Abstract
Proteasomes are a multi-subunit protease complex that produces peptides bound by major histocompatibility complex (MHC) class I molecules. Phylogenetic studies indicate tahat two specialized forms of proteasomes, immunoproteasomes and thymoproteasomes, and the proteasome activator PA28αβ emerged in a common ancestor of jawed vertebrates which acquired adaptive immunity based on the MHC, T cell-receptors and B-cell receptors ~500 million years ago. Comparative genomics studies now provide strong evidence that the genes coding for the immunoproteasome subunits emerged by genome-wide duplication. On the other hand, the gene encoding the thymoproteasome subunit β5t emerged by tandem duplication from the gene coding for the β5 subunit. Strikingly, birds lack immunoproteasomes, thymoproteasomes and the proteasome activator PA28αβ, raising an interesting question of whether they have evolved any compensatory mechanisms.
Keywords: adaptive immune system, genome-wide duplication, immunoproteasome, proteasome activator, thymoproteasome
Introduction
Eukaryotic cells have two major pathways for intracellular protein degradation. One is lysosomal autophagy (Nakatogawa et al. 2009) and the other is the ubiquitin-proteasome system (Tanaka 2009). In the latter, proteins covalently modified with a chain of ubiquitin are specifically recognized by a large protease complex, the 26S proteasome, leading to the degradation of ubiquitin-tagged proteins (Bard et al. 2018; Collins and Goldberg 2017). The ubiquitin-proteasome system is indispensable for cell survival and plays a critical role in many essential cellular processes including protein quality control, regulation of cell cycle progression, transcription, cell trafficking and responses to oxidative stress. The catalytic core of the 26S proteasome, known as the 20S proteasome, has a cylindrical structure composed of a stack of four heptameric rings; the two outer rings are made up of catalytically inactive α subunits, whereas the two inner rings, which are made up of β subunits, contain catalytic subunits, with active sites facing the interior of the cylinder (Unno et al. 2002).
Proteasomes are of ancient origin. They are found not only in eukaryotes but also in all archaea and some eubacteria such as actinobacteria and mycobacteria (Maupin-Furlow 2011). Although the overall structure of the 20S proteasome is highly conserved between eukaryotes and prokaryotes, the complexity of α and β subunits is generally low in prokaryotes. Thus, the archaebacterium Thermoplasma acidophilum has only one kind of α and β subunits, respectively (Lowe et al. 1995). Eubacterial 20S proteasomes usually have one or two kinds of α and β subunits, respectively (Jastrab and Darwin 2015). In contrast, eukaryotic 20S proteasomes contain seven distinct α subunits designated as α1 to α7 and seven distinct β subunits designated as β1 to β7. Among the seven β subunits, only β1 (also known as PSMB6), β2 (also known as PSMB7) and β5 (also known as PSMB5) subunits are proteolytically active and exert caspase-like, trypsin-like and chymotrypsin-like activities, respectively, which confer the ability to cleave peptide bonds at the C-terminal side of acidic, basic and hydrophobic amino acid residues, respectively (Table 1). Archaeal and eubacterial proteasomes efficiently cleave short fluorogenic peptides only after hydrophobic residues whereas eukaryotic proteasomes exhibit three major peptidase activities (Dolenc et al. 1998). Therefore, the chymotrypsin-like activity is most likely the most ancient, fundamental one.
Table 1.
Members of the three catalytic subunit families in jawed vertebrates
| Subunit | Other name | Gene symbol |
|---|---|---|
| β1 | Y, PSMB6 | PSMB6 |
| β1i | LMP2, PSMB9 | PSMB9 |
| PSMB12 | psmb12 | |
| β2 | Z, PSMB7 | PSMB7 |
| β2i | MECL1, PSMB10 | PSMB10 |
| PSMB13 | psmb13 | |
| β5 | X, PSMB5 | PSMB5 |
| β5i | LMP7, PSMB8 | PSMB8 |
| β5t | PSMB11 | PSMB11 |
Gene symbols given are those of humans, except for psmb12 and psmb13, which are found only in fish. In humans and chickens, gene symbols are upper-case italics. Chickens have only PSMB5, PSMB6 and PSMB7. In rodents, gene symbols are italicized, with only the first letter in upper-case. In Xenopus and fish, gene symbols are lower-case italics.
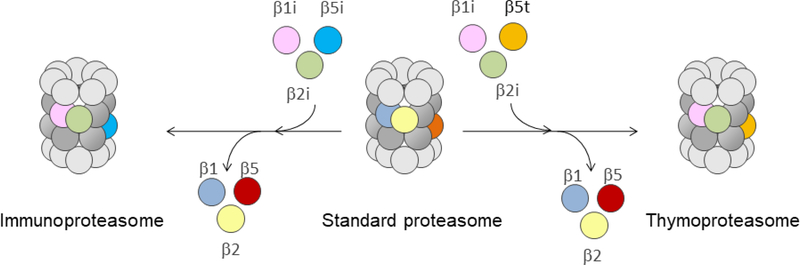
In the immune system, proteasomes play a critical role in the production of antigenic peptides bound by major histocompatibility complex (MHC) class I molecules (Heemels and Ploegh 1995; Kniepert and Groettrup 2014; Tanaka and Kasahara 1998). Notably, jawed vertebrates have two specialized forms of 20S proteasomes named immunoproteasomes (Akiyama et al. 1994) and thymoproteasomes (Murata et al. 2007) in addition to the standard or constitutive 20S proteasome shared by all eukaryotes (Fig. 1). These specialized forms of proteasomes differ from the standard 20S proteasome in the composition of catalytic β subunits (Murata et al. 2018). As a result, immunoproteasomes, which have β1i (also known as LMP2, PSMB9), β2i (also known as MECL1, PSMB10) and β5i (also known as LMP7, PSMB8) subunits instead of β1, β2 and β5 subunits, respectively, have distinct cleavage specificities and produce peptides with C-terminal hydrophobic residues that fit well in the groove of MHC class I molecules more efficiently than standard 20S proteasomes. Recent work has shown that the repertoire of MHC class I-binding peptides differs substantially between wildtype and immunoproteasome-deficient mice and that the presentation of class I epitopes to cytotoxic T cells is greatly impaired in the latter (Kincaid et al. 2011).
Fig. 1.
Subunit composition of immunoproteasomes and thymoproteasomes
Proteasomes have a cylindrical structure made up of four stacked rings: two outer α-rings composed up of α subunits and two inner β-rings composed of β subunits. In eukaryotes, α- and β-rings are made up of seven distinct subunits, respectively. Catalytic β subunits, β1, β2 and β5, are colored. Immunoproteasomes and thymoproteasomes, which occur only jawed vertebrates, differ from standard 20S proteasomes in the composition of catalytic β subunits.
Immunoproteasomes are constitutively expressed by professional antigen presenting cells and medullary thymic epithelial cells, but are induced in response to immune and inflammatory stimuli in other cell types (Kniepert and Groettrup 2014). This induction occurs because the expression of genes coding for β1i, β2i and β5i subunits is induced by exposure to inflammatory cytokines such as interferon γ and tumor necrosis factor. Under inflammatory conditions, immunoproteasomes rather than standard 20S proteasomes are assembled because β1i, β2i and β5i subunits are preferentially incorporated into newly assembled 20S proteasomes in place of β1, β2 and β5 subunits, respectively, partly due to preferential binding of β5i to POMP, a chaperone essential for proteasome assembly (Griffin et al. 1998; Heink et al. 2005).
