheimer's disease [49]. Deficient-C3ar, the complement factor C3 complement receptor that mediates neuroimmune crosstalk critical in network function, reduces expression of Irf8, Spi1, and Runx1 in PS19 mice [6, 49, 50]. This novel finding shows the detrimental effect of activated complement system and how expression of microglial transcription factors can be recovered by inactivation of a complement pathway in a neurodegenerative model [49]. Through unbiased approach using RNA-seq or Nanostring assays, specific pathways such as TREM2-APOE and C3-C3AR can be identified to determine critical regulators in neurodegenerative state in microglia. This approach also identifies key gene markers to identify sub-populations of microglia associated with disease-states, which can be further tested in additional mice models including aging.
PU.1 also plays a role in traumatic brain injury (TBI). Genome-wide methyl binding domain (MBD)-seq and RNA-seq of brain tissues in TBI induced by lateral fluid percussion in adult male Spraque-Dawley rats showed molecular networks and mechanism underlying the chronic dysregulation [51]. Transcription factors including Spi1, Cebpd, Pax6, Tp73 were upregulated at 3 months after TBI [51]. Together, these studies imply that PU.1 and other transcription factors are critical in maintaining homeostatic microglia transcriptome, but can be influenced by neurodegenerative disorder or TBI.
Microglia transcriptome in aging
Transcriptome analysis of aged human microglia reveal specific genes influenced by aging [52-54]. With maturation and aging, expression of pro-inflammatory genes in adult microglia increase, suggesting their reactive state and reduced plasticity in the adult stage possibly due to repetitive exposure to infectious agents and active removal of cell debris [53, 55]. In aging mice, the microglial sensome transcripts for sensing endogenous ligands including Trem2, Gpr34, P2ry12, Fcrl1, and Dap12, are downregulated. Microbe recognition and host defense genes such as Tlr2, Cd74, Ltf, Clec7a, Cxcl16, are also upregulated [12]. Interestingly, Wehrspaun et al. analyzed three transcriptome data sets from postmortem human cortical tissue spanning teens to elderly, and found microglial markers to be assembled into a gene co-expression network regulated by an age-dependent transcriptional module consisting of RUNX1, IRF8, SPI1, and TAL1 [53]. Further analysis revealed that microglia surface receptors for microglia-neuron crosstalk (CX3CR1, P2RY12, and TREM2) and TLRs for communication with pathogen-associated molecular patterns tended to show a negative correlation between gene expression level and age, highlighting the age-dependent change in microglial plasticity [53, 55]. More recently, Olah et al. suggested a protective effect of APOE2 haplotype against increased expression of aged human microglia gene set in human microglia [12, 56]. Their results demonstrated upregulation of CD33, INPP5D, MS4A4A, SORL1, and TREM2 in aged microglia, but no change in PU.1. Compared to previous reports, the mean age of the postmortem tissue donors was greater at 95-years old and their cohort consisted of mostly females, which could explain the discrepancy [56]. However, lack of PU.1-enrichment in aged human microglia gene set might also be due to tissue processing differences including postmortem tissue degradation or microglial isolation. In summary, aged microglia show reduced expression of sensome genes and increased expression of neuroprotection genes, which are modulated by RUNX1, IRF8, PU.1 and TAL1, supporting aged microglia should be not considered dysfunctional but rather have shifting functional roles in aged brain to maintain brain circuit function.
The effect of microbiome on microglia transcriptome
The gut microbiome has emerged as a key regulator of the immune system and microglial physiology. Accumulating evidence indicates that PU.1 expression significantly alters the microglia phenotypes via reshaping the gut microbiota landscape. Microglia are dysregulated in mice with dysbiosis, either due to microbial imbalance or maladaptation [15]: gut symbiosis promoted the maintenance of microglia under homeostasis conditions, while germ-free (GF) condition led to impaired microglia maturation, differentiation and function [15]. RNA-seq analysis of microglia from GF mice revealed that microglia transcription factor PU.1 and survival factor CSF1R were significantly upregulated in microglia from GF mice [1, 15]. The immature phenotype of GF microglia could be corrected by microbiota recolonization [15]. However, the mechanism in which microbiota can influence the maturation and homeostatic state maintenance remains to be determined. These research findings based on mouse models can be helpful in providing shared mechanisms between human and mice on how microbiota can influence microglial phenotype and transcriptome. The human microbiota can be more complex due to changing diet and activities compared to laboratory mice microbiota.
