Abstract
Carboxylesterase1 (CES1) is a primary human hepatic hydrolase involved in hydrolytic biotransformation of numerous medications. Considerable interindividual variability in CES1 expression and activity has been consistently reported. Four isoforms of the CES1 protein are produced by alternative splicing (AS). In the current study, we examined the activity and expression of each CES1 isoform using transfected cell lines, and determined CES1 isoform composition and its impact on CES1 activity in human livers. In transfected cells, isoforms 3 and 4 showed mRNA and protein expressions comparable to isoforms 1 and 2, but had significantly impaired activity when hydrolyzing enalapril and clopidogrel. In individual human liver samples, isoforms 1 and 2 were the major forms, contributing 73%−90% of total CES1 protein expression. In addition, the protein expression ratios of isoforms 1 and 2 to isoforms 3 and 4 were positively associated with CES1 activity in the livers, suggesting that CES1 isoform composition is a factor contributing to the variability in hepatic CES1 function. Further investigations of the regulation of CES1 AS would improve our understanding of CES1 variability and help develop a strategy to optimize the pharmacotherapy of many CES1 substrate medications.
Keywords: carboxylesterase 1 (CES1), alternative splicing (AS), protein isoforms, stable isotope labeling with amino acids in cell culture (SILAC), heavy stable isotope-labeled quantitative concatamer (QconCAT)
Introduction
In humans, carboxylesterase 1 (CES1) is the most abundant hydrolase expressed in the liver [1], contributing 80–95% of total hepatic hydrolytic activity [2]. CES1 is also a predominant hydrolase in human lung, and in monocytes/macrophages, but is absent in human intestine [3]. It catalyzes the hydrolysis of various exogenous and endogenous compounds including carboxylic acid esters, thioesters, amides, and carbamates. Many clinically important medications, such as methylphenidate [4], clopidogrel [5], oseltamivir [6], ACE Inhibitors [7], dabigatran [8, 9], and sacubitril [10], are metabolized by the enzyme. A growing number of studies have demonstrated significant interindividual variability in CES1 activity and expression; this variability is associated with inadequate therapeutic effects and unexpected side effects of CES1 substrate medications [4, 7, 11, 12, 13]. Thus, identifying the factors contributing to CES1 variability is fundamental for improving the therapeutic effect of many medications metabolized by CES1.
CES1 expression and activity are affected by both non-genetic (e.g. age and gender) [14] and genetic factors [7, 9, 10, 15, 16]. CES1 is encoded by the CES1 gene located on chromosome 16. In addition, the non-functional pseudogene CES1P1 coincides with CES1 in the reverse orientation, and has a functional variant CES1P1VAR that can produce functional CES1 proteins. However, CES1P1VAR does not contribute significantly to CES1 expression/activity due to its extremely low transcription efficiency[11, 17]. A variant form of CES1 also exists, CES1VAR, which resulted from a genomic translocation of the 5’ region of CES1P1 to CES1 [17, 18]. It has been demonstrated that CES1/CES1P1 haplotypes or diplotypes have no significant effect on CES1 expression or activity in human liver [7, 19]. In addition, over 2500 single nucleotide polymorphisms (SNPs) have been identified within the CES1 gene (NCBI dbSNP). Some SNPs, such as G143E, D269fs, E220G and L40T, are deleterious to its enzymatic activity, and could alter CES1-mediated drug metabolism [4, 16, 20]. However, these variants only account for a small portion of CES1 variability, leaving the majority unexplained.
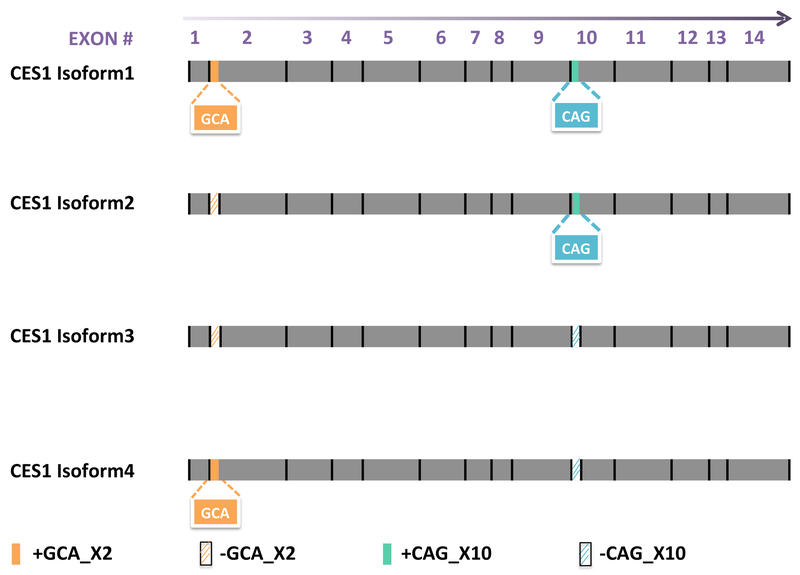
Beyond genetic polymorphisms, alternative splicing (AS), an important transcriptional process, can also affect the expression of a gene and the activity of its product. AS may produce multiple distinct transcripts from the same pre-messenger RNA and yield different protein products [21]. This mechanism is involved in the mRNA processing of approximately 95% of multi-exon genes in humans [22], which not only significantly increases the protein diversity and functional capacity of human genes, but also produces dysfunctional proteins or even causes diseases due to mis-splicing [23]. The CES1 gene features two existing alternative splicing sites and four associated transcripts: isoform1 (NM_001025195.1), isoform2 (NM_001025194.1), isoform3 (NM_001266.4), and isoform 4 (XM_005255774.1), which can be translated into four corresponding protein isoforms. The two CES1 AS events include the retention of three intronic nucleotides “GCA” adjacent to the 5’ -end of exon2 (+GCA_X2) and the deletion of the “CAG” at the 5’- end of exon10 (-CAG_X10). The +GCA_X2 event introduces an “alanine” after amino acid 17 “tryptophan” while the -CAG_X10 event deletes the “glutamine” at amino acid 362. Isoform2 (NM_001025194.1) is considered the reference mRNA, and its protein product has been widely used as the wild-type CES1 in most CES1 pharmacogenetic studies [4–7, 9, 10, 16, 17, 24]. Isoforms 1, 3, and 4 are generated from the AS events +GCA_X2, –CAG_X10, and both +GCA_X2 and -CAG_X10, respectively (Figure 1). It is plausible that these differences in splicing may contribute to the variability in CES1 expression and/or activity observed among individuals, and consequently affect responses to CES1 substrate medications. In the present study, we evaluated the expression and activity of each isoform using transfected cell lines, and further analyzed the endogenous composition of CES1 isoforms and its impact on CES1 activity in individual human liver samples.
Figure 1.