Another specialized form of proteasome, the thymoproteasome, has β1i, β2i and β5t (also known as PSMB11) subunits instead of β1, β2 and β5 (β5i) subunits, respectively (Murata et al. 2007). Because β5t is expressed exclusively in thymic cortical cells (Murata et al. 2007; Tomaru et al. 2009), thymoproteasomes occur only in the thymic cortex. Unlike β5 and β5i whose substrate-binding pockets are mostly composed of hydrophobic amino acids, the substrate-binding pocket of β5t contains many hydrophilic amino acids, thereby producing a distinct set of peptides that presumably bind to MHC class I molecules with low affinity (Kondo et al. 2018; Nitta et al. 2010; Sasaki et al. 2015; Xing et al. 2013). In thymoproteasome-deficient mice, in which cortical thymic epithelial cells express immunoproteasomes instead of thymoproteasomes, CD8+ T cell production is severely impaired (Murata et al. 2007). It is thought that thymoproteasomes play a critical role in thymic positive selection of CD8+ T cells through producing a unique set of peptides involved in positive selection and/or minimizing the overlap of the repertoire of MHC class I-binding peptides displayed to developing T cells in the cortex and medulla where positive and negative selection takes place, respectively (Klein et al. 2014; Murata et al. 2018; Nitta et al. 2010; Takahama et al. 2012).
Here we provide a brief overview of proteasome evolution mainly focusing on immunoproteasomes and thymoproteasomes.
Evolution of adaptive immunity and the origin of specialized forms of proteasomes
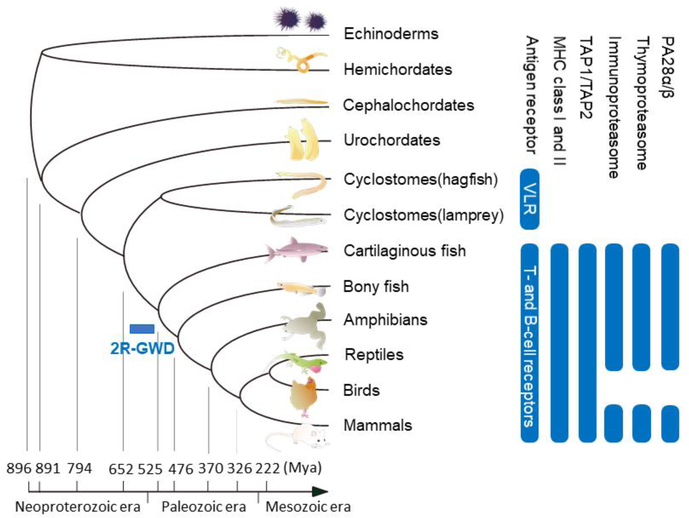
Accumulated evidence indicates that lymphocyte-based adaptive immunity emerged in a common ancestor of vertebrates and once established its basic design has been maintained throughout vertebrate evolution (Flajnik and Kasahara 2001, 2010; Kaufman 2018b) (Fig. 2). Thus, the essential features of lymphocyte-based adaptive immunity, such as the clonal expression of a single type of antigen receptors with allelic exclusion, the clonal proliferation of antigen-stimulated lymphocytes and the dichotomy of lymphocytes into T and B cells are shared by all vertebrates (Boehm et al. 2012; Hirano et al. 2013). There are, however, substantial differences between the adaptive immune systems of jawed and jawless vertebrates (Boehm 2011; Boehm et al. 2018; Flajnik and Kasahara 2010; Kasahara and Sutoh 2014). In jawed vertebrates ranging from cartilaginous fishes to mammals, three cardinal elements involved in antigen recognition are T-cell receptors, B-cell receptors and MHC molecules (Flajnik 2018a). By contrast, jawless vertebrates, represented by lampreys and hagfish, have neither MHC class I nor class II molecules and use variable lymphocyte receptors (VLRs) as antigen receptors (Cooper and Alder 2006; Flajnik 2018b). Unlike T-cell and B-cell receptors, which are both members of the immunoglobulin superfamily, VLRs are members of the leucine-rich repeat (LRR) family and are composed of an N-terminal cap, a variable number of multiple LRR modules, a connecting peptide, a C-terminal cap and an invariant stalk. Because the sequences of LRR modules are diverse, and the number of modules assembled into the VLR gene is variable, a single VLR gene can generate diversity comparable to that of antigen receptors of jawed vertebrates. Because T and B lymphocyte lineages are conserved in all vertebrates, jawed and jawless vertebrates appear to have adopted different molecules for antigen recognition within the context of specialized lymphocyte lineages (Boehm et al. 2012; Flajnik and Kasahara 2010; Kasahara and Sutoh 2014).
Fig. 2.
Evolution of adaptive immunity
Phylogeny of animals is shown on the left along with the divergence times based on molecular data compiled by Blair and Hedges (Blair and Hedges 2005). The phylogenetic distribution of key elements discussed in this review is shown on the right. It is conspicuous that birds lack proteasome elements specialized for antigen presentation by MHC class I molecules. This figure was modified from the previously published one (Murata et al. 2018). Mya, million years ago
Initial evidence that immunoproteasomes occur in non-mammalian vertebrates was obtained in Xenopus laevis, from which the genes coding for β5i and β1i were identified (Namikawa et al. 1995; Nonaka et al. 1997a). Subsequently, the cartilaginous fish was shown to have genes coding for β5i and β5i-like subunits, but in the same study the gene coding for β5i was identified in neither lampreys nor hagfishes, suggesting that immunoproteasomes occur only in jawed vertebrates (Kandil et al. 1996). This suggestion is now confirmed by genome sequence information accumulated in various vertebrates and invertebrates. The gene coding for the β5t subunit, designated PSMB11, also occurs in the cartilaginous fish but not in jawless vertebrates or invertebrates (Sutoh et al. 2012). Considering that immunoproteasomes and thymoproteasomes apparently have functions dedicated to the production of MHC class I-binding peptides, it is reasonable that these specialized forms of proteasomes occur only in jawed vertebrates possessing MHC class I molecules.
