Abstract
Cyanobacteria are photosynthetic prokaryotes that are influential in global geochemistry and are promising candidates for industrial applications. Because the livelihood of cyanobacteria is directly dependent upon light, a comprehensive understanding of metabolism in these organisms requires taking into account the effects of day-night transitions and circadian regulation. These events synchronize intracellular processes with the solar day. Accordingly, metabolism is controlled and structured differently in cyanobacteria than in heterotrophic bacteria. Thus, the approaches applied to engineering heterotrophic bacteria will need to be revised for cyanobacterial chassis. Here, we summarize important findings related to diurnal metabolism in cyanobacteria and present open questions in the field.
Keywords: Circadian clock, Diurnal Physiology, Photosynthesis, Redox regulation, Signaling nucleotides, Light-Dark Cycles
An Introduction to Day-Night Cycles in Cyanobacteria
The daily fluctuation of light is a nearly universal evolutionary pressure for life on Earth. For cyanobacteria, microorganisms that rely almost exclusively on light for energy, the response to these day-night cycles is particularly wide-ranging and includes the redirection of central metabolism [1, 2] and sweeping changes in gene expression [3, 4]. As important primary producers and progenitors (via endosymbiosis) to the other oxygen-evolving photosynthetic organisms [5], cyanobacteria and their responses to light-dark cycles (LDCs) have broad implications for understanding photosynthesis in higher organisms and for characterizing a phylum that has tremendous ecological impact and biotechnological potential [6, 7]. For instance, natural diel cycles influence infection by viruses (see Box 1) and also carry significant economic consequences for industrial-scale growth (see Beyond Cyanobacteria section). However, due to practical experimental considerations, most research, and reviews to date, have focused on the unnaturally static condition of perpetual light. Recent work probing the physiology of cyanobacteria in LDCs has opened up a fresh perspective on the life cycle of this keystone bacterial phylum. In this review we consolidate current knowledge on cyanobacterial growth in LDCs by starting with the cellular functions that are important for the day and night states. Thereafter, we address the current understanding of regulatory processes that are required to coordinate the transitions between the two. Finally, we discuss how research on cyanobacterial biology in LDCs has revealed a paradigm for diurnal growth that generalizes beyond cyanobacteria.
Box 1 : Viral Infections and Light-Dark.
The ocean and freshwater lake ecosystems teem with cyanobacteria as well as viruses that infect them. The ocean is home to upwards of 1030 phage particles, and their interactions with marine life play a critical role in ecosystem dynamics [93] and biogeochemical cycles [94]. Viral particles infecting cyanobacterial cells co-opt and utilize ATP and NADPH produced by the cyanobacterium during photosynthesis. Light and photosynthesis influence the success of the phage infection, including the number of phage particles that are generated during infection and released upon lysis [95, 96]. For example, in S. elongatus light has a strong influence on infection by the contractile phage AS-1, and infection and light absorption are correlated, occurring in a diel pattern under LDCs [97].
Some phage carry metabolism genes that enhance host daytime metabolic flux. For instance, a phage-borne psbA gene, encoding the photosynthetic reaction center D1 protein, may help maintain host photosynthetic capacity during infection. Moreover, genes of the pentose phosphate cycle (talC, gnd, zwf, and even the cp12 gene that encodes the inhibitory factor of the CBBC) have been found in phage and are preferentially expressed during infection in the light [98]. At night overall phage gene expression decreases 100-fold [99].
Surviving the Day
Each day, a cyanobacterium faces the formidable task of turning inorganic carbon into the organic molecules of life via photosynthetic carbon dioxide assimilation. As the sun rises, the cell encounters numerous metabolic challenges. It must perform cellular division by binary fission while also storing energy reserves for the night, a period of photosynthetic quiescence. Daytime activities take place in the background of photosynthesis, a process vital to the cell and one that requires significant resources for efficient function. In the process, damaging reactive oxygen species (ROS) are generated as a byproduct [8]. As a consequence, cyanobacterial metabolism is carefully orchestrated in both space and time [4, 9, 10] (Fig. 1, Key Figure).
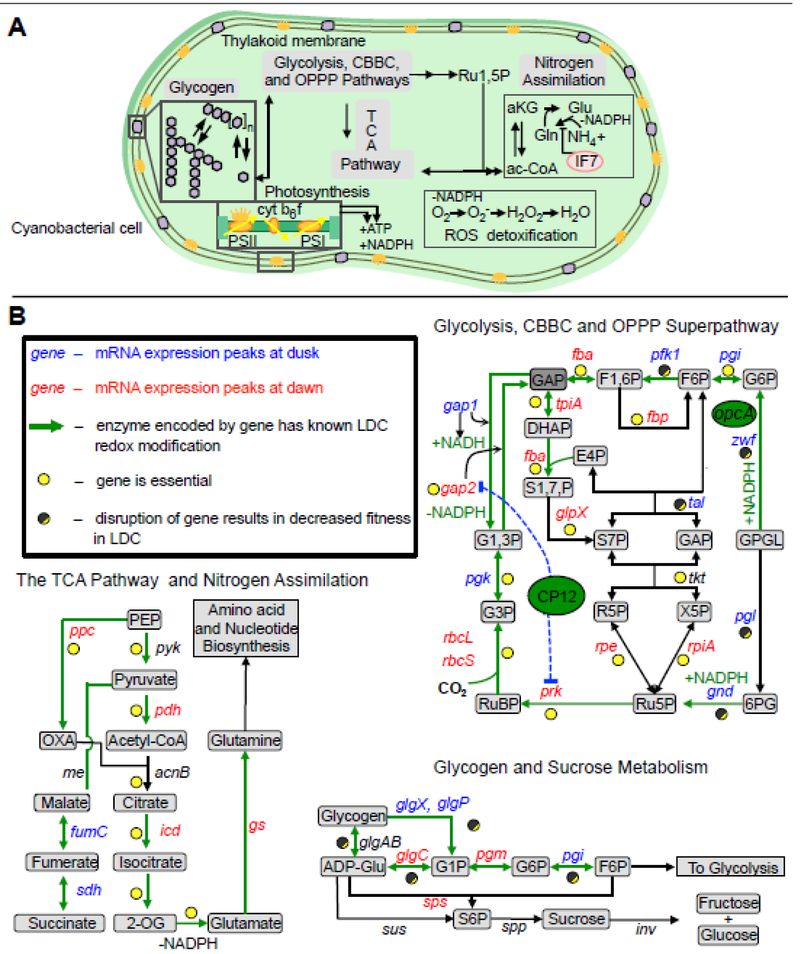
Figure 1. Shift work: snapshot of cellular activities across the day and night.