Illustration of the differences among the four CES1 mRNA isoforms caused by alternative splicing. Compared to isoforms 2 and 3, isoforms 1 and 4 have an insertion of three nucleotides “GCA” at the 5’-end of exon 2 (orange rectangle, +GCA_X2). In addition, three nucleotides “CAG” at the 5’-end of exon 10 are spliced out in isoforms 3 and 4 (diagonal strips in blue, -CAG_X10) relative to that in isoforms 1 and 2.
Materials and Methods
Materials
The QuikChange Lightning Multi Site-Directed Mutagenesis Kit was purchased from Agilent Technologies (Santa Clara, CA, USA). Products purchased from Thermo Fisher Scientific Co. (Waltham, MA, USA) include the Flp-In™-293 cell line, pOG44 plasmid, S.N.A.P.™ Plasmid DNA MiniPrep Kit, Lipofectamine® 2000 Transfection Reagent, Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), dialyzed FBS, hygromycin B in phosphate buffered saline (PBS, 50mg/ml) and antibiotics solution containing penicillin (10,000 IU/mL) and streptomycin (10,000 μg/mL), trypsin-EDTA (0.25%), the stable isotope labeling with amino acids in cell culture (SILAC) Protein Quantitation Kit-DMEM containing SILAC DMEM (deficient in arginine and lysine), 13C6 l-lysine-2HCl, 13C615N4 l-arginine-HCl, urea, dl-dithiothreitol (DTT), trifluoroacetic acid (TFA), TRIzol RNA isolation reagent and acetonitrile. Iodoacetamide (IAA) was the product of Acros Organics (Morris Plains, NJ, USA). TPCK-treated trypsin was obtained from Worthington Biochemical Corporation (Freehold, NJ, USA). Water Oasis HLB columns were from Waters Corporation (Milford, MA, USA). Recombinant CES1 (purity >95%) was the product of R&D Systems (Minneapolis, MN, USA).
Enalapril maleate was purchased from Sigma-Aldrich (St. Louis, MO, USA). The enalapril hydrolytic metabolite enalaprilat dehydrate was the product of Sellechchem (Houston, TX, USA). S-(l)-clopidogrel and clopidogrel carboxylate were obtained from Toronto Research Chemicals, Inc. (Toronto, Canada). The High-Capacity cDNA Reverse Transcription Kit and SYBR Green PCR Master Mix were the products of Applied Biosystems (Foster City, CA, USA).
A total of 52 individual normal human liver samples were obtained from XenoTech LLC (Kansas City, KS, USA) and the Cooperative Human Tissue Network (CHTN, Columbus, OH, USA). These samples came from 23 males and 29 females with ages ranging from 22 to 83 years. The donors included 48 Caucasians, 2 African-Americans, 1 Hispanic, and 1 classified as ‘others’. Based on our genotyping data, none of the samples carried any known functional CES1 variants (data not shown).
The study design is illustrated in Supplementary Figure S1.
Establish cell lines stably expressing four CES1 isoforms
A plasmid with CES1 mRNA isoform2 (NM_001025194.1), which represents the “canonical” CES1 sequence, was generated in our lab previously [16]. Plasmids for the other three CES1 isoforms, including isoform1 (NM_001025195.1), isoform 3 (NM_001266.4), and isoform 4 (XM_005255774.1), were generated using a site-directed mutagenesis assay with the isoform 2 as the template (Supplementary Table S1). The desired sequences of the CES1 plasmids were confirmed by DNA sequencing analysis. The validated CES1 isoform plasmids were then transfected into Flp-In-293 cells following our previously-published protocol [16]. Briefly, CES1 plasmids were co-transfected with pOG44 plasmid at a ratio of 1:10 into Flp-In-293 cells with the Lipofectamine 2000 reagent. Six hours after transfection, cells were gently rinsed to remove the transfection reagents and cultured in complete medium (DMEM containing 10% FBS). After 12 hours, the culture medium was replaced with complete medium supplemented with the selecting antibiotic hygromycin B (100 μg/mL). Four cell lines stably expressing the four different CES1 isoforms were established after culturing with hygromycin B for at least three weeks. All transfected cell lines were further validated by DNA sequencing analysis.
Preparation of S9 fractions from transfected cells and individual human liver samples
S9 fractions were prepared from cell lines and Individual human livers according to our previous publications [7, 16]. Briefly, cultured cells were harvested at approximately 95% confluence and lysed by sonication on ice, while samples from individual human livers were minced and homogenized at 4 °C. The cells and liver lysates were then centrifuged at 9,000 g at 4 °C for 30 min. The supernatant (S9 fraction) was collected and determined for protein concentrations using the Pierce BCA assay (Thermo Fisher, Waltham, USA). The S9 fraction samples were stored at −80 °C until use.
Enzymatic activity assays
Incubation studies were conducted to measure CES1 activity on the hydrolysis of the CES1 substrates enalapril and clopidogrel in the prepared S9 fraction samples. As described in our previous publication[7], incubations for clopidogrel were performed in 1.5 mL Eppendorf tubes, while those of enalapril were carried out in 4 mL silanized glass vials due to enalapril and its hydrolytic metabolite enalaprilat having significant non-specific bindings to plastic Eppendorf tubes. Enalapril (500 μM) and clopidogrel (100 μM) were incubated with S9 fractions at the final protein concentrations of 0.2 mg/ml and 0.1 mg/ml, respectively, at 37°C for 10 min. Hydrolysis reactions were terminated by the addition of either a four-fold volume of methanol containing the internal standard (IS) 5-hydroxy omeprazole (20 ng/mL) for enalapril or a two-fold volume of acetonitrile with the IS clopidogrel-d4 carboxylic acid (25 ng/mL) for clopidogrel. After centrifugation at 17,000 g at 4°C for 20 min, the supernatant was collected and analyzed for the metabolites concentrations using the HPLC-MS/MS methods described below [7, 10, 25]. The S9 fractions prepared from the CES1 isoform2 and vector transfected cells served as the wild-type and blank controls, respectively.
Analysis of CES1 isoform mRNA expressions in transfected cells and CES1 isoforms mRNA levels in individual human liver samples
Transfected cells expressing each of the four CES1 isoforms were cultured in six-well plates until reaching 95% confluence. One μg of total RNA extracted from the cells was reverse-transcribed to cDNA using oligo dT primers. Quantitative real-time polymerase chain reactions (qRT-PCR) were performed on a 7300 Real-Time PCR system (Applied Biosystems, Foster City, CA) using SYBR green fluorescence. Ubiquitin C was used as the internal control for normalization. The mRNA expressions of different CES1 isoforms were determined relative to those in the isoform 2 transfected cells by the 2−ΔΔCT method. The same method was applied to the determination of mRNA transcript levels with (isoforms 3 and 4) or without (isoforms 1 and 2) the -CAG_X10 AS event in individual human liver samples, relative to those in pooled mRNA samples. Additionally, RNA isolated from vector-transfected cells was included as the blank control for CES1 mRNA quantification in transfected cell lines. The primers and experimental conditions for qRT-PCR experiments are described in Supplementary Tables S1 and S2, respectively.