Genome-wide duplications (GWDs) and evolution of immunoproteasomes
Susumu Ohno proposed based mainly on cytological evidence that one or two rounds of GWD took place close to the origin of vertebrates (Ohno 1970). The refined version of this proposal, which became known as the 2R (two round) hypothesis, assumes that the genome of jawed vertebrates underwent two rounds of GWD (2R-GWD) close to the origin of vertebrates (Kasahara 2007, 2013). The 2R hypothesis is now strongly supported by comparative genomics analysis, although the timing of GWD relative to the emergence of jawless vertebrates is still controversial (Mehta et al. 2013; Smith et al. 2018). One of the key observations underpinning the 2R hypothesis is genome paralogy, which refers to the occurrence of closely linked sets of paralogous genes on multiple, typically four different chromosomes. In humans and mice, the genes coding for β1i and β5i (PSMB9 and PSMB8 in humans, and Psmb9 and Psmb8 in mice) are in the class II region of the MHC on chromosomes 6 and 17, respectively. Mapping of the gene coding for the β2 subunit (PSMB7 in humans and Psmb7 in mice) showed that it is located on the regions of human chromosome 9 and mouse chromosome 2 containing a group of genes, the paralogous copies of which are located in the MHC, providing the first indication that the MHC region retains the traces of ancient GWDs (Kasahara et al. 1996). Subsequent studies showed that, along with the HOX gene clusters, the MHC is a prototypical region exhibiting genome paralogy, with an extensive number of gene families sharing paralogous copies among four major paralogous regions (the MHC on chromosome 6 and specific regions of chromosomes 1, 9 and 19) and other minor paralogous regions (Flajnik and Kasahara 2001, 2010; Kasahara 2007). Prompted by this observation, it was realized that other immune gene families such as chemokine, JAK/STAT and B7 have paralogous copies in the regions displaying genome paralogy (Kasahara 1998, 2010).
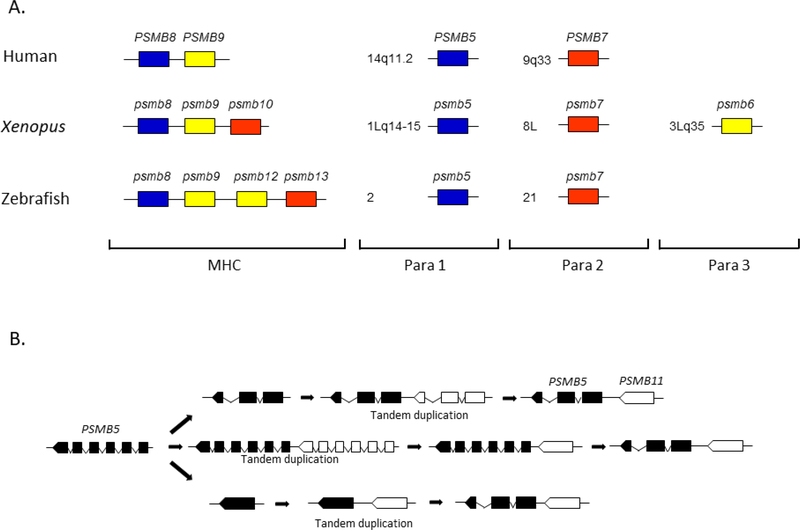
Among the seven β subunits of the 20S proteasome, three catalytic β subunits, β1, β2 and β5, are more closely related to each other than they are to other β subunits. Also, β1i, β2i and β5i, which occur only in jawed vertebrates, are most closely related to β1, β2 and β5, respectively, indicating that each immunoproteasome subunit emerged by gene duplication from the evolutionarily more ancient, corresponding subunit of the standard 20S proteasome. Based on these observations and the finding that the gene coding for the β2 subunit maps to the region paralogous to the MHC, it was proposed that PSMB8, PSMB9 and PSMB10 arose from PSMB6, PSMB5 and PSMB7, respectively, not by individual duplications, but by 2R-GWD (Kasahara et al. 1996)(Fig. 3). However, in mammals, the PSMB10 gene coding for the β2i subunit is not encoded in the MHC; in addition, the genes coding for β1, β2 and β5 are unlinked and do not map to any of the four major paralogous regions initially identified, apparently contradicting the idea that the three immunoproteasome subunit genes arose by GWDs. However, genes often change their locations due to translocations and other mechanisms. It was therefore assumed that the genes coding for β1, β2i and β5 changed their locations secondarily.
Fig. 3.
Evolution of immunoproteasomes and thymoproteasomes
Panel A shows that the majority of genes coding for catalytic β subunits are located in the MHC and MHC-paralogous regions. Para 1, 2 and 3 stand for MHC-paralogous regions 1, 2 and 3. In humans, PSMB10 and PSMB6 map outside of the paralogous regions. In Xenopus tropicalis, all six catalytic β subunit genes are encoded in the MHC and MHC-paralogous regions; psmb10 maps to the class III region of the MHC and psmb6 maps to the region syntenic to human para 3 (19p13.1–13.3). In zebrafish, psmb12 and psmb13, which originated from psmb6 and psmb7, respectively, are in the class I region of the MHC. Panel B shows three possible scenarios accounting for the emergence of the PSMB11 gene (indicated in white) from the PSMB5 gene (indicated in black). In bony fishes, psmb11 and psmb5 are located in a tail-to-head orientation as shown in the figure. In contrast, they are located in a head-to-head orientation in tetrapods (Sutoh et al. 2012). Therefore, PSMB11 appears to have been inverted in a tetrapod lineage (not shown in the figure).
Subsequent studies in non-mammalian vertebrates showed that the gene encoding β2i is encoded in the MHC region in Xenopus, and that the Xenopus β1 subunit is encoded in one of the MHC paralogous regions (Ohta et al. 2006). In zebrafish, the class I region of the MHC was found to contain the psmb13 gene (a member of the β2 subunit family distinct from psmb10), although the teleost psmb10 gene, which was initially mapped to the class I region of the MHC (Clark et al. 2000; Lukacs et al. 2007), actually maps outside of the MHC region on a separate chromosome (McConnell et al. 2016). Furthermore, the region of human chromosome 14 (14q11-q32) containing the gene for β5 is now considered to be part of the MHC paralogous regions (Flajnik and Kasahara 2010; Kasahara 2007). Therefore, it seems that the proteasome subunit genes whose chromosomal localization in mammals initially appeared contradictory to what was expected from 2R-GWD changed their locations secondarily in the course of vertebrate evolution. Considering all the evidence accumulated in various vertebrate species, it is now very likely that the genes coding for immunoproteasome subunits emerged by 2R-GWD.
Class I region in the primordial MHC
An interesting feature of the MHC in nonmammalian vertebrates is the close linkage of the immunoproteasome, transporter associated with antigen processing (TAP) and TAP-binding protein (TAPBP) genes with classical class I genes, whereas in most mammals they are closely linked to class II genes (Michalova et al. 2000; Nonaka et al. 1997b; Ohta et al. 2002; Ohta et al. 2003). These findings suggest that in the primordial MHC, the class I processing and presenting genes co-evolved so that the peptides generated and transported preferentially bound to particular class I allelic products (Flajnik et al. 1999; Kaufman 1999). Consistent with this idea, there are polymorphisms in TAP and immunoproteasome genes that co-segregate with class I polymorphisms in amphibians and some fish (see below). Indeed, chicken TAP genes are polymorphic and very closely linked to the classical class I (BF) genes (Kaufman 1999). Chicken class I allelic products are either ‘permissive’ (generalists) and capable of binding to many peptides or ‘restrictive’ (specialists) binding to peptides with defined anchor residues (Kaufman 2018a); the peptides transported by a particular TAP allelic product have a propensity to bind to the class I allelic product, the coding genes of which are linked to each other (Kaufman 2015; Walker et al. 2011).