A) A representation of an S. elongatus cell with an overview of the major metabolic pathways important for day-night physiology. B) Reactions of glycogen metabolism, TCA pathway, and central metabolism that are present in S. elongatus are provided in more detail. Green arrows in panel B indicate reactions where the respective enzyme has a detectable light/dark-dependent redox modification in Synechocystis sp. PCC 6803 [69]. Data about the peak expression time of circadian genes is indicated using data collected from S. elongatus PCC 7942 [9]. Genes colored in red peak in expression in the subjective morning, genes colored in blue peak in expression in the subjective evening, and genes colored in black have no detectable circadian rhythm in S. elongatus. Essential genes and genes that cause light-dark sensitive phenotypes when mutated are indicated by full yellow and half yellow-half dark grey circles, respectively. Abbreviations: PBS, phycobilisome; PSII, photosystem II; cyt b6f, cytochrome b6f; PSI, photosystem I; Fd(red), ferredoxin (reduced); Ru1,5P, ribulose-1,5-bisphosphate; 3PG, 3-phosphoglycerate; 1,3-BPG, 1,3-bisphosphoglycerate; GAP, glyceraldehyde-3-phosphate; F6P, fructose-6-phosphate; G6P, glucose-6-phosphate; G1P, glucose-1-phosphate; 6PGL, 6-phosphogluconolactone; 6PG, 6-phosphogluconate; Ru5P, ribulose-5-phosphate; ac-CoA, acetyl-CoA; aKG, a-ketoglutarate; Glu, glutamate; Gln, glutamine.
Central Carbon Metabolism.
Much of cyanobacterial metabolism can be described as temporally partitioned, and generalized as anabolic during the day and catabolic at night. Daytime metabolism begins with shifting carbon flux from the oxidative pentose phosphate pathway (OPPP) to the Calvin-Benson-Bassham Cycle (CBBC), and is controlled via products of the photosynthetic light reactions [11-13]. One of the critical steps in this process is inactivation of CP12, a redox-sensitive protein that is a master regulator of the CBBC [14, 15]. During the night, oxidized CP12 structurally sequesters glyceraldehyde 3-phosphate dehydrogenase 2 (Gap2) and phosphoribulokinase (Prk) and inhibits the CBBC. This switch is mediated by the redox state of the cyanobacterial cell, which changes markedly depending on photosynthetic activity. At the onset of light, photosynthetic reducing equivalents are generated, reduced CP12 releases Gap2 and Prk, and CBBC activity resumes. Metabolomic analysis reveals that anabolic metabolism is upregulated during this phase of the day, including pathways related to amino acid, nucleotide, and quinone biosynthesis [1]. Upregulation of amino acid and nucleotide synthesis agrees with physiological observations that protein synthesis and DNA replication occur to a much greater extent during the day [16, 17].
Energy Storage and Electron Sinks.
A principal activity during the daytime is accumulation of excess photosynthate, which is stored as the glucose polymer glycogen. During growth in LDCs, glycogen accumulates during the day and serves two primary purposes: i) as an energy-storage polymer in preparation for night [1, 18-21], and ii) as a “regulatory valve” to assimilate excess reducing power produced under conditions of particularly high light intensity [8, 22-24]. Mutations that target the glycogen biosynthesis genes glgA, glgC, or glgP significantly hinder the ability of cells to grow and remain viable in LDCs, highlighting the importance of glycogen storage [25-27].
Because cyanobacteria cannot rapidly turn off photosynthetic activity, conditions that temporarily impair daytime cell growth can cause a dangerous buildup in membrane redox potential [8, 28, 29]. The role of glycogen as a photosynthetic electron sink has been highlighted by investigations into the cyanobacterial nitrogen-deprivation response [25, 30, 31]. Nitrogen deficiency causes a rapid accumulation of glycogen as the downstream utilization of carbon skeletons via diverse biosynthetic processes is inhibited [30, 32]. In strains unable to synthesize glycogen, nitrogen deprivation causes growth defects and oxidative damage at high-light intensities that otherwise do not affect wild-type (WT) cells [8, 23, 24]. Additionally, under nitrogen deprivation conditions, mutants unable to synthesize glycogen secrete pyruvate and tricarboxylic acid (TCA) pathway intermediates, possibly as an attempt to redirect photosynthetic output [25, 30, 31, 33]. Overall, the buffering of cellular redox state through glycogen synthesis and degradation is an important aspect of regulating cellular physiology.
Surviving the Night
As the day ends and night begins, cyanobacteria face a drastic change of lifestyle. The past few decades of research have taught us a great deal about cyanobacterial processes that occur in the light, but little about cyanobacterial life in the dark. Although a cyanobacterium in darkness is typically viewed as being in a dormant state, the cell is not inactive and many processes still operate dynamically. Studies on transcription, translation, and metabolism have demonstrated specific adaptive responses to darkness in Synechococcus elongatus sp. PCC 7942 (for a historical synopsis of this model organism see Box 2). While overall rates of these processes may be lower than in the light, or even close to zero in the case of DNA replication [34, 35], they are coordinated such that the cell can conserve energy, ensuring its survival until light is available again. With photosynthesis unable to proceed in the dark, a suite of cell-wide changes occurs, ranging from shifts in ATP and reductant levels, to redirection in the flux of carbon compounds, to altered cell division activities.
Box 2 : Brief history of Synechococcus sp. PCC 7942.
Synechococcus elongatus sp. PCC 7942 is the official name of a cyanobacterium that was isolated prior to 1973 from a local freshwater source by students taught by K.W. Floyd at California State University, San Francisco. Several samples were transferred to S.V. Shestakov of Moscow State University, whose lab demonstrated that one of the isolates, termed R-2, was transformable by chromosomal DNA from an antibiotic-resistant strain in their collection called Anacystis nidulans 602 [100]. The California isolate became known for many years as A. nidulans R2. In 1978 C.A.M.J.J. van den Flondel brought the strain from Moscow to the lab of G. van Arkel (University of Utrecht), where he was able to isolate mutants that carried the selectable transposon Tn901 in the small endogenous plasmid of A. nidulans R2. This work began the era of recombinant DNA-based molecular genetics research in cyanobacteria [101]. Drs. van den Flondel and van Arkel deposited the strain in the Pasteur Culture Collection, where it was given the accession number PCC 7942. A re-evaluation of the taxonomic structure of the cyanobacteria in the mid-1980s resulted in a renaming of previous Anacystis strains to the genus Synechococcus. For a period of several years, publications regarding this organism referred to it as Synechococcus sp. strain PCC 7942 without a species designation. A second edition of Bergey’s Manual of Systematic Bacteriology was published in 2001 which included a section on the classification of cyanobacteria. A chapter by M. Herdman, R. Castenholz, J. Waterbury, and R. Rippka described the Synechococcus clade Cluster 1.1, typified by PCC 6301, which is so closely related to PCC 7942 as to be members of the same species [102]. These authors proposed the binomial Synechococcus elongatus, which is a name in keeping with the Botanical Code of Nomenclature. Most papers published since that date refer to the former A. nidulans R2 as S. elongatus PCC 7942. Note that the name Synechococcus elongatus had been used previously with reference to thermophilic cyanobacteria that are phylogenetically distant from PCC 6301 and 7942. The name Thermosynechococcus elongatus is now used for those thermophilic strains, but care is advisable in reading the literature to distinguish the S. elongatus that refers to PCC 7942 and PCC 6301, typically grown at 30 C, from the relatives of T. elongatus, typically grown at 45-50 C [103].
