Abstract
SAMHD1 is a host triphosphohydrolase that degrades intracellular deoxynucleoside triphosphates (dNTPs) to a lower level that restricts viral DNA synthesis, and thus prevents replication of diverse viruses in non-dividing cells. Recent progress indicates that SAMHD1 negatively regulates antiviral innate immune responses and inflammation through interacting with various key proteins in immune signaling and DNA damage repair pathways. SAMHD1 can also modulate antibody production in adaptive immune responses. In this review, we summarize how SAMHD1 regulates antiviral immune responses through distinct mechanisms and discuss the implications of these new functions of SAMHD1. Furthermore, we propose important new questions and future directions that can advance functional and mechanistic studies of SAMHD1-mediated immune regulation during viral infections.
Keywords: SAMHD1, viruses, infection, replication, antiviral immunity
Interplay Between SAMHD1 and Antiviral Immune Responses
Human sterile alpha motif and HD-domain-containing protein 1 (SAMHD1) (see Glossary) was first identified as an ortholog of mouse Mg11 induced by interferon-gamma (IFN-γ)[1],suggesting a link between SAMHD1 and innate immunity. Homozygous mutations in SAMHD1 lead to a genetic encephalopathy named Aicardi-Goutières syndrome (AGS) that mimics congenital viral infection [2]. AGS patients have a rare autosomal recessive neurological condition with high levels of interferon-stimulated gene (ISG) products in peripheral blood [3, 4]. Similarly, SAMHD1-knockout (KO) mice showed spontaneous IFN-I induction, increased intracellular dNTP concentration in lymphocytes, macrophages and dendritic cells (DCs) [5–7]. These data suggest that SAMHD1 modulates the IFN pathway in mammalian cells, and the underlying mechanisms include SAMHD1 inhibition of retrotransposons in SAMHD1-deficient humans or mice [8, 9]. SAMHD1 itself is an ISG and can be induced by interleukin (IL)-12 and IL-18 in human monocyte-derived macrophages (MDM) [10]. Type IFN (IFN-I) treatment increases SAMHD1 expression level in several cell lines, but not in primary CD4+ T-lymphocytes [11]. SAMHD1 is up-regulated by IFN-α in U87-MG cells, but not in MDM or human monocytic THP-1 cells [12]. Moreover, IFN treatment can activate SAMHD1 expression in various cell lines by regulation of micro-RNAs [13, 14]. Thus, IFNs up-regulate SAMHD1 expression in a cell type-dependent manner and the underlying mechanisms may link SAMHD1-mediated antiviral activity.
SAMHD1 is a restriction factor blocking human immunodeficiency virus type 1 (HIV-1) infection in non-dividing cells [15, 16]. Functioning as a dNTP triphosphohydrolase (dNTPase), SAMHD1 hydrolyzes intracellular dNTPs, thereby reducing reverse transcription and HIV-1 cDNA synthesis [17, 18]. Some of SAMHD1’s functions in innate immune responses can be a result of the lowered intracellular dNTP pool. As a multifunctional protein, SAMHD1 plays important roles in regulating cell-cycle progression, cancer development, adaptive immunity, and other viral infections [19]. New functions and mechanisms of SAMHD1 in antiviral immune responses have been recently unveiled [20]. SAMHD1 prevents innate immune response induced by DNA damage during DNA replication [21, 22]. In addition, SAMHD1 promotes hyper-mutation of the immunoglobulin gene in activated B cells and affects antibody production [23]. Since significant progress has been made regarding new antiviral and immune functions of SAMHD1, it is necessary to analyze these new findings and point out the unsolved problems to further advance this research area. To better focus on the role of SAMHD1 in the antiviral immune response, we first provide an overview of the innate and adaptive immune system against virus infection. Then, we summarize the molecular mechanisms of SAMHD1-mediated antiviral function and immune responses regulation. Lastly, we discuss important questions that remain to be addressed in the future.
Overview of Antiviral Immune Responses
Innate antiviral immune responses
The innate immune system is the first cellular line counteracting viral invasion. A variety of molecules in the cell recognize different species of viral nucleic acids, depending on their cellular location. Viral dsRNA in infected cells is recognized by oll-like receptor 3 (TLR3) in the endosome, which triggers TIR-domain-containing adapter-inducing interferon-β (TRIF)-dependent signaling, activating interferon regulatory factor 3 (IRF3) and nuclear factor-κB (NF-κB) [24, 25]. TLR7/8 in the endosome detects viral single-stranded RNA (ssRNA) and induces myeloid differentiation primary response 88 (MyD88)-dependent NF-κB activation [26]. Cytoplasmic viral RNA activates retinoic acid-inducible gene 1 (RIG-I) or melanoma differentiation-associated protein 5 (MDA5), which interact with mitochondrial antiviral-signaling protein (MAVS) [27–30] and activate IRF3/7 and NF-κB [31]. The phosphorylated IRF3/7, through TANK-binding kinase 1 (TBK1) or inhibitor-κB kinase ε (IKKε), forms homo- or hetero-dimers and translocate into the nucleus to induce expression of IFN-I [31]. NF-κB also coordinately modulates antiviral innate immune responses by inducing phosphorylation and degradation of the NF-κB inhibitor IκBα, leading to activation and translocation of NF-κB to the nucleus to bind to the IFN-β promoter. Overall, the activation of IFN-I and NF-κB signaling drives the expression of IFN-I, ISGs, pro-inflammatory cytokines, and/or chemokines, which can limit viral infection [32] (Figure 1).
Figure 1. SAMHD1 down-regulates the IFN-I pathway.
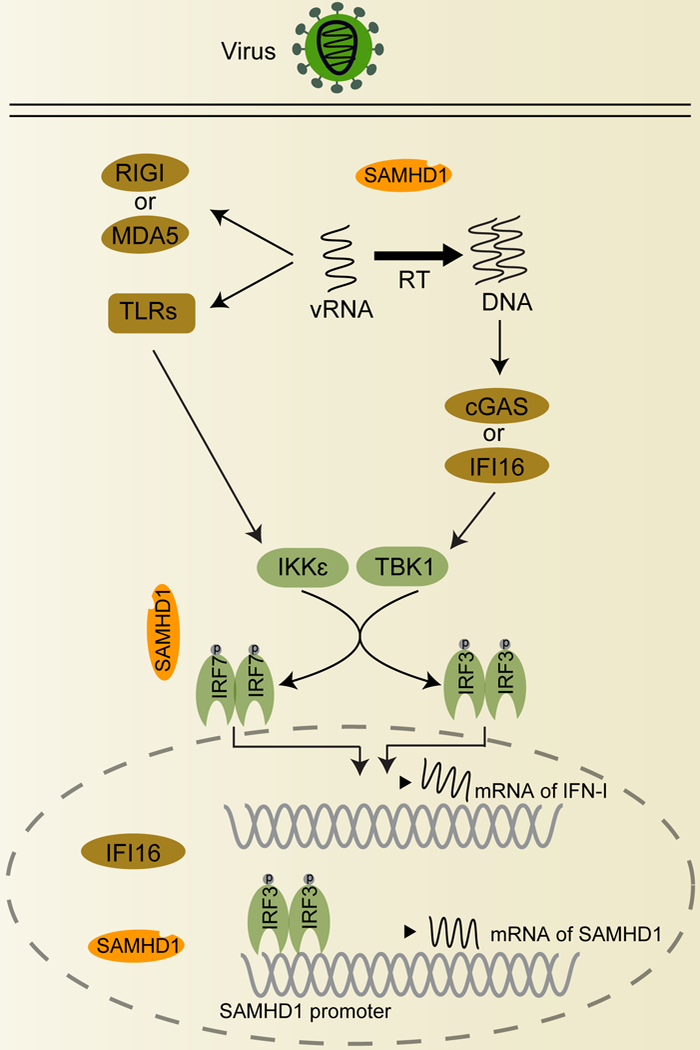
In the IFN-I innate immune pathway, viral RNA released in infected cells is recognized by RIG-I and MDA5 in the cytosol or TLRs in endosome, leading to activation and phosphorylation of both IRF3/7 by TBK1 or IKKε.IRF3/7 forms homo- or hetero-dimers and translocate into the nucleus to induce IFN-I expression. The DNA sensor cGAS in cytoplasm recognizes dsDNA and activates IFN- signaling. IFI16 is expressed in both cytoplasm and nucleus and activates innate immune responses. For retroviruses, SAMHD1 tetramers inhibit viral reverse transcription (RT) by hydrolyzing the intracellular dNTPs, blocking virus replication and preventing subsequent viral DNA-induced innate immune responses. For RNA viruses, SAMHD1 down-regulates RNA sensor-triggered innate immune response by interacting with IRF7 and decreasing IKKε- mediated phosphorylation of IRF7. IRF3 binds to the promoter of SAMHD1 and enhances SAMHD1 mRNA transcription and protein expression. The decreased provirus by SAMHD1 may induce less intensive innate immune response. The circled letter P of IRF3 and IRF7 indicates their phosphorylation. cGAS, cyclic guanosine monophosphate (GMP)-adenosine monophosphate (AMP) synthase; IFI16, gamma-interferon-inducible protein-16; IKKε, inhibitor-κB kinase ε; MDA5, melanoma differentiation-associated protein 5; RIG-I, retinoic acid-inducible gene 1; TBK1, TANK-binding kinase 1; TLRs, Toll-like receptors; vRNA, viral RNA.