psmb8 biallelism in ectotherms
In most ectotherms studied, there are biallelic lineages of psmb8, most conspicuously with modifications in the catalytic site (Kandil et al. 1996; Namikawa et al. 1995; Ohta et al. 2002; Tsukamoto et al. 2012). One lineage with an alanine at position 31 is predicted to be similar to mammalian β5i, which was shown in mammals to produce peptides with hydrophobic C-termini. The catalytic site in the other allele has a conspicuous phenylalanine-31 that is predicted to modify the range of peptides generated (Kandil et al. 1996; Ohta et al. 2002). This biallelism is found in cartilaginous fish, teleosts and amphibians, strongly suggesting that this is the primordial feature of the β5i subunit. Xenopus also shows biallelism of tap, a feature that extends to all Xenopus species, which unfailingly co-segregates with particular psmb8 and class I alleles. Wild frogs of both X. laevis and X. tropicalis maintain these two allelic syntenic sets, demonstrating a balancing selection within populations over 100 million years and likely a block of recombination (i.e. cold spots) between the three genes (Ohta et al. 2003). The paradigm put forward by Kaufman on selection for class I ‘generalists’ and ‘specialists’ in birds may also apply to the class I system in ectotherms (Kaufman 2018a).
This Ala/Phe-31 polymorphism of psmb8 is found in many ectotherms, yet its loss in several fish and amphibians suggests it is under a strong pressure and it can be re-established by convergent evolution in different animal groups (Huang et al. 2013). Additionally, in sharks and some bony fish there has been an expansion of immunoproteasome genes suggesting that: 1) the peptide repertoire can be expanded in some species; and 2) ectotherm classical class I molecules are much more dependent on proteasome-generated peptides than class I molecules of most mammals (McConnell et al. 2016; Michalova et al. 2000; Ohta et al. 2003). We have known of these interesting polymorphisms for a long time, but functional experiments like those done in birds for TAP, e.g. examining peptide generation in vitro or impact on class I expression/function in vivo have not been done for ectotherm immunoproteasomes or TAP. With the ease of crispr usage in teleost models and Xenopus, the experiments should be relatively easy to implement.
Evolution of thymoproteasomes
Like immunoproteasomes, thymoproteasomes are an invention by jawed vertebrates. However, unlike immunoproteasomes, the PSMB11gene coding for the thymoproteasome-specific subunit β5t appears to have emerged by tandem duplication from the evolutionarily more ancient PSMB5 gene coding for the β5 subunit (Sutoh et al. 2012)(Fig. 3). This is indicated by the fact that PSMB11 is located adjacent to the PSMB5 gene in all classes of jawed vertebrates for which information is available, expect for birds that have lost PSMB11. A striking feature of PSMB11 is the complete absence of introns; most β-subunit genes including those coding for β1, β1i, β2, β2i and β5i subunits have six to eight exons, with similar exon–intron organization (Hayashi et al. 1997; Tanaka and Kasahara 1998). The most plausible explanation for this complete absence of introns is that the PSMB5 gene underwent tandem duplication and that the reverse-transcribed, processed mRNA derived from PSMB5 replaced another copy of PSMB5 next to the source gene by homologous recombination (Kaessmann et al. 2009). Interestingly, gnathostome PSMB5 genes have only three exons and do not share any exon-intron boundaries with those of other β subunit genes, whereas lamprey and amphioxus PSMB5 genes have exon-intron organization typical of β subunit genes (Abdulla et al. 1996; Kohda et al. 1997; Sutoh et al. 2012). Therefore, the PSMB5 gene appears to have undergone major alterations in gene structure in a jawed vertebrate lineage. It is unclear whether this change in gene structure has something to do with the emergence of PSMB11. The alteration in gene structure may have taken place prior to the tandem duplication that gave rise to an ancestor of PSMB11 (Fig. 3, panel B, top). Alternatively, PSMB5 may have changed its structure after it had diverged from PSMB11 (Fig. 3, panel B, middle). A third possibility is that the loss of introns in PSMB5 took place before tandem duplication and that PSMB5 acquired introns newly after duplication (Fig. 3, panel B, bottom).
In mammals, reptiles, amphibians and cartilaginous fishes, PSMB11 is a single copy gene. In contrast, teleost fish have two copies of apparently functional psmb11 genes, designated psmb11a and psmb11b, which are thought to have emerged by fish-specific GWD (Sutoh et al. 2012). The β5tb subunit encoded by psmb11b has a typical S1 pocket made up of hydrophilic residues, whereas the S1 pocket of the β5ta subunit encoded by psmb11a is less hydrophilic, suggesting that the cleavage specificities of the thymoproteasomes containing β5ta and β5tb may differ. Whether both β5t subunits are expressed in cortical thymic epithelial cells remains to be investigated.
Evolution of proteasome activators
Two additional proteasome components with a role apparently specialized for MHC class I-mediated antigen presentation are PA28α and PA28β subunits of the PA28 activator complex, also known as the 11S regulator of the 20S proteasome (Cascio 2014; Tanaka and Kasahara 1998). The PA28 activator complex, which is inducible by stimulation with interferon-γ, is a ring-shaped, cytoplasmic heteroheptamer composed of ~28-kDa α and ~28-kDa β subunits with a stoichiometry of α3β4 or a mixture of α3β4 and α4β3 (Zhang et al. 1999); binding of this complex to the outer α-rings of the 20S proteasome stimulates its peptidase activity in an ATP-independent manner and enhances the generation of class I-binding peptides by promoting the assembly of the immunoproteasome and altering its cleavage pattern (de Graaf et al. 2011). Enhanced expression of the PA28α subunit in virus-infected fibroblasts resulted in more efficient presentation of viral peptides to cytotoxic T cells (Groettrup et al. 1996). Furthermore, mice lacking the PA28α subunit (Preckel et al. 1999) or both PA28α and PA28β subunits (Murata et al. 2001) showed impaired cytotoxic T cell responses against some epitopes, demonstrating a critical role of PA28αβ in MHC class I-mediated presentation of some peptides.
The genes coding for PA28α and PA28β subunits (named PSME1 and PSME2) are members of the PA28 family. A third member of this family is PSME3 coding for the PA28γ subunit, which was originally identified as Ki antigen in sera of patients with systemic lupus erythematosus (Nikaido et al. 1990). PA28γ, which forms a homoheptamer, is a nuclear activator of the 20S proteasome that plays an important role in the regulation of cell proliferation, apoptosis, nuclear dynamics and cellular stress response (Wilk et al. 2000).