Glycogen breakdown and the OPPP.
The initiation of glycogen degradation is essential for nighttime survival in those cyanobacteria that are unable to utilize an external fixed carbon source [1, 26]. The majority of the released glucose is shunted directly into the OPPP rather than the glycolytic pathway favored by diverse heterotrophs [11, 13, 36]. The primary function of the OPPP at night is to generate reducing power in the form of reduced nicotinamide adenine dinucleotide phosphate (NADPH) when the main production route during the day, photosynthesis, is not active [2, 26, 37]. This preference for the OPPP over glycolysis is functionally relevant in its production of NADPH over NADH. A number of enzymes in photosynthetic organisms have evolved a preference for NADPH over NADH as a reductant source, including some that are important for detoxifying ROS [38, 39]. Cyanobacteria possess a variety of antioxidant and redox-buffering systems, such as enzymatic defences involving superoxide dismutase and catalyases, and non-enzymatic strategies using glutathione, peroxiredoxin, and caroteniods. Of these ROS responses, reduced glutathione-mediated reactions are critical for protection from the multiple ROS species that cells encounter. Importantly, the reduction of the disulfide in glutathione is mediated by glutathione reductase, which is dependent on NADPH. It is therefore not suprising that NADPH produced through the OPPP appears to be vital to surviving LDCs; many mutations that cause LDC sensitivity result in high oxidative-stress buildup that is not cleared in the dark [2, 27, 40]. Thus, carbon flux through the OPPP is one of the most critical processes for nighttime survival, and inactivation of any of the three core OPPP genes zwf, gap, or gnd causes severely attenuated growth when cells are cultured in LDCs [27, 41-43]. In contrast, the OPPP genes are dispensable when cells are grown under continuous light where NADPH is generated by photosynthesis (Fig. 2). Detoxification of ROS using NADPH produced by the OPPP is a common theme in other higher organisms as well, such as in the hawk moth (see Beyond cyanobacteria).
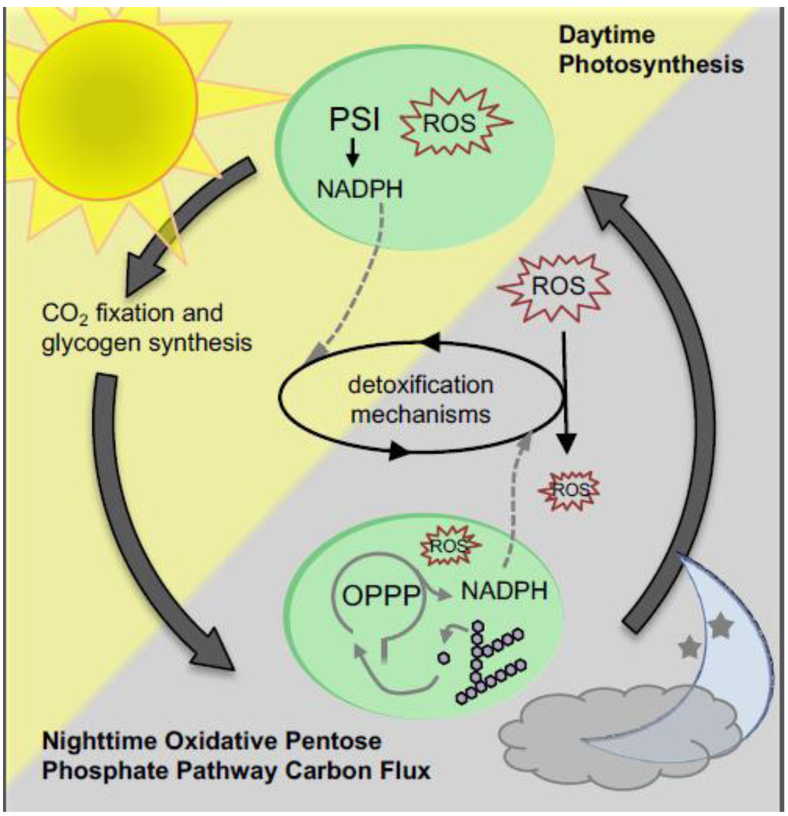
Figure 2. Detoxifying ROS in the diurnal world.
High rates of photosynthetic metabolism in light generate damaging ROS. During the day NADPH produced via photosynthesis can aid in clearing such molecules. At night ROS production ceases, and remaining ROS is cleared by nighttime metabolism. Here, detoxification can be aided by NADPH produced through degradation of glycogen by the OPPP.
Consistent with a paradigm of the OPPP as the primary catabolic route for stored carbon, a genome-wide transposon mutagenesis screen in S. elongatus showed that enzymes of the regenerative phase of the canonical TCA cycle are not essential for viability under either continuous light or in LDC conditions [27]. Metabolic flux studies utilizing 13C isotopic tracing in Synechocystis sp. PCC 6803 show that carbon primarily cycles within the OPPP even when that species grows heterotrophically [12, 44]. These data reveal a fundamental difference in the orchestration of carbon processing pathways between cyanobacteria and most heterotrophs. While many of the reactions that are usually part of a TCA cycle have been shown to be non-essential in S. elongatus, the first three steps leading to 2-oxoglutarate (2-OG) production are mediated by essential genes. It is likely that intermediates produced from other TCA-cycle precursor metabolites can be generated via alternative reactions, but 2-OG is the only known precursor for nitrogen assimilation into glutamine and glutamate in cyanobacteria. Production of 2-OG via the TCA reactions and subsequent nitrogen assimilation strongly influence NADPH reductant balance, because glutamine and glutamate biosynthesis require considerable reductant input via reactions that preferentially utilize NADPH over NADH [45]. Thus, nitrogen assimilation via 2-OG is inhibited in the dark, in part due to the induction of glutamine synthase inactivating factor (IF7) ([46]; also see Supplemental Table S1).
Transcription and Translation.
Several specific enzymatic reactions important for LDCs have been discussed, but an additional layer of regulation should be taken into account: the transcription and translation of the genes that encode these enzymes, and many others, is dynamic across the day-night cycle. Generally, gene expression decreases in the dark, with a few exceptions [3, 47, 48]. Of the genes that are expressed upon dark exposure, some can be classified as being induced by darkness independent of the time of day or circadian phase [49], while others’ expression profile in darkness relies on a functioning circadian clock [3].