Viral DNA is recognized by intracellular DNA sensors, including gamma-interferon-inducible protein-16 (IFI16) [33] and cyclic GMP-AMP (cGAMP) synthase (cGAS) [34, 35]. Upon binding to DNA, cGAS synthesizes cGAMP, which binds to and activates stimulator of IFN genes (STING), leading to IKKε/TBK1-mediated phosphorylation of IRF3/7 (Figure 1). This pathway also induces the activation of NF-κB [36]. IFI16, which is localized in the cytoplasm and the nucleus, signals through STING and promotes productionofIL-1β through the inflammasome and caspase 1 [37]. Innate immune responses triggered by viral RNA or DNA initiate earlier antiviral effects, but clearance of viral infections often relies on adaptive immune responses.
Adaptive antiviral immune responses
The adaptive immune system contains T and B cells and can be triggered by innate immunity to protect against viral infection through antigen-presenting cells (APCs), including macrophages and DCs [38]. The mature, functional T cells include CD4+ and CD8+ T cells [39]. CD8+ T cells have potential to differentiate into cytotoxic T-lymphocytes, which inhibit viral replication and directly kill virus-infected cells by secreting cytokines and effector molecules. CD8+ T cells detect viral antigens presented by major histocompatibility complex (MHC-I). CD4+ T cells recognize antigens processed by lysosomal degradation and presented through MHC-II. Naïve CD4+ T cells can differentiate into effector, regulatory, or follicular helper cells [39]. B cells express receptors that recognize foreign antigens after somatic hypermutation and class switch recombination. These selected and highly specific B cells produce antibodies when B cells differentiate into plasma cells, thus offering humoral immunity to neutralize invading pathogens [40, 41]. Antibody-dependent cell-mediated cytotoxicity of natural killer cells also plays an important role in adaptive antiviral immunity [42, 43]. The adaptive immune system provides immunity during host exposure to pathogens and the process is coordinately regulated by cellular factors and environment-derived signals [44].
SAMHD1-mediated Suppression of Antiviral Innate Immune Responses
SAMHD1 inhibits IFN-I response induced via the cGAS-STING pathway
SAMHD1 restricts HIV-1 infection in non-dividing cells by blocking reverse-transcription [17, 18]. It is known that prematurely terminated reverse transcription products trigger cGAS activation [45, 46]. The low levels of viral cDNAs may inefficiently activate DNA sensing pathways in innate immune responses. This might be a mechanism by which HIV-1 avoids immune surveillance in macrophages and DCs during early stages of infection [47]. The cGAS-STING-IRF3 axis is triggered by HIV-1 and other retroviruses in human cells when SAMHD1 is down-regulated [46]. Both cGAS and IFI16 can detect HIV-1 ssDNA that induces an IFN-β response through the STING-TBK1-IRF3/7 pathway in macrophages [48].singbone-marrow-derived macrophages from SAMHD1-cGAS or SAMHD1-STING double-KO mice, SAMHD1 has been shown to inhibit spontaneous innate responses in a cGAS/STING-dependent manner [49]. By blocking viral cDNA synthesis during lentivirus infection, SAMHD1 also down-regulates IFN induction in myeloid cells [49]. These results suggest that SAMHD1 impairs cGAS triggered innate responses by reducing the cDNA synthesis of retrovirus reverse transcription in the early process of infection (Figure 1).
Potential role of SAMHD1 in the RIG-I/MDA5 or TLR pathways
As a DNA/RNA binding protein without nuclease activity [50, 51], SAMHD1 has a preference for ssRNA over ssDNA [52], and associates with cellular nucleic acids in situ, which is dependent on the HD and C-terminal domains of SAMHD1 [53]. Although SAMHD1 mainly localizes in the nucleus, some AGS-associated mutants with high ssRNA binding activity express in the cytosol, and may affect RIG-I/MDA5 or TLR-mediated sensing of viral RNA. SAMHD1 was reported to have a ribonuclease activity to target retroviral genomic RNA in a phosphorylation-dependent manner [54–56], but its RNase-dependent antiretroviral activity has not been recapitulated [57, 58]. The HD domain of SAMHD1 has been reported to have a 3′−5′ exonuclease activity in vitro [59]; however, the observed nuclease activity of SAMHD1 is likely due to nuclease contamination during the purification of recombinant SAMHD1 [50, 51, 57]. SAMHD1 may act as an ssRNA-binding protein to negatively regulate viral RNA-induced innate immune responses through RIG-I/MDA5 or TLR pathways, but this activity remains to be confirmed (Figure 1).
SAMHD1 interacts with IKKε and IRF7 in the IFN-I pathway
To further understand the mechanisms by which SAMHD1 regulates IFN-I signaling, it is critical to investigate SAMHD1 interaction with key proteins in the IFN-I pathway. IRF3 binds to the SAMHD1 promoter and induces its expression in cells [60], suggesting that SAMHD1 per se is an ISG (Figure 1). Because AGS patients with SAMHD1 mutations present high levels of IFN-I [2] and IRF7 is essential for the induction of IFN-I [61], it is plausible that SAMHD1 interacts with IRF7 to regulate the IFN-I pathway. Indeed, knockdown (KD) of IRF7 in SAMHD1 KO THP-1 cells significantly impairs IFN-I expression, and KD of SAMHD1 in MDM increases IFN-α secretion [20]. SAMHD1 interacts with IRF7 and IKKε in MDM and prevents IKKε-mediated IRF7 phosphorylation, thereby negatively regulating the IFN-I pathway [20]. Furthermore, SAMHD1 does not interact with overexpressed IRF3 or TBK1 in cells, suggesting selective interactions of SAMHD1 with key regulators in IFN-I signaling [20]. Notably, KKα also can phosphorylate IRF7 [62], and whether SAMHD1 interacts with IKKα to suppress IRF7 phosphorylation remains to be investigated.