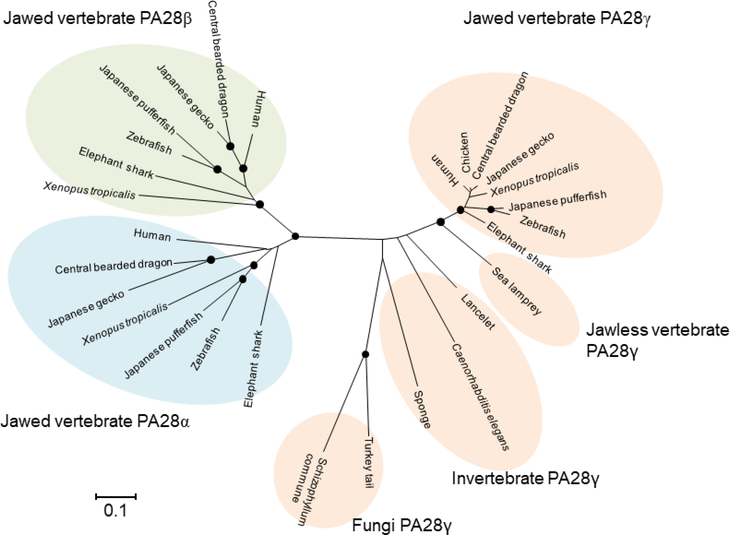
Phylogenetic analysis of the PA28 family of proteins indicates that PA28α and PA28β occur only in jawed vertebrates, whereas PA28γ occurs in wide-ranging multicellular animals including sponges and in some fungi, although it is apparently absent in plants (Fig. 4). Therefore, the PA28γ subunit is clearly of more ancient origin than PA28α and PA28β subunits; it appears that the PSME1 and PSME2 genes emerged from the PSME3 gene in a common ancestor of jawed vertebrates along with other key elements of adaptive immunity including the genes coding for the subunits of immunoproteasomes and thymoproteasomes. In humans and mice, the genes coding for PA28α and PA28β subunits are closely linked to each other in a tail-to-tail orientation, indicating that they arose by tandem duplication (Kandil et al. 1997; Kohda et al. 1998; McCusker et al. 1999). They are also tightly linked in the genomes of Xenopus tropicalis (NCBI Xenopus tropicalis annotation release 103: NC_030677.1) and coelacanth (NCBI Latimeria chalumnae annotation release 101: NW_005821462.1), indicating that the original gene arrangement has been maintained from bony fish to mammals.
Fig. 4.
Evolution of PA28 subunits
For the construction of the phylogenetic tree, amino acid sequences of PA28α, PA28β and PA28γ subunits identified by database searches were aligned using the default Auto setting of the version 7.0 MAFFT program (Kuraku et al. 2013). The tree was constructed using the neighbor-joining algorithm implemented in the MEGA6 program package (Tamura et al. 2013). The distance matrix was obtained by calculating Poisson correction distances for all pairs of sequences. Gaps were excluded using the pairwise-deletion option. The reliability of branching patterns was assessed by bootstrap analysis (1,000 replications). Nodes supported by bootstrap values over 95% are indicated by closed circles. DDBJ/EMBL/NCBI accession numbers are as follows: human PA28α, CAG46459.1; human PA28γ, CAG46543.1; human PA28γ, CAG46545.1; chicken PA28γ, CAG31370.1; central bearded dragon PA28α, XP_020664971.1; central bearded dragon PA28β, XP_020664964.1; central bearded dragon PA28γ, XP_020651679.1; Japanese gecko PA28α, XP_015270891.1; Japanese gecko PA28β, XP_015272197.1; Japanese gecko PA28γ, XP_015282260.1; Xenopus tropicalis PA28α, AAH88020.1; Xenopus tropicalis PA28β, NP_001011494.1; Xenopus tropicalis PA28γ, NP_001096200.1; zebrafish PA28α, NP_571450.1; zebrafish PA28β, NP_571449.1; zebrafish PA28γ, AAF05816.1; Japanese pufferfish PA28α, XP_003967855.1; Japanese pufferfish PA28β, XP_003968209.1; Japanese pufferfish PA28γ, XP_003961088.1; elephant shark PA28α, AFM87012.1; elephant shark PA28β, JK930727; elephant shark PA28γ, XP_007907679.1; sea lamprey PA28γ, CO546357.1; lancelet PA28γ, XP_019630503.1; Caenorhabditis elegans PA28γ, NP_499493.1; sponge Amphimedon queenslandica PA28γ, XP_019856930.1; Turkey tail Trametes versicolor PA28γ, XP_008031792.1; and Schizophyllum commune PA28γ, XP_003038805.1.
Loss of specialized immune proteasomes in birds
The MHC regions of chicken and quail (and most birds) are quite compact, with short introns and intergenic regions, and thus it was relatively easy in the early days of genome analysis to sequence the entire MHC region. Like what was described above for ectotherms, the TAP and TAPBP genes were found closely linked to class I genes (Kaufman et al. 1999). Yet, many other genes found in the mammalian MHC, including the immunoproteasome genes, were not present (Sutoh et al. 2012). Further genomic and transcriptomic studies over the next 20 years also failed to unearth these genes, and a comprehensive proteomic study of proteasome in activated chicken cells revealed only the constitutive subunits (Erath and Groettrup, 2015; Kaufman 2015). Thus, there is little doubt that birds lost their specialized proteasome genes, including PSMB11 and proteasome activator PSME1 and PSME2 genes. It is tempting to speculate that bird class I’s strict dependence on TAP, and the duality of their classical class I system, resulted in the loss of the immune-specialized proteasomes (Kaufman 2018a). However, birds have lost many other immune genes (Magor et al. 2013), and have broken many syntenic groupings outside of the immune system (Lovell et al. 2014), so there may be a general pressure to lose genes rather than something specific to the immune system. Nevertheless, it is doubtful that the loss of both immuno- and thymoproteasome in birds was just a coincidence, since Psmb11 is co-expressed with Psmb9 and Psmb10 in mouse cortical thymic epithelial cells. The lack of the thymoproteasome in birds would call into question the ‘peptide-switch’ hypothesis in which CD8+ T cells are positively selected on a different set of peptides on cortical thymic epithelial cells and thus would escape negative selection on an entirely different set of peptides on medullary thymic epithelial cells. Another head scratcher: while β5t and other immunoproteasome subunits may be co-dependent and generate a different set of peptides compared to constitutive proteasomes, the PA28 subunits have an entirely different function, the rapid production of peptides at the time of viral (or other) infection, as noted above. While the loss of specialized immunoproteasomes can be rationalized by the unusual bird class I system, the loss of enhancing the efficiency of peptide production has no obvious explanation.
There is no doubt that the proteasome system was coopted and modified by gene duplication to service MHC class I antigen presentation. The loss of all of the duplicated genes in birds was either a ‘use it or lose it’ scenario, or the unusual bird class I system could not function well with the duplicates. Regardless of the “reason” for their loss, further studies of CD8+ thymic differentiation and class I antigen presentation in birds are of interest.
Conclusions
Compared to all other 20S proteasome subunits, β5 has a special place in the immune system. As mentioned, β5 has the primordial specificity of the β catalytic proteasome subunit, and one of its paralogues encodes β5i, which is the lynchpin in the formation of 20S immunoproteasomes. As mentioned, the gene coding for β5 has a unique exon/intron organization among β proteasomal subunit genes (and only in jawed vertebrates), which might be related to the generation of the gene encoding β5t early in gnathostome evolution. Interestingly, the immunoproteasome lineages described above, by and large, are found only for β5i, not β1i or β2i. Xenopus speciates by polyploidization with 2n, 4n, 8n, and 12n species, but the MHC (and other genes involved in adaptive immunity) is diploidized, first shown in functional studies such as acute graft rejection (Flajnik et al. 1985; Kobel and Du Pasquier 1986). Later molecular analyses demonstrated that while classical class I, class II and TAP genes are diploidized within a block, other MHC region genes may be found in more copies, i.e. are not diploidized, or the functional gene might be diploidized yet not remain within the MHC. psmb9 and psmb10 genes may fall into this latter category, but the functional psmb8 gene, consistent with its ancient biallelism, always remains linked to class I/TAP/class II genes, i.e. what we would call the ‘true’ MHC (Du Pasquier et al. 2009; Session et al. 2016). Lastly, data suggest that psmb8 and classical class I genes are coexpressed in Xenopus, with little-to-no expression in tadpoles and high expression in many tissues in adults (Salter-Cid et al. 1998).