It is clear that circadian regulation provides a fitness benefit to cyanobacteria when cells are in sync with the environmental LDC [50]. Yet, there is considerable overlap between genes found to be induced during dark exposure at nighttime [3], those induced by dark exposure during the day [49], and those induced by dim light during the late afternoon[51] (Supplemental Table S1). Although many of these genes encode either proteins of unknown function or are annotated simply as dig for dark-induced gene, some gene annotations provide clues to important functions during dark exposure. For example, a protein with high sequence similarity to CP12 (synpcc7942_0252) [14], a regulator of ribosomal status hpf (IrtA; synpcc7942_2352) [49], and a probable chaperone protein (hspA, synpcc7942-0241) suggest that redox regulation and control of protein synthesis and degradation are key processes in the dark.
Examination of the protein landscape tells a different, less dramatic story. Ansong et al.[52] showed that only 4% of proteins in Synechococcus sp. PCC 7002 change in abundance between light and dark. This trend is also seen in other cyanobacteria; for example, few proteins were found to have >2 fold change across light and dark phases in Cyanothece, a N2-fixing cyanobacterium [53]. However, while the overall abundance of protein is relatively constant, the rate of translation of proteins has been observed to decrease in the dark [17, 49], and post-translational modifications and intracellular localizations may act as strong regulatory components at the protein level in LDCs.
Orchestration of Cell Physiology Under Light-Dark Cycles
The many metabolic changes that occur during the day-night transition are managed through a complex network of interconnected regulatory processes. The majority of the genome in S. elongatus is differentially transcribed in LDCs. A number of factors drive both transcriptional and protein levels including: circadian clock output, chromosome topology, signaling nucleotides, and changing concentrations of metabolites, such as NADPH and ATP (Fig. 3).
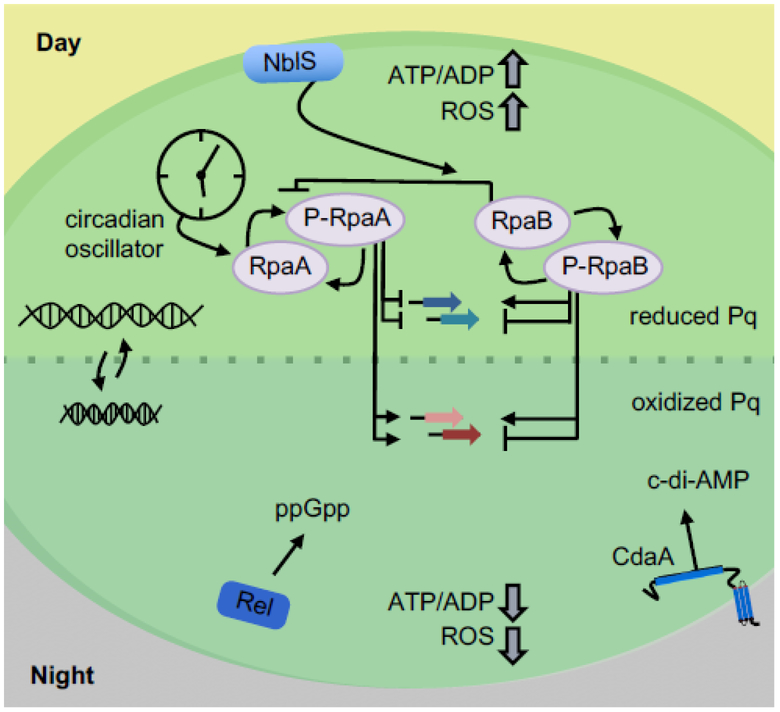
Figure 3. Getting the message: signaling pathways important for the day-night transition.
The signaling pathways effective during the day-to-night transition consist of environmental sensing via RpaB and its cognate histidine protein kinase NblS, circadian status via RpaA, intracellular redox and energy status through changes in concentrations of NADPH and ATP and redox state of the plastoquinone (Pq) pools, chromosome compaction, and nucleotide signaling molecules such as ppGpp and cyclic-di-AMP (synthesized by RelA and CdaA, respectively).
Transcription Factors.
While multiple transcription factors affect transcription in cyanobacteria, two with wide-ranging influence stand out. The response regulators RpaA and RpaB are master transcription factors in cyanobacteria that act as control hubs of LDC physiology. RpaA provides the key output mechanism to convey temporal information from the circadian clock, and is responsible for the clock-dependent regulation of hundreds of genes [54, 55].
The link between the circadian clock – whose oscillator comprises proteins KaiA, KaiB, and KaiC – and activity of the transcription factor RpaA lies with two histidine protein kinases, SasA and CikA, which engage with the oscillator complex at different times of day [56]. Association of SasA or CikA with the oscillator, stimulates an activity that either phosphorylates (SasA) or dephoshorylates RpaA (CikA) [57]. Phosphorylated RpaA (P-RpaA) activates genes important for nighttime metabolism, and in the absence of RpaA (or KaiA [27]) the clock is locked into a daytime transcriptional regime that leads to metabolic imbalance, increased oxidative stress, and death in LDCs [2, 27, 54, 58]. In contrast, a strain that is locked in a “nighttime” mode through elimination of KaiC experiences constitutive expression of the OPPP and other nighttime metabolic pathways. This situation is generally permissive for growth in LDCs, although the lack of a timing mechanism exacts a fitness cost that can be observed when WT and KaiC-null strains are grown together in competition [59].
Less investigated is the activity of RpaB. It acts as a light-responsive regulator of gene expression that is independent of the clock [60, 61]. Manipulating RpaB expression level or phosphorylation state affects growth in LDCs, and its output overlaps with that of RpaA in ways that are not yet understood [62]. While RpaA acts as the sole output signal of the clock, RpaB feeds in signals of environmental light status to the cell, and integration of both signals is important for fitness in LDCs.
Chromosome Topology.
One striking cellular change in cyanobacteria that occurs over a daily cycle and is circadian-controlled is that of chromosome topology. The extent of chromosome or plasmid compaction in S. elongatus varies depending on the time of day, being compact and highly supercoiled at some times and relatively relaxed at others [9, 63-65]. When supercoiling is relaxed by addition of an inhibitor of DNA gyrase, changes in gene expression patterns are observed. Expression of genes that are normally expressed when the genome is in a highly supercoiled state decrease, and those normally expressed when the genome is relaxed increase, upon the addition of the inhibitor, consistent with expectations for the time-of-day peak expression of a given gene and the topological state. While mostly correlative, this relationship between DNA compaction and supercoiling with transcriptional outputs suggests another possible mechanism for global regulation. The details of how genome organization, compaction, and accessibility to interacting proteins may impact gene expression, protein localization, and cell division [16, 66, 67] is an area ripe for further study.