SAMHD1 inhibits activation of the NF-κB pathway
NF-κB activation by viral infections can lead to inflammatory responses and modulate innate immunity [63]. Recent studies demonstrated that SAMHD1 plays a role in regulating the NF-κB pathway [20]. SAMHD1 expression can be induced by tumor necrosis factor alpha (TNF-α) treatment, which activates NF-κB [64] (Figure 2), and significantly higher levels of serum TNF-α were detected in SAMHD1-defective patients [65]. A case report indicated that intravenous infusion of IL-6 antibodies relieved disease progression in a SAMHD1-defective patient [66], highlighting an important function of SAMHD1 in down-regulating diseases associated with abnormal pro-inflammatory response. Consistently, loss of SAMHD1 in THP-1 cells significantly up-regulates NF-κB signaling and the inflammatory response. SAMHD1 interacts with endogenous p50, p100/p52 and IκBα in THP-1 cells and MDM [20]. Treatment with pro-inflammatory cytokines, lipopolysaccharide (LPS), or Sendai virus (SeV) infection decreases the levels of endogenous IκBα, thereby reducing IκBα interaction with SAMHD1. The LPS derivative, monophosphoryl lipid , induces more TNF-α positive macrophages from SAMHD1 KO mice than those from heterozygous mice. LPS treatment of THP-1 cells also increases SAMHD1 expression levels, suggesting an innate response of SAMHD1 to inflammation through NF-κB activation [20]. Thus, SAMHD1 acts as an immune modulator that inhibits the NF-κB pathway and inflammatory response to viral infection or inflammatory stimuli (Figure 2) [20]. However, the detailed mechanisms remain unclear. Future structural analyses of SAMHD1/NF-kB complexes can help clarify the functional significance of these interactions.
Figure 2. SAMHD1 prevents the activation of NF-κB signaling.
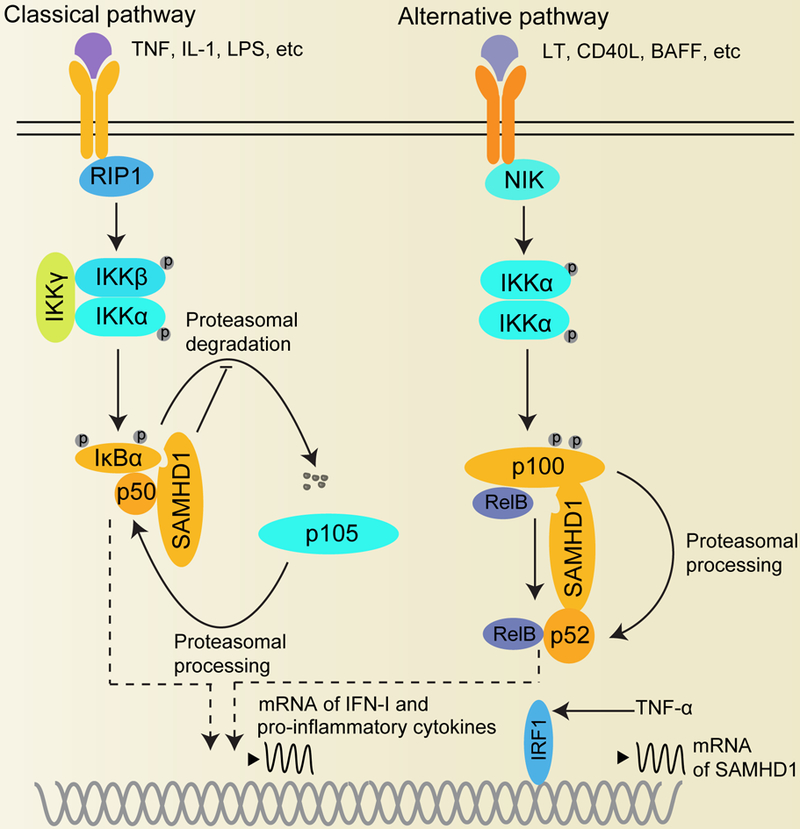
The NF-κB signaling contains the classical (canonical) and alternative (noncanonical) pathways. The classical pathway is induced by factors such as TNF, IL-1, or LPS, and is activated by the phosphorylation of IKKα/IKKβ heterodimers of the IKK complex and targets p50/p65 dimers. The alternative pathway is activated by factors including LT, CD40L, or BAFF, and requires NIK induced phosphorylation of IKKα homodimers, leading to proteasomal cleavage of phosphorylated p100 and causes activation of RelB/p52 heterodimers. In the classical pathway, SAMHD1 interacts with p50 and IκBα to inhibit IκBα phosphorylation and degradation, thus negatively regulates NF-κB activation in human macrophages and cell lines. In the alternative pathway, SAMHD1 binds to p100/p52 and prevents NF-κB signaling in cells. TNF-α promotes SAMHD1 protein expression through activation of the transcription factor IRF1. The circled letter P of indicated proteins indicates their phosphorylation. BAFF, B-cell activating factor; CD40L, cluster of differentiation 154; IκBα, nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor alpha; LPS, Lipopolysaccharide; LT, Lymphotoxin; NIK, NF-κB inducing kinase; RIP1, Receptor-interacting serine/threonine-protein kinase 1; TNF, tumor necrosis factor superfamily.
SAMHD1 in cell cycle regulation and the PI3K-AKT-IRF3 pathway
The expression and activity of SAMHD1 can be affected by cell cycle status [67, 68]. SAMHD1 interacts with CDK1/2 [69–73] and impairs the regulation of the cell cycle and apoptosis in THP-1 cells [74]. Because cell cycle checkpoints are critical for controlling proliferation and activation of immune cells, SAMHD1 deficiency may lead to aberrant cell division and cause immune dysfunction [19]. The anti-retroviral function of SAMHD1 is negatively regulated by cell cycle-dependent phosphorylation at threonine 592 (T592) [69, 70, 75, 76]. IFNs treatment reduces T592 phosphorylation of SAMHD1, thus enhancing HIV-1 restriction in MDM [70, 76]. This suggests a crosstalk between IFN-I signaling and SAMHD1 T592 phosphorylation in a cell cycle-dependent manner (Figure 3).
Figure 3. SAMHD1 in cell cycle regulation and PI3K-AKT-IRF3 innate immune signaling.
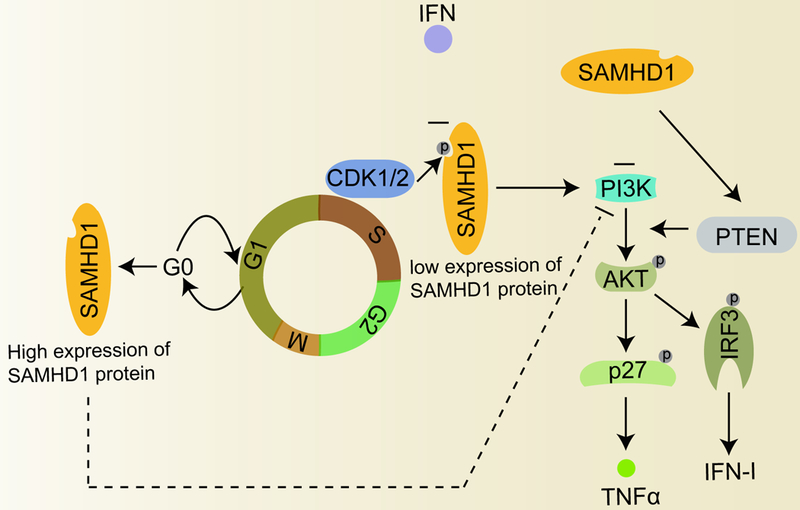
SAMHD1 protein expression is high in G0 phase and low in S-phase during the cell cycle. SAMHD1 also impairs apoptosis through PI3K-AKT-p27 axis and thus regulates p27 induced protein expression of TNF-α in NF-κB signaling. SAMHD1 interacts with CDK1/2, which are kinases involved in cell cycle checkpoints. CDK1 and CDK2 are important for regulation of phosphorylation and expression of SAMHD1 during cell cycle. IFNs can reduce SAMHD1 phosphorylation. SAMHD1 controls IFN-I signaling through PI3K-AKT-IRF3 axis. The circled letter P of indicated proteins indicates their phosphorylation. AKT, protein kinase B; CDK1, cyclin dependent kinase 1; CDK2, cyclin dependent kinase 2; PI3K, phosphoinositide 3-kinase.