HPLC-MS/MS analysis
The HPLC-MS/MS analysis for CES1 activity and total protein expression was performed on a Shimadzu HPLC system (Shimadzu, Tokyo, Japan) coupled with an Applied Biosystems API 4000 triple quadruple mass spectrometer (Foster City, CA, USA). In addition, the quantitative analysis of CES1 protein isoforms in human livers was conducted on a TripleTOF 5600+ mass spectrometer (AB Sciex, Framingham, MA) coupled with an Eksigent 2D plus LC system (Eksigent Technologies, Dublin, CA).
LC-MS/MS method for assaying CES1 activity
The hydrolytic metabolites enalaprilat and clopidogrel carboxylate were quantitated using previously described assays, with some modifications [5, 7, 10]. The analytes were isolated on a Shimadzu VP-ODS column (5 μm, 150 × 2.0 mm, Shimadzu, Japan) with mobile phase delivered at a constant flow rate (0.3 mL/min for enalaprilat and 0.25 mL/ min for clopidogrel carboxylate). The gradient conditions are summarized in Supplementary Table S3. Column temperature was set at 50 °C for enalaprilat and 40 °C for clopidogrel carboxylate. An injection volume of 10 μL was used for all analytes. Positive electrospray ionization mode was applied, and ions were monitored by multiple reaction monitoring (MRM) with the following m/z transitions: enalaprilat (349.0 > 206.0), clopidogrel carboxylate (308.0 > 197.9), 5-hydroxy omeprazole (362.34 > 213.9), and clopidogrel-d4 carboxylic acid (312.1 > 202.0).
Quantification of CES1 protein in cells transfected with CES1 isoforms
Relative CES1 protein expression levels in the S9 fractions from transfected cells were quantified using the SILAC-based LC-MS/MS assay that we established previously[26] without calibration curves. In brief, the intensities of six selected CES1 unique peptides were normalized by their SILAC counterpart peptides (i.e. IS), and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal reference protein to further control for potential sample loading variations. The relative CES1 protein expression in each transfected cell line was calculated by comparison to the CES1 level in cells transfected with CES1 isoform 2. Six CES1 unique peptides (i.e. AISESGVALTSVLVK, FLSLDLQGDPR, TAMSLLWK, SYPLVC[CAM]IAK, ELIPEATEK, FWANFAR), one GAPDH unique peptide (GALQNIIPASTGAAK), and the corresponding heavy isotope-labeled SILAC peptides were separated on a ZORBAX 300SB-C18 column (5 μm, 150 × 2.1 mm, Agilent Technologies, Santa Clara, CA, USA). The column temperature was set at 40 °C. Mobile phase was delivered at a constant flow rate of 0.2 mL/min with the gradient conditions listed in Supplementary Table S3. Analytes were monitored under positive ionization mode at the transitions described previously [26]. The data were analyzed using Skyline software (University of Washington, Seattle, WA, USA).
Quantification of CES1 isoforms and total protein levels in individual human livers
To determine the CES1 isoform compositions in human liver samples, the wild-type peptide “QEFGWLIPMQLMSYPLSEGQLDQK” and the corresponding AS peptide “QEFGWLIPMLMSYPLSEGQLDQK” were utilized to quantitate the expression levels of CES1 isoforms with (i.e. isoforms 3 and 4) and without (i.e. isoforms 1 and 2) the -CAG_X10 splicing event. Total CES1 expression in human liver samples was determined using the four CES1 unique peptides, including AISESGVALTSVLVK, TAMSLLWK, SYPLVC[CAM]IAK, and ELIPEATEK. The heavy stable isotope-labeled quantitative concatamer (QconCAT) technique was used to produce the stable heavy isotope-labeled internal CES1 protein standard (IS) for the calibration of both CES1 isoforms and total CES1 protein relative quantification (Supplementary Figure S2).
Samples were prepared according to our previous publication with some modifications[27] Briefly, 80 μg human liver S9 protein was mixed with 10-fold acetone and incubated at −20 °C overnight, followed by centrifugation at 17,000 g for 15 min at 4 °C. After being washed twice with ice-cold 80% ethanol, the precipitated protein pellet was resuspended and mixed with 0.28 μg heavy stable isotope-labeled QconCAT CES1 IS. The mixture was then subjected to reduction and alkylation using dithiothreitol (4mM DTT in 8 M urea/100 mM ammonium bicarbonate solution) and iodoacetamide (20mM IAA in 8 M urea/100 mM ammonium bicarbonate solution), respectively. Next, a two-step incubation was carried out for protein digestion, which consists of a first digestion with Lysyl endopeptidase (Wako Chemicals, Richmond, VA) at 37 °C for 6 hrs (protein: Lysyl endopeptidase= 100:1) and a second digestion with trypsin at 37 °C overnight (protein: trypsin=50:1). Following a cleaning step using Waters Oasis HLB columns, the desalted peptides were dried in a SpeedVac SPD1010 concentrator (Thermo Scientific, Hudson, NH), and reconstituted in 80 μL of 3% acetonitrile solution with 0.1% formic acid.
A high resolution scheduled multiple-reaction monitoring (sMRM-HR) analysis was performed on a TripleTOF 5600+ mass spectrometer (AB Sciex, Framingham, MA) for quantification of the isoform-specific peptides “QEFGWLIPMQLMSYPLSEGQLDQK” and “QEFGWLIPMLMSYPLSEGQLDQK”. The analytes were trapped on a trapping column and cleaned with water containing 0.1% formic acid (phase A) at a flow rate of 10 μL/min for 3 min before being separated on an analytical column (ChromXP C18-CL, 120 Å, 150 × 0.3 mm, 5 μm, Eksigent Technologies, Dublin, CA) using the gradient listed in Supplementary Table S3 at a flow rate of 5 μL/min.
The sMRM-HR acquisition consisted of one 200 ms TOF-MS scan from 400 to 1250 Da and subsequent MS/MS scans from 100 to 1500 Da of the inclusion precursors with rolling collision energy and the scheduled retention times. Retention times were obtained from preliminary non-scheduled MRM-HR runs. The sMRM-HR data were analyzed using the Skyline software (version 3.7.1.11271, University of Washington, Seattle, WA) [28]. The precursor to product ion transitions extracted for quantitative analysis were listed in Supplemental Table S4. To compare the relative CES1 isoform expression levels among individual human liver samples, the ratios of the peak areas of light to heavy peptides were further calibrated by the corresponding standard curves. The standard curve calibrators were a serial mixture of CES1 isoforms 1 and 4 transfected cell S9 fractions at the amounts of 5, 10, 20, 30, 50, and 80 μg S9 proteins. The coefficients of determination of the standard curves of all quantitative peptides varied from 0.986 to 0.999, with the coefficients of variation ranging from 0.1% to 15.8%. When a signal-to-noise ratio of ten is used to define the lower limit of quantification (LOQ), the lowest concentration of the standard curve calibrators and the lowest CES1 concentration detected in our liver samples were approximately 37.5-fold and 175-fold higher than the LOQ, respectively, indicating the assay had adequate sensitivity for CES1 quantification. The impact of different CES1 isoforms on CES1 function was determined by linear regression of CES1 activities against the protein expression of CES1 isoforms.