Cellular Energy Levels
Shifts in ATP/ADP and NADPH/NADP+ ratios unavoidably occur in LDCs due to temporal division of photosynthesis and catabolism. The dynamics of ATP changes in LDCs have been measured by many researchers over decades, but results vary. In S. elongatus ATP levels were reported to fall precipitously within the first 2 minutes after a shift to darkness and then recover to near pre-dark levels within 20 to 60 minutes [48, 68]. More recently, Rust et al. made similar measurements, but over a longer duration of dark exposure. They found that, despite this quick recovery, ATP levels gradually decrease overall during dark exposure, reaching ~50% of pre-dark level and remaining low until light is reintroduced, after which ATP concentration rapidly recovers [69]. The physiological implication of these changes are likely to be global for the cell. For instance, ATP concentration directly affects the status of the circadian oscillator and its stimulation of the RpaA kinase, SasA [69].
Cellular Redox.
Cellular redox homeostasis is of critical importance for growth in LDCs [52, 70, 71]. Mutants that are unable to funnel carbon metabolites through the OPPP produce an insufficient amount of NADPH at night [2]. This deficit results in diminished ability to detoxify cellular ROS accumulated during the day, which requires NADPH reducing equivalents [2, 8, 72]. Thus, limiting nighttime NADPH production by compromising OPPP activity likely has severe redox consequences for broad metabolic and regulatory systems in photoautotrophic microorganisms.
One common theme between cyanobacteria and plant chloroplasts is the redox-dependent regulation of many proteins [73]. Although both photosynthetic reactions and the OPPP generate NADPH, reductant levels drop in the dark as the photosynthetic reactions cease [14, 68]. Compounding this redox change, NADH levels rise in the dark, leading to a further decrease in the NADPH / NADH ratio [14]. Overall, the concentrations of NADPH and NADP+ vary to an even greater extent than that of ATP over the course of LDCs, and have been correlated with changes in the transcription of genes that encode enzymes of photosynthetic and metabolic homeostasis processes [71]. In addition, many metabolic enzymes are redox-modified. As LDCs drive the changes in oxidation state that control these modifications the subsequent enzymatic activities also follow suit [14, 52, 70, 73-76] (Fig. 1).
In the absence of notable changes in protein levels, redox modifications of metabolic enzymes likely play a major role in dictating metabolic flux when cells are in the dark. Indeed, many of the critical enzymes required for survival in darkness are redox modified, including Zwf, Gnd, GlgC, GlgA, GlgP, and enzymes that regulate nitrogen assimilation [70, 77, 78]. Redox regulation directly mediates the critical shift between CBBC and OPPP activity at light-to-dark transitions through the direct inhibition of the CBBC enzymes Gap2 and Prk by CP12 in a redox-controlled and light-dependent manner [14, 15]. Moreover, numerous redox-active proteins and small molecules – thioredoxins, ferredoxins, peroxiredoxins, and glutathione – can directly modify target protein thiols [77-81] and impact enzymatic activity [52, 70]. While many questions remain regarding the mechanisms of redox regulation during growth under LDCs, the maintenance of redox homeostasis is unquestionably important for cyanobacterial metabolic processes at night.
Signaling nucleotides.
Evidence is accumulating that signaling nucleotides act as intracellular messengers of LDCs. Levels of cAMP, c-di-AMP, c-di-GMP, and ppGpp are all light dependent in cyanobacteria [40, 49, 82, 83]. ppGpp in particular is a potent effector of transcription in S. elongatus that is synthesized after a light-to-dark transition and is critical for maintaining fitness during dark-induced stress [49]. Viability in cells unable to synthesize ppGpp is impaired after exposure to darkness, although the mechanisms behind this phenotype are not yet known.
c-di-AMP, a newly discovered signaling nucleotide in cyanobacteria [40, 84], is also important for survival of S. elongatus during darkness. Inactivation of its cyclase, cdaA, leads to increased oxidative stress and decreased survival of the night periods of LDCs [40]. c-di-AMP and ppGpp levels are linked in Firmicutes [85, 86], bringing up the possibility that their activity is coordinated in cyanobacteria grown in LDCs; however, this potential connection remains unexplored.
Beyond Cyanobacteria
The need for metabolic shifts as an adaptation to diel cycles is also a dominating force for plants and eukaryotic algae, and influences global biogeochemical cycles such as CO2 balance. In plants, constant adjustments to physiology in response to changes in light quality, intensity, and duration are made through the use of photoreceptors [87]. Changes in the photoperiod of LDCs cue plants to undergo different phases of growth, development, and metabolism. In Arabidopsis, for example, darkness elicits the expression of over 80 genes that code for functions involved in photosystem II inhibition, starch degradation, chloroplastic translation inhibition, and redox regulation, similar to what we observe in cyanobacteria [88].
The responses of plants to darkness and LDCs are also important from an economic and agricultural point of view. Post-harvest storage of green leafy vegetables, such as kale and cabbage, in LDCs results in significantly improved appearance and health value of crops compared to constant-condition controls due to increased tissue integrity, chlorophyll content, and levels of glucosinolates [89, 90].
Moreover, photosynthetic organisms are not alone in struggling with oxidative stress during LDCs, and some of the preventative mechanisms they utilize may be conserved in heterotrophs. For instance, hovering flight in nectarivores is an immensely energetic endeavor that comes with high metabolic turnover that generates ROS. In hawkmoths, like cyanobacteria, this oxidative stress is likely detoxified by activity of the OPPP during rest, which by producing NADPH maintains sufficient quantities of reduced glutathione to act as an antioxidant [91] (Fig. 2). This strategy for oxidative stress management after intense exercise may be of importance in other animals as well [92].
Concluding Remarks
Lessons from the genetically tractable and evolutionarily ancient cyanobacteria can educate us on the metabolic strategies that have evolved to enable organisms to deal with LDC stress, a phenomenon that is difficult to study in many other phyla. Insights from cyanobacteria can also aide in developing strategies to harness photosynthetic organisms for real-world industrial, biotech, and agricultural applications.
In this review, we have attempted to condense much of the knowledge that now exists on the cyanobacterial response to LDCs. There remain, though, many areas for growth in this respect (see Outstanding Questions). Many variables such as protein and metabolite levels appear surprisingly constant in LDCs on average. However, this balance seems possible only in the context of large shifts in transcription, primary metabolism, and glycogen levels that occur during the day. These shifts, in turn, are caused by large alterations in ratios of electron carriers, redox poise, nucleotide signaling, and chromosome structure, all compelled by signals from the internal circadian clock and external environment. Thus, maintaining physiological balance in a world of potentially jarring LDCs is an exhaustive task that requires a vigilant sensing of external and internally generated signals, and appropriate cellular responses.
Outstanding Questions Panel.
Increased ROS has been correlated with growth defects in all of the LDC-sensitive mutants where it has been assayed. However, is this truly a causal relationship? Is ROS the dominant stress that must be mitigated in LDCs, and what are the important pathways in resisting this stress?
How can the direct response to LDCs be perturbed in a controlled way (through mutant or condition) to enable observations, similar to the manipulations that have proven so fundamental to research on the circadian clock?