SAMHD1 deficiency induces activation of IFN- I signaling through PI3K-AKT-IRF3 axis in SAMHD1 KO cells, which showed increased expression of genes involved in the PI3K/AKT pathway and activated STAT1 and AKT phosphorylation [77]. In addition, SAMHD1 and AKT1 double KO cells failed to produce IFN-I induced by endogenous RNA substrates, suggesting a direct link between SAMHD1 function and RNA-mediated activation of the PI3K-AKT-IRF3 axis (Figure 3) [77]. Consistent with this report, we identified that SAMHD1 KO in THP-1 cells increased PI3K activity. Mechanistically, SAMHD1 KO reduced expression of the tumor suppressor PTEN, increased p27 phosphorylation and caused p27 mis-localization to the cytoplasm, suggesting that SAMHD1 regulates the PI3K-AKT-p27 signaling axis [78]. Since PTEN potentiates IRF3-mediated induction of IFN-I during RNA virus infection [79], it is possible that SAMHD1 modulates PTEN-mediated IFN-I signaling. Significantly increased expression of TNF-α was found in SAMHD1 KO cell-derived tumors in xenografted immunodeficient mice [78], indicating that SAMHD1 regulates TNF-α-mediated inflammation response in vivo. These data also implicate that SAMHD1 regulation of TNF-α may also affect innate immunity.
SAMHD1 in DNA damage repair induced IFN-I pathway
Genome stability, often regulated by DNA methylation, maintains normal cell behavior in healthy individuals [80]. Because mutations of SAMHD1 and other nucleic acid-degrading enzymes cause AGS, it is unsurprising that fibroblasts from AGS patients display atypical genomic phenotypes, including global loss of DNA methylation and increased RNA:DNA hybrids in DNA hypomethylated regions [81]. These RNA:DNA hybrids may activate the cGAS-STING pathway or TLR9-mediated IFN response [81]. Multiple double-stranded DNA (dsDNA) break repair proteins such as ataxia-telangiectasia mutated (ATM) and meiotic recombination protein 11 (MRE11) can detect damaged DNAs and induce IFN-I responses [82, 83]. SAMHD1 can be recruited to sites of DNA damage, as evidenced by an increased co-localization of SAMHD1 with 53BP1 foci after camptothecin treatment [84]. DNA damage activates IFN-I in SAMHD1 mutated cells, rendering the cells more sensitive to genotoxic stress [85]. Furthermore, DNA damage induced by etoposide in human MDM does not trigger IFN-I, but mediates SAMHD1 inhibition of HIV-1 infection through the p53-p21-CDK1-MCM2 pathway [86]. Thus, the function of SAMHD1 in DNA damage response induced IFN-I expression is likely dependent on treatment with different DNA instability chemicals and different cell types.
Recently, a dNTPase-independent role of SAMHD1 in homologous recombination (HR)-mediated DNA double-strand break (DSB) repair was identified [22]. Residue K484 in SAMHD1 was responsible for interaction with c-terminal binding protein 1-interacting protein (CtIP) and recruitment of SAMHD1 to DSB sites to facilitate DNA end resection in HR (Figure 4 ) [22]. SAMHD1 degradation by Vpx of HIV-2 or simian immunodeficiency viruses (SIVs) or SAMHD1 deficiency by KD or KO renders cancer cells hypersensitive to DSB-inducing agents, consistent with previous results showing that Vpx sensitizes acute myelogenous leukemia cells to cytarabine chemotherapy [87, 88]. he byproducts of DNA repair induced by SAMHD1 dysfunction also contribute to the production of IFN-I and pro-inflammatory factors [21]. DNA damage and inflammation are connected by the cGAS-cGAMP-STING pathway [89], which affects innate antiviral immunity. However, it is unclear whether and how SAMHD1 regulates DNA damage-triggered innate immune response to various virus infections, which represents an important new area to explore.
Figure 4. SAMHD1 negatively regulates IFN-I response during DNA damage repair.
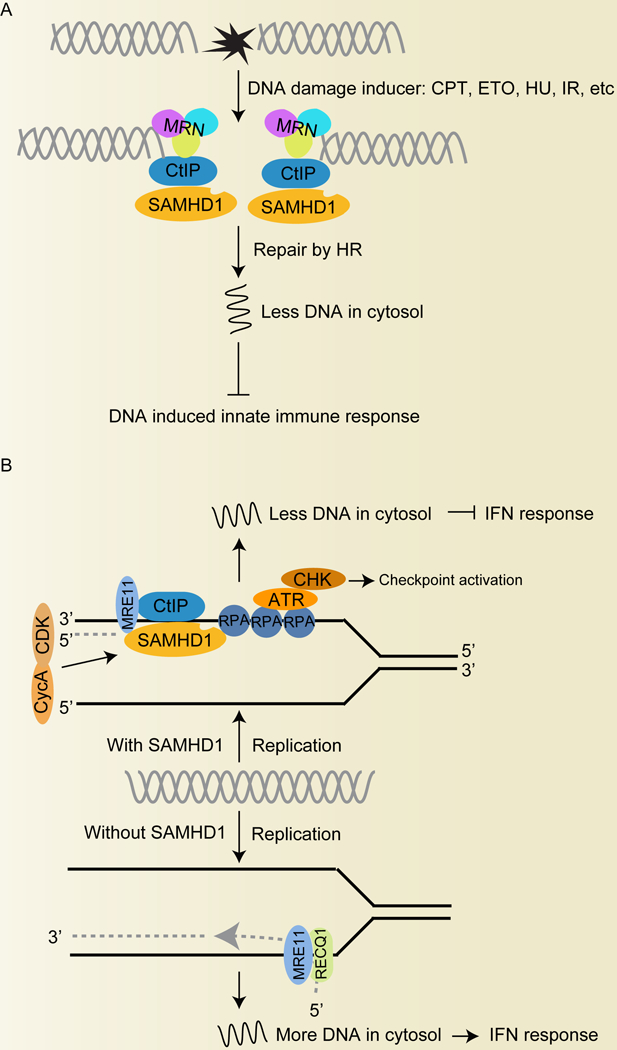
(A). SAMHD1 regulates DNA damage response induced by DNA instability chemicals such as CPT, ETO, HU or ionizing radiation (IR). SAMH 1 co-localized with MRN after DNA damage induced by IR or CPT in cells. SAMHD1 interacts and recruits CtIP to DSB sites to facilitate DNA end resection in HR process. SAMHD1 inhibits DNA damage-induced innate immune response. (B). SAMHD1 prevents the release of ssDNA from stalled replication forks and inhibits activation of the cGAS-STING-IRF3 pathway, leading to block of IFN-I production. SAMHD1 forms foci at replication forks and interacts with nascent DNA. SAMHD1 binds RPA and CtIP to promote fork progression and resection. SAMHD1 acts with MRE11 and promotes its exonuclease activity to accelerate resection at forks and DSBs, resulting in degradation of nascent DNA at stalled replication forks. SAMHD1 activates the ATR-CHK1 checkpoint for the reboot of CPT-arrested forks. Mutation or loss of SAMHD1 increases the accumulation of ssDNA and activate the DNA sensor-dependent IFN pathway. ATR, ataxia telangiectasia and Rad3-related protein; CDK, cyclin-dependent kinase; CHK, ATM serine/threonine kinase; CPT, Camptothecin; CtIP, C-terminal binding protein 1 interacting protein; CycA, Cyclin A; ETO, etoposide; HR, homologous recombination; HU, hydroxyurea; IR, ionizing radiation; MRE11, double-strand break repair protein; MRN, a protein complex consisting of MRE11, Rad50 and Nbs1; RECQ1, ATP-dependent DNA helicase Q1; RPA, replication protein A.