Data Analysis
Data are presented as mean ± standard deviation (S.D.). Significant differences of expression and activity between the CES1 reference isoform (i.e. isoform 2) and other isoforms were identified by unpaired, two-tailed t-test. A two-tailed Pearson correlation analysis was conducted to analyze the correlation between CES1 activity and the protein levels of total CES1 and CES1 isoforms, and isoform compositions (GraphPad Prism software version 6.0; GraphPad Software, San Diego, CA, USA). The data from cellular experiments were obtained from three independent replicates (n=3), and p < 0.05 was considered statistically significant.
Results
CES1 isoforms 3 and 4 showed significantly impaired CES1 activity
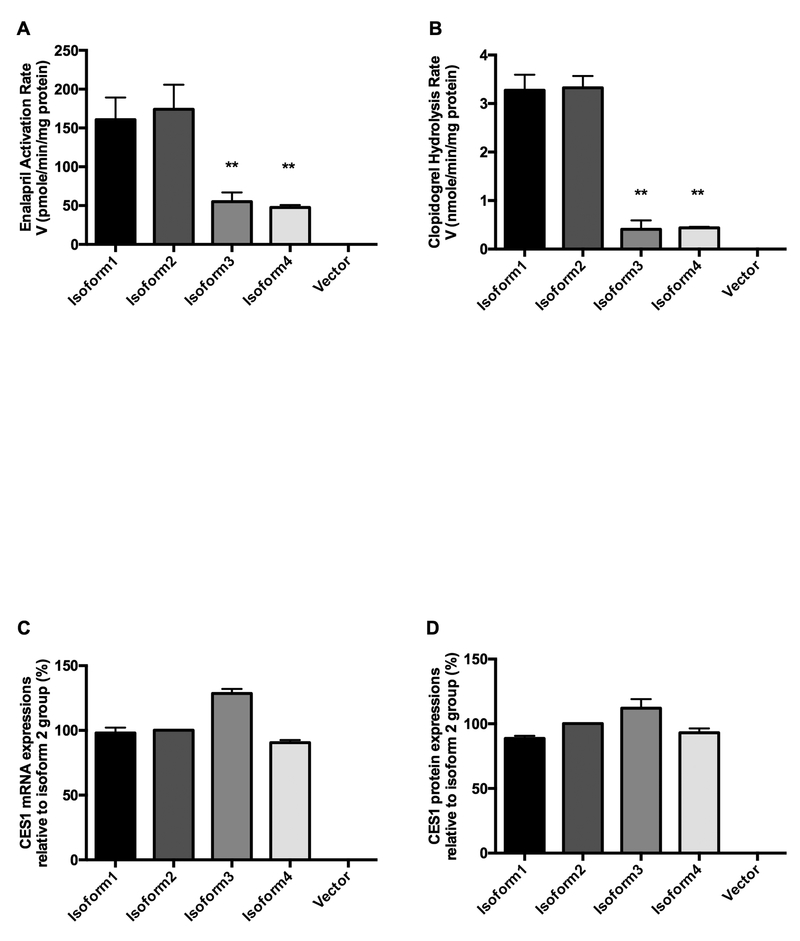
Four cell lines stably expressing CES1 isoforms were established to determine the hydrolysis activities of different isoforms on the two CES1-specific substrate drugs enalapril (Figure 2A) and clopidogrel (Figure 2B). Both substrates were efficiently hydrolyzed by the wild-type form isoform2, and no appreciable metabolites were detected in the vector control. Hydrolytic activities of isoform 1 on enalapril and clopidogrel were comparable to those of isoform 2, while CES1 isoforms3 and 4 showed significantly decreased activities for both substrates (Figure 2A & B). Compared to CES1 isoforms 1 and 2, both isoform 3 and 4 have a deletion of three nucleotides “CAG” at the 5’- end of exon 10 (i.e. -CAG_X10).
Figure 2.
CES1 activity on hydrolyzing enalapril (A) and clopidogrel (B), CES1 mRNA (C) and protein (D) expressions of each isoform in transfected cells. Enalapril (500 μM) and clopidogrel (100 μM) were incubated with the S9 fractions prepared from four cell lines transfected with different CES1 isoforms. Hydrolytic activity of CES1 was determined based on the formation rates of respective hydrolytic products enalaprilat and clopidogrel carboxylic acid. CES1 mRNA and protein levels of isoforms 1, 3, and 4 relative to that of isoform 2 were measured using quantitative real-time PCR and targeted proteomics, respectively. Isoform 2- and vector- transfected cells were used as wild-type and blank controls, respectively. Data are presented as means ± S.D. (n=3), *p<0.05, **p<0.001 (isoforms 1, 3, and 4 vs. isoform 2).
CES1 mRNA and protein levels of the four different isoforms were comparable in transfected cells
The mRNA and protein levels of the CES1 isoforms were measured in the transfected cell lines with vector-transfected cell line included as the negative control. As expected, neither CES1 mRNA nor protein was detected in the vector controls. Significant expression was found for both CES1 mRNA and protein in the transfected cells with levels being comparable among all four CES1 isoforms (Figure 2C & D).