ATAC-seq analysis of embryonic microglia from specific pathogen-free (SPF) and GF mice have shown that several regions of the differentially accessible regions are more accessible in embryonic microglia of SPF but not GF mice during development [57]. More recently, Thion et al. demonstrated that microglia acquire sexually dimorphic transcriptome profiles and different response to the GF environment in C57BL/6J SPF and GF mice [57]. Guneykaya et al. also reported sex-dependent differences in transcriptomic and proteomic profiles in microglia [58]. However, PU.1 was not among the top transcription regulators, supporting the idea that PU.1 is a critical regulator in microglial identity and survival that can be regulated by the presence of microbiota in sex-independent manner. These studies suggest that gut microbiota can induce subtle, but global chromatin accessibility changes during embryonic microglia development and regulates several key targets involved in microglial maturation in sex and stage-specific manner.
Early life environmental perturbation of microglia
Maternal immune activation (MIA) by administration of polyinosinic:polycytidylic acid (polyI:C), which is a synthetic analog of double-stranded RNA used to simulate viral infections via interaction with TLRs, can disrupt pre-microglial development in offspring, leading to a more advanced developmental stage. Hippocampal microglial cells from polyI:C offspring mice showed dysregulation of genes associated with inflammatory response, cell migration and phagocytosis [13, 59, 60]. Microglia from polyI:C offspring had reduced expression of PU.1 and its target genes, which were restored after administration of anti-inflammatory drug minocycline [26, 59]. Downregulated genes included Fcgr1, Itgav, P2ry6, Sirpa, Siglece and Cx3cr1 that constitute the sensome. This study demonstrates the significant environmental effect by maternal immune activation on microglial gene expression phenotype, which are regulated by PU.1, and show similarity to those of microglia derived from AD mouse models including APP/PS1 mice (Table 3). Future studies are needed to validate the function of PU.1 in dysregulated microglia in both germ-free and MIA mice.
Table 3.
Epigenetic regulation of PU.1 expression in microglia
| Model | Tissue treatment | Isolation method | PU.1 expression change | Dynamic changes | Ref |
|---|---|---|---|---|---|
| Maternal immune activation (MIA) | C57BL/6 5mg/kg PolyIC intraperitoneal injection at G15-Hippocampal tissue-> anti-inflammatory minocycline | Percoll gradient-> MACS beads with CD11b | ↓PU.1 | Downregulated genes included Fcgr1, Itgav, P2ry6, Sirpa, Siglece, Cx3cr1 that constitute the sensome in which microglia-neuron communicates | [59] |
| Early life stress | BALB/cByj mice separation procedure occurred daily from PND1-21; Hippocampus collected from P28 mice | Percoll gradient-> FACS CD11b, CD45 | ↓PU.1 | reduced expression of pro-inflammatory genes such as Tlr3, IL17ra, increased expression of IL6, high rate of phagocytic activity, reduced expression of many developmental genes | [64] |
| Germ-free (GF) | 6-20 weeks adult GF C57BL/6 mice, SPF mice treated with antibiotics (cefoxitin, gentamicin, metronidazole-> Recolonization of the gut with complex microbiota | density gradient-> FACS for CD11b and CD45 | ↑PU.1 | Immature phenotype of GF microglia, reduced innate immune response, GF microglia failed to display activated morphology after LPS stimulation | [15] |
| Alzheimers Disease (AD) | CK-p25 3-month old female mice-hippocampus | IHC | ↑PU.1 | N/a | [89] |
| PS19 tau transgenic mice-> C3aR KO mice | Myelin removal-> FACS with CD11b and CD45 | ↑PU.1 | Increased volumes and reduced branching and complexity in microglia and astrocytes of PS19, which were corrected by C3aR-KO in mice. | [49] | |
| Human postmortem hippocampal; Human primary microglial-enriched culture-> Vorinostat (HDAC inhibitor) | IHC | ↑PU.1 | N/a | [30] | |
| Huntingtons Disease (FID) | BV2 cell line; R6/2 mouse line with mHTT knock in Hdh175/175 mice -> siRNA – PU.1 | Percoll gradient→ FACS with CD11b, CD45, CD16/32 | Age/stage dependent ↑PU.1 | Increased proinflammatory phenotype including IL6 expression | [23] |
| Traumatic Brain Injury (TBI) | adult male Spraque-Dawley rats lateral fluid percussion | IHC | ↑PU.1 | N/a | [51] |
More recent studies have linked microbiota with behavioral deficits associated with MIA [61, 62]. The MIA phenotypes require maternal intestinal bacteria that promote Th17 cell differentiation [63]. In addition, when polyI:C offspring are orally treated with human commensal Bacteroides fragilis, the gut microbiota composition is altered and ameliorates MIA phenotypes including behavioral abnormalities [61]. Other environmental factors such as early life stress may also affect microglia development. Early life stress caused by maternal separation can induce pro-inflammatory microglia phenotype in offspring and reduced PU.1 transcriptional activity [64]. Together, these studies suggest microbiota have a large impact on the host metabolome and behavior as observed in microglia development and MIA studies. Further research is necessary to identify additional environmental factors and determine how different combinations of factors can influence microglia development and homeostasis.