SAMHD1 in DNA replication-induced interferon pathway
Lower dNTP pool reduces DNA synthesis, leading to fork stalling and accumulation of ssDNA in cells, thus causing genomic instability and chromosomal rearrangements in tumorigenesis [90, 91]. Consistent with these observations, SAMHD1 mutations in colon cancers have been shown to increase gene mutation rates because of the abnormal DNA replication fidelity [92]. Treatment of MDM with the DSB inducer, neocarzinostatin, enhances anti-HIV-1 activity of unphosphorylated SAMHD1, activates the innate immune response and up-regulates ISG expression [93]. These data indicate that SAMHD1 is important for suppressing DNA replication-induced IFN-I expression. A recent study reported a phosphorylation-dependent and dNTPase-independent role of SAMHD1 in maintaining DNA replication stability and regulating IFN-Iinduction[21].SAMHD1KD in cells increases accumulation of cytosolic ssDNA, which triggers the cGAS-STING-IRF3 pathway and leads to higher mRNA levels of IFN-I and TNF-α. SAMHD1 forms foci at replication forks and interacts with nascent DNA and binds replication protein A and CtIP to promote fork progression and resection. In addition, SAMHD1 acts with MRE11 and promotes its exonuclease activity to accelerate degradation of nascent DNA [21]. Therefore, SAMHD1 mutation may increase the accumulation of ssDNA from stalled replication forks and promote ssDNA accumulation to activate the DNA sensor-dependent IFN pathway (Figure 4B). However, it remains unknown whether and how SAMHD1 regulates D A replication-induced innate immune and inflammation responses to virus infections.
SAMHD1 Affects Adaptive Antiviral Immune Responses
Because induction of innate immune responses results in the activation of adaptive immunity, the role of SAMHD1 in the innate immune pathway can affect adaptive immune signaling (Figure 5). A few studies revealed mechanisms of SAMHD1 in restricting HIV-1 infection in CD4+ T cells [88, 94–96]. SAMHD1 also promotes abortive HIV-1 infection of CD4+ memory stem cells [97]. Vpx-induced degradation of SAMHD1 enables cytotoxic T lymphocyte killing of monocyte-derived dendritic cells (MDDCs) and inhibits transmission of HIV-1 to CD4+ T cells [98], thus emphasizing the importance of SAMHD1 on adaptive immune signaling during HIV-1 infection. In addition, SAMHD1 depletion increases DCs maturation [99]. Moreover, co-culture of autologous CD4+ lymphocytes from HIV-1 infected patients with SAMHD1-deficient MDDCs displayed enhancement of both viral infection and IFN-I response [100], suggesting that SAMHD1 controls immune signaling from producer cell transmission to target MDDCs and prevents their ability to sense the virus and produce ISGs in innate immune responses. Furthermore, the crosstalk between DCs and lymphocytes down-regulates SAMHD1 expression, which stimulates HIV-1 production and induces the innate sensing of HIV-1 and DC maturation [101].
Figure 5. Role of SAMHD1 in the crosstalk between innate and adaptive immune responses.
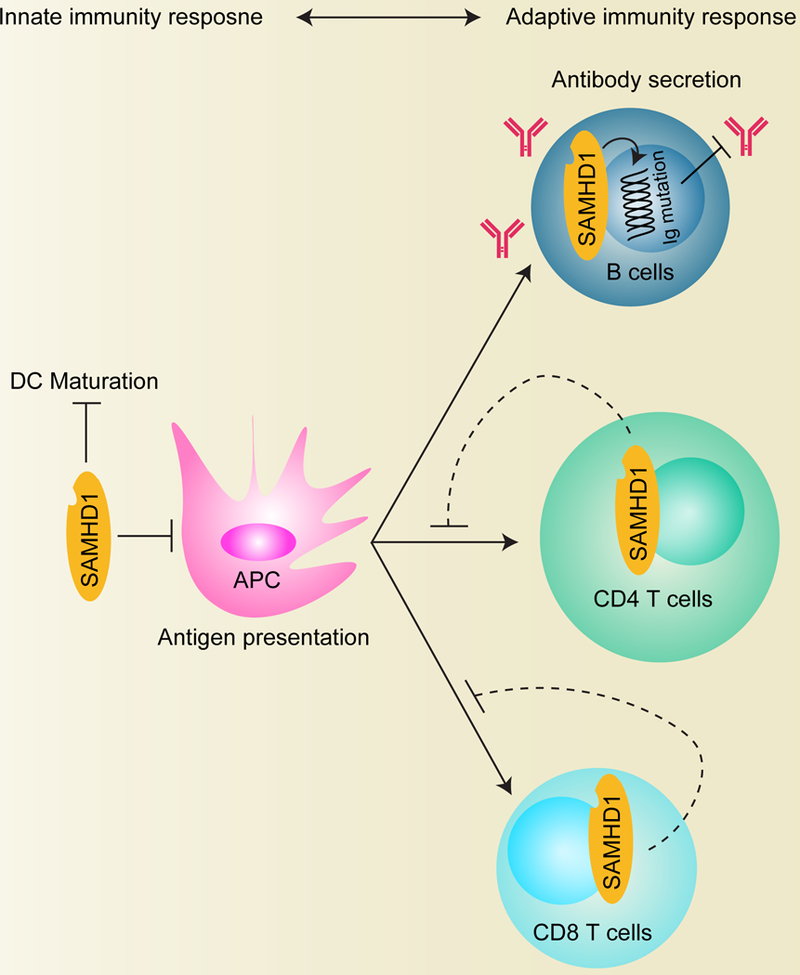
Because of the crosslink between innate immune responses and adaptive immunity, the function of SAMHD1 in the regulation of innate immune pathway may affect adaptive immune responses. SAMHD1 prevents DC maturation induced by HIV-1. SAMHD1 blocks HIV-1 infection in resting CD4+ T cells, non-dividing DCs and macrophages. SAMHD1 also reduces HIV-1 antigen-specific CD8+ T cell response ex vivo. In germinal center B cells, SAMHD1 increases immunoglobulin mutation and reduces antibody production. DC, dendritic cells; APC, antigen-presenting cells.
SAMHD1 affects HIV-1 infection and cell-cell transmission by interacting with CD81, a protein important for syncytia formation [102]. SAMHD1 also reduces HIV-1 antigen-specific CD8+ T cell response in vivo, suggesting that SAMHD1 may function in antigen presentation in adaptive immune responses (Figure 5) [49]. In addition to T cells, CRISPR-Cas9-mediated SAMHD1 KO in germinal center B cells exacerbated immunoglobulin hypermutation by activation-induced deaminase, thus altering antibody binding characteristics and influencing virus restriction. Furthermore, the mutation causes G1-phase base excision repair and mismatch repair (MMR), which can also act as an antiviral mechanism because MMR evolved to attack viral genomes and trigger apoptosis [23]. In conclusion, SAMHD1 provides a preexisting set of intracellular antivirus mechanisms in adaptive immunity of both T- and B-cells (Figure 5).
SAMHD1 Regulation of Immunity: Broad Implications
Because the immune response is a conserved mechanism and indicator of defense against pathogenic infection and disease progression, our understanding of SAMHD1 function in immune regulation can help design novel therapies against multiple diseases. Loss of SAMHD1 causes the pathogenic cytosolic accumulation of DNA likely generated by endogenous retroelements, which induces innate immune activation and leads to inflammatory responses. SAMHD1 has been reported to inhibit human endogenous retroelements in cells [8, 9, 103].
The findings of SAMHD1-mediated down-regulation of pro-inflammatory responses in cells through NF-κB and IFN-I pathways provide a basis for treating SAMHD1-defective AGS patients. This hypothesis has been supported by a case study showing that IL-6 antibodies relieved cerebral vasculopathy in a AGS patient [66]. It is speculated that combined therapy based on IL-6, TNF-α and IFN-I antibodies may have a better effect on treatment of AGS. It is conceivable that NF-κB-targeting drugs could be an alternative or effective way to relieve autoimmune disorders including AGS. When using nucleoside analogues for chemotherapy of certain cancers such as leukemia, the role of SAMHD1 should be considered regarding degradation of nucleic acid, anti-viral function, cell cycle regulation and the DNA damage response pathway [84, 87, 88]. While a number of agents have been reported to inhibit the dNTPase activity of SAMHD1 [104–106], it is intriguing to know whether these SAMHD1 inhibitors can be developed into therapeutic drugs for clinical use.