Clearly, there is still much to learn about the immunoproteasomes, nearly 30 years after their discovery. Peptides have yet to be purified from class I molecules on cortical thymic epithelial cells, which would aid in distinguishing between various models for CD8+ T cell development. A longstanding issue is determining the types of peptides generated by ectotherm immunoproteasomes, and the specificity of those peptides for particular tap and class I allelic products. Lastly, the literature suggests that there are immunoproteasome responsibilities besides the production of peptides (e.g. NF-κB activation), and such functions are poorly studied (Yewdell 2005).
Acknowledgements
We thank Dr. Yoichi Sutoh, Iwate Tohoku Medical Megabank Organization, Iwate Medical University, for his help in preparing Figure 2.
Funding information Experimental work from the authors’ laboratories has been supported by grants from The Ministry of Education, Culture, Sports, Science and Technology, Japan and The National Institutes of Health, USA (RO1AI140326)
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no competing financial interests.
References
- Abdulla S, Beck S, Belich M, Jackson A, Nakamura T, Trowsdale J (1996). Divergent intron arrangement in the MB1/LMP7 proteasome gene pair. Immunogenetics 44:254–258 [DOI] [PubMed] [Google Scholar]
- Akiyama K, Yokota K, Kagawa S, Shimbara N, Tamura T, Akioka H, Nothwang HG, Noda C, Tanaka K, Ichihara A (1994) cDNA cloning and interferon γ down-regulation of proteasomal subunits X and Y. Science 265:1231–1234 [DOI] [PubMed] [Google Scholar]
- Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A (2018) Structure and function of the 26S proteasome. Annu Rev Biochem 87:697–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JE, Hedges SB (2005) Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol 22:2275–2284 [DOI] [PubMed] [Google Scholar]
- Boehm T (2011) Design principles of adaptive immune systems. Nat Rev Immunol 11:307–317 [DOI] [PubMed] [Google Scholar]
- Boehm T, Hirano M, Holland SJ, Das S, Schorpp M, Cooper MD (2018) Evolution of alternative adaptive immune systems in vertebrates. Annu Rev Immunol 36:19–42 [DOI] [PubMed] [Google Scholar]
- Boehm T, McCurley N, Sutoh Y, Schorpp M, Kasahara M, Cooper MD (2012) VLRbased adaptive immunity. Annu Rev Immunol 30:203–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascio P (2014) PA28αβ: the enigmatic magic ring of the proteasome? Biomolecules 4:566–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MS, Pontarotti P, Gilles A, Kelly A, Elgar G (2000) Identification and characterization of a β proteasome subunit cluster in the Japanese pufferfish (Fugu rubripes). J Immunol 165:4446–4452 [DOI] [PubMed] [Google Scholar]
- Collins GA, Goldberg AL (2017) The logic of the 26S proteasome. Cell 169:792–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MD, Alder MN (2006) The evolution of adaptive immune systems. Cell 124:815–822 [DOI] [PubMed] [Google Scholar]
- de Graaf N, van Helden MJ, Textoris-Taube K, Chiba T, Topham DJ, Kloetzel PM, Zaiss DM, Sijts AJ (2011) PA28 and the proteasome immunosubunits play a central and independent role in the production of MHC class I-binding peptides in vivo. Eur J Immunol 41:926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolenc I, Seemuller E, Baumeister W (1998) Decelerated degradation of short peptides by the 20S proteasome. FEBS Lett 434:357–361 [DOI] [PubMed] [Google Scholar]
- Du Pasquier L, Wilson M, Sammut B (2009) The fate of duplicated immunity genes in the dodecaploid Xenopus ruwenzoriensis. Front Biosci (Landmark Ed) 14:177–191 [DOI] [PubMed] [Google Scholar]
- Erath S, Groettrup M (2015) No evidence for immunoproteasomes in chicken lymphoid organs and activated lymphocytes. Immunogenetics 67:51–60 [DOI] [PubMed] [Google Scholar]
- Flajnik MF (2018a) A cold-blooded view of adaptive immunity. Nat Rev Immunol 18:438–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF (2018b) A convergent immunological holy trinity of adaptive immunity in lampreys: discovery of the variable lymphocyte receptors. J Immunol 201:13311335. [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M (2001) Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity 15:351–362 [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M (2010) Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet 11:47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF, Kaufman JF, Du Pasquier L (1985) Studies on the Xenopus major histocompatibility complex. Dev Comp Immunol 9:777–781 [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Ohta Y, Namikawa-Yamada C, Nonaka M (1999) Insights into the primordial MHC from studies in ectothermic vertebrates. Immunol Rev 167:59–67 [DOI] [PubMed] [Google Scholar]
- Griffin TA, Nandi D, Cruz M, Fehling HJ, Kaer LV, Monaco JJ, Colbert RA (1998) Immunoproteasome assembly: cooperative incorporation of interferon γ (IFN-γ)inducible subunits. J Exp Med 187:97–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groettrup M, Soza A, Eggers M, Kuehn L, Dick TP, Schild H, Rammensee H-G, Koszinowski UH, Kloetzel P-M (1996) A role for the proteasome regulator PA28α in antigen presentation. Nature 381:166–168 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Ishibashi T, Tanaka K, Kasahara M (1997) The mouse genes encoding the third pair of β-type proteasome subunits regulated reciprocally by IFN-γ: structural comparison, chromosomal localization, and analysis of the promoter. J Immunol 159:2760–2770 [PubMed] [Google Scholar]
- Heemels M-T, Ploegh H (1995) Generation, translocation, and presentation of MHC class I-restricted peptides. Annu Rev Biochem 64:463–491 [DOI] [PubMed] [Google Scholar]
- Heink S, Ludwig D, Kloetzel PM, Kruger E (2005) IFN-γ-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc Natl Acad Sci U S A 102:9241–9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Guo P, McCurley N, Schorpp M, Das S, Boehm T, Cooper MD (2013) Evolutionary implications of a third lymphocyte lineage in lampreys. Nature 501:435–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Tanaka Y, Fujito NT, Nonaka M (2013) Dimorphisms of the proteasome subunit β type 8 gene (PSMB8) of ectothermic tetrapods originated in multiple independent evolutionary events. Immunogenetics 65:811–821 [DOI] [PubMed] [Google Scholar]
- Jastrab JB, Darwin KH (2015) Bacterial proteasomes. Annu Rev Microbiol 69:109–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M (2009) RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet 10:19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandil E, Kohda K, Ishibashi T, Tanaka K, Kasahara M (1997) PA28 subunits of the mouse proteasome: primary structures and chromosomal localization of the genes. Immunogenetics 46:337–344 [DOI] [PubMed] [Google Scholar]