PU.1 in transcriptomic and epigenetic alterations in vitro
Microglia culture models have been developed to recapitulate microglia in vivo, but have multiple limitations. Microglia lose their distinct expression patterns once cultured in vitro [5, 22]. Gosselin et al. reported that majority of transcript levels between human and mouse microglia were similar, but there are genes with species-specific bias in expression magnitudes [22]. Expression levels for complement system genes such as C2 and C3 and brain structure developmental genes including SYNDIG1 and GLDN were higher in human microglia, showing distinct differences between the human and microglia transcriptome [22]. Another limitation of microglia culture is that it develops immature microglia with amoeboid morphology, rapid proliferation, and heightened phagocytic activity [65]. It can be reversed by engraftment of isolated cells into brains lacking microglia, suggesting that additional CNS-specific cues are required to sustain microglia specification [18, 66].
Primary human microglia remain poorly characterized due to limited accessibility, lack of standardized isolation methods and culturing conditions. Recently, protocols for induce human iPSC-derived microglial-like cells (iMG) have been developed [67-71]. These protocols use induced pluripotent stem cell (iPSC) to differentiate them into iMG. iMG exhibited a transcriptomic phenotype more similar to human microglia in vitro than ex vivo microglia. (Table 4) [22, 67, 68]. iMG can integrate into organotypic 3D culture [67]. When co-cultured with iPSC-derived neurons, iMG display a more motile phenotype and dampened secretion of cytokines compared to monoculture [67, 68]. Numerous protocol used to culture microglia require complex recipes that induces key microglial regulators including PU.1 to recapitulate microglia in vivo. Further improvements are necessary for the differentiation of iPSC to more microglia-like cells for functional studies.
Table 4.
Maintenance of microglial gene signature in human iPSC-derived microglia-like cells
| Cell type | Key Factors in Maintaining Microglia |
Microglia Protein Expression | Method | Ref |
|---|---|---|---|---|
| Pluripotent stem cell-derived microglia-like cells (pMGLs) | IL34 and CSF1 | CD45, IBA1, P2RY12, TMEM119 | IHC, qPCR, and RNAseq showing pMGLs resemble primary fetal human and mouse microglia | [67] |
| Human microglial-like cells (iMGLs) differentiated from human iPSC | TGFβ, M-CSF, IL-34, insulin, CD200, CX3CL1 | CD45, CX3CR1, TREM2, TGFβR1, P2RY12, MERTK, PROS1, and ITGB5 | IHC, RNAseq PCA analysis showing close resemblance to human fetal and adult microglia | [68] |
| Human iPSC-derived microglia (iPSC-MG) | IL34 and GM-CSF | CD11b CD11c, CX3CR1, IBA1, P2RY12, TMEM119 | IHC, Flow cytometry, and RNAseq revealing close similarities between iPSC-derived (iPSC-MG) and human primary microglia as well as clear distinctions from macrophages. mRNA levels of TMEM119 were very low in iPSC-MG compared with hMG, even though the protein was detected by immunofluorescence. | [69] |
| Human IPSC-derived Microglia co-culture them with iPSC-derived cortical neurons (co-pMG) | Astrocyte-derived GM-CSF, M-CSF, and TGFβ, IL-34 | CD11b, CD14, CD45, IBA1, MERTK | IHC, Flow cytometry, qPCR, and RNAseq PCA showed weak neural cell signature in the co-cultured pMG isolated with CD11b beads, but close similarity of these cells to fetal microglia | [70] |
| Human and murine iPS-MG | Subsequent culture of iPS-HPC on astrocyte monolayers supplemented with GM-CSF, M-CSF and IL-3 | CD11b and IBA1, HLA-DR, CD45, TREM-2 and CX3CR1 | IHC, microarray hybridization and qPCR show human iPS-MG resembled those of human fetal microglia including expression of P2RY12, GPR34, MERTK, C1QA, PROS1 and GAS6 as well as those of DC and Mac human iPS-MG did not cluster as tightly with the commercially obtained human fetal microglia compared to murine analysis | [71] |
Most of the mRNAs encoding transcription factors that recognize enriched motifs in microglia enhancers exhibited significant reductions in vitro [22]. De novo analysis of ATAC-seq peaks associated with a significant decrease in H3K27ac-recognition elements for PU.1, IRF, CTCF, AP-1, SMAD, and MEF2 motifs, revealing epigenetic regulation. Tissue micro-environmental factors activate cascades resulting in co-operative binding of certain transcription factors with PU.1, leading to a unique enhancer profile enabling microglial dynamic response to its surrounding micro-environment [24, 72]. Several groups have attempted different cell culture conditions to maintain homeostatic microglia signature and ramified morphology In vitro using a variety of cytokines, such as CSF1, GM-CSF, TGFβ, and IL-34 [73-76]. Butovsky et al. showed TGFβ act as a crucial upstream regulator in microglia survival, differentiation and maintenance of microglia signature In vitro [24]. Further improvements such as overexpression of PU.1 in microglia to enhance CSF1R signaling can be considered as well in future studies [31]. However, the addition of key factors for microglial maintenance can hinder the interpretation of In vitro disease models, since factors such as CSF1 can promote microglia viability and increased PU.1 expression that can also influence microglial function in disease states [76].