Structural analyses of SAMHD1 in complex with its interacting partners of the NF-κB and IFN-I pathways can help further understand the molecular mechanism of SAMHD1 in immunity. he crystal structure of the catalytic region of human SAMHD1 with dNTPs has been reported [107, 108]. Distinct from human SAMHD1, the crystal structure and functional studies of full-length mouse SAMHD1 shows that the SAM domain is required to exert its hydrolysis activity [109]. The SAMHD1 domains among different species might be functionally distinct. Previous studies showed that Vpx drives the evolution of primate SAMHD1 [110] and fish SAMHD1 exhibits a positive role in IFN-I expression [111]. Thus, the function of SAMHD1 is complicated and needs further study. A better understanding of the structural changes of SAMHD1 during viral infection should be achieved by using a complex containing human SAMHD1 and its interacting partners.
The balance between immune responses and viral replication is important to avoid excessive responses that might be detrimental to the host. By down-regulating the activation of NF-κB and IFN-I signaling pathways upon viral infection or inflammatory stimuli, SAMHD1 may contribute to the maintenance of this equilibrium. Conversely, it is plausible that SAMHD1-mediated HIV-1 restriction prevents activation of innate and adaptive immune responses, indicating competition between cell-autonomous virus control and viral replication. A recent study reported that SAMHD1-mediated suppression of HIV-1 gene expression negatively affects reactivation of viral latency in CD4+ T cells, suggesting a novel role for SAMHD1 in regulating HIV-1 latency [112]. The function of SAMHD1 in anti-viral immune regulation is complex and connected to multi-cellular pathways. How SAMHD1 maintains the equilibrium between virus replication and immune responses requires more investigation.
Concluding Remarks and Future Perspectives
In this review, we provide an updated summary of SAMHD1 in restricting viral infections and modulating antiviral immunity by various strategies. Based on the detailed discussion on functions and mechanisms of SAMHD1 in modulating immuno-pathogenesis and antiviral immune responses, we expect that more rapid progress (see Outstanding Questions) will be obtained to broaden our understanding of SAMHD1 regulation of these responses. The expected progress will Be fundamental for future development of more effective therapeutic strategies against immune disorders and viral infections.
Outstanding Questions Box.
What are structural and functional changes of SAMHD1 and its interacting proteins (such as NF-κB and IRF7) in the context of viral infections?
In addition to SAMHD1 phosphorylation, do other post-translational modifications and/or gene polymorphisms of SAMHD1 affect the innate immune response to viral infections, and how?
How does SAMHD1 balance restriction of virus replication and inhibition of immune responses in infected cells and hosts?
Does nuclear or cytoplasmic localization of SAMHD1 affect its ability to regulate antiviral immune responses?
In addition to SAMHD1-knockout mouse models, could other SAMHD1-knockout animal models be generated to study SAMHD1 physiological antiviral immune functions and the development of drugs to treat autoimmune diseases?
Could human SAMHD1 protein be a drug target for development of new and more effective therapies against viral infections, certain cancers, and autoimmune disorders?
Could SAMHD1 inhibitors be developed into therapeutic drugs for clinical use?
Do multiple functions and mechanisms of SAMHD1 in regulating antiviral immune responses exist among different mammalian species?
Could human or animal SAMHD1 affect immune responses to infections of pathogenic microbes other than viruses?
Highlights.
SAMHD1 negatively modulates innate and adaptive immune responses to HIV-1 infection via the cGAS/STING pathway.
SAMHD1 suppresses the innate immune responses to viral infections and inflammatory stimuli by interacting with key proteins in the NF-κB and type I interferon pathways.
The PI3K/AKT/IRF3 signaling pathway is important for type I interferon expression induced by SAMHD1-defeciency.
SAMHD1 prevents interferon induction by promoting nascent DNA degradation during cellular DNA replication.
SAMHD1 increases immunoglobulin hypermutation in activated B cells, thereby contributing to antibody diversity and antiviral immunity.
Acknowledgements
We thank members of the Wu laboratory for discussions. The work in the Wu laboratory was supported by NIH grants AI104483, AI120209, and GM128212 (to L.W.). S C and Z.Q. were supported partially by scholarships from the China Scholarship Council. Z.Q. is supported by C. Glenn Barber Funds from the Ohio State University.
Glossary
- DNA damage response
Deoxyribonucleic acid mutation or breakage induced by various chemical drugs or stimulation, such as camptothecin (CPT), etoposide (ETO), hydroxyurea (HU), or ionizing radiation (IR). The DNA damage causes genome instability and is detrimental to human. It can be repaired by cellular repair system or homologous recombination in cells.
- dNTPs
deoxyribonucleoside triphosphates, substrates for DNA synthesis including dATP, dGTP, dCTP and dTTP, which are important in cellular genome replication, retrovirus genome reverse transcription, and DNA damage repair process in cells.
- dsRNA and ssRNA
Double-stranded RNA and single-stranded RNA, respectively, which are assembled as double chains or single chain of nucleotides including guanine (G), uracil (U), adenine (A), and cytosine (C). It often refers to the genome of RNA viruses; for example, a rotavirus harbors a dsRNA genome and HIV-1 has an ssRNA genome.
- SAMHD1
Sterile alpha motif and HD-domain-containing protein 1, which is a host triphosphohydrolase that hydrolyzes intracellular dNTPs to deoxynucleosides and inorganic triphosphates. Loss of human SAMHD1 due to genetic mutation can lead to Aicardi-Goutières syndrome (AGS), a genetic encephalopathy mimicking congenital viral infection. The patients have rare autosomal recessive neurological condition with high levels of interferon-stimulated gene products and inflammatory cytokines in peripheral blood.
- IRF3 and IRF7
Interferon regulatory factor (IRF) 3/7, which are major transcription factors belonging to IRF family members that contain an N-terminal DNA-binding domain specifically binding to a conserved IFN-stimulated response element and a carboxyl-terminal IRF association domain. Upon virus infection, IRF3/7 are phosphorylated by TBK1/IKKε or IKKα, form homo or hetero dimmers and translocate to the nucleus to bind promoters of IFN-I, thus inducing innate antiviral immune responses.
- IFN-I
Type I interferon, including interferon α/β that are induced by IRF3/7. IRF3 specially induces interferon β and IRF7 is responsible for both interferon α and β. In the late stage of the innate immune response, IFN-I bind to IFN receptors on adjacent cells to promote phosphorylation of tyrosine kinase 2 and Janus kinase 1, which further phosphorylate signal transducer and activator of transcription 1 (STAT1) and STAT2. The phosphorylated hetero-dimers of STAT1 and STAT2 then bind to IRF9 and translocate into the nucleus to activate expression of interferon-stimulated genes.