Correlations between CES1 activity and CES1 isoform mRNA levels in individual human liver samples
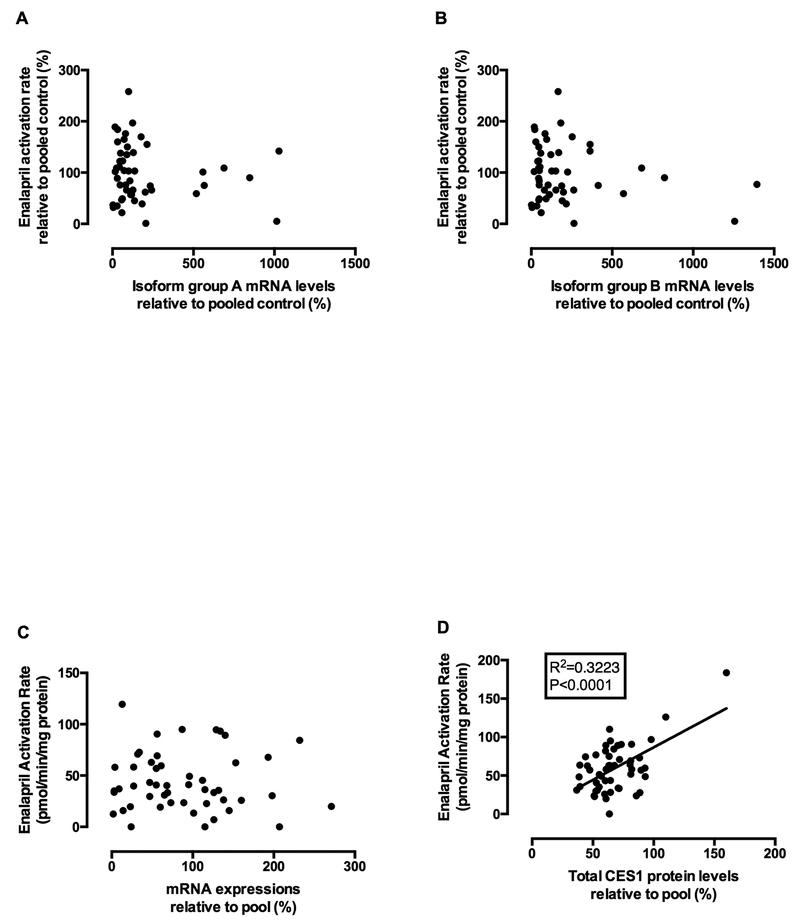
In the transfected cells, the AS event -CAG_X10 (i.e. isoform 3 and 4) was found to significantly impair CES1 activity, while the AS event +GCA_X2 did not impose significant effect on CES1 activity. Accordingly, we classified the isoforms into two groups, group A consisting of isoforms 1 and 2 with normal CES1 function and group B containing isoforms 3 and 4 with significantly impaired activity. We then evaluated the correlation between CES1 activity on enalapril hydrolysis and the mRNA levels of these two CES1 isoform groups in 52 individual human livers. No significant correlation was observed between CES1 activity and the mRNA levels of either group A (Figure 3A) or group B (Figure 3B). Importantly, we also noticed an overall poor correlation between CES1 mRNA level and CES1 activity (Figure 3C), while the correlation between CES1 protein levels and activity was statistically significant (Figure 3D).
Figure 3.
Correlations between CES1 activity and mRNA expressions of CES1 isoform group A (isoforms 1 and 2) (A) and group B (isoforms 3 and 4) (B), total CES1 mRNA (C), and CES1 protein expressions (D) in 52 individual human liver samples. CES1 activity was determined based on the formation rates of enalapril hydrolytic product enalaprilat following incubation of enalapril with individual human liver S9 prepared from human livers. mRNA expressions of total CES1 and isoform groups A & B were analyzed by qRT-PCR using primers for total CES1 and specific CES1 isoform groups, respectively. Total CES1 protein expression was determined by targeted proteomics. Neither isoform group A nor group B mRNA expressions were significantly correlated to CES1 activity in the human liver samples. CES1 activity on enalapril hydrolysis was significantly correlated to CES1 protein expression but not mRNA level.
Correlation analysis between CES1 activity and CES1 isoform protein levels in individual human liver samples
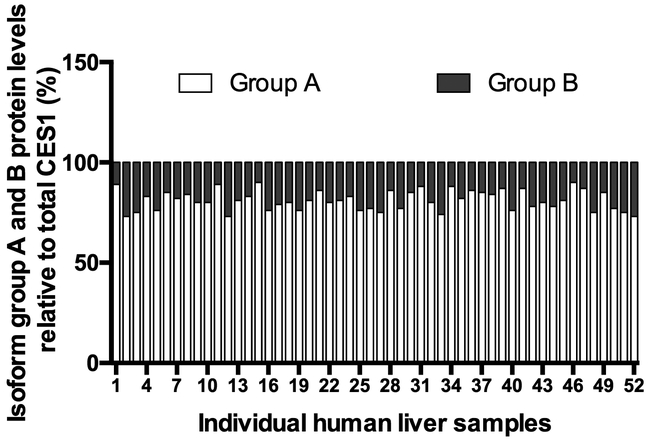
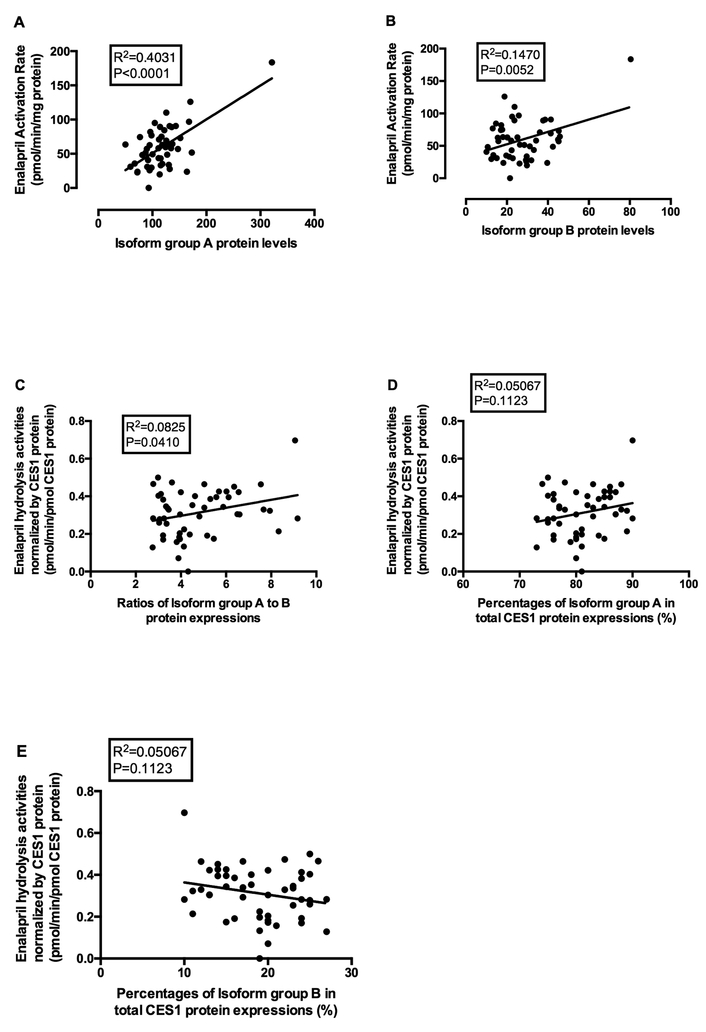
Given that the activity of CES1 isoforms 3 and 4 (group B) is significantly lower than that of isoforms 1 and 2 (group A) in the transfected cells (Figure 2A & B), we further measured protein expression to determine the effects of isoform composition on CES1 activity in human livers. We developed a quantitative proteomics assay to differentiate the isoforms group A and B. The isoform group A was found to be the major CES1 isoform, contributing approximately 74–90% of total CES1 protein expression in the liver samples (Figure 4). The correlation between CES1 activity and the protein expression levels of isoform group A (R2=0.4031, p<0.0001) was much greater than that for isoform group B (R2=0.1470, p=0.0052) (Figure 5A and 5B). In addition, the specific abundance of isoforms group A was found to be superior to total CES1 expression for the prediction of CES1 activity, based on their squared correlation coefficients (R2, 0.4031 vs. 0.3223) (Figure 5A and 3D). Moreover, the protein expression ratio of isoform group A to group B was positively correlated with CES1 activity normalized by total CES1 expression (R2=0.0825, p=0.0410), indicating that varied CES1 isoform compositions may affect CES1 activity in the liver (Figure 5C). As expected, the CES1 unit activity (i.e. CES1 protein-normalized CES1 activity) showed positive and negative trends of correlation with the compositions of CES1 isoform group A (Figure 5D) and B (Figure 5E), respectively, although the correlations were not statistically significant.