Concluding remarks
The tight regulation of microglia transcription involves multiple key transcriptional factors including PU.1, SALL1, MEF2, and MAFB (Figure 1). Among these factors, PU.1 is one of the more specific factors critical to microglial identity and survival and is sensitive to micro-environmental cues. Current studies show that PU.1 can interact with the other specific transcription factors and epigenetic regulators such as HDAC, highlighting the importance of PU.1 in regulating microglial homeostatic and reactive state at transcriptional and epigenetic levels. Dysregulation of transcription factors in disease states are likely responsible for altering the microglial gene expression landscape, which may contribute to the disease progression. Identification of key transcription factors and epigenetic factors will lead to development of effective therapeutic strategies via modulation of MGnD in the future. Immune training and induction of immune memory of microglia can be potentially beneficial in suppressing their pro-inflammatory response in disease states. Additional studies will be necessary to address that (se Outstanding Questions), which would direct to a better understanding of how transcriptional regulators orchestrate homeostatic and neurodegenerative microglia.
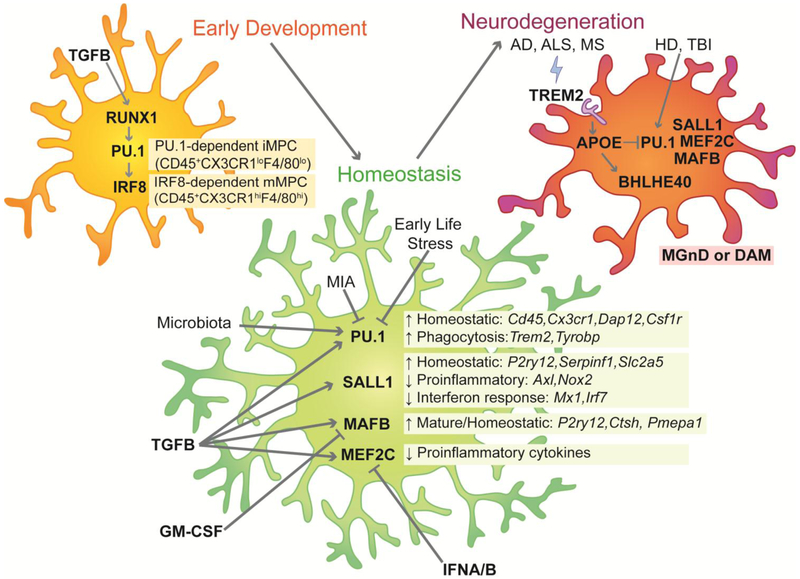
Figure 1: Key transcription factors in early development, homeostasis and neurodegeneration.
In early development microglia (yellow) the transcription factors RUNX1, PU.1 and IRF8 are very active. In homeostatic microglia (green) PU.1, SALL1, MAFB, MEF2c regulate certain homeostatic genes, external factors can epigenetically regulate the expression of these transcription factors. Neurodegenerative microglia (red), also referred as disease-associated microglia (DAM) or microglia neurodegenerative phenotypes DAM (MGnD) display increased signaling of APOE pathway via TREM2 (purple) that inhibits the expression of key transcription factors and activates BHLHE40 that is associated with the neurodegenerative state of microglia. (AD: Alzheimer’s disease, ALS: amyotrophic lateral sclerosis, MS: multiple sclerosis, HD: Huntington’s disease, TBI: traumatic brain injury)
Outstanding Questions:
How do transcription factors work with one another to regulate microglia in different disease states at a single cell level?
How can we develop therapeutic approach that target modulation of epigenetics via immune training in microglia?
Which of the transcriptional factors are potential therapeutic targets for health and disease?
What improvements can be made to study microglia in vitro to resemble In vivo mature microglia?
How can microglia isolation methods and RNA-seq processing methods be standardized for reproducibility and comparison between datasets?
RNA-seq and ChiP-seq provide powerful platforms to characterize microglia in homeostatic and neurodegenerative state, but face certain limitations (Table 5). Future work should focus on single-cell transcriptomics since the recent accumulation of data based on RNA-seq and ChIP-seq are dependent on bulk sequencing. Single-cell RNA and ChiP-sequencing can provide characterization of subpopulations, distinct transcriptomic phenotype, but may have significant limitation in sequencing depth. Focusing on individual cells across ages and brain regions to refine key regulators in different states and evaluate the variability in cell identity, response and location in the CNS may constitute the next level of microglial transcriptomic characterization (see Clinician’s Corner). Further research should also focus on the discrepancy between human and mouse microglia transcriptome regulation, which can provide targets that shared between mouse and human microglia for therapeutic strategies.