- NF-κB
The mammalian NF-κB family includes five retroviral oncoprotein (v-Rel)-like monomers, NF-κB1 (p105/p50), NF-κB2 (p100/p52), RelA/p65, RelB and c-Rel. Among these Rel family proteins, p50 and p52 are from -terminal region of precursors p105 (NF-κB1) and p100 (NF-κB2), respectively, from which the -terminal domain containing ankyrin repeats is posttranslationally and proteolytically processed by the proteasome. However, RelA, RelB, and c-Rel are translated as mature proteins with transcription transactivation domains (TADs). The NF-κB family members function as two types of dimers, of which the p50 or p52 homodimers are transcriptional repressors, while other TAD-containing heterodimers are activators.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li N et al. (2000)Identification of human homologue of mouse IFN-gamma induced protein from human dendritic cells. Immunol Lett 74 (3), 221–4. [DOI] [PubMed] [Google Scholar]
- 2.Rice GI et al. (2009) Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet 41 (7), 829–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crow YJ and Rehwinkel J (2009) Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum Mol Genet 18 (R2), R130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crow YJ and Manel N (2015) Aicardi-Goutieres syndrome and the type I interferonopathies. Nat Rev Immunol 15 (7), 429–40. [DOI] [PubMed] [Google Scholar]
- 5.Behrendt R et al. (2013) Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Rep 4 (4), 689–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rehwinkel J et al. (2013) SAMHD1-dependent retroviral control and escape in mice. EMBO J 32 (18), 2454–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu L (2013) SAMHD1 knockout mice: modeling retrovirus restriction in vivo. Retrovirology 10, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao K et al. (2013) Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell Rep 4 (6), 1108–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrmann A et al. (2018) The SAMHD1-mediated block of LINE-1 retroelements is regulated by phosphorylation. Mob DNA 9, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pauls E et al. (2013) Restriction of HIV-1 replication in primary macrophages by IL-12 and IL-18 through the upregulation of SAMHD1. J Immunol 190 (9), 4736–41. [DOI] [PubMed] [Google Scholar]
- 11.St Gelais C et al. (2012) SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goujon C et al. (2013) Evidence for IFNalpha-induced, SAMHD1-independent inhibitors of early HIV-1 infection. Retrovirology 10, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riess M et al. (2017) Interferons Induce Expression of SAMHD1 in Monocytes through Down-regulation of miR-181a and miR-30a. J Biol Chem 292 (1), 264–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin C et al. (2014) MicroRNA-181 expression regulates specific post-transcriptional level of SAMHD1 expression in vitro. Biochem Biophys Res Commun 452 (3), 760–7. [DOI] [PubMed] [Google Scholar]
- 15.Hrecka K et al. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474 (7353), 658–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laguette N et al. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474 (7353), 654–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstone DC et al. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480 (7377), 379–82. [DOI] [PubMed] [Google Scholar]
- 18.Lahouassa H et al. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol 13 (3), 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ballana E and Este JA (2015) SAMHD1: at the crossroads of cell proliferation, immune responses, and virus restriction. Trends Microbiol 23 (11), 680–692. [DOI] [PubMed] [Google Scholar]
- 20.Chen S et al. (2018) SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-kappaB and interferon pathways. Proc Natl Acad Sci U S A 115 (16), E3798–E3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coquel F et al. (2018) SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557 (7703), 57–61. [DOI] [PubMed] [Google Scholar]
- 22.Daddacha W et al. (2017) SAMHD1 Promotes DNA End Resection to Facilitate DNA Repair by Homologous Recombination. Cell Rep 20 (8), 1921–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thientosapol ES et al. (2018) SAMHD1 enhances immunoglobulin hypermutation by promoting transversion mutation. Proc Natl Acad Sci U S A 115 (19), 4921–4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blasius AL and Beutler B (2010) Intracellular toll-like receptors. Immunity 32 (3), 305–15 [DOI] [PubMed] [Google Scholar]
- 25.Alexopoulou L et al. (2001) Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413 (6857), 732–8. [DOI] [PubMed] [Google Scholar]
- 26.Kawai T and Akira S (2009) The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol 21 (4), 317–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seth RB et al. (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122 (5), 669–82. [DOI] [PubMed] [Google Scholar]
- 28.Kawai T et al. (2005) IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6 (10), 981–8. [DOI] [PubMed] [Google Scholar]
- 29.Meylan E et al. (2005) Cardif is an adaptor protein in the RIG- antiviral pathway and is targeted by hepatitis C virus. Nature 437 (7062), 1167–72. [DOI] [PubMed] [Google Scholar]
- 30.Xu LG et al. (2005) VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19 (6), 727–40. [DOI] [PubMed] [Google Scholar]
- 31.Chow KT et al. (2018) RIG-I and Other RNA Sensors in Antiviral Immunity. Annu Rev Immunol 36, 667–694. [DOI] [PubMed] [Google Scholar]
- 32.Jensen SandThomsen AR(2012) Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol 86 (6), 2900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unterholzner L et al. (2010) IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11 (11), 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L et al. (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339 (6121), 786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao P et al. (2013) Cyclic [G(2’,5’)pA(3’,5’)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153 (5), 1094–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan YK and Gack MU (2016) Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol 14 (6), 360–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin T et al. (2012) Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36 (4), 561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evavold CL and Kagan JC (2018) How Inflammasomes Inform Adaptive Immunity. J Mol Biol 430 (2), 217–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laidlaw BJ et al. (2016) The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat Rev Immunol 16 (2), 102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodnow CC et al. (2010) Control systems and decision making for antibody production. Nat Immunol 11 (8), 681–8. [DOI] [PubMed] [Google Scholar]
- 41.Nutt SL et al. (2015) The generation of antibody-secreting plasma cells. Nat Rev Immunol (3), 160–71. [DOI] [PubMed] [Google Scholar]
- 42.Fujii SI et al. (2018) Vaccine Designs Utilizing Invariant NKT-Licensed Antigen-Presenting Cells Provide NKT or T Cell Help for B Cell Responses. Front Immunol 9, 1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams NM et al. (2018) Transcription Factor IRF8 Orchestrates the Adaptive Natural Killer Cell Response. Immunity 48 (6), 1172–1182 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rankin LC and Artis D (2018) Beyond Host Defense: Emerging Functions of the Immune System in Regulating Complex Tissue Physiology. Cell 173 (3), 554–567. [DOI] [PubMed] [Google Scholar]
- 45.Lahaye X et al. (2013) The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity 39 (6), 1132–42. [DOI] [PubMed] [Google Scholar]
- 46.Gao D et al. (2013) Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341 (6148), 903–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antonucci JM et al. (2017) The Dynamic Interplay between HIV-1, SAMHD1, and the Innate Antiviral Response. Front Immunol 8, 1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jakobsen MR et al. (2013)IFI16senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci U S A 110 (48), E4571–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maelfait J et al. (2016) Restriction by SAMHD1 Limits cGAS/STING-Dependent Innate and Adaptive Immune Responses to HIV-1. Cell Rep 16 (6), 1492–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seamon KJ et al. (2015) SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res 43 (13), 6486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seamon KJ et al. (2016) Single-Stranded Nucleic Acids Bind to the Tetramer Interface of SAMHD1 and Prevent Formation of the Catalytic Homotetramer. Biochemistry 55 (44), 6087–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goncalves,. et al. (2012) SAMHD1 is a nucleic-acid binding protein that is mislocalized due to aicardi-goutieres syndrome-associated mutations. Hum Mutat 33 (7), 1116–22. [DOI] [PubMed] [Google Scholar]
- 53.Tungler V et al. (2013) Single-stranded nucleic acids promote SAMHD1 complex formation. J Mol Med (Berl) 91 (6), 759–70. [DOI] [PubMed] [Google Scholar]
- 54.Ryoo J et al. (2014) The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med 20 (8), 936–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi J et al. (2015) SAMHD1 specifically restricts retroviruses through its RNase activity. Retrovirology 12, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryoo J et al. (2016) Reply to SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nat Med 22 (10), 1074–1075. [DOI] [PubMed] [Google Scholar]