While transcript and protein data can provide insight into metabolism during LDCs, we currently do not have a good picture of what the total metabolome looks like as cells transition between day and night growth phases. Is the flux of carbon and accumulation of metabolites indicative of a restorative process or an active process to prevent light-induced redox stress?
What differences are detected in physiological responses when cells are exposed to abrupt (square-wave) versus sinusoidal LDC (more like that found in nature)?
While the activities of the circadian clock output transcription factor RpaA when it is in its phosphorylated active state have been detailed as being important for LDC viability, what is its role, if any, when it is in its “inactive” unphosphorylated state during the day?
Cyanobacteria are pervasive across latitudinal space and must contend with large seasonal variations in day and night lengths, but how the cyanobacterial clock functions when driven by LDCs of different photoperiods, as would be present in different seasons is still a relatively unexplored area in the field.
Supplementary Material
Highlights.
A cyanobacterium integrates signals from the environment and from an internal circadian clock to orchestrate diurnal physiology.
Large datasets from genomic, proteomic, and metabolomic analyses have elucidated daytime and nighttime programs that cyanobacterial cells employ during diurnal growth.
A critical aspect of metabolism in the dark is the production of NADPH by the oxidative pentose phosphate pathway when photosynthesis is inactive, which drives the suppression of potentially lethal reactive oxygen species.
Understanding diurnal physiology in cyanobacteria may help to harness these organisms for biotechnology applications, where outdoor growth may be desirable.
Acknowledgments
We thank Drs. R. Rippka, L.A. Sherman, S.V. Shestakov, and C.A.M.J.J. van den Hondel for their input on the history of S. elongatus PCC 7942. The authors were supported by grants from the National Institutes of Health (R35GM118290 to SSG; training grant T32GM007240 to BER and SD); the US Department of Energy Office of Science Early Career Research Program, Office of Basic Energy Sciences (DE-SC0006394 to DFS); National Science Foundation Graduate Research Fellowship Program (RDH), and a University of California, San Diego CRES postdoc award (DGW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Diamond S, et al. (2015) The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc. Natl. Acad. Sci. U. S. A 112, E1916–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diamond S, et al. (2017) Redox crisis underlies conditional light-dark lethality in cyanobacterial mutants that lack the circadian regulator, RpaA. Proc. Natl. Acad. Sci. U. S. A. 114, E580–E589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hosokawa N, et al. (2011) Circadian transcriptional regulation by the posttranslational oscillator without de novo clock gene expression in Synechococcus. Proc. Natl. Acad. Sci. U. S. A 108, 15396–15401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito H, et al. (2009) Cyanobacterial daily life with Kai-based circadian and diurnal genomewide transcriptional control in Synechococcus elongatus. Proc. Natl. Acad. Sci. U. S. A. 106, 14168–14173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries J and Archibald JM (2017) Endosymbiosis: Did plastids evolve from a freshwater cyanobacterium? Curr. Biol. 27, R103–R105 [DOI] [PubMed] [Google Scholar]
- 6.Flombaum P, et al. (2013) Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U. S. A 110, 9824–9829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliver NJ, et al. (2016) Cyanobacterial metabolic engineering for biofuel and chemical production. Curr. Opin. Chem. Biol 35, 43–50 [DOI] [PubMed] [Google Scholar]
- 8.Latifi A, et al. (2009) Oxidative stress in cyanobacteria. FEMS Microbiol. Rev 33, 258–278 [DOI] [PubMed] [Google Scholar]
- 9.Vijayan V, et al. (2009) Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proceedings of the National Academy of Sciences 106, 22564–22568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsby AE (2007) Cyanobacterial heterocysts: terminal pores proposed as sites of gas exchange. Trends Microbiol 15, 340–349 [DOI] [PubMed] [Google Scholar]
- 11.Knoop H, et al. (2013) Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6803. PLoS Comput. Biol 9, e1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.You L, et al. (2015) Photoheterotrophic fluxome in Synechocystis sp. strain PCC 6803 and its implications for cyanobacterial bioenergetics. J. Bacteriol 197, 943–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young JD, et al. (2011) Mapping photoautotrophic metabolism with isotopically nonstationary (13)C flux analysis. Metab. Eng. 13, 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamoi M, et al. (2005) The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J 42, 504–513 [DOI] [PubMed] [Google Scholar]
- 15.Wedel N and Soll J (1998) Evolutionary conserved light regulation of Calvin cycle activity by NADPH-mediated reversible phosphoribulokinase/CP12/glyceraldehyde-3-phosphate dehydrogenase complex dissociation. Proc. Natl. Acad. Sci. U. S. A 95, 9699–9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori T, et al. (1996) Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc. Natl. Acad. Sci. U. S. A 93, 10183–10188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singer RA and Doolittle WF (1975) Control of gene expression in blue-green algae. Nature 253, 650–651 [DOI] [PubMed] [Google Scholar]
- 18.Gaudana SB, et al. (2013) Diurnal rhythm of a unicellular diazotrophic cyanobacterium under mixotrophic conditions and elevated carbon dioxide. Photosynth. Res 118, 51–57 [DOI] [PubMed] [Google Scholar]
- 19.Hanai M, et al. (2014) The effects of dark incubation on cellular metabolism of the wild type cyanobacterium Synechocystis sp. PCC 6803 and a mutant lacking the transcriptional regulator cyAbrB2. Life 4, 770–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki E, et al. (2007) Role of the GlgX protein in glycogen metabolism of the cyanobacterium, Synechococcus elongatus PCC 7942. Biochim. Biophys. Acta 1770, 763–773 [DOI] [PubMed] [Google Scholar]
- 21.Wyman M and Thom C (2012) Temporal orchestration of glycogen synthase (glgA) gene expression and glycogen accumulation in the oceanic picoplanktonic cyanobacterium Synechococcus sp. strain WH8103. Appl. Environ. Microbiol. 78, 4744–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, et al. (2014) Isobutanol production as an alternative metabolic sink to rescue the growth deficiency of the glycogen mutant of Synechococcus elongatus PCC 7942. Photosynth. Res. 120, 301–310 [DOI] [PubMed] [Google Scholar]
- 23.Miao X, et al. (2003) Changes in photosynthesis and pigmentation in an agp deletion mutant of the cyanobacterium Synechocystis sp. Biotechnol. Lett. 25, 391–396 [DOI] [PubMed] [Google Scholar]
- 24.Work VH, et al. (2015) Lauric acid production in a glycogen-less strain of Synechococcus sp. PCC 7002. Front Bioeng Biotechnol 3, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grundel M, et al. (2012) Impaired glycogen synthesis causes metabolic overflow reactions and affects stress responses in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 158, 3032–3043 [DOI] [PubMed] [Google Scholar]