Table 5.
Technical limitations in transcriptomic and epigenetic analysis of microglia
| Limitations | Impact | Current trend | Future directions |
|---|---|---|---|
| Different microglia isolation methods | Difficulty in reproducing data and reaching consensus of microglial subpopulation function. | Identifying subpopulations using single-cell RNAseq | Provide standardized method using updated microglial markers. |
| RNAseq processing differences | The difference in reading depth can result in missing specific genes that are not highly expressed in microglia. | Provide optimal reading depth to analyze all genes possible. | |
| Different preparation methods of DNA libraries can lead to different reads of transcript due to fragmentation of mRNA. | SmartSeq, designed for small starting amounts, involves PCR amplification before the final fragmentation of the sequencing library TruSeq method using heat-fragmentation of mRNA and the only amplifies the sequencing library | Provide consensus of most efficient method for RNA preparation | |
| Difference between protein expression and mRNA transcript level | Difference between transcriptional and protein level can lead to misinterpretation of gene expression changes. | Flow cytometry or IHC confirmation Mass spectrometry | Provide protein validation of identified genes. |
Clinician’s Corner:
Non-specific anti-inflammatory drugs such as minocycline can negate the balance of neuroprotective and neurotoxic function of microglia.
Disease-specific microglial transcription factors are new therapeutic targets of neuroinflammation
Complement system and microglia-specific immune molecules are novel therapeutic targets of neurodegenerative disorders.
Analysis of disease-specific microglial markers will facilitate evaluation of the clinical outcome of anti-neuroinflammatory therapy
The information obtained from murine microglia may differ from those from human microglia. The transcriptomic and epigenetic results will require careful validation using freshly isolated human microglia.
Highlights:
Microglia transcriptome is tightly regulated by a combination of transcription factors, such as PU.1, for their role in the development and homeostatic function of microglia.
Environmental factors including microbiome, maternal immune activation, and early life stress can influence the transcription factor network and epigenetic modification of target genes.
Distinct clusters of transcription factors regulate the transcription network in early development, homeostatic and disease conditions
The PU.1-regulated genes are influenced by epigenetic modifications, which are altered by disease and tissue culture environment
Acknowledgement
We would like to thank JC Delpech, Ashley Comer, and Seiko Ikezu for critical review of the manuscript. This work is in funded in part by National Institute of Health grants 5T32GM008541 (HY), 1RF1AG05419, R01AG054672, R56AG057469, R01AG056318, BrightFocus Foundation A2016551S, Cure Alzheimer’s Fund, Nancy Lurie Marks Family Fund and Landreth Family Foundation (TI).
Glossary
- Apolipoprotein E (APOE):
A class of lipid-binding proteins involved in transportation and metabolism of lipids, lipid-soluble vitamins and cholesterol.
- Assay for transposase accessible chromatin (ATAC) sequencing:
A method for mapping chromatin accessibility genome-wide using Tn5 transposase that inserts sequencing adapters into accessible regions of chromatin.
- Binding motif:
A collection of DNA binding sites that can be represented by a consensus sequence in which are bound by DNA-binding proteins to regulate gene transcription.
- Chromatin immunoprecipitation (ChIP)-seq:
A method to identify the genome-wide DNA binding sites for specifically modified chromatins by combining chromatin immunoprecipitation (ChIP) with DNA sequencing.
- Dystrophic neurites:
Abnormal neuronal processes characterized by aberrant sprouting, dystrophic expansion, and accumulation of various cellular organelles and cytoskeletal proteins, such as microtubule-associated tau proteins.
- Enhancer:
Enhancers are short regions of DNA that form complexes with transcriptional factors to regulate the transcription of targeted genes
- Epigenome:
The study of change in phenotypes such as post-translational modifications of chromatins and DNA, which does not involve alteration in DNA sequence.
- ETS domain transcription factor:
Characterized by the presence of a conserved DNA-binding domain and interacts with multitude of co-regulatory partners to elicit gene-specific responses and drive distinct biological processes.
- Fluorescence-activated cell sorting (FACS):
Fluorescence intensity-based sorting for collection of specific cell populations
- Hematopoietic stem cell:
A subset of stem cells found in peripheral blood and bone marrows that can develop into all types of blood cells.
- Histone deacetylase:
A class of enzyme that removes the acetyl group from histone proteins on DNA, making the DNA less accessible to transcription factors.
- Immune memory:
A cellular immune function for specific recognition of an antigen that the body has previously encountered. In microglia, there are two types of immune memory: training, which augments innate immune responses; and tolerance, which dampens them.
- Lineage-determining transcription factor:
Transcription factors that bind to enhancer regions and recruit secondary transcription factors for cell-type-specific gene expression.