Figure 4.
Compositions of CES1 isoform group A and B proteins in 52 individual human livers. The heights of white (bottom) and grey (top) bars represent the percentages of isoform groups A and B in total CES1 protein expressions, respectively. The sum of group A and B protein expression was the total CES1 protein (100%) in each individual liver. The numbers on x-axis indicate different individual liver samples (1–52).
Figure 5.
Correlation analysis between CES1 activity and protein levels of isoform group A (A) and isoform group B (B), and the correlation between CES1 protein expression-normalized activity on enalapril hydrolysis (pmol/min/pmol CES1 protein) and the ratios of CES1 protein isoform group A to group B (C) and the percentages of the group A (D) and B (E) in 52 individual human liver samples. CES1 activity was evaluated based on the enalapril hydrolysis rates. The protein levels of CES1 isoform groups A and B were determined using a QconCAT-based sMRM-HR proteomics assay. CES1 activity exhibited a stronger correlation with isoform group A protein levels relative to those of group B. CES1 protein normalized activity was found to be positively correlated to the protein expression ratios of isoforms group A to group B. The normalized CES1 activity also showed positive and negative trends of correlation with the compositions of CES1 isoform group A (D) and B (E), respectively, although the correlations were not statistically significant.
Discussion
A recent proteomics study has shown that CES1 is the most abundant drug-metabolizing enzyme in human liver microsomes [1]. As the predominant hepatic hydrolase, CES1 plays a critical role in the activation/deactivation of various medications. Importantly, previous studies have shown hepatic CES1 expression and activity to vary among individuals by 11.3-fold [26] and 47.8-fold [29], respectively. Additionally, significant interindividual variability has been consistently reported in responses to CES1 substrate medications, which is in part associated with CES1 genetic polymorphisms[30]. Many SNPs have been identified in the coding regions of CES1, some of these have been demonstrated to significantly affect CES1 function. The first two recognized loss-of-function CES1 variants, G143E (rs71647871) and D269fs (rs71647872), were identified in a study subject who exhibited markedly impaired CES1 activity towards the hydrolysis of methylphenidate [31]. The deleterious effect of the G143E variant on CES1 activity has been consistently demonstrated by several in vitro and clinical investigations for other CES1 substrate drugs, such as oseltamivir [31], trandolapril [32], clopidogrel [5], ACE inhibitors [7], dabigatran [9] and sacubitril [10]. Additionally, we recently conducted a comprehensive functional study of CES1 nonsynonymous variants, and revealed a few CES1 SNPs which impaired CES1 activity in the hydrolysis of enalapril, clopidogrel, and sacubitril; these include L40Ter (rs151291296), A158V (rs202121317), R199H (rs2307243), E220G (rs200707504), and T290M (rs202001817) [16]. Aside from nonsynonymous variants, two SNPs −816A>C (rs3785161) and −75T>G (rs3815583), located in the promoter regions of CES1P1 and CES1, respectively, have been reported to be associated with responses to CES1 substrates, such as imidapril [33], clopidogrel [34] and methylphenidate [13], although not all studies have supported this association [29]. Taken together, these findings have established an important role for CES1 genetic polymorphisms in regulating CES1 function; however, it is worth noting that a substantial portion of CES1 variability remains unexplained.
In humans, four different CES1 protein isoforms are generated by AS, but the functions of each isoform and how isoform composition affects the metabolism of CES1 substrates have not been explored. In the current study, we made an initial attempt to characterize the expression and activity of the four CES1 isoforms produced by AS using isoform-transfected cells, and to further analyze the expression patterns of CES1 isoforms at both mRNA and protein levels, as well as the consequent impact on CES1 activity in individual human liver samples. Two well-established CES1 specific substrate drugs, enalapril [7] and clopidogrel [5], were utilized to evaluate CES1 activity in the transfected cell lines and human livers. To avoid potential interference from functional CES1 SNPs, liver samples with any known functional CES1 variants, such as the G143E, were excluded from the study.
The results from the transfected cell line study suggest that the CES1 AS events do not affect mRNA and protein expressions. However, compared to the CES1 reference isoform 2, the hydrolysis activities of isoforms 3 and 4 on enalapril and clopidogrel were decreased by 60–90%, while the activity of isoform 1 remained the same. The activities of isoforms 3 and 4 were comparable, and both isoforms had a deletion of “CAG” at the beginning of exon 10 (i.e. -CAG_X10), suggesting the amino acid “glutamine” at 362 (Q362) encoded by those three nucleotides is critical for CES1 function (Figure 2A & B). It has been reported that L363 is present in the flexible pocket of the active site of CES1 [35]. We speculate that the deletion of Q362, which is adjacent to L363, might affect the structure of the enzyme active site and consequently impair CES1 activity for isoforms 3 and 4. As this deletion is likely to influence the flexible pocket rather than the rigid pocket of the CES1 active site, its effect on different CES1 substrates might vary, e.g. the decrease in CES1 activity was more significant for clopidogrel hydrolysis than enalapril activation (enalapril vs. clopidogrel: 70% vs. 87% decrease). Given the substrate-dependent effect observed for the Q362 deletion, further studies of its impact on other CES1 substrate medications are warranted.
As CES1 isoforms 3 and 4 showed markedly impaired activity, varied composition of CES1 isoforms among individuals might be a factor contributing to observed CES1 variability. To evaluate the impact of different CES1 isoform compositions on CES1 activity in human livers, we assigned the isoforms to two groups based on their activities, i.e. group A (isoforms 1 and 2) with normal CES1 activity and group B (isoforms 3 and 4) with decreased CES1 activity. We suspected CES1 activity would correlate better with the isoforms of group A than group B. No significant correlation was found between CES1 activity and isoform mRNA levels. This could be attributed to the fact that correlation is overall lacking between CES1 activity and CES1 mRNA levels. Poor mRNA-protein and mRNA-activity correlations have also been documented for many other proteins [36, 37]. We further analyzed the correlations between CES1 protein isoform groups and CES1 activity in individual human liver samples. Isoforms 1 and 2 were found to be the major CES1 protein forms in human livers, contributing to ~80% of total CES1 expression (Figure 4). As expected, CES1 activities correlated better with CES1 isoform group A protein expressions (Figure 5A, R2=0.4031) than those with isoform group B (Figure 5B, R2=0.1470) or with total CES1 protein expressions (Figure 3D, R2=0.3223). According to previous publications, the R2 of the correlations between protein expression and activity of different hepatic drug-metabolizing enzymes varied from 0.26 to 0.97 [37, 38], which is comparable to what we observed for CES1 in the present study. In addition, a positive correlation was observed between the CES1 unit activities (i.e. CES1 protein expression-normalized activity: CES1 activity/CES1 protein expression) and the protein expression ratios of isoform group A to group B in human livers (Figure 5C, R2=0.0825). The CES1 unit activities also showed positive and negative correlation trends with the percentages of isoform group A (Figure 5D, R2=0.05067) and group B (Figure 5E, R2=0.05067), respectively, although the correlations were not statistically significant. Overall, it was indicated that the CES1 isoform composition is a factor contributing to CES1 activity variability.