- 26.Osanai T, et al. (2007) Sugar catabolism regulated by light- and nitrogen-status in the cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. Sci. 6, 508–514 [DOI] [PubMed] [Google Scholar]
- 27.Welkie DG, et al. (2018) Genome-wide fitness assessment during diurnal growth reveals an expanded role of the cyanobacterial circadian clock protein KaiA. Proc Natl Acad Sci U S A DOI: 10.1073/pnas.1802940115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klotz A, et al. (2015) Nitrogen starvation acclimation in Synechococcus elongatus: Redoxcontrol and the role of nitrate reduction as an electron sink. Life 5, 888–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh AK, et al. (2008) Integration of carbon and nitrogen metabolism with energy production is crucial to light acclimation in the cyanobacterium Synechocystis. Plant Physiol 148, 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hickman JW, et al. (2013) Glycogen synthesis is a required component of the nitrogen stress response in Synechococcus elongatus PCC 7942. Algal Research 2, 98–106 [Google Scholar]
- 31.Jackson SA, et al. (2015) Dynamics of photosynthesis in a glycogen-deficient glgC mutant of Synechococcus sp. strain PCC 7002. Appl. Environ. Microbiol. 81, 6210–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen MM and Smith AJ (1969) Nitrogen chlorosis in blue-green algae. Arch. Mikrobiol. 69, 114–120 [DOI] [PubMed] [Google Scholar]
- 33.Davies FK, et al. (2014) Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front Bioeng Biotechnol 2, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohbayashi R, et al. (2013) DNA replication depends on photosynthetic electron transport in cyanobacteria. FEMS Microbiol Lett 344, 138–144 [DOI] [PubMed] [Google Scholar]
- 35.Watanabe S, et al. (2012) Light-dependent and asynchronous replication of cyanobacterial multi-copy chromosomes. Mol. Microbiol. 83, 856–865 [DOI] [PubMed] [Google Scholar]
- 36.Knowles VL and Plaxton WC (2003) From genome to enzyme: analysis of key glycolytic and oxidative pentose-phosphate pathway enzymes in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 44, 758–763 [DOI] [PubMed] [Google Scholar]
- 37.Yang C, et al. (2002) Integration of the information from gene expression and metabolic fluxes for the analysis of the regulatory mechanisms in Synechocystis. Appl. Microbiol. Biotechnol 58, 813–822 [DOI] [PubMed] [Google Scholar]
- 38.Flores E and Herrero A (2005) Nitrogen assimilation and nitrogen control in cyanobacteria. Biochem. Soc. Trans 33, 164–167 [DOI] [PubMed] [Google Scholar]
- 39.Ohashi Y, et al. (2011) Regulation of nitrate assimilation in cyanobacteria. J. Exp. Bot 62, 1411–1424 [DOI] [PubMed] [Google Scholar]
- 40.Rubin BE, et al. (2018) High-throughput interaction screens illuminate the role of c-di-AMP in cyanobacterial nighttime survival. PLoS Genet 14, e1007301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broedel SE Jr. and Wolf RE Jr. (1990) Genetic tagging, cloning, and DNA sequence of the Synechococcus sp. strain PCC 7942 gene (gnd) encoding 6-phosphogluconate dehydrogenase. J. Bacteriol 172, 4023–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doolittle WF and Singer RA (1974) Mutational analysis of dark endogenous metabolism in the blue-green bacterium Anacystis nidulans. J. Bacteriol 119, 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scanlan DJ, et al. (1995) Characterization of a zwf mutant of Synechococcus sp. strain PCC 7942. J. Bacteriol. 177, 2550–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.You L, et al. (2014) 13C-MFA delineates the photomixotrophic metabolism of Synechocystis sp. PCC 6803 under light- and carbon-sufficient conditions. Biotechnol J 9, 684–692 [DOI] [PubMed] [Google Scholar]
- 45.Muro-Pastor MI, et al. (2005) Ammonium assimilation in cyanobacteria. Photosynth Res 83, 135–150 [DOI] [PubMed] [Google Scholar]
- 46.Marques S, et al. (1992) Light-mediated regulation of glutamine synthetase activity in the unicellular cyanobacterium Synechococcus sp. PCC 6301. Planta 187, 247–253 [DOI] [PubMed] [Google Scholar]
- 47.Doolittle WF (1979) The cyanobacteril genome, its expression, and the control of that expression. Adv. Microb. Physiol. 20, 1–102 [DOI] [PubMed] [Google Scholar]
- 48.Takano S, et al. (2015) The initiation of nocturnal dormancy in Synechococcus as an active process. BMC Biol 13, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hood RD, et al. (2016) The stringent response regulates adaptation to darkness in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. U. S. A 113, E4867–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woelfle MA, et al. (2004) The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr. Biol. 14, 1481–1486 [DOI] [PubMed] [Google Scholar]
- 51.Piechura JR, et al. (2017) Natural changes in light interact with circadian regulation at promoters to control gene expression in cyanobacteria. Elife 6, DOI: 10.7554/eLife.32032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ansong C, et al. (2014) Characterization of protein redox dynamics induced during light-to-dark transitions and nutrient limitation in cyanobacteria. Front. Microbiol. 5, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welkie D, et al. (2014) Transcriptomic and proteomic dynamics in the metabolism of a diazotrophic cyanobacterium, Cyanothece sp. PCC 7822 during a diurnal light-dark cycle. BMC Genomics 15, 1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markson JS, et al. (2013) Circadian control ^of global gene expression by the cyanobacterial master regulator RpaA. Cell 155, 1396–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shultzaberger RK, et al. (2015) Giving time purpose: The Synechococcus elongatus clock in a broader network context. Annu. Rev. Genet 49, 485–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen SE and Golden SS (2015) Circadian Rhythms in Cyanobacteria. Microbiol Mol Biol Rev 79, 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gutu A and O'Shea EK (2013) Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol Cell 50, 288–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paddock ML, et al. (2013) Active output state of the Synechococcus Kai circadian oscillator. Proc. Natl. Acad. Sci. U. S. A. 110, E3849–3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ouyang Y, et al. (1998) Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A 95, 8660–8664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seino Y, et al. (2009) The response regulator RpaB binds to the upstream element of photosystem I genes to work for positive regulation under low-light conditions in Synechocystis sp. Strain PCC 6803. J. Bacteriol 191, 1581–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilde A and Hihara Y (2016) Transcriptional and posttranscriptional regulation of cyanobacterial photosynthesis. Biochim. Biophys. Acta 1857, 296–308 [DOI] [PubMed] [Google Scholar]