- Methyl-CpG binding protein 2 (MECP2):
This protein is capable of binding to methylated DNA, further condensing the chromatin structure to repress or activate gene transcription depending on the context.
- Promoter regions:
DNA sequences that define where transcription of a gene by RNA polymerase begins and initiates transcription of a particular gene
- Toll-like receptors (TLRs):
Evolutionally conserved innate immunity receptors for detecting pathogen- and damage-associated pattern molecules for host-defense response.
- Transcription factor:
A protein that controls the transcription from DNA to messenger RNA.
- Transcriptomics:
The study of genome-wide set of all RNA molecules, including mRNA, non-coding RNA.
- Triggering receptor expressed on myeloid cells 2 (TREM2):
A transmembrane molecule expressed in microglia and myeloid cells, which is functionally implicated in phagocytosis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kierdorf K, et al. , Microglia emerge from erythromyeloid precursors via Pu.1- and irf8-dependent pathways. Nature Neuroscience, 2013. 16: p. 273. [DOI] [PubMed] [Google Scholar]
- 2.Crotti A and Ransohoff RM, Microglial Physiology and Pathophysiology: Insights from Genome-wide Transcriptional Profiling. Immunity, 2016. 44(3): p. 505–515. [DOI] [PubMed] [Google Scholar]
- 3.Nimmerjahn A, Kirchhoff F, and Helmchen F, Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science, 2005. 308. [DOI] [PubMed] [Google Scholar]
- 4.Zhan Y, et al. , Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nature Neuroscience, 2014. 17: p. 400. [DOI] [PubMed] [Google Scholar]
- 5.Gosselin D, et al. , Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell, 2014. 159(6): p. 1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong S, et al. , Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science (New York, N.Y.), 2016. 352(6286): p. 712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekar A, et al. , Schizophrenia risk from complex variation of complement component 4. Nature, 2016. 530(7589): p. 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen DV, Hanson JE, and Sheng M, Microglia in Alzheimer’s disease. The Journal of Cell Biology, 2018. 217(2): p. 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prata J, et al. , Bridging Autism Spectrum Disorders and Schizophrenia through inflammation and biomarkers - pre-clinical and clinical investigations. Journal of Neuroinflammation, 2017. 14(1): p. 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu IM, et al. , A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep, 2013. 4(2): p. 385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wendeln A-C, et al. , Innate immune memory in the brain shapes neurological disease hallmarks. Nature, 2018. 556(7701): p. 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickman SE, et al. , The microglial sensome revealed by direct RNA sequencing. Nat Neurosci, 2013. 16(12): p. 1896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matcovitch-Natan O, et al. , Microglia development follows a stepwise program to regulate brain homeostasis. Science, 2016. 353(6301): p. aad8670. [DOI] [PubMed] [Google Scholar]
- 14.Ginhoux F and Prinz M, Origin of microglia: current concepts and past controversies. Cold Spring Harb Perspect Biol, 2015. 7(8): p. a020537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erny D, et al. , Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci, 2015. 18(7): p. 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ransohoff RM, A polarizing question: do M1 and M2 microglia exist? Nat Neurosci, 2016. 19(8): p. 987–991. [DOI] [PubMed] [Google Scholar]
- 17.Cronk James C., et al. , Methyl-CpG Binding Protein 2 Regulates Microglia and Macrophage Gene Expression in Response to Inflammatory Stimuli. Immunity, 2015. 42(4): p. 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett FC, et al. , A Combination of Ontogeny and CNS Environment Establishes Microglial Identity. Neuron, 2018. 98(6): p. 1170–1183.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett ML, et al. , New tools for studying microglia in the mouse and human CNS. Proceedings of the National Academy of Sciences of the United States of America, 2016. 113(12): p. E1738–E1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldmann T, et al. , Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol, 2016. 17(7): p. 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buttgereit A, et al. , Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol, 2016. 17(12): p. 1397–1406. [DOI] [PubMed] [Google Scholar]
- 22.Gosselin D, et al. , An environment-dependent transcriptional network specifies human microglia identity. Science, 2017. 356(6344). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crotti A, et al. , Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat Neurosci, 2014. 17(4): p. 513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butovsky O, et al. , Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci, 2014. 17(1): p. 131–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dakic A, et al. , PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. The Journal of Experimental Medicine, 2005. 201(9): p. 1487–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satoh J, et al. , A Comprehensive Profile of ChIP-Seq-Based PU.1/Spi1 Target Genes in Microglia. Gene Regul Syst Bio, 2014. 8: p. 127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beers DR, et al. , Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A, 2006. 103(43): p. 16021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenbauer F, et al. , Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nature Genetics, 2004. 36: p. 624. [DOI] [PubMed] [Google Scholar]