- 57.Antonucci JM et al. (2016) SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nat Med 22 (10), 1072–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wittmann S et al. (2015) Phosphorylation of murine SAMHD1 regulates its antiretroviral activity. Retrovirology 12, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beloglazova N et al. (2013) Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J Biol Chem 288 (12), 8101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang S et al. (2016) Interferon regulatory factor 3 is a key regulation factor for inducing the expression of SAMHD1 in antiviral innate immunity. Sci Rep 6, 29665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tamura T et al. (2008) The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 26, 535–84. [DOI] [PubMed] [Google Scholar]
- 62.Honda K et al. (2005) IRF-7 is the master regulator of type- interferon-dependent immune responses. Nature 434 (7034), 772–7. [DOI] [PubMed] [Google Scholar]
- 63.Rahman MM and McFadden G (2011) odulation of NF-kappaB signalling by microbial pathogens. Nat Rev Microbiol 9 (4), 291–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao W et al. (2008) Dendritic cell-derived interferon-gamma-induced protein mediates tumor necrosis factor-alpha stimulation of human lung fibroblasts. Proteomics 8 (13), 2640–50. [DOI] [PubMed] [Google Scholar]
- 65.Xin B et al. (2011) Homozygous mutation in SAMHD1 gene causes cerebral vasculopathy and early onset stroke. Proc Natl Acad Sci U S A 108 (13), 5372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henrickson M and Wang H (2017) Tocilizumab reverses cerebral vasculopathy in a patient with homozygous SAMHD1 mutation. Clin Rheumatol 36 (6), 1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Franzolin E et al. (2013) The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc Natl Acad Sci U S A 110 (35), 14272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan J et al. (2015) CyclinA2-Cyclin-dependent Kinase Regulates SAMHD1 Protein Phosphohydrolase Domain. J Biol Chem 290 (21), 13279–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.White TE et al. (2013) The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13 (4), 441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cribier A et al. (2013) Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep 3 (4), 1036–43. [DOI] [PubMed] [Google Scholar]
- 71.St Gelais C et al. (2014) Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J Virol 88 (10), 5834–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pauls E et al. (2014) Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J Immunol 193 (4), 1988–97. [DOI] [PubMed] [Google Scholar]
- 73.St Gelais C et al. (2016) A Putative Cyclin-binding Motif in Human SAMHD1 Contributes to Protein Phosphorylation, Localization, and Stability. J Biol hem 291 (51), 26332–26342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bonifati S et al. (2016) SAMHD1 controls cell cycle status, apoptosis and HIV-1 infection in monocytic THP-1 cells. Virology 495, 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.St Gelais C et al. (2018) A Cyclin-Binding Motif in Human SAMHD1 Is Required for Its HIV-1 Restriction, dNTPase Activity, Tetramer Formation, and Efficient Phosphorylation. J Virol 92 (6), e01787–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szaniawski MA et al. (2018) SAMHD1 Phosphorylation Coordinates the Anti-HIV-1 Response by Diverse Interferons and Tyrosine Kinase Inhibition. MBio 9 (3), e00819–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oh C et al. (2018) A central role for PI3K-KT signaling pathway in linking SAMHD1-deficiency to the type I interferon signature. Sci Rep 8 (1), 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kodigepalli KM et al. (2018) SAMHD1 modulates in vitro proliferation of acute myeloid leukemia-derivedTHP-1cellsthroughthe PI3K-Akt-p27 axis. Cell Cycle 17 (9), 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li S et al. (2016) The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat Immunol 17 (3), 241–9. [DOI] [PubMed] [Google Scholar]
- 80.Jasiulionis MG (2018) Abnormal Epigenetic Regulation of Immune System during Aging. Front Immunol 9, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim YW et al. (2015) Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutieres syndrome. elife 4, 10.7554/eLife.08007. [DOI] [PMC free article] [PubMed]
- 82.Hartlova,. et al. (2015) DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 42 (2), 332–343. [DOI] [PubMed] [Google Scholar]
- 83.Kondo T et al. (2013) DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci U S A 110 (8), 2969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Clifford R et al. (2014) SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood 123 (7), 1021–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kretschmer S et al. (2015) SAMHD1 prevents autoimmunity by maintaining genome stability. Ann Rheum Dis 74 (3), e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mlcochova P et al. (2018) DNA damage induced by topoisomerase inhibitors activates SAMHD1 and blocks HIV-1 infection of macrophages. EMBO J 37 (1), 50–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herold N et al. (2017) Targeting SAMHD1 with the Vpx protein to improve cytarabine therapy for hematological malignancies. Nat Med 23 (2), 256–263. [DOI] [PubMed] [Google Scholar]
- 88.Schneider C et al. (2017) SAMHD1 is a biomarker for cytarabine response and a therapeutic target in acute myeloid leukemia. Nat Med 23 (2), 250–255. [DOI] [PubMed] [Google Scholar]
- 89.Li T and Chen ZJ (2018) The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med 215 (5), 1287–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bester AC et al. (2011) Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145 (3), 435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kohnken R et al. (2015) Regulation of deoxynucleotide metabolism in cancer: novel mechanisms and therapeutic implications. Mol Cancer 14, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rentoft M et al. (2016) Heterozygous colon cancer-associated mutations of SAMHD1 have functional significance. Proc Natl Acad Sci U S 113 (17), 4723–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jauregui P and Landau NR (2018) DNA damage induces a SAMHD1-mediated block to the infection of macrophages by HIV-1. Sci Rep 8 (1), 4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Coiras M et al. (2016) IL-7 Induces SAMHD1 Phosphorylation in CD4+ Lymphocytes, Improving Early Steps of HIV-1 Life Cycle. Cell Rep 14 (9), 2100–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Descours B et al. (2012) SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 9, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baldauf HM et al. (2012) SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med 18 (11), 1682–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tabler CO et al. (2014) CD4+ memory stem cells are infected by HIV-1 in a manner regulated in part by SAMHD1 expression. J Virol 88 (9), 4976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ayinde D et al. (2015) SAMHD1 Limits HIV-1 Antigen Presentation by Monocyte-Derived Dendritic Cells. J Virol 89 (14), 6994–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hertoghs N et al. (2015) SAMHD1 degradation enhances active suppression of dendritic cell maturation by HIV-1. J Immunol 194 (9), 4431–7. [DOI] [PubMed] [Google Scholar]
- 100.Puigdomenech I et al. (2013) SAMHD1 restricts HIV-1 cell-to-cell transmission and limits immune detection in monocyte-derived dendritic cells. J Virol 87 (5), 2846–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Su B et al. (2014) Dendritic cell-lymphocyte cross talk downregulates host restriction factor SAMHD1 and stimulates HIV-1 replication in dendritic cells. J Virol 88 (9), 5109–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rocha-Perugini V et al. (2017) CD81 association with SAMHD1 enhances HIV-1 reverse transcription by increasing dNTP levels. Nat Microbiol 2 (11), 1513–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu S et al. (2015) SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation. PLoS Genet 11 (7), e1005367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Seamon KJ et al. (2014) Small molecule inhibition of SAMHD1 dNTPase by tetramer destabilization. J Am Chem Soc 136 (28), 9822–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Seamon KJ and Stivers JT (2015) High-Throughput Enzyme-Coupled Assay for SAMHD1 dNTPase. J Biomol Screen 20 (6), 801–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hollenbaugh JA et al. (2017) Substrates and Inhibitors of SAMHD1. PLoS One 12 (1), e0169052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ji X et al. (2013) Mechanism of allosteric activation of SAMHD1 by dGTP. Nat Struct Mol Biol 20 (11), 1304–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ji X et al. (2014) Structural basis of cellular dNTP regulation by SAMHD1. Proc Natl Acad Sci U S A 111 (41), E4305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Buzovetsky O et al. (2018), The SAM domain of mouse SAMHD1 is critical for its activation and regulation. Nat Commun 9 (1)411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang C et al. (2012)Co-evolution of primate SAMHD1 and lentivirus Vpx leads to the loss of the vpx gene in HIV-1 ancestor. PLoS One 7 (5), e37477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li M et al. (2018) Fish SAMHD1 performs as an activator for IFN expression. Dev Comp Immunol 86, 138–146. [DOI] [PubMed] [Google Scholar]
- 112.Antonucci JM et al. (2018) SAMHD1 Impairs HIV-1 Gene Expression and Negatively Modulates Reactivation of Viral Latency in CD4(+) T Cells. J Virol 92 (15), e00292–18. [DOI] [PMC free article] [PubMed] [Google Scholar]