- 62.Espinosa J, et al. (2015) Cross-talk and regulatory interactions between the essential response regulator RpaB and cyanobacterial circadian clock output. Proc. Natl. Acad. Sci. U. S. A 112, 2198–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murata K, et al. (2016) Ultrastructure of compacted DNA in cyanobacteria by high-voltage cryo-electron tomography. Sci. Rep. 6, 34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith RM and Williams SB (2006) Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. U. S. A 103, 8564–8569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woelfle MA, et al. (2007) Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 104, 18819–18824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cohen SE, et al. (2014) Dynamic localization of the cyanobacterial circadian clock proteins. Curr. Biol 24, 1836–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dong G, et al. (2010) Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell 140, 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ihlenfeldt MJA and Gibson J (1975) CO2 fixation and its regulation in Anacystis nidulans (Synechococcus). Arch. Microbiol 102, 13–21 [DOI] [PubMed] [Google Scholar]
- 69.Rust MJ, et al. (2011) Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science 331, 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo J, et al. (2014) Proteome-wide light/dark modulation of thiol oxidation in cyanobacteria revealed by quantitative site-specific redox proteomics. Mol. Cell. Proteomics 13, 3270–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saha R, et al. (2016) Diurnal Regulation of Cellular Processes in the Cyanobacterium Synechocystis sp. Strain PCC 6803: Insights from Transcriptomic, Fluxomic, and Physiological Analyses. MBio 7, DOI: 10.1128/mBio.00464-00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cameron JC and Pakrasi HB (2010) Essential role of glutathione in acclimation to environmental and redox perturbations in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 154, 1672–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lindahl M and Kieselbach T (2009) Disulphide proteomes and interactions with thioredoxin on the track towards understanding redox regulation in chloroplasts and cyanobacteria. J. Proteomics 72, 416–438 [DOI] [PubMed] [Google Scholar]
- 74.Diaz-Troya S, et al. (2014) Redox regulation of glycogen biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803: analysis of the AGP and glycogen synthases. Mol Plant 7, 87–100 [DOI] [PubMed] [Google Scholar]
- 75.Schürmann P (2003) Redox signaling in the chloroplast: the ferredoxin/thioredoxin system. Antioxid. Redox Signal 5, 69–78 [DOI] [PubMed] [Google Scholar]
- 76.Singh AK, et al. (2004) Microarray analysis and redox control of gene expression in the cyanobacterium Synechocystis sp. PCC 6803. Physiol. Plant 120, 27–35 [DOI] [PubMed] [Google Scholar]
- 77.Chardonnet S, et al. (2015) First proteomic study of S-glutathionylation in cyanobacteria. J. Proteome Res 14, 59–71 [DOI] [PubMed] [Google Scholar]
- 78.Li M, et al. (2007) Identification of novel targets of cyanobacterial glutaredoxin. Arch. Biochem. Biophys 458, 220–228 [DOI] [PubMed] [Google Scholar]
- 79.Dietz K-J (2011) Peroxiredoxins in plants and cyanobacteria. Antioxid. Redox Signal 15, 1129–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lindahl M and Florencio FJ (2003) Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc. Natl. Acad. Sci. U. S. A 100, 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zaffagnini M, et al. (2012) Redox regulation in photosynthetic organisms: focus on glutathionylation. Antioxid. Redox Signal 16, 567–586 [DOI] [PubMed] [Google Scholar]
- 82.Agostoni M and Montgomery B (2014) Survival strategies in the aquatic and terrestrial world: The impact of second messengers on cyanobacterial processes. Life 4, 745–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Puszynska AM and O'Shea EK (2017) ppGpp Controls Global Gene Expression in Light and in Darkness in S. elongatus. Cell Rep 21, 3155–3165 [DOI] [PubMed] [Google Scholar]
- 84.Agostoni M, et al. (2018) Homeostasis of second messenger cyclic-di-AMP Is critical for cyanobacterial fitness and acclimation to abiotic stress. Front Microbiol 9, 1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Corrigan RM, et al. (2015) Cross-talk between two nucleotide-signaling pathways in Staphylococcus aureus. J. Biol. Chem 290, 5826–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Whiteley AT, et al. (2015) The PAMP c-di-AMP Is Essential for Listeria monocytogenes Growth in Rich but Not Minimal Media due to a Toxic Increase in (p)ppGpp. Cell Host Microbe 17, 788–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seluzicki A, et al. (2017) Dancing in the dark: darkness as a signal in plants. Plant Cell Environ 40, 2487–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang J, et al. (2016) Proteomic insight into the response of Arabidopsis chloroplasts to darkness. PLoS One 11, e0154235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Graf A and Smith AM (2011) Starch and the clock: the dark side of plant productivity. Trends Plant Sci 16, 169–175 [DOI] [PubMed] [Google Scholar]
- 90.Liu JD, et al. (2015) Keeping the rhythm: light/dark cycles during postharvest storage preserve the tissue integrity and nutritional content of leafy plants. BMC Plant Biol 15, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Levin E, et al. (2017) Hawkmoths use nectar sugar to reduce oxidative damage from flight. Science 355, 733–734 [DOI] [PubMed] [Google Scholar]
- 92.Del Rio CM and Dillon ME (2017) Sweet relief for pollinators. Science 355, 686–687 [DOI] [PubMed] [Google Scholar]
- 93.Suttle CA (2007) Marine viruses - major players in the global ecosystem. Nat. Rev. Microbiol 5, 801–812 [DOI] [PubMed] [Google Scholar]
- 94.Jover LF, et al. (2014) The elemental composition of virus particles: implications for marine biogeochemical cycles. Nat. Rev. Microbiol. 12, 519–528 [DOI] [PubMed] [Google Scholar]
- 95.Adolph KW and Haselkorn R (1972) Photosynthesis and the development of blue-green algal virus N-1. Virology 47, 370–374 [DOI] [PubMed] [Google Scholar]
- 96.Sherman LA (1976) Infection of Synechococcus cedrorum by the cyanophage AS-1M. III. Cellular metabolism and phage development. Virology 71, 199–206 [DOI] [PubMed] [Google Scholar]
- 97.Jia Y, et al. (2010) Light-dependent adsorption of photosynthetic cyanophages to Synechococcus sp. WH7803. FEMS Microbiol. Lett 310, 120–126 [DOI] [PubMed] [Google Scholar]
- 98.Thompson LR, et al. (2011) Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. U. S. A 108, E757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thompson LR, et al. (2016) Gene expression patterns during light and dark infection of Prochlorococcus by cyanophage. PLoS One 11, e0165375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grigorieva GA and Shestakov SV (1976) Application of the genetic transformation method for taxonomic analysis of unicellular blue-green algae. In Proc 2nd Int Symp Photosynthetic Prokaryotes, pp. 220–221 [Google Scholar]
- 101.van den Hondel CA, et al. (1980) Introduction of transposon Tn901 into a plasmid of Anacystis nidulans: preparation for cloning in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A 77, 1570–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Herdman (2001) Form-genus XIII. Synechococcus. Bergey's Manual of Systematic Bacteriology 1, 508–512 [Google Scholar]
- 103.Michel KP, et al. (1998) Immunocytochemical localization of IdiA, a protein expressed under iron or manganese limitation in the mesophilic cyanobacterium Synechococcus PCC 6301 and the thermophilic cyanobacterium Synechococcus elongatus. Planta 205, 73–81 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.