We applied different proteomic approaches to the quantification of total CES1 protein in transfected cells and different CES1 isoforms in individual human liver samples. CES1 expressions in the transfected cells were quantified using a SILAC method we previously developed [26], which involves five CES1 signature peptides shared by all four CES1 isoforms. As only a single CES1 isoform was expressed in each transfected cell line, the total CES1 expression in any one transfected cell line would represent the expression of the respective isoform. In contrast, human livers may contain all four CES1 isoforms, which requires a quantitative method that is able to differentiate between them. According to the results from the study of isoform-transfected cell lines, the Q362 deletion at the 5’-end of exon 10 common to isoforms 3 and 4 (group B) was associated with significantly lower CES1 activity, while the insertion at exon 2 had no effect on CES1 activity. Accordingly, two surrogate peptides “QEFGWLIPMQLMSYPLSEGQLDQK” and “QEFGWLIPMLMSYPLSEGQLDQK”, which are respectively unique to isoform groups A and B, were used to quantify CES1 isoforms in liver samples. The QconCAT IS we generated contains both surrogate peptides, allowing simultaneous quantification of both isoform groups. Additionally, all quantitative QconCAT peptides were flanked by at least 15 native CES1 amino acids in order to ensure identical digestion efficiencies for the QconCAT peptides and their corresponding endogenous peptides.
The major hepatic CES1 isoforms were shown to be isoforms 1 and 2, which in the present study constituted over 70% of total CES1 protein expression in normal human livers (Figure 4). Nonetheless, it has been widely reported that alternative splicing could be affected by a variety of diseases [39]. It is possible that the CES1 AS patterns that we observed in normal liver tissues might be disrupted in patients with certain diseases, which may result in an altered composition of CES1 isoforms and consequently affect the metabolism of CES1 substrate drugs.
In summary, this study demonstrated for the first time that CES1 isoforms 3 and 4 exhibit significantly lower CES1 activity than isoforms 1 and 2, while the AS event itself does not affect gene expression. Furthermore, CES1 isoform composition may be associated with CES1 activity in human livers. Therefore, variation in CES1 AS may contribute to the documented interindividual variability in response to CES1 substrate medications. Further studies of CES1 AS regulation would improve our understanding of CES1 variability and help develop a strategy to optimize the pharmacotherapy of many clinically important drugs metabolized by the enzyme.
Supplementary Material
Statement of Significance.
Carboxylesterase1 (CES1), the primary hepatic hydrolase in humans, plays a critical role in the metabolism of numerous medications. CES1 expression and activity vary markedly among individuals, resulting in significant interindividual variability in the pharmacokinetics and pharmacodynamics of CES1 substrate drugs. CES1 protein consists of four isoforms due to alternative splicing; however, the activities and expression profiles of these isoforms remain unknown. This study is the first to examine the functions of CES1 isoforms in cell lines transfected with different CES1 isoforms and individual human livers using a targeted proteomics approach. The activities of CES1 isoforms 3 and 4 were found to be significantly lower than the isoforms 1 and 2. The study further revealed that CES1 isoforms 1 and 2 are the major protein forms in human livers, and different CES1 isoform composition may be associated with CES1 activity variation in human livers. The data suggest that CES1 alternative splicing may contribute to the interindividual variability in response to many clinically important medications metabolized by CES1.
Acknowledgements
This work was partially supported by the National Institutes of Health National Heart, Lung, and Blood Institute [Grant R01HL126969, Hao-Jie Zhu], and the Michigan Institute of Clinical Health Research (MICHR) Pre-K Award [Grant UL1TR002240, Xinwen Wang].
Footnotes
Conflict of Interest
The authors have declared no conflict of interest.
Supporting information
All protein LC-MS/MS data have been deposited to the PeptideAtlas with the dataset identifier PASS01246. The following supporting information is available in the supplementary materials in Wiley Online Library.
Study design scheme; the amino acid sequences of CES1 QconCAT protein; the primer pairs, templates and thermocycling conditions used in Site Directed Mutagenesis and Real-Time PCR; the gradient conditions of HPLC-MS/MS analysis and the target peptides information for both the MRM acquisition and sMRM-HR data analysis.
References
- [1].Achour B, Al Feteisi H, Lanucara F, Rostami-Hodjegan A, Barber J, Drug Metab Dispos 2017, 45, 666. [DOI] [PubMed] [Google Scholar]
- [2].Ross MK, Crow JA, J Biochem Mol Toxicol 2007, 21, 187. [DOI] [PubMed] [Google Scholar]
- [3].Imai T, Drug Metab Pharmacokinet 2006, 21, 173. [DOI] [PubMed] [Google Scholar]
- [4].Zhu H-J, Patrick KS, Yuan H-J, Wang J-S, Donovan JL, DeVane CL, Malcolm R, Johnson JA, Youngblood GL, Sweet DH, Langaee TY, Markowitz JS, Am J Hum Genet 2008, 82, 1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhu H-J, Wang X, Gawronski BE, Brinda BJ, Angiolillo DJ, Markowitz JS, J Pharmacol Exp Ther 2013, 344, 665. [DOI] [PubMed] [Google Scholar]
- [6].Zhu HJ, Markowitz JS, Eur J Clin Pharmacol 2013, 69, 733. [DOI] [PubMed] [Google Scholar]
- [7].Wang X, Wang G, Shi J, Aa J, Comas R, Liang Y, Zhu HJ, Pharmacogenomics J 2015, 6, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dimatteo C, D’Andrea G, Vecchione G, Paoletti O, Cappucci F, Tiscia GL, Buono M, Grandone E, Testa S, Margaglione M, Thromb Res 2016, 144, 1. [DOI] [PubMed] [Google Scholar]