- Kandil E, Namikawa C, Nonaka M, Greenberg AS, Flajnik MF, Ishibashi T, Kasahara M (1996) Isolation of low molecular mass polypeptide complementary DNA clones from primitive vertebrates: Implications for the origin of MHC class I-restricted antigen presentation. J Immunol 156:4245–4253 [PubMed] [Google Scholar]
- Kasahara M (1998) What do the paralogous regions in the genome tell us about the origin of the adaptive immune system? Immunol Rev 166:159–175 [DOI] [PubMed] [Google Scholar]
- Kasahara M (2007) The 2R hypothesis: an update. Curr Opin Immunol 19:547–552 [DOI] [PubMed] [Google Scholar]
- Kasahara M (2010) Genome duplication and T cell immunity. Prog Mol Biol Transl Sci 92:7–36 [DOI] [PubMed] [Google Scholar]
- Kasahara M (2013) Impact of whole-genome duplication on vertebrate development and evolution. Sem Cell Dev Biol 24:81–82 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Hayashi M, Tanaka K, Inoko H, Sugaya K, Ikemura T, Ishibashi T (1996) Chromosomal localization of the proteasome Z subunit gene reveals an ancient chromosomal duplication involving the major histocompatibility complex. Proc Natl Acad Sci U S A 93:9096–9101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Sutoh Y (2014) Two forms of adaptive immunity in vertebrates: similarities and differences. Adv Immunol 122:59–90 [DOI] [PubMed] [Google Scholar]
- Kaufman J (1999) Co-evolving genes in MHC haplotypes: the “rule” for nonmammalian vertebrates? Immunogenetics 50:228–236 [DOI] [PubMed] [Google Scholar]
- Kaufman J (2015) What chickens would tell you about the evolution of antigen processing and presentation. Curr Opin Immunol 34:35–42 [DOI] [PubMed] [Google Scholar]
- Kaufman J (2018a) Generalists and specialists: a new view of how MHC class I molecules fight infectious pathogens. Trends Immunol 39:367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman J (2018b) Unfinished business: evolution of the MHC and the adaptive immune system of jawed vertebrates. Annu Rev Immunol 36:383–409 [DOI] [PubMed] [Google Scholar]
- Kaufman J, Milne S, Gobel TW, Walker BA, Jacob JP, Auffray C, Zoorob R, Beck S (1999) The chicken B locus is a minimal essential major histocompatibility complex. Nature 401:923–925 [DOI] [PubMed] [Google Scholar]
- Kincaid EZ, Che JW, York I, Escobar H, Reyes-Vargas E, Delgado JC, Welsh RM, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, Rock KL (2011) Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol 13:129–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein L, Kyewski B, Allen PM, Hogquist KA (2014) Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol 14:377–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniepert A, Groettrup M (2014) The unique functions of tissue-specific proteasomes. Trends Biochem Sci 39:17–24 [DOI] [PubMed] [Google Scholar]
- Kobel HR, Du Pasquier L (1986) Genetics of polyploid Xenopus. Trends Genet 2:310–315 [Google Scholar]
- Kohda K, Matsuda Y, Ishibashi T, Tanaka K, Kasahara M (1997) Structural analysis and chromosomal localization of the mouse Psmb5 gene coding for the constitutively expressed β-type proteasome subunit. Immunogenetics 47:77–87 [DOI] [PubMed] [Google Scholar]
- Kohda K, Ishibashi T, Shimbara N, Tanaka K, Matsuda Y, Kasahara M (1998) Characterization of the mouse PA28 activator complex gene family: complete organizations of the three member genes and a physical map of the ~150-kilobase region containing the α- and β-subunit genes. J Immunol 160:4923–4935 [PubMed] [Google Scholar]
- Kondo K, Ohigashi I, Takahama Y (2018) Thymus machinery for T-cell selection. Int Immunol 31: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S, Zmasek CM, Nishimura O, Katoh K (2013) aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res 41:W22–W28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Wirthlin M, Wilhelm L, Minx P, Lazar NH, Carbone L, Warren WC, Mello CV (2014) Conserved syntenic clusters of protein coding genes are missing in birds. Genome Biol 15:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R (1995) Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science 268:533–539 [DOI] [PubMed] [Google Scholar]
- Lukacs MF, Harstad H, Grimholt U, Beetz-Sargent M, Cooper GA, Reid L, Bakke HG, Phillips RB, Miller KM, Davidson WS, Koop BF (2007) Genomic organization of duplicated major histocompatibility complex class I regions in Atlantic salmon (Salmo salar). BMC Genomics 8:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magor KE, Miranzo Navarro D, Barber MR, Petkau K, Fleming-Canepa X, Blyth GA, Blaine AH (2013) Defense genes missing from the flight division. Dev Comp Immunol 41:377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin-Furlow J (2011) Proteasomes and protein conjugation across domains of life. Nat Rev Microbiol 10:100–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SC, Hernandez KM, Wcisel DJ, Kettleborough RN, Stemple DL, Yoder JA, Andrade J, de Jong JLO (2016) Alternative haplotypes of antigen processing genes in zebrafish diverged early in vertebrate evolution. Proc Natl Acad Sci U S A 113:E5014–E5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker D, Wilson M, Trowsdale J (1999) Organization of the genes encoding the human proteasome activators PA28α and β. Immunogenetics 49:438–445 [DOI] [PubMed] [Google Scholar]
- Mehta TK, Ravi V, Yamasaki S, Lee AP, Lian MM, Tay BH, Tohari S, Yanai S, Tay A, Brenner S, Venkatesh B (2013) Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). Proc Natl Acad Sci U S A 110:16044–16049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalova V, Murray BW, Sultmann H, Klein J (2000) A contig map of the Mhc class I genomic region in the zebrafish reveals ancient synteny. J Immunol 164:5296–5305 [DOI] [PubMed] [Google Scholar]
- Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K (2007) Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316:1349–1353 [DOI] [PubMed] [Google Scholar]
- Murata S, Takahama Y, Kasahara M, Tanaka K (2018) The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat Immunol 19:923–931 [DOI] [PubMed] [Google Scholar]
- Murata S, Udono H, Tanahashi N, Hamada N, Watanabe K, Adachi K, Yamano T, Yui K, Kobayashi N, Kasahara M, Tanaka K, Chiba T (2001) Immunoproteasome assembly and antigen presentation in mice lacking both PA28α and PA28β. EMBO J 20:5898–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10:458–467 [DOI] [PubMed] [Google Scholar]