- 29.Smith AM, et al. , The transcription factor PU.1 is critical for viability and function of human brain microglia. Glia, 2013. 61(6): p. 929–42. [DOI] [PubMed] [Google Scholar]
- 30.Rustenhoven J, et al. , PU.1 regulates Alzheimer's disease-associated genes in primary human microglia. Mol Neurodegener, 2018. 13(1): p. 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang KL, et al. , A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer's disease. Nat Neurosci, 2017. 20(8): p. 1052–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaikkonen MU, et al. , Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Molecular cell, 2013. 51(3): p. 310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Atalaya JP, et al. , Development and maintenance of the brain's immune toolkit: Microglia and non-parenchymal brain macrophages. Dev Neurobiol, 2018. 78(6): p. 561–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lavin Y, et al. , Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell, 2014. 159(6): p. 1312–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laribee RN and Klemsz MJ, Histone H4 HDAC activity is necessary for expression of the PU.1 gene. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression, 2005. 1730(3): p. 226–234. [DOI] [PubMed] [Google Scholar]
- 36.Krämer OH, et al. , The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. The EMBO journal, 2003. 22(13): p. 3411–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson KL, et al. , PU.1 Is a Lineage-specific Regulator of Tyrosine Phosphatase CD45. Journal of Biological Chemistry, 2001. 276(10): p. 7637–7642. [DOI] [PubMed] [Google Scholar]
- 38.Gibbons HM, et al. , Valproic acid induces microglial dysfunction, not apoptosis, in human glial cultures. Neurobiology of Disease, 2011. 41(1): p. 96–103. [DOI] [PubMed] [Google Scholar]
- 39.Rustenhoven J, et al. , Isolation of highly enriched primary human microglia for functional studies. Sci Rep, 2016. 6: p. 19371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin L-W, et al. , Dysregulation of Glutamine Transporter SNAT1 in Rett Syndrome Microglia: A Mechanism for Mitochondrial Dysfunction and Neurotoxicity. The Journal of Neuroscience, 2015. 35(6): p. 2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keren-Shaul H, et al. , A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell, 2017. 169(7): p. 1276–1290.e17. [DOI] [PubMed] [Google Scholar]
- 42.Ayata P, et al. , Epigenetic regulation of brain region-specific microglia clearance activity. Nature Neuroscience, 2018. 21(8): p. 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freilich RW, Woodbury ME, and Ikezu T, Integrated Expression Profiles of mRNA and miRNA in Polarized Primary Murine Microglia. PLoS ONE, 2013. 8(11): p. e79416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ponomarev ED, et al. , MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α–PU.1 pathway. Nature medicine, 2011. 17(1): p. 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krasemann S, et al. , The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity, 2017. 47(3): p. 566–581 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mrdjen D, et al. , High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity, 2018. 48(2): p. 380–395 e6. [DOI] [PubMed] [Google Scholar]
- 47.Bouvier DS, et al. , High Resolution Dissection of Reactive Glial Nets in Alzheimer’s Disease. Scientific Reports, 2016. 6: p. 24544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heppner FL, Ransohoff RM, and Becher B, Immune attack: the role of inflammation in Alzheimer disease. Nature Reviews Neuroscience, 2015. 16: p. 358. [DOI] [PubMed] [Google Scholar]
- 49.Litvinchuk A, et al. , Complement C3aR Inactivation Attenuates Tau Pathology and Reverses an Immune Network Deregulated in Tauopathy Models and Alzheimer’s Disease. Neuron, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lian H, et al. , NFκB-Activated Astroglial Release of Complement C3 Compromises Neuronal Morphology and Function Associated with Alzheimer’s Disease. Neuron, 2015. 85(1): p. 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lipponen A, et al. , Transcription factors Tp73, Cebpd, Pax6, and Spi1 rather than DNA methylation regulate chronic transcriptomics changes after experimental traumatic brain injury. Acta Neuropathol Commun, 2018. 6(1): p. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galatro TF, et al. , Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat Neurosci, 2017. 20(8): p. 1162–1171. [DOI] [PubMed] [Google Scholar]
- 53.Wehrspaun CC, Haerty W, and Ponting CP, Microglia recapitulate a hematopoietic master regulator network in the aging human frontal cortex. Neurobiology of Aging, 2015. 36(8): p. 2443.e9–2443.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanamsagar R, et al. , Generation of a microglial developmental index in mice and in humans reveals a sex difference in maturation and immune reactivity. Glia, 2017. 65(9): p. 1504–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kierdorf K and Prinz M, Factors regulating microglia activation. Frontiers in Cellular Neuroscience, 2013. 7: p. 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olah M, et al. , A transcriptomic atlas of aged human microglia. Nat Commun, 2018. 9(1): p. 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thion MS, et al. , Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell, 2018. 172(3): p. 500–516.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guneykaya D, et al. , Transcriptional and Translational Differences of Microglia from Male and Female Brains. Cell Reports, 2018. 24(10): p. 2773–2783.e6. [DOI] [PubMed] [Google Scholar]