- [9].Shi J, Wang X, Nguyen JH, Bleske BE, Liang Y, Liu L, Zhu HJ, Biochem Pharmacol 2016, 119, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shi J, Wang X, Nguyen J, Wu AH, Bleske BE, Zhu HJ, Drug Metab Dispos 2016, 44, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sai K, Saito Y, Tatewaki N, Hosokawa M, Kaniwa N, Nishimaki-Mogami T, Naito M, Sawada J-I, Shirao K, Hamaguchi T, Yamamoto N, Kunitoh H, Tamura T, Yamada Y, Ohe Y, Yoshida T, Minami H, Ohtsu A, Matsumura Y, Saijo N, Okuda H, Br J Clin Pharmacol 2010, 70, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shi D, Yang J, Yang D, LeCluyse EL, Black C, You L, Akhlaghi F, Yan B, J Pharmacol Exp Ther 2006, 319, 1477; [DOI] [PubMed] [Google Scholar]; Nemoda Z, Angyal N, Tarnok Z, Gadoros J, Sasvari-Szekely M, Neuropharmacology 2009, 57, 731. [DOI] [PubMed] [Google Scholar]
- [13].Bruxel EM, Salatino-Oliveira A, Genro JP, Zeni CP, Polanczyk GV, Chazan R, Rohde LA, Hutz MH, Pharmacogenomics J 2013, 13, 476. [DOI] [PubMed] [Google Scholar]
- [14].Zhu H-J, Appel DI, Jiang Y, Markowitz JS, Drug Metab Dispos 2009, 37, 1819; [DOI] [PubMed] [Google Scholar]; Shi J, Wang X, Eyler RF, Liang Y, Liu L, Mueller BA, Zhu HJ, Basic Clin Pharmacol Toxicol 2016, 119, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tarkiainen EK, Backman JT, Neuvonen M, Neuvonen PJ, Schwab M, Niemi M, Clin Pharmacol Ther 2012, 92, 68. [DOI] [PubMed] [Google Scholar]
- [16].Wang X, Rida N, Shi J, Wu A, Bleske B, Zhu HJ, Drug Metab Dispos 2017, 45, 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fukami T, Nakajima M, Maruichi T, Takahashi S, Takamiya M, Aoki Y, McLeod HL, Yokoi T, Pharmacogenet Genomics 2008, 18, 911. [DOI] [PubMed] [Google Scholar]
- [18].Holmes RS, Wright MW, Laulederkind SJ, Cox LA, Hosokawa M, Imai T, Ishibashi S, Lehner R, Miyazaki M, Perkins EJ, Potter PM, Redinbo MR, Robert J, Satoh T, Yamashita T, Yan B, Yokoi T, Zechner R, Maltais LJ, Mamm Genome 2010, 21, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sanford JC, Wang X, Shi J, Barrie ES, Wang D, Zhu H-J, Sadee W, Pharmacogenet Genomics 2016, 26, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Oh J, Lee S, Lee H, Cho JY, Yoon SH, Jang IJ, Yu KS, Lim KS, PLoS One 2017, 12, e0176320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ryu JY, Kim HU, Lee SY, Mol Biosyst 2015, 11, 2798. [DOI] [PubMed] [Google Scholar]
- [22].Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ, Nat Genet 2008, 40, 1413. [DOI] [PubMed] [Google Scholar]
- [23].Lopez-Bigas N, Audit B, Ouzounis C, Parra G, Guigo R, FEBS Lett 2005, 579, 1900; [DOI] [PubMed] [Google Scholar]; Kim E, Goren A, Ast G, Trends Genet 2008, 24, 7. [DOI] [PubMed] [Google Scholar]
- [24].Zschunke F, Salmassi A, Kreipe H, Buck F, Parwaresch MR, Radzun HJ, Blood 1991, 78, 506. [PubMed] [Google Scholar]
- [25].Zhu HJ, Wang X, Gawronski BE, Brinda BJ, Angiolillo DJ, Markowitz JS, J Pharmacol Exp Ther 2013, 344, 665. [DOI] [PubMed] [Google Scholar]
- [26].Wang X, Liang Y, Liu L, Shi J, Zhu HJ, Rapid Commun Mass Spectrom 2016, 30, 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shi J, Wang X, Zhu H, Jiang H, Wang D, Nesvizhskii A, Zhu HJ, J Proteome Res 2018, 17, 3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ, Bioinformatics 2010, 26, 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhu H-J, Langaee TY, Gong Y, Wang X, Pepine CJ, Cooper-DeHoff RM, Johnson JA, Markowitz JS, Eur J Clin Pharmacol 2016, 72, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rasmussen HB, Bjerre D, Linnet K, Jurgens G, Dalhoff K, Stefansson H, Hankemeier T, Kaddurah-Daouk R, Taboureau O, Brunak S, Houmann T, Jeppesen P, Pagsberg AK, Plessen K, Dyrborg J, Hansen PR, Hansen PE, Hughes T, Werge T, Consortium I, Pharmacogenomics 2015, 16, 649. [DOI] [PubMed] [Google Scholar]
- [31].Zhu H-J, Markowitz JS, Drug Metab Dispos 2009, 37, 264. [DOI] [PubMed] [Google Scholar]
- [32].Zhu H-J, Appel DI, Johnson JA, Chavin KD, Markowitz JS, Biochem Pharmacol 2009, 77, 1266. [DOI] [PubMed] [Google Scholar]
- [33].Geshi E, Kimura T, Yoshimura M, Suzuki H, Koba S, Sakai T, Saito T, Koga A, Muramatsu M, Katagiri T, Hypertens Res 2005, 28, 719. [DOI] [PubMed] [Google Scholar]
- [34].Xie C, Ding X, Gao J, Wang H, Hang Y, Zhang H, Zhang J, Jiang B, Miao L, Pharmacogenet Genomics 2014, 24, 204; [DOI] [PubMed] [Google Scholar]; Zou JJ, Chen SL, Fan HW, Tan J, He BS, Xie HG, J Cardiovasc Pharmacol 2014, 63, 178; [DOI] [PubMed] [Google Scholar]; Bjerre D, Ferrero L, Madsen M, Hansen PR, Rasmussen HB, Consortium I, Pharmacogenet Genomics 2015, 25, 147. [DOI] [PubMed] [Google Scholar]
- [35].Nzabonimpa GS, Rasmussen HB, Brunak S, Taboureau O, Consortium I, Drug Metab Pers Ther 2016. [DOI] [PubMed] [Google Scholar]
- [36].Maier T, Guell M, Serrano L, FEBS Lett 2009, 583, 3966. [DOI] [PubMed] [Google Scholar]
- [37].Ohtsuki S, Schaefer O, Kawakami H, Inoue T, Liehner S, Saito A, Ishiguro N, Kishimoto W, Ludwig-Schwellinger E, Ebner T, Terasaki T, Drug Metab Dispos 2012, 40, 83. [DOI] [PubMed] [Google Scholar]
- [38].Achour B, Russell MR, Barber J, Rostami-Hodjegan A, Drug Metab Dispos 2014, 42, 500. [DOI] [PubMed] [Google Scholar]
- [39].Caceres JF, Kornblihtt AR, Trends Genet 2002, 18, 186; [DOI] [PubMed] [Google Scholar]; Hasler R, Kerick M, Mah N, Hultschig C, Richter G, Bretz F, Sina C, Lehrach H, Nietfeld W, Schreiber S, Rosenstiel P, Eur J Cell Biol 2011, 90, 603; [DOI] [PubMed] [Google Scholar]; Faustino NA, Cooper TA, Genes Dev 2003, 17, 419; [DOI] [PubMed] [Google Scholar]; Musunuru K, Trends Cardiovasc Med 2003, 13, 188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.