- Namikawa C, Salter-Cid L, Flajnik MF, Kato Y, Nonaka M, Sasaki M (1995) Isolation of Xenopus LMP-7 homologues. Striking allelic diversity and linkage to MHC. J Immunol 155:1964–1971 [PubMed] [Google Scholar]
- Nikaido T, Shimada K, Shibata M, Hata M, Sakamoto M, Takasaki Y, Sato C, Takahashi T, Nishida Y (1990) Cloning and nucleotide sequence of cDNA for Ki antigen, a highly conserved nuclear protein detected with sera from patients with systemic lupus erythematosus. Clin Exp Immunol 79:209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta T, Murata S, Sasaki K, Fujii H, Ripen AM, Ishimaru N, Koyasu S, Tanaka K, Takahama Y (2010) Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity 32:29–40 [DOI] [PubMed] [Google Scholar]
- Nonaka M, Namikawa-Yamada C, Sasaki M, Salter-Cid L, Flajnik MF (1997a) Evolution of proteasome subunits δ and LMP2: complementary DNA cloning and linkage analysis with MHC in lower vertebrates. J Immunol 159:734–740 [PubMed] [Google Scholar]
- Nonaka M, Namikawa C, Kato Y, Sasaki M, Salter-Cid L, Flajnik MF (1997b) Major histocompatibility complex gene mapping in the amphibian Xenopus implies a primordial organization. Proc Natl Acad Sci U S A 94:5789–5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S (1970) Evolution by Gene Duplication. Springer-Verlag, New York [Google Scholar]
- Ohta Y, Goetz W, Hossain MZ, Nonaka M, Flajnik MF (2006) Ancestral organization of the MHC revealed in the amphibian Xenopus. J Immunol 176:3674–3685 [DOI] [PubMed] [Google Scholar]
- Ohta Y, McKinney EC, Criscitiello MF, Flajnik MF (2002) Proteasome, transporter associated with antigen processing, and class I genes in the nurse shark Ginglymostoma cirratum: Evidence for a stable class I region and MHC haplotype lineages. J Immunol 168:771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Powis SJ, Lohr RL, Nonaka M, Pasquier LD, Flajnik MF (2003) Two highly divergent ancient allelic lineages of the transporter associated with antigen processing (TAP) gene in Xenopus: further evidence for co-evolution among MHC class I region genes. Eur J Immunol 33:3017–3027 [DOI] [PubMed] [Google Scholar]
- Preckel T, Fung-Leung WP, Cai Z, Vitiello A, Salter-Cid L, Winqvist O, Wolfe TG, Von Herrath M, Angulo A, Ghazal P, Lee JD, Fourie AM, Wu Y, Pang J, Ngo K, Peterson PA, Fruh K, Yang Y (1999) Impaired immunoproteasome assembly and immune responses in PA28−/− mice. Science 286:2162–2165 [DOI] [PubMed] [Google Scholar]
- Salter-Cid L, Nonaka M, Flajnik MF (1998) Expression of MHC class Ia and class Ib during ontogeny: high expression in epithelia and coregulation of class Ia and lmp7 genes. J Immunol 160:2853–2861 [PubMed] [Google Scholar]
- Sasaki K, Takada K, Ohte Y, Kondo H, Sorimachi H, Tanaka K, Takahama Y, Murata S (2015) Thymoproteasomes produce unique peptide motifs for positive selection of CD8(+) T cells. Nat Commun 6:7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, van Heeringen SJ, Quigley I, Heinz S, Ogino H, Ochi H, Hellsten U, Lyons JB, Simakov O, Putnam N, Stites J, Kuroki Y, Tanaka T, Michiue T, Watanabe M, Bogdanovic O, Lister R, Georgiou G, Paranjpe SS, van Kruijsbergen I, Shu S, Carlson J, Kinoshita T, Ohta Y, Mawaribuchi S, Jenkins J, Grimwood J, Schmutz J, Mitros T, Mozaffari SV, Suzuki Y, Haramoto Y, Yamamoto TS, Takagi C, Heald R, Miller K, Haudenschild C, Kitzman J, Nakayama T, Izutsu Y, Robert J, Fortriede J, Burns K, Lotay V, Karimi K, Yasuoka Y, Dichmann DS, Flajnik MF, Houston DW, Shendure J, DuPasquier L, Vize PD, Zorn AM, Ito M, Marcotte EM, Wallingford JB, Ito Y, Asashima M, Ueno N, Matsuda Y, Veenstra GJ, Fujiyama A, Harland RM, Taira M, Rokhsar DS (2016) Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Timoshevskaya N, Ye C, Holt C, Keinath MC, Parker HJ, Cook ME, Hess JE, Narum SR, Lamanna F, Kaessmann H, Timoshevskiy VA, Waterbury CKM, Saraceno C, Wiedemann LM, Robb SMC, Baker C, Eichler EE, Hockman D, Sauka-Spengler T, Yandell M, Krumlauf R, Elgar G, Amemiya CT (2018) The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat Genet 50:270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutoh Y, Kondo M, Ohta Y, Ota T, Tomaru U, Flajnik MF, Kasahara M (2012) Comparative genomic analysis of the proteasome β5t subunit gene: implications for the origin and evolution of thymoproteasomes. Immunogenetics 64:49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama Y, Takada K, Murata S, Tanaka K (2012) β5t-containing thymoproteasome: specific expression in thymic cortical epithelial cells and role in positive selection of CD8+ T cells. Curr Opin Immunol 24:92–98 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K (2009) The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci 85:12–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Kasahara M (1998) The MHC class I ligand-generating system: roles of immunoproteasomes and the interferon-γ-inducible proteasome activator PA28. Immunol Rev 163:161–176 [DOI] [PubMed] [Google Scholar]
- Tomaru U, Ishizu A, Murata S, Miyatake Y, Suzuki S, Takahashi S, Kazamaki T, Ohara J, Baba T, Iwasaki S, Fugo K, Otsuka N, Tanaka K, Kasahara M (2009) Exclusive expression of proteasome subunit β5t in the human thymic cortex. Blood 113:5186–5191 [DOI] [PubMed] [Google Scholar]
- Tsukamoto K, Miura F, Fujito NT, Yoshizaki G, Nonaka M (2012) Long-lived dichotomous lineages of the proteasome subunit beta type 8 (PSMB8) gene surviving more than 500 million years as alleles or paralogs. Mol Biol Evol 29:3071–3079 [DOI] [PubMed] [Google Scholar]
- Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T (2002) The structure of the mammalian 20S proteasome at 2.75 Å resolution. Structure 10:609–618 [DOI] [PubMed] [Google Scholar]
- Walker BA, Hunt LG, Sowa AK, Skjodt K, Gobel TW, Lehner PJ, Kaufman J (2011) The dominantly expressed class I molecule of the chicken MHC is explained by coevolution with the polymorphic peptide transporter (TAP) genes. Proc Natl Acad Sci U S A 108:8396–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk S, Chen WE, Magnusson RP (2000) Properties of the nuclear proteasome activator PA28γ (REGγ). Arch Biochem Biophys 383:265–271 [DOI] [PubMed] [Google Scholar]
- Xing Y, Jameson SC, Hogquist KA (2013) Thymoproteasome subunit-β5T generates peptide-MHC complexes specialized for positive selection. Proc Natl Acad Sci U S A 110:6979–6984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell JW (2005) Immunoproteasomes: regulating the regulator. Proc Natl Acad Sci U S A 102:9089–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Krutchinsky A, Endicott S, Realini C, Rechsteiner M, Standing KG (1999) Proteasome activator 11S REG or PA28: Recombinant REGα/REGβ hetero-oligomers are heptamers. Biochemistry 38:5651–5658 [DOI] [PubMed] [Google Scholar]