- 59.Mattei D, et al. , Maternal immune activation results in complex microglial transcriptome signature in the adult offspring that is reversed by minocycline treatment. Transl Psychiatry, 2017. 7(5): p. e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsiao EY, et al. , Modeling an autism risk factor in mice leads to permanent immune dysregulation. Proc Natl Acad Sci USA, 2012. 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsiao EY, et al. , Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell, 2013. 155(7): p. 1451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hsiao EY, et al. , The microbiota modulates gut physiology and behavioral abnormalities associated with autism. Cell, 2013. 155(7): p. 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim S, et al. , Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature, 2017. 549(7673): p. 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delpech J-C, et al. , Early life stress perturbs the maturation of microglia in the developing hippocampus. Brain, behavior, and immunity, 2016. 57: p. 79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kettenmann H, et al. , Physiology of microglia. Physiol Rev, 2011. 91. [DOI] [PubMed] [Google Scholar]
- 66.Bohlen CJ, et al. , Diverse Requirements for Microglial Survival, Specification, and Function Revealed by Defined-Medium Cultures. Neuron, 2017. 94(4): p. 759–773 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muffat J, et al. , Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med, 2016. 22(11): p. 1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abud EM, et al. , iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron, 2017. 94(2): p. 278–293 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Douvaras P, et al. , Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Reports, 2017. 8(6): p. 1516–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haenseler W, et al. , A Highly Efficient Human Pluripotent Stem Cell Microglia Model Displays a Neuronal-Co-culture-Specific Expression Profile and Inflammatory Response. Stem Cell Reports, 2017. 8(6): p. 1727–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pandya H, et al. , Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci, 2017. 20(5): p. 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gautier EL, et al. , Gene expression profiles and transcriptional regulatory pathways underlying mouse tissue macrophage identity and diversity. Nature immunology, 2012. 13(11): p. 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ginhoux F, et al. , Fate Mapping Analysis Reveals That Adult Microglia Derive from Primitive Macrophages. Science (New York, N.Y.), 2010. 330(6005): p. 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, et al. , IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nature immunology, 2012. 13(8): p. 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Greter M, et al. , Stroma-Derived Interleukin-34 Controls the Development and Maintenance of Langerhans Cells and the Maintenance of Microglia. Immunity, 2012. 37(6): p. 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith AM, et al. , M-CSF increases proliferation and phagocytosis while modulating receptor and transcription factor expression in adult human microglia. Journal of Neuroinflammation, 2013. 10(1): p. 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gomez Perdiguero E, et al. , Tissue-resident macrophages originate from yolk sac-derived erythro-myeloid progenitors. Nature, 2015. 518(7540): p. 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barbosa AC, et al. , MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proceedings of the National Academy of Sciences, 2008. 105(27): p. 9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deczkowska A, et al. , Mef2C restrains microglial inflammatory response and is lost in brain ageing in an IFN-I-dependent manner. Nat Commun, 2017. 8(1): p. 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zusso M, et al. , Regulation of Postnatal Forebrain Amoeboid Microglial Cell Proliferation and Development by the Transcription Factor Runx1. The Journal of Neuroscience, 2012. 32(33): p. 11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Samokhvalov IM, Samokhvalova NI, and Nishikawa S.-i. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature, 2007. 446: p. 1056. [DOI] [PubMed] [Google Scholar]
- 82.Huang G, et al. , PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nature Genetics, 2007. 40: p. 51. [DOI] [PubMed] [Google Scholar]
- 83.Harrison SJ, et al. , Sall1 regulates cortical neurogenesis and laminar fate specification in mice: implications for neural abnormalities in Townes-Brocks syndrome. Disease Models & Mechanisms, 2012. 5(3): p. 351–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Koshida R, et al. , MafB antagonizes phenotypic alteration induced by GM-CSF in microglia. Biochem Biophys Res Commun, 2015. 463(1-2): p. 109–15. [DOI] [PubMed] [Google Scholar]
- 85.Kelly LM, et al. , MafB is an inducer of monocytic differentiation. The EMBO Journal, 2000. 19(9): p. 1987–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Santambrogio L, et al. , Developmental plasticity of CNS microglia. Proceedings of the National Academy of Sciences of the United States of America, 2001. 98(11): p. 6295–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aziz A, et al. , MafB/c-Maf Deficiency Enables Self-Renewal of Differentiated Functional Macrophages. Science, 2009. 326(5954): p. 867. [DOI] [PubMed] [Google Scholar]
- 88.Kim H and Seed B, The transcription factor MAFB antagonizes anti-viral responses by blockade of coactivator recruitment to IRF3. Nature immunology, 2010. 11(8): p. 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gjoneska E, et al. , Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. Nature, 2015. 518(7539): p. 365–9. [DOI] [PMC free article] [PubMed] [Google Scholar]